Abstract
To examine the relationship between folding and export competence by the twin-arginine translocation (Tat) pathway we analyzed the subcellular localization of fusions between a set of eight putative Tat leader peptides and alkaline phosphatase in isogenic Escherichia coli strains that either allow or disfavor the formation of protein disulfide bonds in the cytoplasm. We show that export by the Tat translocator is observed only in strains that enable oxidative protein folding in the cytoplasm. Further, we show that other disulfide-containing proteins, namely single-chain Fv and heterodimeric FAB antibody fragments, are export-competent only in strains having an oxidizing cytoplasm. Functional, heterodimeric FAB protein was exported from the cytoplasm by means of a Tat leader peptide fused to the heavy chain alone, indicating that the formation of a disulfide-bonded dimer preceeds export. These results demonstrate that in vivo only proteins that have attained the native conformation are exported by the Tat translocator, indicating that a folding quality-control mechanism is intrinsic to the export process. The ability to export proteins with disulfide bonds and the folding proofing feature of the Tat pathway are of interest for biotechnology applications.
Two distinct pathways exist for the export of proteins across the cytoplasmic membrane of Escherichia coli. The bulk of protein translocation across the cytoplasmic membrane occurs by the general secretory (Sec) pathway (1, 2). However, a fundamentally different mechanism for periplasmic protein localization was recently discovered, first in the thylakoid membranes of photosynthetic organisms and subsequently in bacteria and archaea (3, 4). In the latter, this mechanism has been named the twin-arginine translocation (Tat) pathway because of the signature Arg-Arg motif found near the N terminus of the leader peptide of proteins that are engaged in this mode of export from the cytoplasm (5).
The bacterial Tat export machinery is structurally and mechanistically similar to the ΔpH-driven protein import pathway found in plant thylakoids (6–8). For this reason, bacterial Tat leader peptides are functionally interchangeable with thylakoid import signals (9–11). In general, the Tat pathway appears to be responsible for the membrane translocation of polypeptides that are incompatible with the Sec machinery such as those requiring the incorporation of cofactors (e.g., GFOR; ref. 12) or subunit association (e.g., HybO; ref. 13) before export and also proteins that can not be exported by the Sec pathway in a functional form, for unknown reasons (e.g., GFP; ref. 14). Processes such as cofactor incorporation or the assembly of protein subunits hinge on the formation of secondary or tertiary structure. In vitro experiments in the thylakoid system have shown that: (i) bovine pancreatic trypsin inhibitor (BPTI) fused to a ΔpH-dependent pathway precursor is transported into thylakoids, even when BPTI was permanently folded by using irreversible disulfide cross-linkers that prevented any unfolding during the translocation process (15); and (ii) dihydrofolate reductase (DHFR) bound to methotrexate, a ligand that greatly stabilizes the folded state of the protein, is specifically targeted to the thylakoid lumen by the ΔpH-dependent pathway (16). However, BPTI and DHFR are significantly smaller (56 and 187 aa, respectively) than typical ΔpH-dependent polypeptide substrates.
In vivo the export of cofactor-containing proteins by the bacterial Tat pathway is abolished when the incorporation of the cofactor in the cytoplasm is blocked (17). The attachment of cofactors is a complex process that involves interactions with other cellular components including specific chaperones. Whether on cofactor attachment the protein assumes a fully native conformation before export, and if so, whether partial unfolding occurs during translocation are far from clear. Moreover, the folding state of Tat-competent proteins that do not contain cofactors has not been established. An equally intriguing question is whether the Tat pathway is able to accept both folded and unfolded polypeptides as substrates, or if only proteins that have reached a substantially native state in the cytoplasm are competent for translocation. Earlier in vitro data regarding the import of DHFR by the ΔpH system provide some support for the former model (16). The alternative model, whereby the Tat pathway can export only native-like proteins, implies the existence of a folding quality-control mechanism inherent to this pathway. Because the Tat translocon is necessary and sufficient for protein export, the ability to assess the folding competence of exported proteins would then have to reside with the membrane TatABCE components.
We examined the relationship between protein folding and export competence by analyzing the subcellular localization of proteins with structural disulfide bonds in strains that either allow or disfavor oxidative protein folding in the cytoplasm. Tat-dependent accumulation of active alkaline phosphatase (AP), as well as other multidisulfide proteins including single-chain and heterodimeric FAB antibody fragments, into the periplasm was observed only in strains that enable the formation of disulfide bonds in the cytoplasm. Thus in vivo only proteins with substantially native-like conformation can be exported by the Tat translocator, demonstrating the presence a folding quality-control mechanism that is intrinsic to the export process and is unrelated to cofactor association.
Experimental Procedures
Bacterial Strains, Growth, and Induction Conditions.
The bacterial strains and plasmids used in this study are described in Table 1. For monitoring the export of AP, cells grown in LB media overnight were subcultured into M9 salts supplemented with 0.2% glucose/1 μg/ml vitamin B1/1 mM MgSO4/50 μg/ml 18 aa (excluding methionine and cysteine) at a 100-fold dilution, and then incubated at 30°C. For the expression of single-chain antibody (scFv) and Fab antibody fragments, cells were subcultured from overnight cultures into fresh LB medium (5% vol/vol) and then incubated at 30°C. Protein synthesis was induced by adding isopropyl β-d-thiogalactoside to a final concentration of 0.1 mM when the cells reached an OD600 ≈ 0.5. Where appropriate, the coexpression of DsbC from pBAD-ΔssDsbC was induced by using 0.2% arabinose. Antibiotic selection was maintained for all markers on plasmids at the following concentrations: ampicillin, 100 μg/ml; chloramphenicol, 25 μg/ml; kanamycin, 50 μg/ml; and spectinomycin, 100 μg/ml.
Table 1.
Bacterial strains and plasmids used in this study
| E. coli strain or plasmid | Relevant phenotype or features | Source |
|---|---|---|
| DHB4 | MC1000 phoR Δ(phoA) PvuII Δ(malF)3 F′[lacIqZYA pro] | Laboratory stock |
| DR473 | DHB4 ΔtrxB gor552 Tn10Tet ahpC* Tn10Cm (araC Para-trxB) | J. Beckwith |
| FA113 | DHB4 trxB gor552 Tn10tetrahpC* | J. Beckwith |
| DHBA, DRA | DHB4 dsbA∷kan, DR473 dsbA∷kan | This work |
| DRB | DR473 tatB∷kan | This work |
| DRC | DR473 tatC∷spec | This work |
| pTrc99A | trc promoter, ColE1 ori, ampr | Amersham Biosciences |
| pAID135 | Δ(2–22)AP | Ref. 20 |
| pTorA-AP | ssTorA-Δ(1–22)AP cloned in pTrc99A | This work |
| pKK-AP | as pTorA-AP with R11K;R12K mutation in ssTorA | This work |
| pFdnG-AP | ssFdnG-Δ(1–22)-AP in pTrc99A | This work |
| pTrc99-Fab | Ref. 22 | |
| pssTorA-Fab | ssTorA-VH-CH1 and V1-C1 dicistronic operon in Trc99 | This work |
| pKK-Fab | as above with ssTorA(R11K;R12K) | This work |
| pBAD-ΔssdsbC | Ref. 22 |
Enzyme Activity Assays.
Cells expressing AP were harvested 3 h after induction, treated with 100 mM iodoacetamide as above, pelleted by centrifugation, and fractionated by the cold osmotic shock procedure (18, 19). Soluble protein was quantified by the Bio-Rad protein assay, with BSA as standard. AP activity and β-galactosidase activity assays were performed as described (20). Only data from fractionation experiments in which ≥95% of the β-galactosidase activity was in the cytoplasmic fraction are reported. The trypsin resistance of AP was assessed as described (21), except that samples were first treated with iodoacetamide.
ELISA was performed according to Levy et al. (22). Western blotting was performed as described by Chen et al. (23).
Results
Alkaline Phosphatase Can Be Exported by Tat only in Strains with an Oxidizing Cytoplasm.
In bacteria, the thioredoxin and glutaredoxin pathways maintain the cytoplasm in a highly reducing state that strongly disfavors the oxidation of protein thiols (reviewed in ref. 24). For this reason, proteins requiring disulfide bonds have to be exported into a more oxidizing environment. AP is usually secreted by a Sec-specific leader peptide into the periplasmic space where it is rapidly oxidized by DsbA to form its two disulfide bonds that are critical for the stability and catalytic activity of the protein (21). Earlier studies had demonstrated that fusions of AP to Tat-specific leader peptides are not exported by the Tat pathway (25–29). We reasoned that the inability of AP fusions to be exported by the Tat pathway might be due to the fact that such proteins are usually unfolded in the cytoplasm, which is maintained in a strongly reducing state that precludes the formation of disulfide bonds. To investigate the above-mentioned hypothesis and to examine the relationship between folding and Tat export competence, we exploited the availability of mutant E. coli that allow the formation of disulfide bonds in the cytoplasm or, conversely, disable protein oxidation in the periplasm.
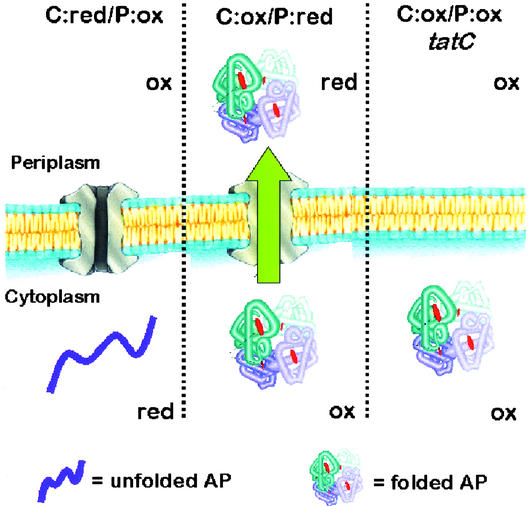
Beckwith and coworkers (30) had demonstrated that disulfide bonds can form readily when AP is expressed without its signal sequence in the cytoplasm of oxidizing mutant strains such as E. coli DR473 (trxB gor ahpC*). For convenience, this strain background is designated C:ox/P:ox because both the cytoplasm and the periplasm are oxidizing (Fig. 1). In a dsbA derivative of DR473, protein oxidation in the periplasm is impaired (31), and therefore, such a strain is designated C:ox/P:red. In DR473 dsbA cells, the formation of disulfide bonds in AP can only occur within the cytoplasm.
Figure 1.
AP folding and export in different strain backgrounds. Ox and red refer to oxidizing and reducing redox potentials in the specified subcellular compartment. In C:ox (DR473) cells, AP is able to fold in the cytoplasm and thus can serve as a substrate for the Tat pathway. The localization of AP is not impaired in the C:ox/P:red strain DRA (DR473 dsbA). Deletion of tatC (or tatB) in the C:ox/P:ox strain blocks AP export.
A set of eight leader peptides that have been shown to direct export by the Tat pathway were fused to AP and expressed in DHB4, DR473, DR473 dsbA, and finally in DR473 tatB∷kan and in DR473 tatC∷spec. In the latter two strain backgrounds, the mutational inactivation of tatB or tatC is expected to impair export through the Tat pathway. Cells were grown in minimal media and the subcellular distribution of AP was determined. After cell harvesting, cell samples were treated with 100 mM iodoacetamide to prevent the formation of disulfide bonds during fractionation by osmotic shock and the AP enzymatic activities in the cytoplasmic and periplasmic fractions were then determined. The degree of leakage of cytoplasmic components during fractionation was <5% as determined by the subcellular distribution of β-galactosidase activity and of GroEL ptotein (detected by Western blotting).
The fusion proteins were classified into two classes on the basis of whether the formation of active AP was strictly dependent on an oxidizing cytoplasm (Table 1). The first class of Tat leader peptides (class I: ssFdnG, ssFdoG, ssHyaA, signal sequence trimethylamine N-oxide reductase [ssTorA]) was found to export AP to the periplasm in a C:ox-dependent and Tat-dependent manner. In other words, accumulation of AP activity in the periplasm only occurred in strains having both an oxidizing cytoplasm and an intact Tat apparatus. Specifically, when class I leaders were expressed in the parental strain DHB4, which has a reducing cytoplasm (i.e., it is C:red/P:ox) only background AP activity was observed. However, in a C:ox strain, a significant fraction of the AP activity (25–50% of total) was found in the periplasm. The periplasmic AP activity was not dependent on DsbA and was thus virtually the same in C:ox/P:ox cells relative to C:ox/P:red cells (i.e., in DR473 dsbA). This finding indicates that the oxidation of AP occurs only in the cytoplasm before export. The accumulation of active protein in the periplasm was abolished when the signature RR motif in the leader peptide was changed to a KK sequence (data not shown), a substitution known to completely block export (5). Likewise, inactivation of either tatB or tatC in a C:ox/P:ox strain resulted in near complete loss of AP enzymatic activity in the periplasm, whereas the cytoplasmic activity remained virtually unchanged. For one of the four class I Tat-leader peptides (ssHyaA), export was only partially blocked in a tatB strain.
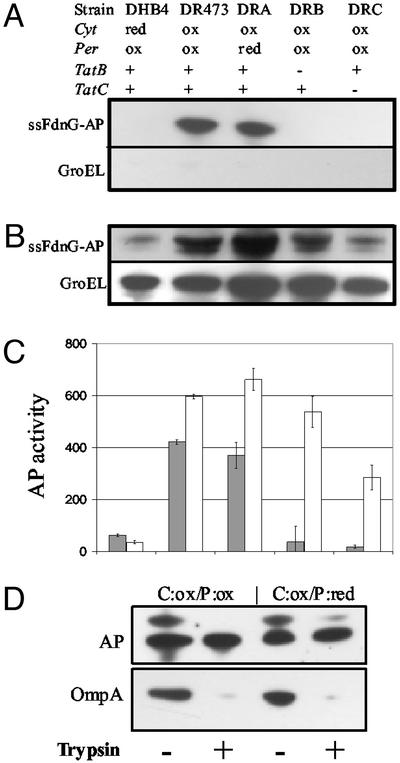
Fig. 2 A–C shows the subcellular distribution of one class I leader peptide fusion, ssFdnG-AP, in the different strain backgrounds. Inactivation of tatC in the C:ox strain background resulted in a lower level of cytoplasmic fusion protein. Similarly, a smaller amount of ssFdnG-AP fusion was observed in the cytoplasm of C:red cells. A higher-molecular-weight species corresponding to the ssFdnG-AP precursor could be resolved in 4–20% acrylamide gradient gels (see Fig. 4D). This high-molecular-weight species could be processed to a band with an electrophoretic mobility identical to the mature protein by incubation with trypsin, because of the presence of trypsin-sensitive sites in the leader peptide (26). The mature protein was completely resistant to trypsin under conditions that result in the disappearance of full-length OmpA (Fig. 2D). Further, no loss of AP enzymatic activity was detected in these experiments consistent with the observation of Sone et al. (21) that only the native, fully oxidized form of AP is trypsin-resistant.
Figure 2.
Subcellular localization of ssFdnG-AP. Immunoblotting of periplasmic (A) and cytoplasmic (B) fractions from cells expressing ssFdnG-AP. Samples were normalized on the basis of the amount of total cell protein and resolved on SDS-12% polyacrylamide gels. GroEL was used as a fractionation marker by probing with anti-GroEL serum. (C) AP activity for the same periplasmic (filled bars) and cytoplasmic (open bars) fractions as in A and B. (D) Trypsin sensitivity analysis of periplasmic fractions collected from C:ox/P:ox and C:ox/P:red cells. Samples were separated on SDS-4–20% polyacrylamide gels and probed with anti-AP and anti-OmpA serum.
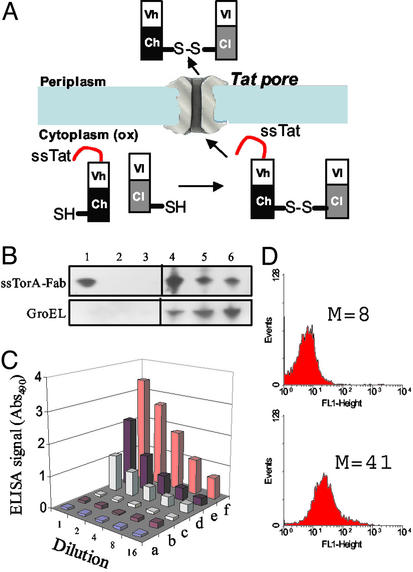
Figure 4.
Tat export of an antidigoxin antibody fragment (FAB). (A) Tat export of FAB antibodies. (B) Western blotting of periplasmic (lanes 1–3) and cytoplasmic (lanes 4–6) fractions collected from cells coexpressing TorA-Fab fusion and ΔssDsbC. Strains used were (1, 4) C:ox/P:ox; (2, 5) C:ox/P:ox/tatC, and (3, 6) C:red/P:ox. All lanes were loaded with the same amount of total protein and anti-mouse IgG F(ab′)2 antibody was used to detect the FAB light chain. GroEL was used as a fractionation marker for the spheroplast fractions. (C) ELISA of periplasmic (a, c, and e) and cytoplasmic (b, d, and f) fractions collected from C:red/P:ox (a and b), C:ox/P:ox/tatC (c and d), and C:ox/P:ox (e and f) cells. (D) Flow cytometric analysis of C:red/P:ox cells (Upper) and C:ox/P:ox cells (Lower) cells expressing TorA-Fab and ΔssDsbC and labeled with FITC-digoxin.
Four of the eight Tat leader peptides (class II: ssDmsA, ssSufI, ssYacK, and ssYcbK) exhibited high AP activity in the WT strain DHB4 (C:red/P:ox). Further analysis indicated that the class II leader peptides can engage both the Tat and the Sec pathways. Specifically the ability to engage the Sec pathway was evident by: (i) significant reduction of periplasmic AP activity in dsbA mutants relative to dsbA+ cells [compare DHB4 (C:red/P:ox) to DHB4 dsbA (C:red/P:red), and also DR473 (C:ox/P:ox) to DR473 dsbA (C:ox/P:red)]. For all four class II leader peptides, the level of periplasmic AP activity in the dsbA mutant strains cells was ≈60% of what was obtained in the isogenic dsbA+ E. coli. (ii) The appearance of significant periplasmic AP activity in tatB or tatC mutants. (iii) Loss of AP activity in a secA conditional mutant (strain MM52 secA51(ts), C:red/P:ox) after upshift to the nonpermissive temperature of 42°C relative to WT cells also shifted to 42°C (D.T., M.P.D., Y. K. Kawarasaki, and G.G., unpublished work). The ability of class II leader peptides to also engage the Tat pathway is revealed by the presence of high AP activity in the periplasmic fraction of DR473 dsbA (C:ox/P:red) cells.
Tat Export of Antibody Fragments.
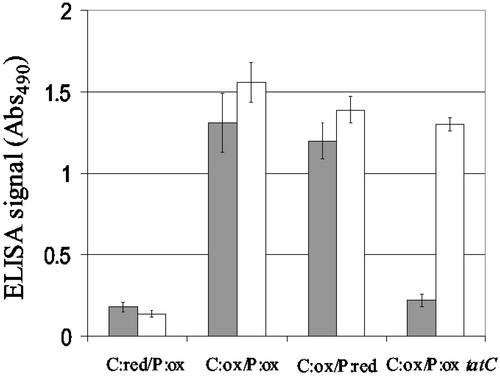
In addition to AP, class I leader peptides could mediate the export of other proteins with intra- or intermolecular disulfide bonds from the cytoplasm of C:ox strains. scFvs contain two intermolecular disulfide bonds, one in the VH and one in the Vl chain. The 26–10 antidigoxin scFv (22) was fused to the class I Tat leader peptide ssTorA, and the accumulation of antigen-binding protein in the periplasmic space of C:ox/P:ox cells was monitored by ELISA (Fig. 3). Nearly 45% of the digoxin-binding activity was localized in the periplasmic space. The accumulation of active antidigoxin scFv in the periplasm was not affected in DR473 dsbA (C:ox/P:red) but reduced to background levels in a tatC mutant.
Figure 3.
Tat export of a scFv. Detection of scFv by ELISA in the periplasmic and cytoplasmic fractions. Periplasmic (filled bars) and cytoplasmic (open bars) samples were serially diluted and started from the same amount of total protein. Data reported are from a 4-fold dilution and are the average of two independent experiments.
FAB antibodies are heterodimeric proteins in which the heavy chain (VH-CH1) and light chain (Vl-Cl) are linked by an intermolecular disulfide bond. In addition, FAB proteins contain four more intrachain disulfides, two within each of the heavy and light chains. A dicistronic operon consisting of a gene encoding a ssTorA-VH-CH1 fusion followed by the light chain (Vl-Cl) was constructed (Fig. 4A; ref. 22). In this construct the light chain does not contain a leader peptide and therefore can not be exported from the cytoplasm by itself. Induction of protein synthesis in DR473 dsbA (i.e., C:ox/P:red) was found to result in the accumulation of a small but significant amount of light-chain polypeptide in the osmotic shock fraction. Western blot analysis by using an anti-mouse IgG F(ab′)2 antibody specific for the FAB light chain indicated that the intensity of the Vl-Cl band in the osmotic shock fraction was ≈15–20% of that in the cytoplasm (data not shown). Neither light chain nor FAB protein could be detected in the osmotic shock fraction of a tatC mutant strain or from DHB4 dsbA (C:red/P:ox) cells.
We reasoned that if preassociation of the heavy and light chains is required for secretion by the Tat pathway, then conditions that enhance the yield of correctly folded FAB may increase the export efficiency. The yield of the antidigoxin FAB expressed in the cytoplasm of a trxB gor aphC* mutant (the C:ox strain FA113 was used for these experiments to enable coexpression of DsbC from the ara promoter) is increased markedly on coexpression of a signal-sequenceless version of the periplasmic disulfide isomerase DsbC (ΔssDsbC) (22). Consistent with this hypothesis, in cells coexpressing ΔssDsbC the intensity of the light-chain band in the osmotic shock fraction was 70% of that in the cytoplasm (compared with 15–20% without ΔssDsbC, as noted above). A similar partitioning of the FAB digoxin-binding activity was detected by ELISA (Fig. 4C). In the FA113 tatC∷spec strain, no FAB could be detected in the periplasmic fraction and a significant reduction in the amount of protein remaining in the cytoplasm was also observed. We and others have observed that depletion of the tat genes often results in inactivation or degradation of Tat substrate proteins in the cytoplasm (32, 33). Similarly, no light-chain protein or FAB-binding activity could be detected in E. coli DHB4 (Fig. 4 B and C) or when the RR dipeptide in ssTorA was substituted by KK (data not shown).
As a final test we showed that functional FAB protein capable of binding to the hapten digoxin is formed in vivo. Incubation of E. coli in a hypertonic solution (5× PBS) increases the outer membrane permeability such that fluorescent ligands can equilibrate within the periplasm whereas periplasmic proteins are unable to leak out of the cell (23). Cells binding the fluorescent ligand can thus be detected by flow cytometry. C:ox/:P:ox cells expressing ssTorA-VH-CH1 and Vl-Cl and incubated with a digoxin-fluorescein complex exhibited a marked increase in cell fluorescence relative to control cells (C:red/P:ox; Fig. 4D). The detection of digoxin-binding activity in intact cells in situ further rules out the possibility that formation of disulfide bonds in the FAB protein may have occurred during cell fractionation.
Discussion
The hallmark of the Tat pathway that sets it apart form all other modes of protein translocation across lipid bilayer membranes is the ability to export polypeptides that have already assumed a degree of stable secondary, or perhaps even tertiary, structure. The relationship between the folding state of a protein and Tat secretion competence is poorly understood. Export of folded proteins had only been demonstrated experimentally in vitro for the plant equivalent of Tat, the ΔpH-dependent import system of thylakoids. Clark and Theg (15) showed that prefolded and chemically crosslinked BPTI could be imported into thylakoids when linked to the 17-kDa subunit of the oxygen-evolving complex of photosystem II (prOE17). However, BPTI is a very small protein (56 residues, 6.5 kDa) and therefore not typical of native Tat and ΔpH substrates. Furthermore, crosslinked BPTI could also be transported across the chloroplast envelope membrane, a process that usually involves polypeptides that are largely unfolded (34). Hynds et al. (16) found that DHFR, bound to methotrexate, could be transported in a ΔpH-dependent manner. The incorporation of amino acid analogues, which destabilize DHFR and render it protease-sensitive, did not inhibit transport. Such “malfolded” DHFR may (i) be able to fold into a nonnative compact conformation that is accepted by the ΔpH system, (ii) remain unfolded and able to engage the thylakoid Sec pathway for import, in analogy with the Tat class II leader peptides described in the present study, or (iii) be transported in an unfolded state by the Tat pathway. Hynds et al. (16) favored the third scenario, but the other two possibilities could not be ruled out on the basis of their data.
The ability to promote genetically oxidative protein folding selectively in the cytoplasm or in the periplasmic space enabled us to examine the requirements for export competence of proteins that (i) do not contain cofactors and (ii) exhibit well characterized folding kinetics. Comparison of AP accumulation in the periplasmic fraction of WT E. coli mutant strains that enable the formation of disulfide bonds in the cytoplasm and their dsbA derivatives (Table 2) revealed that for all eight leader peptides oxidative folding in the cytoplasm is a prerequisite for Tat export. Sone et al. (21) demonstrated that partially oxidized AP lacking the Cys-168-Cys-178 disulfide is proteolytically unstable and is subject to rapid degradation by DegP in vivo. Thus, the stable periplasmic accumulation of the AP fusions in our studies (in a degP+ strain background) as well as the trypsin stability of the AP domain of ssFdnG-AP in the osmotic shock fraction (Fig. 2) indicate that the exported form of the AP fusion must be fully oxidized. On the basis of these considerations, we conclude that the Tat pore is capable of translocating AP that is fully oxidized and contains the native disulfide bonds. At present it is not possible to ascertain whether the protein is exported as a dimer or whether the association of the oxidized, folded monomers occurs after translocation into the periplasmic space. However, because in vitro dimerization represents a slow step in the folding of AP (35), the latter mechanism seems more plausible.
Table 2.
Periplasmic alkaline phosphatase activity in cells expressing Tat leader peptide-AP fusions*
| FdnG | MDVSRRQFFKICAGGMAGTTVAALGFAPKQALAQ |
| FdoG | MQVSRRQFFKICAGGMAGTTAAALGFAPSVALAE |
| HyaA | MNNEETFYQAMRRQGVTRRSFLKYCSLAATSLGLGAGMAPKIAWAL |
| TorA | MNNNDLFQASRRRFLAQLGGLTVAGMLGPSLLTPRRATAA |
| DmsA | MKTKIPDAVLAAEVSRRGLVKTTAIGGLAMASSALTLPFSRIAHAV |
| SufI | MSLSRRQFIQASGIALCAGAVPLKASAA |
| YacK | MQRRDFLKYSVALGVASALPLWSRAVFAA |
| YcbK | MDKFDANRRKLLALGGVALGAAILPTPAFAT |
| Leader peptide | Periplasmic AP activity
|
|||||
|---|---|---|---|---|---|---|
| DHA C:red/P:red | DHB4 C:red/P:ox | DR473 C:ox/P:ox | DRA C:ox/P:red | DRB (DR473 tatB∷kan) C:ox/P:ox | DRC (DR473 tatC∷spec) C:ox/P:ox | |
| Δ2–20† | 43 | 56 (52) | 69 (7) | 57 (8) | 56 (8) | 47 (9) |
| FdnG‡ | 32 | 45 (48) | 420 (42) | 371 (38) | 30 (5) | 6 (2) |
| FdoG | 55 | 62 (54) | 446 (53) | 385 (50) | 17 (2) | 11 (2) |
| HyaA‡ | 17 | 28 (46) | 187 (61) | 185 (59) | 65 (23) | 20 (4) |
| TorA‡ | 42 | 51 (51) | 437 (26) | 403 (28) | 38 (3) | 17 (1) |
| DmsA‡ | 35 | 123 (>90) | 280 (23) | 175 (14) | 112 (12) | 74 (9) |
| SufI‡ | 75 | 323 (>90) | 459 (30) | 381 (19) | 263 (15) | 308 (18) |
| YacK‡ | 25 | 102 (>90) | 153 (18) | 135 (12) | 185 (19) | 80 (9) |
| YcbK | 27 | 95 (>90) | 178 (22) | 137 (13) | 170 (18) | 81 (10) |
All signal sequences were fused in frame to E. coli AP(Δ1–22), and the fusions were inserted into vector pTrc99A (see Experimental Procedures).
Cells were treated with 100 mM iodoacetamide immediately after harvesting to prevent formation of disulfide bonds during subsequent sampling processing. AP activity was calculated as the amount of p-nitrophenyl phosphate hydrolyzed (in micromoles) per minute at 25°C and pH 8.0. Reported values for AP activity are the average of three separate measurements from two independent experiments (n = 6). Standard error is <10% for all reported data. Values in parentheses indicate the percentage of the total enzymatic activity in the periplasmic fraction.
AP with a Δ2–20 in the native leader peptide.
Leader peptide carries a c-region positive charge.
For 4/8 leader peptides (class I: ssFdnG, ssFdoG, ssHyaA, and ssTorA) export occurs exclusively by the Tat pathway as evidenced by the fact that (i) AP enzymatic activity in the osmotic shock fraction is unaffected by the presence or absence of DsbA and (ii) export was completely abolished by the substitution of the invariant RR in the leader peptide by KK, in tatC mutants, and, with the exception of ssHyaA, also in tatB mutants. The role of TatB in the export of hydrogenases has been the subject of some controversy (36, 37). TatB is dispensable in the export of the nonphysiological substrate colicin (38), and the TatB plant homologue, Hcf106, has also been reported to be nonessential for the secretion of the chloroplast 16- and 23-kDa subunits of the oxygen-evolving complex (39).
In cells expressing AP fusions to class I leader peptides, ≈26% (for ssTorA) to ≈60% (for ssHyaA) of the total enzymatic activity in cell lysates was found in the periplasmic fraction. The observed efficiency of AP export is typical of native Tat substrates and of Tat fusions to heterologous proteins (14, 29, 32, 40, 41). If indeed the Tat translocator exports preferentially oxidized monomeric AP, as discussed above, then the enzymatically active protein remaining in the cytoplasm could represent dimerized protein that is incompatible with export. In addition, we noticed the accumulation of various amounts of inactive, export-incompetent AP in the cytoplasm. The inactive cytoplasmic AP in C:ox cells was trypsin-sensitive, and its abundance was elevated at higher growth temperatures and on expression of the fusions from high-copy-number plasmids and dependent on the leader peptide used (data not shown). In contrast, we found no evidence of inactive AP fusions in the periplasmic fraction for any of the constructs tested. In some cases (e.g., for ssFdnG-AP in Fig. 2D), a portion of the AP fusion protein in the periplasm migrated as a higher-molecular-weight band having the expected electrophoretic mobility of the precursor. This high-molecular-weight species was processed by trypsin to the mature band, presumably because of the presence of sensitive sites within Tat leader peptides (26).
Class II Tat leader peptides (ssDmsA, ssSufI, ssYacK, and ssYcbK) afforded some AP export from cells with a reducing cytoplasm. This AP activity was not abolished in C:red tatB or tatC mutants, indicating that export of the protein to the periplasm did not involve the Tat machinery. When class II leader peptide fusions were expressed in C:ox/P:ox cells, the amount of active AP in the periplasm increased by between 42% (for ssSufI-AP) and >100% (for ssDmsA). However, inactivation of dsbA in the C:ox strain DR473 resulted in a significant reduction in periplasmic AP. The two possible explanations for these results are either that translocation of misfolded or unfolded AP occurs by the Tat apparatus or that class II leader peptides can engage both the Sec and Tat translocons. The finding that the export of AP activity in C:red cells is eliminated in a secA51(ts) mutant strain grown at the nonpermissive temperature (E. coli MM52 at 42°C) supports the latter hypothesis. The ability of class II leader peptides to engage both the Sec and the Tat secretion apparatus is in agreement with the recent studies by Sanders et al. (42), who found that the export of heterologous cytochrome c fused to the HyaA leader peptide could proceed by the Sec pathway in the absence of the heme cofactor, whereas when the heme was enzymatically ligated in the cytoplasm, Tat became the predominant export route. Earlier studies had indicated that the presence of a positive charge at the C terminus of Tat leader peptides can serve as a “Sec-avoidance” signal (5, 43). Although ssDmsA, ssSufI, and ssYacK all possess a charged amino acid in this region it appears that the C-domain-positive charge alone is insufficient for preventing promiscuous export by the Sec pathway, at least when these leaders are fused to AP.
Several earlier efforts to export AP by the Tat pathway had been unsuccessful (25, 27–29). In some cases AP activity could be detected in the periplasmic space but export was independent of the Tat pathway (29). The analysis presented here now explains these previous observations. Although the export of AP by Tat depends on oxidative folding in the cytoplasm, certain leader peptides are able to engage both the Sec and Tat pathways. The export flux among these two pathways presumably depends on the folding kinetics of the polypeptide, with folded molecules exported through the Tat system and proteins that have not yet reached a critical state in folding able to access the Sec pathway.
In addition to AP, we find that fusions to other multidisulfide proteins, such as scFv and FAB antibody fragments, become competent for export by the Tat pathway only when expressed in strains where the cytoplasm promotes oxidative protein folding. The accumulation of FAB antibody in the periplasm could only be observed in C:ox strains and depended on an intact Tat apparatus and on the presence of a functional leader peptide on the heavy chain. The simplest explanation for these findings is that the FAB first assembles in the cytoplasm where the two chains become linked by an intermolecular disulfide followed by Tat export of the folded protein. Increasing the folding efficiency of FAB in the cytoplasm through the coexpression of ΔssDsbC greatly enhanced the export efficiency of FAB. The export of fully folded Fab molecules into the periplasm, even though only one of the two chains contains a leader peptide, is representative of the “hitchhiker” mode of export whereby a leaderless polypeptide is exported by its association with a second polypeptide that can engage the secretion apparatus (13).
The results outlined above provide conclusive evidence that (i) in the absence of folding in the cytoplasm there is no protein export by the Tat pathway and that (ii) folded proteins of at least ≈43 kDa (the size of monomeric AP), as well as proteins that must assemble subunits in the cytoplasm, are exported in a folded conformation by the bacterial Tat pathway in vivo. The inability to export AP, scFv, and FAB proteins in the absence of disulfide bonds indicates the existence of a quality-control mechanism that is an integral part of Tat translocation machinery. Because only the Tat proteins are required for translocation, the folding quality-control mechanism must be inherent to the TatABCE proteins in the membrane. For example, a cytoplasmic domain of a Tat component may be able to function as a chaperone by binding to exposed hydrophobic regions in unfolded protein intermediates. Alternatively, interactions with aberrantly folded proteins could inhibit either the assembly of the Tat translocation pore or the gating of the pore once formed. The isolation of Tat mutations that enable the export of unfolded AP fused to class I leader peptides in C:red strains will help distinguish between these two mechanisms and is currently underway.
Finally, we note that the results of the present study have significant ramifications for biotechnology. First, we have demonstrated that proteins with structural disulfide bonds can be efficiently exported by the Tat pathway. Many industrially important secreted proteins including enzymes contain multiple disulfide bonds that are required for proper folding. Such proteins may be secreted by the Tat pathway in C:ox strains. The advantage of using the Tat pathway is that it may alleviate problems with cell toxicity often encountered when proteins exported by the Sec pathway become “stuck” in the translocon. Second, the folding quality control feature of the Tat pathway may be exploited in combinatorial library-screening experiments to “weed out” mutant polypeptides that can not fold properly.
Acknowledgments
We thank Tracy Palmer and Jon Beckwith for providing E. coli strains, and Tracy Palmer, Steve Theg, and Lluis Masip for critical review of the manuscript. This work was supported by grants from the Foundation for Research and by National Institutes of Health Grant GM-55090 (to G.G.). D.T. was supported by a National Science Foundation graduate fellowship.
Abbreviations
- Tat
twin-arginine translocation
- Sec
general secretory pathway
- ss
signal sequence
- TorA
trimethylamine N-oxide reductase
- AP
alkaline phosphatase
- scFv
single-chain antibody
- BPTI
bovine pancreatic trypsin inhibitor
- DHFR
dihydrofolate reductase
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Pugsley A P. Microbiol Rev. 1993;57:50–108. doi: 10.1128/mr.57.1.50-108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stuart R A, Neupert W. Nature. 2000;406:575–577. doi: 10.1038/35020668. [DOI] [PubMed] [Google Scholar]
- 3.Settles A M, Yonetani A, Baron A, Bush D R, Cline K, Martienssen R. Science. 1997;278:1467–1470. doi: 10.1126/science.278.5342.1467. [DOI] [PubMed] [Google Scholar]
- 4.Weiner J H, Bilous P T, Shaw G M, Lubitz S P, Frost L, Thomas G H, Cole J A, Turner R J. Cell. 1998;93:93–101. doi: 10.1016/s0092-8674(00)81149-6. [DOI] [PubMed] [Google Scholar]
- 5.Cristobal S, de Gier J W, Nielsen H, von Heijne G. EMBO J. 1999;18:2982–2990. doi: 10.1093/emboj/18.11.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dalbey R E, Robinson C. Trends Biochem Sci. 1999;24:17–22. doi: 10.1016/s0968-0004(98)01333-4. [DOI] [PubMed] [Google Scholar]
- 7.Keegstra K, Cline K. Plant Cell. 1999;11:557–570. doi: 10.1105/tpc.11.4.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Settles A M, Martienssen R. Trends Cell Biol. 1998;8:494–501. doi: 10.1016/s0962-8924(98)01387-7. [DOI] [PubMed] [Google Scholar]
- 9.Halbig D, Hou B, Freudl R, Sprenger G A, Klosgen R B. FEBS Lett. 1999;447:95–98. doi: 10.1016/s0014-5793(99)00269-0. [DOI] [PubMed] [Google Scholar]
- 10.Mori H, Cline K. J Biol Chem. 1998;273:11405–11408. doi: 10.1074/jbc.273.19.11405. [DOI] [PubMed] [Google Scholar]
- 11.Wexler M, Bogsch E G, Klosgen R B, Palmer T, Robinson C, Berks B C. FEBS Lett. 1998;431:339–342. doi: 10.1016/s0014-5793(98)00790-x. [DOI] [PubMed] [Google Scholar]
- 12.Halbig D, Wiegert T, Blaudeck N, Freudl R, Sprenger G A. Eur J Biochem. 1999;263:543–551. doi: 10.1046/j.1432-1327.1999.00536.x. [DOI] [PubMed] [Google Scholar]
- 13.Rodrigue A, Chanal A, Beck K, Muller M, Wu L F. J Biol Chem. 1999;274:13223–13228. doi: 10.1074/jbc.274.19.13223. [DOI] [PubMed] [Google Scholar]
- 14.Thomas J D, Daniel R A, Errington J, Robinson C. Mol Microbiol. 2001;39:47–53. doi: 10.1046/j.1365-2958.2001.02253.x. [DOI] [PubMed] [Google Scholar]
- 15.Clark S A, Theg S M. Mol Biol Cell. 1997;8:923–934. doi: 10.1091/mbc.8.5.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hynds P J, Robinson D, Robinson C. J Biol Chem. 1998;273:34868–34874. doi: 10.1074/jbc.273.52.34868. [DOI] [PubMed] [Google Scholar]
- 17.Robinson C, Bolhuis A. Nat Rev Mol Cell Biol. 2001;2:350–356. doi: 10.1038/35073038. [DOI] [PubMed] [Google Scholar]
- 18.Sargent F, Bogsch E G, Stanley N R, Wexler M, Robinson C, Berks B C, Palmer T. EMBO J. 1998;17:3640–3650. doi: 10.1093/emboj/17.13.3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Derman A I, Beckwith J. J Bacteriol. 1995;177:3764–3770. doi: 10.1128/jb.177.13.3764-3770.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Derman A I, Prinz W A, Belin D, Beckwith J. Science. 1993;262:1744–1747. doi: 10.1126/science.8259521. [DOI] [PubMed] [Google Scholar]
- 21.Sone M, Kishigami S, Yoshihisa T, Ito K. J Biol Chem. 1997;272:6174–6178. doi: 10.1074/jbc.272.10.6174. [DOI] [PubMed] [Google Scholar]
- 22.Levy R, Weiss R, Chen G, Iverson B L, Georgiou G. Protein Expression Purif. 2001;23:338–347. doi: 10.1006/prep.2001.1520. [DOI] [PubMed] [Google Scholar]
- 23.Chen G, Hayhurst A, Thomas J G, Harvey B R, Iverson B L, Georgiou G. Nat Biotechnol. 2001;19:537–542. doi: 10.1038/89281. [DOI] [PubMed] [Google Scholar]
- 24.Ritz D, Beckwith J. Annu Rev Microbiol. 2001;55:21–48. doi: 10.1146/annurev.micro.55.1.21. [DOI] [PubMed] [Google Scholar]
- 25.Berks B C. Mol Microbiol. 1996;22:393–404. doi: 10.1046/j.1365-2958.1996.00114.x. [DOI] [PubMed] [Google Scholar]
- 26.Kebir M O, Kendall D A. Biochemistry. 2002;41:5573–5580. doi: 10.1021/bi015798t. [DOI] [PubMed] [Google Scholar]
- 27.Reinartz M, Tschape J, Bruser T, Truper H G, Dahl C. Arch Microbiol. 1998;170:59–68. doi: 10.1007/s002030050615. [DOI] [PubMed] [Google Scholar]
- 28.Sambasivarao D, Scraba D G, Trieber C, Weiner J H. J Bacteriol. 1990;172:5938–5948. doi: 10.1128/jb.172.10.5938-5948.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stanley N R, Sargent F, Buchanan G, Shi J, Stewart V, Palmer T, Berks B C. Mol Microbiol. 2002;43:1005–1021. doi: 10.1046/j.1365-2958.2002.02797.x. [DOI] [PubMed] [Google Scholar]
- 30.Bessette P H, Aslund F, Beckwith J, Georgiou G. Proc Natl Acad Sci USA. 1999;96:13703–13708. doi: 10.1073/pnas.96.24.13703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Belin P, Boquet P L. C R Acad Sci Ser III. 1993;316:469–473. [PubMed] [Google Scholar]
- 32.Angelini S, Moreno R, Gouffi K, Santini C, Yamagishi A, Berenguer J, Wu L. FEBS Lett. 2001;506:103–107. doi: 10.1016/s0014-5793(01)02890-3. [DOI] [PubMed] [Google Scholar]
- 33.Santini C L, Bernadac A, Zhang M, Chanal A, Ize B, Blanco C, Wu L F. J Biol Chem. 2001;276:8159–8164. doi: 10.1074/jbc.C000833200. [DOI] [PubMed] [Google Scholar]
- 34.Schnell D J, Blobel G. J Cell Biol. 1993;120:103–115. doi: 10.1083/jcb.120.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Walker K W, Gilbert H F. J Biol Chem. 1994;269:28487–28493. [PubMed] [Google Scholar]
- 36.Chanal A, Santini C, Wu L. Mol Microbiol. 1998;30:674–676. doi: 10.1046/j.1365-2958.1998.01095.x. [DOI] [PubMed] [Google Scholar]
- 37.Sargent F, Stanley N R, Berks B C, Palmer T. J Biol Chem. 1999;274:36073–36082. doi: 10.1074/jbc.274.51.36073. [DOI] [PubMed] [Google Scholar]
- 38.Ize B, Gerard F, Zhang M, Chanal A, Voulhoux R, Palmer T, Filloux A, Wu L F. J Mol Biol. 2002;317:327–335. doi: 10.1006/jmbi.2002.5431. [DOI] [PubMed] [Google Scholar]
- 39.Roy L M, Barkan A. J Cell Biol. 1998;141:385–395. doi: 10.1083/jcb.141.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.DeLisa M P, Samuelson P, Palmer T, Georgiou G. J Biol Chem. 2002;277:29825–29831. doi: 10.1074/jbc.M201956200. [DOI] [PubMed] [Google Scholar]
- 41.Yahr T L, Wickner W T. EMBO J. 2001;20:2472–2479. doi: 10.1093/emboj/20.10.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sanders C, Wethkamp N, Lill H. Mol Microbiol. 2001;41:241–246. doi: 10.1046/j.1365-2958.2001.02514.x. [DOI] [PubMed] [Google Scholar]
- 43.Bogsch E, Brink S, Robinson C. EMBO J. 1997;16:3851–3859. doi: 10.1093/emboj/16.13.3851. [DOI] [PMC free article] [PubMed] [Google Scholar]