Abstract
The biosynthesis of most secondary metabolites in different bacteria is strongly depressed by inorganic phosphate. The two-component phoR-phoP system of Streptomyces lividans has been cloned and characterized. PhoR showed all of the characteristics of the membrane-bound sensor proteins, whereas PhoP is a member of the DNA-binding OmpR family. Deletion mutants lacking phoP or phoR-phoP, were unable to grow in minimal medium at low phosphate concentration (10 μM). Growth was fully restored by complementation with the phoR-phoP genes. Both S. lividans ΔphoP and ΔphoR-phoP deletion mutants were unable to synthesize extracellular alkaline phosphatase (AP) as shown by immunodetection with anti-AP antibodies and by enzymatic analysis, suggesting that the PhoR-PhoP system is required for expression of the AP gene (phoA). Synthesis of AP was restored by complementation of the deletion mutants with phoR-phoP. The biosynthesis of two secondary metabolites, actinorhodin and undecylprodigiosin, was significantly increased in both solid and liquid medium in the ΔphoP or ΔphoR-phoP deletion mutants. Negative phosphate control of both secondary metabolites was restored by complementation with the phoR-phoP cluster. These results prove that expression of both phoA and genes implicated in the biosynthesis of secondary metabolites in S. lividans is regulated by a mechanism involving the two-component PhoR-PhoP system.
Phosphate control of the biosynthesis of antibiotics and many other types of secondary metabolites is a well known phenomenon (1–5), although the molecular mechanism by which this control is exerted is unknown (6, 7). Expression of genes encoding enzymes for the biosynthesis of secondary metabolites is negatively regulated by phosphate, and formation of the corresponding transcripts occurs only under phosphate-limiting conditions (8–10); but, surprisingly, nothing is known about the molecular mechanism of phosphate control of expression of the corresponding biosynthetic genes (11).
In Escherichia coli and Bacillus subtilis the genes belonging to the pho regulon, including the alkaline phosphatase (AP) gene (phoA) and the phosphate-specific transport (pst) genes, are regulated by a two-protein system consisting of a phosphate-sensor protein, PhoR, and a transcriptional activator protein, PhoB (named PhoP in B. subtilis; refs. 12–14). The sensor kinase PhoR is self-phosphorylated under phosphate starvation conditions (forming PhoR-P) that transfer its phosphate group to dephosphorylated PhoB. The phosphorylated PhoB transcriptional factor (PhoB-P) activates the expression of ≈30 different genes by binding to the pho boxes located upstream of the phosphate-regulated genes (12). Expression of phoA and other members of the pho regulon occurs under phosphate limitation when the transcriptional activator is available in its phosphorylated form.
An important question is whether the control of the biosynthesis of secondary metabolites in actinomycetes is exerted by the same mechanism as the control of AP and other genes involved in phosphate metabolism, or whether control of secondary metabolism proceeds by an entirely different molecular mechanism.
Recently, we cloned the AP gene (phoA, a member of the pho regulon) of Streptomyces griseus IMRU 3570 (the producer of the polyene macrolide candicidin) as a model to study the phosphate control of primary metabolism in actinomycetes (15), but this strain is not amenable to genetic manipulation. It was, therefore, of great interest to elucidate the presence in the model actinomycetes Streptomyces lividans and Streptomyces coelicolor A3 (2) of a two-component signal transduction system similar to phoR-phoB of E. coli and its possible involvement in the control of the biosynthesis of either primary metabolites, secondary metabolites, or both.
Experimental Procedures
Bacterial Strains and Plasmids.
The bacterial strains and plasmids used are listed in Table 1. The phoR-phoP genes of S. coelicolor were cloned from cosmid D8A (16) and those of S. lividans were cloned by PCR (see below). The phoR-phoP cluster from S. coelicolor was subcloned in a 2.9-kb EcoRI–StuI fragment into the EcoRI–SmaI site of pBluescript KS+. This plasmid, named pBSphoRP, was digested with EcoRV–Eco72I and religated, obtaining plasmid pBSphoP (Fig. 1). pBSΔphoRP is a derivative of pBSphoRP in which a 1,651-bp SalI–SphI fragment from phoR and phoP genes was deleted and replaced by the aphII (kanamycin resistance) gene in the opposite orientation (Fig. 1). Similarly, pBSΔphoP was derived from pBSphoP by deleting a 434-bp NruI–SphI fragment from the phoP gene and replacing it by the aphII gene. Plasmids pHZΔphoRP and pHZΔphoP contain the DNA inserts from pBSΔphoRP and pBSΔphoP, respectively, cloned into the recombination-prone unstable vector pHZ1351 (17). The plasmid pIJphoRP contains pBSphoRP cloned into the positive selection vector pIJ699 (17).
Table 1.
Bacterial strains, cosmids, and plasmids
| Strains/plasmids | Characteristics* | Ref. |
|---|---|---|
| Strains | ||
| Streptomyces lividans JI 1326 | Wild type | John Innes† |
| S. lividans ΔphoP | JI 1326, phoP∷aphII | This work |
| S. lividans ΔphoRP | JI 1326, phoRP∷aphII | This work |
| S. lividans phoP+ | ΔphoP, harboring the plasmid pIJphoRP | This work |
| S. lividans phoRP+ | ΔphoRP, harboring the plasmid pIJphoRP | This work |
| E. coli DH5α | F′ φ80 dlacZ ΔM15 Δ(lacZYA-argF) U169 recA1 endA1 hsdR17 (r− m+) supE44 medλ− thi-1 gyrA relA1 | 33 |
| Plasmids/cosmids | ||
| D8A | 41.8-kb DNA fragment from S. coelicolor cloned into Supercos-1 | 16 |
| pBluescript KS+ | Cloning vector, ColE1 origin, Ampr | Stratagene |
| pBSphoP | Eco72I (1,690)–StuI (2,887) fragment carrying phoP gene, cloned into pBluescript KS+ (EcoRV–SmaI) | This work |
| pBSphoRP | EcoRI (from the cosmid D8A)–StuI (2,887) fragment carrying phoR-phoP genes, cloned into pBluescript KS+ (EcoRI–SmaI) | This work |
| pBSΔphoP | Like pBSphoP, with aphII (SphI–Ecl136II) insertion in SphI (2,433)–NruI (1,999) | This work |
| pBSΔphoRP | Like pBSphoRP, with aphII (SphI–SalI) insertion in SphI (2,433)–SalI (782) | This work |
| pGEM-T-easy | Vector system for the cloning of PCR products | Promega |
| pHZ1351 | Highly unstable Sti+ vector, useful for gene replacement in Streptomyces | 17 |
| pHZΔphoP | Insert from pBSΔphoP (SacI–HindIII), cloned into pHZ1351 (SacI–HindIII) | This work |
| pHZΔphoRP | Insert from pBSΔphoRP (SacI–HindIII), cloned into pHZ1351 (SacI–HindIII) | This work |
| pIJ699 | High-copy number cloning vector for Streptomyces | 17 |
| pIJphoRP | Streptomyces–E. coli bifunctional plasmid, with pBSphoRP cloned into pIJ699 | This work |
Numbers in parentheses represent positions in the sequence of cosmid D8A of S. coelicolor.
Collection of microorganisms of the John Innes Institute.
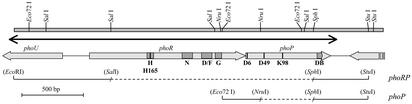
Figure 1.
Restriction map and organization of the DNA region containing the phoR-phoP cluster of S. lividans. The same organization was found in the genome of S. coelicolor A3(2). The double-headed arrow indicates the sequenced DNA fragment. Thick arrows show the size and orientation of the phoU, phoR, and phoP genes. The H, N, D/F, and G boxes in phoR and the DNA-binding (DB) domain in phoP are indicated as shaded regions, and the highly conserved residues His-165 in phoR and Asp-6, Asp-49, and Lys-98 in phoP are shown as a vertical solid line. Horizontal solid lines represent DNA fragments cloned into different plasmids. The discontinuous line indicates the deleted DNA fragment replaced by aphII. phoRP refers to the DNA insert into plasmids pBSphoRP, pBSΔphoRP, pHZΔphoRP, and pIJphoRP. phoP refers to the insert in plasmids pBSphoP, pBSΔphoP, and pHZΔphoP.
S. lividans Deletion Mutants.
The S. lividans ΔphoP and ΔphoRP mutants were constructed by gene replacement using pHZΔphoP and pHZΔphoRP plasmids, respectively. Protoplasts of S. lividans 1326 were transformed and kanamycin-resistant thiostrepton-sensitive transformants were selected. For complementation studies the ΔphoP and ΔphoRP mutants were transformed with pIJphoRP yielding S. lividans phoP+ and phoRP+ strains, respectively.
Culture Conditions.
All strains were grown in solid asparagine-minimal medium (18) supplemented with KH2PO4 in low (10 μM; phosphate starvation conditions) or high concentration (5 mM) and in R5 solid medium with low (0.37 mM) or high (1.85 mM) phosphate concentrations (17).
For studies on the production of secondary metabolites and on AP secretion, seed cultures were made in 100 ml of YED medium [(10 g/liter yeast extract/10 g/liter d-glucose (pH 7.0)] in 500-ml baffled flasks inoculated with ≈107 spores. Five milliliters of a seed culture grown for 30 h at 30°C and 250 rpm were centrifuged, washed twice with NaCl 0.9%, and used to inoculate 100 ml of R5 liquid medium (17).
Thiostrepton was added at final concentrations of 5 and 50 μg/ml in liquid and solid media, respectively, to the cultures of S. lividans phoP+ and phoRP+ strains.
DNA Procedures.
Genomic DNA was isolated by standard procedures (17). For cloning the S. lividans phoR-phoP genes, a 2.7-kb DNA fragment was amplified by PCR using two primers based in S. coelicolor genome sequence: O1, 5′-GGTCCATCGCGTCGTCGTCCTGCTC-3′, and O2, 5′-GGTCCCGCCCTTCGCCGTGTCC-3′. This fragment was cloned into pGEM-T-easy (Promega) vector and sequenced by using pUC/M13 forward and reverse primers in an ABI PRISM 310 genetic analyzer (Perkin–Elmer).
Assay and Immunodetection of AP.
The AP activity in culture supernatants was measured as described (15). In addition, AP was observed by Western blot analysis. The proteins in liquid R5 culture filtrates of 72 h were precipitated with ammonium sulfate at a final concentration of 80% saturation at 4°C (15). The protein pellet was collected by centrifugation, resuspended in 10 mM Tris⋅HCl (pH 8.0), applied to a desalting Sephadex G-25 column, and eluted with the same buffer. Samples were analyzed by SDS/PAGE and blotted to a PVDF membrane. The AP protein band was detected with anti-AP rabbit antibodies raised against pure S. griseus AP (15).
Actinorhodin and Undecylprodigiosin Determination.
Actinorhodin and undecylprodigiosin were extracted and determined spectrophotometrically as described by Kieser et al. (17).
Phosphate Uptake.
S. lividans cultures were grown in liquid YED medium for 20 h (30°C, 250 rpm). The collected cells were washed twice with NaCl 0.9% and transferred to asparagine-minimal medium without inorganic phosphate. After stabilization of the cell suspension for 6 h, 32P-labeled Na2HPO4 (Amersham Biosciences) was added (2 × 105 cpm/ml). The uptake of phosphate was quantified as described (1).
Results
Cloning of the phoU-phoR-phoP Genes of S. lividans.
The S. lividans phoU-phoR-phoP region was cloned by PCR using oligonucleotides O1 and O2 as primers. Sequence analysis of a 2.7-kb region of the S. lividans cloned DNA fragment revealed that this region has the same organization as the homologous region described in the S. coelicolor genome. The S. lividans phoR-phoP cluster is arranged in a head-to-tail organization and phoU is located in opposite orientation (Fig. 1). The three genes appear to be expressed from a bidirectional promoter region both in S. lividans and in S. coelicolor. This particular two-component system has been designated PhoR-PhoP (see below) by using the B. subtilis designation, rather than that of E. coli, to avoid confusion, because the phoB name has previously been used to nominate one of the four AP genes (phoA, phoB, phoC, and phoD) occurring in S. coelicolor (15). The nucleotide sequence and deduced amino acid sequences for the PhoR and PhoP proteins were very similar in S. lividans and S. coelicolor. All of the amino acid changes in S. lividans with respect to the S. coelicolor sequence, Leu-24 to Pro, Ala-62 to Ser, Gly-86 to Ser, and Ala-384 to Pro, were found in PhoR. No amino acid changes were observed in the S. lividans PhoP with respect to the S. coelicolor homologous protein. The separation of phoR and phoP genes was only 5 bp in both Streptomyces species, strongly suggesting that both genes are cotranscribed.
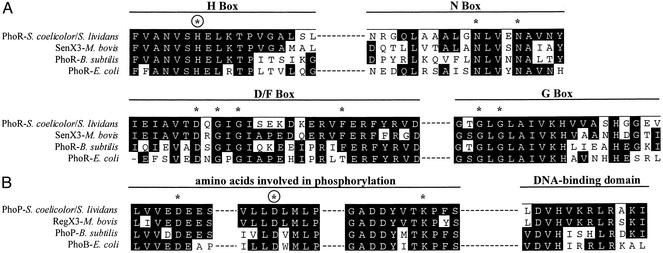
The phoR product of S. lividans is a 426-aa protein with a deduced molecular mass of 45.4 kDa. It contains a hypothetical transmembrane domain in its amino-terminal region that may serve as an anchor to the cytoplasmic membrane. The deduced protein shows all of the characteristics of the sensor proteins of the PhoR family, including the H box (residues His-159 to Leu-176, His-165 being the putative phosphorylation site) and motifs N (residues Asn-267 to Tyr-285), D/F (residues Ile-307 to Asp-334) and G (residues Gly-343 to Val-362) (ref. 19; Fig. 2).
Figure 2.
(A) Conserved motifs in the H box, N box, D/F box, and G box of S. lividans PhoR as compared with the homologous boxes of sensor proteins of two-component systems of M. bovis, B. subtilis, and E. coli. Identical amino acids are shown as white on black letters. Many differences in the four sequences correspond to functionally conserved amino acids. (B) Conserved amino acids involved in phosphorylation of PhoP of S. lividans or S. coelicolor and in the DNA-binding domain. Asterisks indicate essential amino acids for phosphorylation. Encircled asterisks show the amino acid (H or D) that is phosphorylated.
The phoP gene product is a protein of 223 aa with a deduced molecular weight of 24.7 kDa and belongs to the OmpR family (20). It shows the conserved amino acids in the amino-terminal region of PhoB(PhoP)-like proteins including Asp-6, Asp-49 (the putative phosphorylation site), and Lys-98 (21). The carboxyl terminal region includes a DNA-binding motif (residues 190–201 in the S. lividans PhoP protein; Fig. 2).
Deletion of the phoP and phoR-phoP Genes.
Two different deletion mutants, ΔphoP and ΔphoRP, were constructed by in vivo gene replacement in S. lividans (Fig. 3). The ΔphoP mutant was obtained by using the construction pHZΔphoP. After recombination, transformants resistant to kanamycin and sensitive to thiostrepton were selected. The recombination was confirmed by Southern hybridization using probes corresponding to phoR-phoP and aphII (Fig. 3). The DNA fragment deleted from phoP in the ΔphoP mutant removed most of the conserved motifs of PhoP (except Asp-6), including the putative phosphorylation site Asp-49 (Fig. 1).
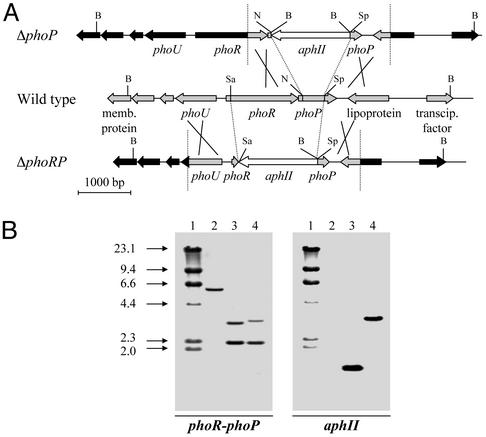
Figure 3.
(A) Strategies for obtaining the S. lividans deletion mutants ΔphoP and ΔphoRP by gene replacement with the kanamycin resistance gene (aphII) (white arrow). The organization of genes in the wild-type strain is shown in the center, and the crossing over resulting in ΔphoP and ΔphoRP mutants are in the upper and lower lines, respectively. DNA fragments originating from the integrating plasmid are shown in gray, and the remaining genes (in black) correspond to the wild-type chromosome. B, BamHI; N, NruI; Sa, SalI; Sp, SphI. (B) Hybridization patterns of BamHI-digested DNA from S. lividans deletion mutants ΔphoP and ΔphoRP (lanes 3 and 4, respectively) as compared with that of the parental wild-type S. lividans strain (lane 2), using the phoR-phoP (Left) and the aphII (Right) probes. Note the lack of hybridization of the wild type with the aphII probe and the change in size of the hybridizing bands in the deletion mutants. DNA molecular size markers (lane 1; lambda DNA/HindIII digested) in kilobases are shown on the left.
Similarly, a 1,651-bp DNA fragment was removed from the phoR-phoP region by in vivo recombination with plasmid pHZΔphoRP. The deleted region in the ΔphoRP mutant comprised all motifs of the PhoR protein involved in the self-phosphorylation, including site His-165, and all of the residues involved in the phosphorylation of PhoP.
Southern analysis of both ΔphoP and ΔphoRP mutants revealed the presence of a single copy of the aphII marker integrated at the expected location in the phoR-phoP locus (Fig. 3).
The S. lividans Mutants ΔphoP and ΔphoRP Do Not Grow at Low Phosphate Concentrations.
In E. coli, several inorganic- and organic-phosphate transport proteins (e.g., the pst system) are known to be regulated by PhoR-PhoP. As shown in Fig. 4, both the ΔphoP and ΔphoRP mutants were unable to grow in Streptomyces asparagine-minimal medium containing 10 μM inorganic phosphate, whereas the parental S. lividans strain grew well at this low phosphate concentration. At high phosphate concentration (5 mM) the deletion mutants were able to grow, showing more intense red pigmentation than the parental strain.
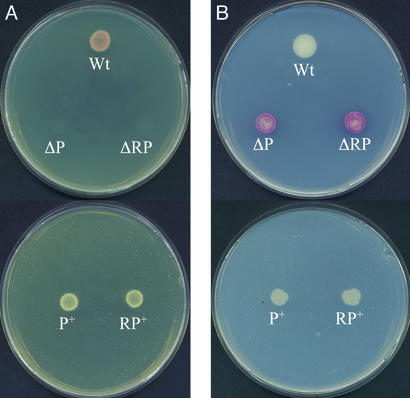
Figure 4.
(A) Lack of growth of the S. lividans ΔphoP and ΔphoRP mutants in asparagine-minimal medium at low (10 μM) phosphate concentration. (B) Growth in the same medium at 5 mM inorganic phosphate. Note that transformants phoP+ and phoRP+ grow well at either low or high phosphate concentrations. The ΔphoP and ΔphoRP deletion mutants overproduce red pigment, whereas the phoP+ and phoRP+ complemented transformants do not. Wt, wild type; ΔP, ΔphoP; ΔRP, ΔphoRP; P+, phoP+; RP+, phoRP+. Strains phoP+ and phoRP+ were grown in the presence of thiostrepton.
Growth of both S. lividans ΔphoP and ΔphoRP mutants was restored by complementation with plasmid pIJphoRP containing both phoR-phoP genes and its complete promoter region. Transformation of ΔphoP and ΔphoRP with this plasmid gave rise to S. lividans strains phoP+ and phoRP+ that were perfectly able to grow on asparagine-minimal medium. The complemented S. lividans phoP+ and phoRP+ transformants did not show the red color of the ΔphoP or ΔphoRP mutants in asparagine-minimal medium with 5 mM phosphate (Fig. 4B).
The ΔphoRP Mutant Is Partially Defective in Inorganic Phosphate Uptake.
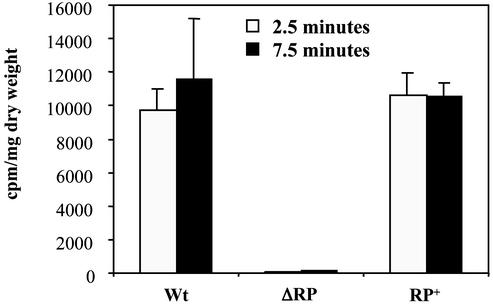
To confirm that the lack of growth in asparagine-minimal medium with reduced phosphate concentration was due to a defect in phosphate uptake, the cellular incorporation of 32P-labeled phosphate during 2.5 or 7.5 min was determined. The parental strain takes inorganic phosphate rapidly and reaches an equilibrium level at 7.5 min, whereas the ΔphoRP mutant showed a drastic reduction in the uptake of inorganic phosphate (Fig. 5). The complemented mutant phoRP+ showed the same behavior as the wild-type strain, although the uptake of phosphate at short times (e.g., 2.5 min) was higher, probably because of the positive effect on transport of the oversynthesis of the phosphate regulatory proteins in the transformant containing the phoR-phoP genes in a multicopy plasmid.
Figure 5.
Uptake of 32P-labeled phosphate after 2.5 min (open vertical bars) or 7.5 min (black bars) in the wild-type S. lividans (Wt), the ΔphoRP deletion mutant (ΔRP), and the complemented transformant phoRP+ (RP+). Thin vertical bars indicate SD of the mean.
The PhoR-PhoP System Regulates AP.
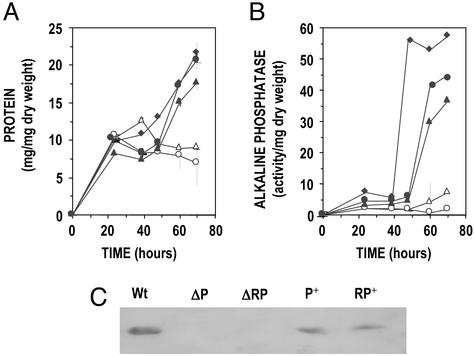
When the parental S. lividans and the deletion mutants ΔphoP and ΔphoRP were compared on R5 liquid medium that supports a good AP formation, the total amount of extracellular protein was similar until 48 h of cultivation, but it was reduced in cultures at 60 and 72 h when extracellular enzymes are known to accumulate (Fig. 6A). The extracellular AP was drastically reduced in the S. lividans ΔphoP and ΔphoRP mutants when compared with the parental strain (Fig. 6B). Complementation of each deletion mutant with the phoR-phoP cluster restored the formation of AP.
Figure 6.
Total extracellular protein (A) and AP activity (B) in R5 liquid cultures of wild-type S. lividans (filled diamonds), the ΔphoP (open triangles), and ΔphoRP (open circles) mutants and the complemented strains phoP+ (filled triangles) and phoRP+ (filled circles). Experiments were done in triplicate. Thin vertical bars indicate SD of the mean. phoP+ and phoRP+ strains were grown in the presence of thiostrepton. (C) Western blot analysis using anti-AP antibodies of extracellular proteins of S. lividans wild-type (Wt), ΔphoP (ΔP), ΔphoRP (ΔPR), phoP+ (P+), and phoRP+ (RP+) strains grown in R5 liquid medium. Note that the AP is restored by complementation of the deletion mutants with the phoR-phoP cluster (transformants P+ and RP+).
These results were confirmed by Western analysis using anti-AP antibodies. The immunoblot analysis clearly indicated that the extracellular AP is absent in the ΔphoP and ΔphoRP mutants (Fig. 6C) and is restored by complementation with the phoR-phoP cluster, i.e., formation of AP depends on the PhoR-PhoP regulatory system.
The Production of Actinorhodin and Undecylprodigiosin Is Strongly Regulated by the PhoR-PhoP System.
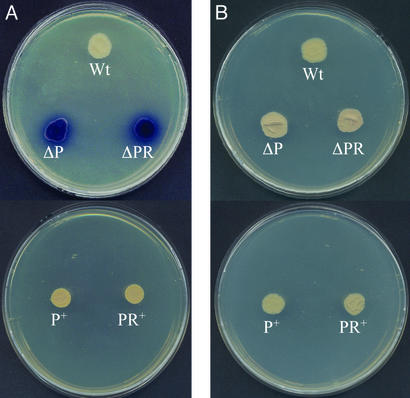
S. lividans contains the actinorhodin gene cluster but, unlike S. coelicolor, it does not synthesize actinorhodin efficiently. When the S. lividans ΔphoP and ΔphoRP mutants were compared with the parental S. lividans strain, it was observed that both mutants were actinorhodin hyperproducers in solid R5 medium with low phosphate (0.37 mM; Fig. 7A). The overexpression of the actinorhodin genes was suppressed in both deletion mutants by complementation with the phoR-phoP cluster (strains P+ and RP+ in Fig. 7). When excess phosphate was added to the R5 medium the production of actinorhodin was still controlled by phosphate in the ΔphoP and ΔphoRP mutants, as shown by the reduction in blue pigment production (Fig. 7B), indicating that the regulatory effect of PhoR-PhoP is bypassed at high phosphate concentrations.
Figure 7.
Growth and actinorhodin production on R5 solid medium containing low (0.37 mM; A) or high (1.85 mM; B) phosphate concentration of the wild-type S. lividans, the ΔphoP and ΔphoRP deletion mutants, and the complemented transformants S. lividans phoP+ and phoRP+. Note that there is a very high actinorhodin (blue pigment) production in the ΔphoP and ΔphoRP deletion mutants (A) growing under a low phosphate concentration but not in the plate with a high phosphate level. The complemented transformants phoP+ and phoRP+ did not overproduce actinorhodin, even under low phosphate concentrations. Abbreviations in the photograph are as in Fig. 4. phoP+ and phoRP+ strains were grown in the presence of thiostrepton.
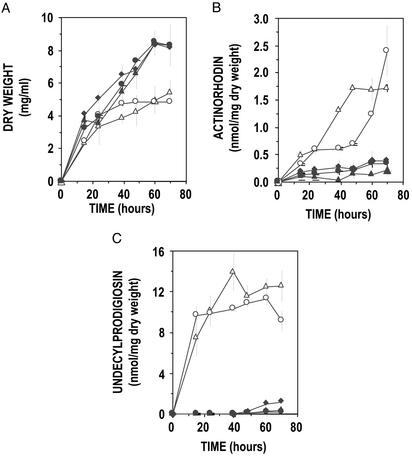
The biosynthesis of actinorhodin and undecylprodigiosin was quantified in R5 liquid cultures. As shown in Fig. 8, both the S. lividans ΔphoP and ΔphoRP overproduced actinorhodin and undecylprodigiosin, reaching levels up to 2.5 nmol (1.59 μg) of actinorhodin and 14 nmol (5.49 μg) of undecylprodigiosin per milligram of dry weight. The oversynthesis of both antibiotics in the deletion mutants in liquid R5 medium was again suppressed by complementation with the phoR-phoP regulatory genes (Fig. 8).
Figure 8.
Growth (A), actinorhodin (B), and undecylprodigiosin (C) production in R5 liquid cultures of S. lividans wild type (filled diamonds), ΔphoP (open triangles), ΔphoRP (open circles), and the complemented transformants phoP+ (filled triangles) and phoRP+ (filled circles). Note that high levels of actinorhodin and undecylprodigiosin are synthesized in the ΔphoP and ΔphoRP deletion mutants but not in the complemented phoP+ and phoRP+ strains. Experiments were done in triplicate. Thin vertical bars indicate SD of the mean. phoP+ and phoRP+ strains were grown in the presence of thiostrepton.
Discussion
Many secondary metabolites, including antibiotics, pigments, immunomodulators, and siderophores, produced by actinomycetes and other Gram-positive and -negative bacteria are strongly repressed by high inorganic phosphate concentrations in the culture medium (2). Production of these compounds has to be performed under phosphate-limiting conditions (4).
Two-component systems (22) provide a conserved mechanism for the coordinate control of gene expression in response to different environmental factors (19, 23). Three putative phoR-phoP-like systems, were found by computer search in the genome of S. coelicolor (24). We have found that the phoR-phoP system showing the best match to the homologous E. coli system is linked to a putative phoU gene (encoding a modulator of phosphate transduction; ref. 12) and that the three genes are expressed from a bidirectional promoter region. The phoU-phoR-phoP organization is very similar in S. lividans and S. coelicolor. The best match of the S. lividans and S. coelicolor PhoR and PhoP proteins were, respectively, with the Mycobacterium bovis two-component SenX3 (41.6% identical amino acids) and the RegX3 (72.7% identity) system of unknown function (25).
PhoR of S. lividans corresponds to the sensor protein of two-component systems. Sensor proteins autophosphorylate in a conserved histidine (corresponding to His-165 in the S. lividans protein; ref. 25). In addition to His-165, PhoR also contains all of the consensus motifs of the kinase domains (26).
PhoP is closely related to the ROII subfamily of response regulators (26). It contains a helix-turn-helix motif in its C-terminal region similar to that identified in the OmpR protein (27, 28) that is also present in the RegX3 protein of M. bovis (25). In the two-component systems the mechanisms of signal transduction involve, first, autophosphorylation of the sensor protein (PhoR), followed by transfer of the phosphate group onto a conserved aspartate residue of the response regulator (Asp-49 in the S. lividans PhoP; Figs. 1 and 2).
The AP encoded by phoA is a prototype member of the pho regulon and is known to be repressed by phosphate in Streptomyces species (15). As shown in this article, PhoA is not formed in the S. lividans ΔphoP and ΔphoRP deletion mutants and its synthesis is restored by complementation of the deletion mutants with the phoR-phoP cluster. These results indicate that formation of PhoA is under positive control of the PhoR-PhoP system as occurs in E. coli (12, 29) and B. subtilis (30, 31). The drastic reduction in phosphate uptake in the S. lividans ΔphoRP deletion mutant suggests that some components of a high-affinity phosphate transport system also depend on the PhoR-PhoP system. However, growth of the deletion mutants occurs at high phosphate concentration, indicating that alternative phosphate transport is still possible in the absence of the PhoR-PhoP system at high phosphate concentrations.
An interesting finding of this work was the observation that production of both actinorhodin and undecylprodigiosin is greatly increased in the ΔphoP or ΔphoRP mutants either in solid or liquid medium. These results clearly indicate that the biosynthesis of secondary metabolites is under the control of the PhoR-PhoP system. The biosynthesis of actinorhodin is known to be regulated negatively by phosphate (5). The wild-type S. lividans, unlike S. coelicolor, produces very low levels of actinorhodin and undecylprodigiosin. The large increase in the production of those secondary metabolites in the ΔphoP and ΔphoRP mutants indicates that these secondary metabolites are repressed by a form of PhoP (i.e., a negative regulation) that accumulates in response to high phosphate concentration in the culture medium. Alternatively, the PhoR-PhoP system may activate the formation of a specific repressor protein for phosphate-controlled promoters by a cascade mechanism. In either case, the ΔphoP and ΔphoRP deletion mutants are largely derepressed in secondary metabolite biosynthesis because they lack a functional PhoP protein.
These regulatory proteins may serve to integrate different input signals (32) that affect secondary metabolite biosynthesis, e.g., the negative role exerted by polyphosphate through autophosphorylation of the polyphosphate glucokinase (7).
The kinetics of expression of different regulatory proteins in S. coelicolor has been reported recently (11). One of the genes studied SCD8A.03 corresponds to phoP in S. lividans. The pattern of expression of this gene coincides with that of SCD8A.01c that corresponds to phoU in our cloned fragment. Both genes are induced (derepressed) at the middle of the growth phase coinciding with the depletion of phosphate in the growth medium.
In summary, all available evidence suggest that inorganic phosphate concentrations control the pattern of expression of a series of genes involved in the biosynthesis of different types of antibiotics in S. lividans and probably in S. coelicolor and other Streptomyces species. The PhoR-PhoP system may serve as a general transduction system for the expression of genes involved in secondary metabolism.
Acknowledgments
We thank D. Hopwood and H. Kieser for providing S. coelicolor cosmid D8A. This work was supported by grants from the Ramón Areces Foundation (Madrid) and the Agencia de Desarrollo Económico de la Junta de Castilla y León, Spain (Proyecto Genérico 10-2/98/LE/0003).
Abbreviation
- AP
alkaline phosphatase
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AJ544582).
References
- 1.Liras P, Villanueva J R, Martín J F. J Gen Microbiol. 1977;102:269–277. doi: 10.1099/00221287-102-2-269. [DOI] [PubMed] [Google Scholar]
- 2.Martín J F, Demain A L. Microbiol Rev. 1980;44:230–251. doi: 10.1128/mr.44.2.230-251.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Masuma R, Tanaka Y, Tanaka H, Omura S. J Antibiot (Tokyo) 1986;39:1557–1564. doi: 10.7164/antibiotics.39.1557. [DOI] [PubMed] [Google Scholar]
- 4.Martín J F. In: Regulation of Secondary Metabolism in Actinomycetes. Shapiro S, editor. Boca Raton, FL: CRC; 1989. pp. 213–237. [Google Scholar]
- 5.Doull J L, Vining L C. Appl Microbiol Biotechnol. 1990;32:449–454. doi: 10.1007/BF00903781. [DOI] [PubMed] [Google Scholar]
- 6.Liras P, Asturias J A, Martín J F. Trends Biotechnol. 1990;8:184–189. doi: 10.1016/0167-7799(90)90170-3. [DOI] [PubMed] [Google Scholar]
- 7.Chouayekh H, Virolle M-J. Mol Microbiol. 2002;43:919–930. doi: 10.1046/j.1365-2958.2002.02557.x. [DOI] [PubMed] [Google Scholar]
- 8.Asturias J A, Liras P, Martín J F. Gene. 1990;93:79–84. doi: 10.1016/0378-1119(90)90139-i. [DOI] [PubMed] [Google Scholar]
- 9.Martín J F, Marcos A T, Martín A, Asturias J A, Liras P. In: Phosphate in Microoganisms. Torriani-Gorini A, Yagil E, Silver S, editors. Washington, DC: Am. Soc. Microbiol.; 1994. pp. 140–147. [Google Scholar]
- 10.McDowall K J, Thamchaipenet A, Hunter I S. J Bacteriol. 1999;181:3025–3032. doi: 10.1128/jb.181.10.3025-3032.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang J, Lih C J, Pan K H, Cohen S N. Genes Dev. 2001;15:3183–3192. doi: 10.1101/gad.943401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Torriani-Gorini A. In: Phosphate in Microorganisms. Torriani-Gorini A, Yagil E, Silver S, editors. Washington, DC: Am. Soc. Microbiol.; 1994. pp. 1–4. [Google Scholar]
- 13.Hulett F M. Mol Microbiol. 1996;19:933–939. doi: 10.1046/j.1365-2958.1996.421953.x. [DOI] [PubMed] [Google Scholar]
- 14.Pragai Z, Harwood C R. Microbiology. 2002;148:1593–1602. doi: 10.1099/00221287-148-5-1593. [DOI] [PubMed] [Google Scholar]
- 15.Moura R S, Martín J F, Martín A, Liras P. Microbiology. 2001;147:1525–1533. doi: 10.1099/00221287-147-6-1525. [DOI] [PubMed] [Google Scholar]
- 16.Redenbach M, Kieser H M, Denapaite D, Eichner A, Cullum J, Kinashi H, Hopwood D A. Mol Microbiol. 1996;21:77–96. doi: 10.1046/j.1365-2958.1996.6191336.x. [DOI] [PubMed] [Google Scholar]
- 17.Kieser T, Bibb M J, Buttner M J, Chater K F, Hopwood D A. Practical Streptomyces Genetics. Norwich, U.K.: The John Innes Foundation; 2000. [Google Scholar]
- 18.Martín J F, McDaniel L E. Biochim Biophys Acta. 1975;411:186–194. doi: 10.1016/0304-4165(75)90298-6. [DOI] [PubMed] [Google Scholar]
- 19.Stock J B, Surette M G, Levit M, Park P. In: Two-Component Signal Transduction. Hoch J A, Silhavy T J, editors. Washington, DC: Am. Soc. Microbiol.; 1995. pp. 25–52. [Google Scholar]
- 20.Mizuno T, Tanaka I. Mol Microbiol. 1997;24:665–667. doi: 10.1046/j.1365-2958.1997.3571723.x. [DOI] [PubMed] [Google Scholar]
- 21.Stock J B, Ninfa A J, Stock A M. Microbiol Rev. 1989;53:450–490. doi: 10.1128/mr.53.4.450-490.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoch J A, Silhavy T J. Two-Component Signal Transduction. Washington, DC: Am. Soc. Microbiol.; 1995. [Google Scholar]
- 23.Alex L A, Simon M I. Trends Genet. 1994;10:133–138. doi: 10.1016/0168-9525(94)90215-1. [DOI] [PubMed] [Google Scholar]
- 24.Bentley S D, Chater K F, Cerdeno-Tarraga A M, Challis G L, Thomson N R, James K D, Harris D E, Quail M A, Kieser H, Harper D, et al. Nature. 2002;417:141–147. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- 25.Himpens S, Locht C, Supply P. Microbiology. 2000;146:3091–3098. doi: 10.1099/00221287-146-12-3091. [DOI] [PubMed] [Google Scholar]
- 26.Parkinson J S, Kofoid E C. Annu Rev Genet. 1992;26:71–112. doi: 10.1146/annurev.ge.26.120192.000443. [DOI] [PubMed] [Google Scholar]
- 27.Martínez-Hackert E, Stock A M. J Mol Biol. 1997;269:301–312. doi: 10.1006/jmbi.1997.1065. [DOI] [PubMed] [Google Scholar]
- 28.Kondo H, Nakagawa A, Nishihira J, Nishimura Y, Mizuno T, Tanaka I. Nat Struct Biol. 1997;4:28–31. doi: 10.1038/nsb0197-28. [DOI] [PubMed] [Google Scholar]
- 29.Makino K, Shinagawa H, Amemura M, Kawamoto T, Yamada M, Nakata A. J Mol Biol. 1989;210:551–559. doi: 10.1016/0022-2836(89)90131-9. [DOI] [PubMed] [Google Scholar]
- 30.Hulett F M, Lee J, Shi L, Sun G, Chesnut R, Sharkova E, Duggan M F, Kapp N. J Bacteriol. 1994;176:1348–1358. doi: 10.1128/jb.176.5.1348-1358.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu W, Hulett M. J Bacteriol. 1997;179:6302–6310. doi: 10.1128/jb.179.20.6302-6310.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kang S G, Jin W, Bibb M, Lee K J. FEMS Microbiol Lett. 1998;168:221–226. doi: 10.1111/j.1574-6968.1998.tb13277.x. [DOI] [PubMed] [Google Scholar]
- 33.Hanahan D. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]