Abstract
Amyloid-β peptide (Aβ) accumulation in the brain is an early, toxic event in the pathogenesis of Alzheimer's disease (AD). Aβ is produced by proteolytic processing of a transmembrane protein, β-amyloid precursor protein (APP), by β- and γ-secretases. Mounting evidence has demonstrated that alterations in APP cellular trafficking and localization directly impact its processing to Aβ. Recent studies have shown that members of the low-density lipoprotein receptor family, including LRP, LRP1B, SorLA/LR11, and apolipoprotein E (apoE) receptor 2, interact with APP and regulate its endocytic trafficking. Another common feature of these receptors is their ability to bind apoE, which exists in three isoforms in humans and the presence of the ε4 allele represents a genetic risk factor for AD. In this review, we summarize the current understanding of the function of these apoE receptors with a focus on their role in APP trafficking and processing. Knowledge of the interactions between these distinct low-density lipoprotein receptor family members and APP may ultimately influence future therapies for AD.
Background
Alzheimer's disease (AD) is the most common cause of dementia among people age 65 and older. A diagnosis of AD is confirmed upon autopsy by the presence of characteristic lesions in specific regions of the brain, notably the hippocampus, amygdala, and association cortices of the frontal, temporal and parietal lobe of the cortex [1]. Fittingly, these affected regions are responsible for memory, emotion and decision making abilities, which are impaired in AD dementia. Lesions found in AD are deposits of amyloid plaques in the cerebrovasculature and parenchyma of the brain and intracellular neurofibrillary tangles. Amyloid plaques are either dense/fibrillar or diffuse in nature; fibrillar plaques are surrounded by dystrophic neurites, activated microglia, and reactive astrocytes, while diffuse plaques lack fibrils and are associated with few or no dystrophic neurites or altered glia.
A major component of the amyloid plaques found in AD is the ~4 kDa amyloid-β peptide (Aβ) [2], which is a cleavage product of the β-amyloid precursor protein (APP) [3]. Aβ ranges in size from 37 to 43 amino acids; however, Aβ42(43) may act as a pathogenic seed for fibrillar plaque formation since it is found in insoluble cores of fibrillar and diffuse plaques [4]. One current hypothesis known as the "amyloid hypothesis" postulates that increased Aβ production or reduced Aβ metabolism results in the formation of aggregated Aβ deposits leading to AD dementia (for review see [5]). In support of this idea, in vitro studies have demonstrated that Aβ42 aggregates and forms fibrils more rapidly and is more neurotoxic than Aβ40 [6-8]. In vivo, studies in mice demonstrate that expression of only human Aβ42 not Aβ40 results in overt amyloid pathology indicating a requirement for Aβ42 in Aβ plaque deposition and AD pathogenesis [9]. It is possible that aggregation of Aβ into fibrils is not the principal cause of AD dementia. Recent studies have also associated non-fibrillar assemblies of Aβ with neuronal injury, synaptic loss and dementia associated with AD. These Aβ assemblies, including soluble Aβ oligomers and intraneuronal Aβ deposits, have been hypothesized to act as an early, causal factor in the pathogenesis AD [1,10].
Genetic studies have confirmed that the processing of APP to Aβ is important for AD pathogenesis. Mapping of genes that segregate within families that develop early onset AD dementia (<65 years of age) led to the identification of a mutation in the APP gene on chromosome 21 [11]. Twenty-five separate pathogenic mutations within the APP gene have been described in familial cases of AD [12]. Several of these mutations increase APP processing to Aβ. Furthermore, persons affected by Down's syndrome (trisomy-21), who have three copies of chromosome 21 and therefore the APP gene, inevitably develop AD. Individuals who have Down's syndrome but lack the region of chromosome 21 containing the APP gene do not develop AD [13]. Together, these findings imply that a gain-of-function mechanism for APP is an important factor in the development of AD. Although genetic mutations in APP, have enhanced our understanding of the biology of AD, they only account for <1% of known AD cases [12]. For this reason, it is of interest to study proteins that interact with APP and modulate its processing to Aβ.
APP biology and processing
The APP gene is alternatively spliced to produce three major isoforms of 695, 751, and 770 amino acids in length. The two longer APP isoforms, APP751 and APP770, both contain a 56 amino acid Kunitz Protease Inhibitor (KPI) homology domain within their extracellular regions. APP is ubiquitously expressed throughout the body, but APP695, which lacks the KPI domain, is the predominant form found in neurons [14,15], and may play a role in neurite outgrowth and axonal sprouting (for review see [16]). Targeted deletion of the APP gene in mice produces no apparent phenotype, suggesting that other members of the APP family, such as amyloid precursor like proteins-1 (APLP1) and 2 (APLP2), can compensate for its function [17]. Triple knockout mice lacking APP, APLP1 and APLP2 die shortly after birth and have cranial abnormalities or cortical dysplasia, suggesting an essential function of this gene family in neuronal migration and brain development [18]. APP and APLP2 may have functional redundancy in development since a double knockout mouse of both genes displays a post-natal lethal phenotype while a double knockout mouse of APP and APLP1 is viable [19]. APP, however, is the only member of the family to contain the Aβ region and produce the AD-associated Aβ peptide.
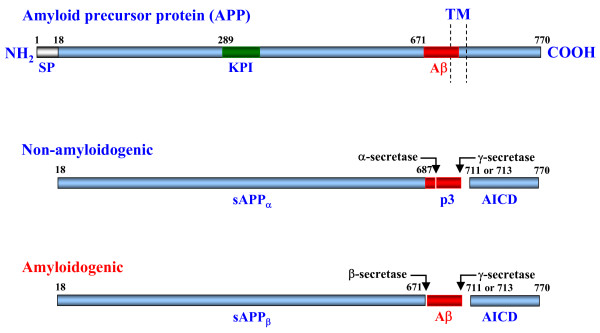
During its trafficking to the cell surface and in the endocytic pathway, APP can undergo proteolysis by secretase enzymes to release either the Aβ peptide (amyloidogenic pathway) or a shorter, non-toxic peptide known as sAPP (non-amyloidogenic pathway) [20] (Fig. 1). In the amyloidogenic pathway, APP is first cleaved at a β-secretase site by the enzyme BACE (β-site APP cleaving enzyme), which releases a soluble β-cleaved APP fragment (sAPPβ) and leaves a 99 amino acid C-terminal fragment (CTF) known as C99 attached to the membrane. C99 is subsequently cleaved by a γ-secretase/presenilin complex within its intramembrane region to release the Aβ peptide [1]. In the non-amyloidogenic pathway, APP is processed by an α-secretase that clips within the Aβ region, which results in the release of a soluble ~110–120 kDa α-cleaved APP fragment (sAPPα). This pathway also releases a CTF that is 83 amino acids in length known as C83. C83 can also be cleaved by γ-secretase to release p3. In both the amyloidogenic and non-amyloidogenic pathway, the γ-secretase cleavage of APP can also release an APP intracellular domain fragment (AICD). The processing of APP to these separate components may have important consequences in both diseased and normal physiology (for review see [21]). sAPP, which contains a KPI domain, has been identified as the serine protease inhibitor, protease nexin II (PNII), which inhibits the serine protease factor XIa in the blood coagulation cascade [22,23]. In addition, the C-terminal cleavage products of APP may activate gene transcription in concert with other proteins such as FE65 [24].
Figure 1.
Schematic representation of APP processing. APP is a type I transmembrane protein that can undergo two separate proteolytic pathways. In the non-amyloidogenic pathway, APP is processed by an α-secretase that clips within the Aβ region (thus precluding its formation), resulting in the release of a soluble ~110–120 kDa N-terminal APP fragment (sAPPα). This pathway also releases a CTF that is 83 amino acids in length (C83). C83 can also be cleaved by a γ-secretase to release a small, non-toxic 3 kDa fragment known as p3 and a γCTF known as APP intracellular domain (AICD). In the amyloidogenic pathway, APP is cleaved first by β-secretase releasing a sAPPβ fragment and leaving a 99 amino acid CTF attached to the membrane (C99). C99 is subsequently cleaved by a γ-secretase, within its intramembrane region to release the Aβ peptide and AICD. SP, Signal Peptide; KPI, Kunitz-type Proteinase Inhibitor domain.
Endocytic trafficking of APP
The presence of APP and APP cleavage products in clathrin-coated vesicles first suggested that the amyloidogenic processing of APP could occur in the endocytic pathway [25]. In 1994, Koo and Squazzo showed that cell surface radiolabeled APP releases Aβ, and that endocytosis of APP is also necessary for Aβ production. Inhibiting endocytosis of cell surface APP by potassium depletion, which disrupts the formation of clathrin lattices, or by C-terminal deletions of the APP tail, which removes important internalization motifs, leads to a decrease in Aβ production along with an increase in cell surface APP and sAPPα secretion [26].
The cytoplasmic tail of APP contains two motifs, YENPTY and YTSI, which are similar to the tyrosine-based NPXY and YXXØ (where X can be any amino acid and Ø is any amino acid with a bulky hydrophobic group) consensus endocytic motifs found in other well-known endocytic receptors such as the low-density lipoprotein and epidermal growth factor receptors [27]. Fusing the APP tail to the ectodomain of the transferrin receptor resulted in a functional endocytic receptor [28]. However, the endocytosis rate mediated by the APP tail is relatively slow at ~6%/minute [29]. Using metabolic labeling followed by cell surface biotinylation, Lai et al. (1995) demonstrated that deletion of the YENPTY motif in full length APP both decreased endocytosis and increased sAPPα secretion. Using a radioiodinated monoclonal antibody against APP to monitor its trafficking, it was also shown that deletion of the entire APP tail increased APP retention at the cell surface and sAPPα production by 2.5-fold when compared to wild-type APP [30]. Further mutational analyses indicated that the dominant endocytosis motif within the APP tail was the tetrapeptide YENP [31].
These studies establish a tight correlation between APP endocytosis and Aβ secretion. Substantial research efforts have examined the localization of the secretases involved in APP processing. The recent identification of the endosomally localized β-secretase, BACE, further supports the idea that Aβ is formed in the endocytic pathway. BACE localizes to the Golgi and endosomes and has optimal activity at the acidic pH found within endosomal compartments [32]. Components of the γ-secretase complex have been localized to the endoplasmic reticulum (ER), lysosomes and cell surface [33,34], whereas α-secretase activity is found primarily at the cell surface [35]. Since the secretases responsible for APP proteolysis have optimal enzymatic activity or distribution within specific cellular compartments, shifting APP to these compartments leads to an increased probability that APP will be cleaved by that secretase.
When cell surface APP is internalized to the endosomes, it is cleaved at the β-secretase site by BACE, and then returned to the cell surface or trafficked to the lysosome where it is cleaved by γ-secretase to produce Aβ. On the other hand, if APP accumulates at the cell surface it has a greater availability for α-secretase interaction and is cleaved to sAPPα via the non-amyloidogenic pathway. Although a large amount of work has been devoted to the study of APP trafficking within the endocytic pathway, there is only emerging evidence that APP-interacting receptors can affect its trafficking and processing. This review focuses on several members of the low density lipoprotein receptor family that have been shown to interact and influence the cellular localization and processing of APP to Aβ.
The low density lipoprotein (LDL) receptor family
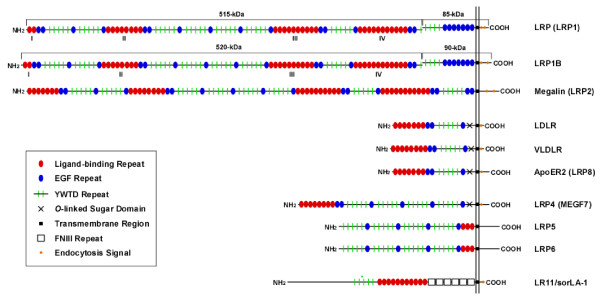
The LDL receptor family consists of a large class of cell surface receptors of diverse function. The family currently consists of the LDL receptor, LDL receptor-related protein (LRP, also known as LRP1), LDL receptor-related protein 1B (LRP1B), megalin/LRP2, the very low density lipoprotein receptor (VLDLR), apoE receptor 2 (apoER2), LRP4/MEGF7, LRP5, LRP6, and sorting protein-related receptor containing LDLR class A repeats (sorLA) or LR11 (Fig. 2). Although members of the LDL receptor family perform a variety of functions from cholesterol metabolism to cellular signaling, they share several features and structural motifs: 1) ligand-binding complement-type cysteine-rich repeats, 2) epidermal growth factor (EGF) receptor-like repeats, 3) YWTD β-propeller domains, 4) one or more endocytic motifs within their cytoplasmic domains such as NPXY, YXXØ or di-leucine motifs [36], and 5) binding of apolipoprotein E (apoE), a protein involved in cholesterol transport. Interestingly, apoE exists in three isoforms (E2, E3, and E4) in humans, and the presence of an ε4 allele represents a genetic risk factor for late-onset AD [37].
Figure 2.
Schematic representation of members of the LDLR family. Members of the LDLR family have diverse functions from cholesterol metabolism, Reelin and Wnt signalling, to intracellular transport. Despite multiple functions, they share common structural motifs, including ligand-binding repeats, epidermal growth factor (EGF) repeats, YWTD spacer domains, a single transmembrane domain and a short cytoplasmic domain containing conserved endocytic motifs. FN, fibronectin.
LRP
LRP is a multi-functional endocytic receptor that is highly expressed in the brain. At ~600 kDa in size, LRP is one of the largest receptors of the LDL receptor family. LRP is synthesized as a single polypeptide precursor and is cleaved by furin in the trans-Golgi network to produce a non-covalently associated heterodimer: a heavy chain (~515 kDa) containing the extracellular and ligand binding domains of LRP and a light chain (85 kDa) containing the transmembrane domain and cytoplasmic tail of LRP. The 515 kDa subunit contains four putative ligand binding domains (designated by Roman numerals I, II, III, and IV) [38] consisting of 2, 8, 10 and 11 cysteine-rich complement-type repeats, respectively. Each of these clusters is interspersed with EGF precursor homology repeats and YWTD repeats that form β-propeller modules (Fig. 2).
Two approaches have been successfully employed to identify which cluster of ligand binding repeats within LRP is responsible for binding to ligands. In one approach, LRP minireceptors were generated by fusing various clusters of ligand-binding repeats to the LRP light-chain and measuring their ability to mediate ligand internalization [39,40]. Another approach has been the creation of soluble recombinant receptor fragments that can be individually tested for ligand binding [41]. These studies have also been aided by the discovery of the ~39 kDa receptor-associated protein (RAP) that has high affinity for LRP (for review see [42]). RAP is normally an ER resident protein that serves as a molecular chaperone for members of the LDL receptor family to prevent premature binding of ligands and aid in their proper folding. Purified RAP is an excellent pharmacological tool because its exogenous application was found to universally antagonize binding of ligands to LRP as well as other members of the LDL receptor family. The majority of LRP ligands, including RAP, have been shown to bind to domains II and IV with equal affinity. No ligand has been demonstrated to bind domain I and only RAP and apoE were found to bind Domain III [40,43,44]. LRP was initially identified as a receptor for activated alpha-2-macroglobulin (α2M) [45]. Since then, LRP has been shown to bind and endocytose over 30 structurally and functionally diverse ligands. The function of these ligands can be divided into several classes, including lipoprotein metabolism, proteinases, proteinase-inhibitor complexes, blood coagulation factors, growth factors, extracellular matrix proteins, chaperones, and bacteria/viral proteins. Numerous ligands may bind LRP by interactions with either a combination of repeats within a single ligand-binding domain or several repeats from separate ligand-binding domains [46].
The 100 amino acid cytoplasmic tail of LRP contains two NPXY motifs, one YXXØ motif, and two di-leucine motifs. A unique feature of LRP is its rapid rate of endocytosis, with half of the receptors at the cell surface able to internalize within 30 seconds (t1/2 < 0.5 min) [47]. Other members of the LDL receptor family endocytose at much slower rates, e.g., megalin has a t1/2 ~ 4.8 min and the apoER2 has a t1/2 ~ 8.1 min [48]. Using site directed mutagenesis, Li et al. (2000) defined the YXXL motif and distal di-leucine repeat as the major endocytic motifs within the LRP tail. Although the ability of LRP to rapidly endocytose a wide variety of ligands suggests a primary function as a cargo transporter, several studies have found that the LRP cytoplasmic domain interacts with proteins involved in cell signaling, axonal transport, and glutamate receptor scaffolding. These adaptor proteins include disabled-1 (Dab1), FE65, JIP-1 and 2, and PSD-95 [49,50]. These findings indicate that LRP may have dual roles as both a signal transduction receptor and a major cargo transport receptor. LRP may also function in synaptic plasticity and memory via association with tissue-type plasminogen activator (tPA) [51], by influencing calcium influx via NMDA receptors [52], or interactions with ApoE and KPI-containing APP in the dentate gyrus [53].
LRP and Alzheimer's disease
Since LRP is a neuronal receptor for apoE, a well-known AD risk factor, LRP was investigated for its significance in AD pathology. From these studies, LRP has been linked to AD in several ways. First, LRP mediates the clearance of Aβ in vitro either by binding to Aβ itself or Aβ complexed to apoE, activated α2M, or lactoferrin [54-57]. Second, LRP and its ligands are found in amyloid plaques in AD brains and also in fibrillar amyloid plaques in a mouse model of AD [58-60]. Finally, several polymorphisms within the LRP gene on chromosome 12 have been associated with AD: a 5' tetranucleotide repeat, a single base pair change within exon 3 (C766T), and a weakly protective polymorphism in exon 6 [61-63]. It must be stated however, that a number of recent papers have discounted an association between the 5' tetranucleotide repeat of LRP and AD [64-66]. Conversely, the C→T change in exon 3 has been confirmed in additional studies and in distinct ethnic groups [65,67-69]. Although this silent polymorphism does not affect protein structure, an analysis of AD patients revealed that carriers of the T allele had a later age of onset than non-carriers. AD cases with C/T or T/T genotypes also had significantly higher levels of cortical LRP compared to carriers of the C/C genotype, indicating a possible protective effect of higher levels of LRP and/or the T allele [70]. Additional studies may be required as this polymorphism has been criticized as being only weakly correlated with AD [71]. Overall, these findings suggest that LRP could play a role in the development of AD pathology.
Interaction between LRP and APP
Although it is possible that LRP plays a role in the clearance of Aβ, it also alters the metabolism of Aβ via extracellular and intracellular interactions with the Aβ precursor, APP. Several studies have indicated that APP processing to Aβ is modified by LRP expression. In 1995, Kounnas et al. reported that LRP binds and internalizes secreted sAPPα, which contains a KPI domain. Soon after, it was demonstrated that cell surface KPI-containing APP complexed with epidermal growth factor binding protein (EGFBP) is internalized by LRP [72]. This internalization of APP was inhibited by the LRP antagonist, RAP, indicating that cell surface and secreted APP are degraded by a mutual pathway that requires LRP.
An intracellular interaction also exists between LRP and non-KPI containing APP through the cytoplasmic adaptor protein, FE65. FE65 contains a WW domain and two phosphotyrosine binding domains (PTB1 and PTB2), similar to the adaptor protein Shc. The PTB domains of FE65 specifically bind ψXNPXpY (where ψ is a hydrophobic residue and pY is phosphotyrosine) motifs within receptor tails [73]. APP binding to FE65 is not dependent on phosphorylation, but can be abolished by mutation of the first tyrosine within the YENPTY motif of APP [74]. Pull down experiments demonstrated that the amino-terminal PTB1 of FE65 binds to LRP and the carboxyl-terminal PTB2 of FE65 binds APP [50], suggesting that FE65 could act as an adaptor to complex these two proteins. A cytoplasmic interaction between APP and LRP, bridged by FE65, could further strengthen the association between LRP and KPI-containing forms of APP and also account for an association between non-KPI containing APP and LRP. These interactions between APP and LRP at cell surface and in the Golgi apparatus have been substantiated with cell surface biotinylation, co-immunoprecipitation, and fluorescence resonance energy transfer (FRET) experiments in cells overexpressing APP, LRP and FE65 [75,76].
To determine if disrupting the interaction between LRP and APP could influence the processing of APP to Aβ, Ulery et al. (2000) antagonized the extracellular interaction between cell surface APP and LRP with RAP. Cells expressing APP751 were incubated with RAP for five days. Remarkably, long-term treatment of cells with RAP caused an increase in cell surface APP and a decrease in Aβ production. In the same study, co-transfection of APP and LRP in LRP-deficient cells, led to a ~3-fold increase in Aβ levels in the media compared to media from cells transfected with APP alone. These data demonstrate that LRP expression can influence APP processing to Aβ, possibly via an extracellular interaction between LRP and APP.
In a study utilizing LRP+/- or LRP-/- mouse fibroblasts expressing APP751, Pietzrik et al. (2002) demonstrated that cells endogenously expressing LRP or transfected with an LRP C-terminal fragment have increased Aβ levels and decreased sAPPα levels compared to LRP-null cells. These studies indicate that the cytoplasmic domain of LRP alone is sufficient for its effect on APP processing. In the same study, it was demonstrated that cells expressing LRP have a higher ratio of intracellular to cell surface APP compared to LRP-null cells, suggesting that the internalization rate of APP is enhanced with LRP expression. Interestingly, expression of an LRP C-terminal fragment bearing a Y→A mutation within the distal NPXY motif did not decrease sAPPα levels. Since this tyrosine is also important for LRP endocytosis [47], it is possible that mutation of this residue influenced the endocytosis of this LRP fragment, which would also affect APP endocytosis and processing. Studies in our laboratory found that mutations within this distal NPXY motif of LRP as well as the leucine within the YXXL endocytic motif increased cell surface levels of LRP and also resulted in an accumulation cell surface APP [77]. Interestingly, we found that only when both LRP and APP were overexpressed together that there was a net accumulation of extracellular Aβ. When LRP alone was overexpressed, endogenous Aβ levels in the media were lower likely due to the ability of LRP to bind and endocytose Aβ.
Ye et al. (2005) recently reported that application of ApoE4 to cells expressing KPI-lacking APP increases APP endocytosis and Aβ levels [78]. This effect was abolished when cells were co-incubated with RAP or when expression of LRP was reduced using small interference RNA. These results suggest that the binding of ApoE to LRP cause levels of Aβ to increase. Although this study provides an interesting link between the pathogenic allele of ApoE, LRP, and APP endocytosis and processing, it is unclear how the binding of ApoE4 to LRP influences APP processing. Future studies should determine if ApoE binding to LRP alters LRP endocytosis, localization, or its ability to interact with APP. Since these studies were performed with APP which lacks an extracellular interaction with LRP, it is possible that ApoE4 binding would enhance an intracellular interaction between LRP and APP.
Our laboratory has also demonstrated a significant link between LRP expression and Aβ levels in vivo [79]. Expression of a functional LRP minireceptor in neurons of an amyloid mouse model of AD was associated with an increase in soluble Aβ levels and memory deficits in aged mice. These changes in APP trafficking and processing appear to be linked to the rapid rate of LRP endocytosis. Altogether these findings indicate that interactions between LRP and APP have the ability to modulate Aβ levels.
Recent findings that LRP interacts with presenilin 1 and BACE and is a substrate for γ-secretase and β-secretase [80-82] suggest that the alterations in APP processing by LRP could be more complex than originally considered. LRP may influence APP access to secretases through interactions with the secretases themselves or by changing the compartmentalization of APP. In the case of γ-secretase activity, LRP C-terminal fragments may compete with APP as a substrate for cleavage [82]. In the case of β-secretase activity, LRP may co-operatively aid interactions between APP and BACE possibly in lipid raft domains [80,83]. The recent generation of a mouse that selectively lacks LRP in differentiated neurons may provide a useful model to further analyze the affect of LRP on amyloid deposition [84].
LRP1B
LRP1B was first characterized in 2000 as a novel LDL receptor family member with extensive homology to LRP [85]. LRP1B shares 59% amino acid identity with LRP. The overall structure of LRP1B is like LRP with similar spacing of its 32 cysteine-rich ligand binding repeats into four clusters of putative ligand binding domains, eight EGF-precursor domains and two NPXY motifs within its cytoplasmic tail. There are two major structural differences between LRP and LRP1B. LRP1B contains one additional ligand binding repeat within ligand binding domain IV and also has a unique 33 amino acid sequence within in its cytoplasmic tail [85,86] (See Fig. 2).
LRP1B was initially named LRP-deleted in tumors (LRP-DIT) because in a study of non-small cell lung cancer cell lines (NSCLC) the LRP1b gene was deleted or inactivated in 40% of the cell lines [85]. Since then, inactivation of the LRP1b gene has also been described in grade 3 (G3) urothelial cancers [87] and esophageal squamous cell carcinomas [88]. Due to its loss of function in cancer cell lines, it is hypothesized that LRP1B acts as a tumor suppressor. Additionally, the expression pattern of LRP1B suggests that it may have an important function in the brain. Tissue expression analysis established that LRP1B was expressed primarily in the brain, thyroid and salivary gland [86] and in situ hybridization of tissue sections determined that LRP1B mRNA expression in the brain was highest in the dentate gyrus of the hippocampus and ventral to the fourth ventricle [89].
In order to determine the trafficking and function of LRP1B, our laboratory created a minireceptor consisting of its fourth putative ligand binding domain, full transmembrane domain, and cytoplasmic tail [86]. This LRP1B minireceptor, designated mLRP1B4, contains both of the main structural differences between LRP and LR1B – an extra ligand binding repeat and a cytoplasmic 33 amino acid repeat. RAP binds both mLRP1B4 and the analogous LRP minireceptor (mLRP4). Several other LRP ligands also bind LRP1B, including complexes of urokinase plasminogen activator and plasminogen activator inhibitor type-1 [86]. Utilizing 125I-labeled RAP, Liu et al. (2001) measured the endocytosis rate of the LRP1B minireceptor. LRP1B exhibits a much slower rate of endocytosis (t1/2 > 10 min) compared to LRP (t1/2 < 0.5 min) [86], which may influence the cellular distribution and catabolism of ligands [86,90].
Since LRP1B shares several ligands with LRP, we sought to determine whether LRP1B could also interact with APP. If the fast endocytosis rate of LRP is responsible for facilitating APP processing to Aβ [91,92], we hypothesized that an interaction between APP and LRP1B, which has a much slower rate of endocytosis, would lead to decreased Aβ production. Using an LRP1B minireceptor, we found that mLRP1B4 and APP form an immunoprecipitable complex [93]. Furthermore, mLRP1B4 bound and facilitated the degradation of a soluble isoform of APP containing a Kunitz proteinase inhibitor (KPI) domain, but not soluble APP lacking a KPI domain. A functional consequence of mLRP1B4 expression was a significant accumulation of APP at the cell surface, which is likely related to the slow endocytosis rate of LRP1B. More importantly, mLRP1B4 expressing cells that accumulated cell surface APP produced less Aβ and secreted more soluble APP. Consistent with our finding of decreased Aβ levels, mLRP1B4 transfected cells also had 40% less β-CTF to full-length APP compared to empty vector transfected cells [93].
To determine whether β-secretase processing was a limiting factor to the production of Aβ in mLRP1B4 expressing cells, we transiently transfected the β-cleaved APP fragment, C99, into mLRP1B4 or empty vector transfected cells. We still detected less Aβ in the media of mLRP1B4 cells compared to empty vector transfected cells, indicating that alterations of APP/C99 trafficking rather than changes in β-secretase activity likely contributed to the decreased levels of Aβ found in mLRP1B4 expressing cells [93].
Using an antibody against the C-terminus of LRP1B, we confirmed the expression LRP1B at the protein level in the cortex, hippocampus, and cerebellum. Interestingly, we detected the highest levels of LRP1B in the cerebellum which is a region that is relatively unaffected in AD [93]. Future studies are still needed to address if LRP1B levels are altered in human brain during normal and pathological states. Since these studies suggest its expression may decrease the extracellular release of Aβ, examination of the regulation of LRP1B may have important applications to AD therapy.
SorLA/LR11
SorLA/LR11 was first described in 1996 as a ~250 kDa receptor containing 11 putative ligand-binding complement-type repeats, 5 YWTD domains, and a vacuolar protein sorting 10 protein (vps10p) domain, which is homologous to a yeast receptor that transports proteins between the late Golgi and a prevacuolar endosome-like compartment [94]. Abundant mRNA expression of sorLA/LR11 was found in human brain, spinal cord, and testis [95]. The functions of SorLA/LR11 are not entirely known. It shares structural and functional similarities with the LDL receptor family in its ability to bind and internalize RAP, ApoE, and lipoprotein lipase [95,96]; however, its endocytosis rate is much slower than LRP. A chimeric receptor of the cytoplasmic and transmembrane domains of SorLA/LR11 endocytosed only 60% of bound ligand after 15 min of incubation at 37°C [96]. Since SorLA/LR11 is also considered to be part of the family of VPS10 domain containing receptors, its main role may be to chaperone proteins as an intracellular sorting receptor. SorLA/LR11 does not appear to play an important role in development since SorLA/LR11 receptor-deficient mice were viable and fertile [97,98].
It was hypothesized that SorLA/LR11 expression may play a preventative role in AD dementia because SorLA/LR11 transcripts were down-regulated in lymphoblasts from AD patients. Also less SorLA/LR11 protein was found in neurons from AD brains by immunocytochemistry and Western blotting [99]. To determine the relevance of SorLA/LR11 expression for Aβ processing, Andersen et al. (2005) examined if SorLA/LR11 could interact with APP and affect its cellular localization [97]. Using several methods, including surface plasmon resonance analysis, immunocytochemistry and co-immunoprecipitation, they showed that SorLA/LR11 and APP interact. Further studies elucidated that APP interacts with the extracellular cluster of ligand-binding complement-type repeats in SorLA/LR11 similar to APP binding to the ligand-binding repeats in LRP [77,100]. Unlike the interaction between LRP and LRP1B, the KPI domain of APP was not necessary for an extracellular interaction with SorLA/LR11 [100].
Expression of SorLA/LR11 shifted the localization of APP in membrane fractions from the ER and plasma membrane to the cis-Golgi and early endosomes [97]. In a neuronal cell line, SorLA/LR11 expression also reduced surface-localized APP and resulted in an accumulation of mature, glycosylated APP [97]. These alterations in APP trafficking were associated with a decrease in APP processing to Aβ. In this study, APP was overexpressed with SorLA/LR11 and levels of full-length APP appeared to be stable. In another study in which APP was not overexpressed, SorLA/LR11 expression also reduced levels of full-length endogenous APP although the authors note that APP mRNA levels were unchanged [101]. Whether APP was overexpressed or not, SorLA/LR11 expression significantly decreased Aβ levels [97,101]. These findings suggest that expression of SorLA/LR11 alters APP trafficking through the secretory pathway, prevents APP trafficking to the cell surface and may influence its turnover. In agreement with the correlative studies in humans and in vitro findings, ablation of SorLA/LR11 expression in mice increased endogenous murine Aβ levels in the cerebral cortex by ~30% compared to controls [97].
Although SorLA/LR11 expression altered the trafficking of APP within intracellular compartments, it did not change the endocytosis rate of APP as in the case of LRP [92,102]. Interestingly, SorLA/LR11 was found to interact with BACE and appeared to compete with interactions between APP and BACE in the Golgi apparatus [102]. Additionally, it has been reported that SorLA/LR11 is also proteolytically processed by γ-secretase [103] and proposed to act as a competitive substrate with APP for γ-secretase activity. Altogether these current findings indicate that SorLA/LR11 regulates APP trafficking into discrete intracellular compartments and also influences its interactions with secretases. Decreased levels of SorLA/LR11 as found in AD could result in increased interactions between APP and BACE which would enhance its processing to C99. With less SorLA/LR11 expression, C99 would have a greater chance to be cleaved by γ-secretase resulting in higher levels of Aβ. Since SorLA/LR11 limits APP processing to Aβ it would be important to know how this receptor is regulated. It has been previously demonstrated that the expression and proteolysis of SorLA/LR11 can be enhanced by binding to its ligands, such as the neuropeptide head activator [104]. Other ligands of SorLA/LR11 e.g. ApoE, or possible ligands e.g. sAPP, that are linked to AD pathogenesis are potential candidates and should be investigated.
ApoER2
ApoER2 is primarily known for its role in cortical development and neuronal migration. Together with VLDLR, ApoER2 acts as a co-receptor for Reelin, an important extracellular signaling protein that regulates positioning of cortical neurons [105]. Recently, several studies have suggested that Reelin binding to ApoER2 may also influence synaptic function, learning, and memory in the adult brain. ApoER2 knock-out mice have deficits in learning and memory and hippocampal slice preparations from these mice are deficient in long term potentiation (LTP), a phenomenon associated with synaptic plasticity [106]. The reason for these deficits could be due to the latest finding that ApoER2 interacts extracellularly with the NR1, NR2A and NR2B subunits of NMDA receptors and also associates intracellularly with PSD-95, proteins which both have important roles in synaptic plasticity [107,108]. Also, alternative splicing of the gene encoding ApoER2 results in a 59 amino acid cytoplasmic insert that is selectively upregulated during periods of high activity in mice. The presence of this insert in ApoER2 was necessary for the propagation of LTP by Reelin and also in NMDA receptor phosphorylation after Reelin binding [107].
In addition to its interactions with the NMDA receptor, it has recently been found that ApoER2 can associate with APP through F-spondin, a protein associated with the extracellular matrix [109]. F-spondin expression alone was previously shown to inhibit BACE-dependent APP cleavage and decrease APP β-CTF levels [110]. An interaction between APP and ApoER2 was demonstrated by co-immunoprecipitation of lysates from primary neuronal cultures and COS7 cells. Incubation with F-spondin-containing medium increased this interaction by over 100%, suggesting an important role of F-spondin in the clustering of these proteins [109]. Interestingly concurrent expression of APP, ApoER2 and F-spondin results in an increase in cell surface levels of both APP and ApoER2.
The clustering of ApoER2 and APP with F-spondin alters the processing of both of these cell surface proteins. With increased cell surface levels of ApoER2 and APP, the authors found an increase in secreted ApoER2 and sAPPα and an increase in their CTFs [109]. ApoER2 expression with F-spondin was also associated with reduced Aβ and β-CTF levels, indicating a decrease in APP processing by β-secretase. The ability of F-spondin and ApoER2 to decrease APP processing by β-secretase was inhibited by preincubation of cells with RAP, indicating the extracellular domain of ApoER2 is important for F-spondin and APP interactions [109]. It would be interesting to know if other extracellular ApoER2 ligands, such as ApoE, could also disrupt ApoER2 interactions with APP and alter its influence on APP processing.
Utilizing a well known FE65-dependent APP luciferase transactivation assay [24] in which luciferase transactivation is dependent on FE65 and γ-secretase cleavage of APP, it was found that application of soluble F-spondin decreased luciferase transactivation [109]. This decrease in luciferase transactivation by the APP fragment and the accumulation of APP CTFs with ApoER2 expression suggest that ApoER2 could also influence downstream transcriptional activity of APP. Future studies are needed to address whether these processes could influence synaptic activity and AD pathogenesis.
A novel finding is that like LRP, ApoER2 is proteolytically processed by a metalloproteinase at the cell surface which releases an extracellular soluble fragment, and subsequently by γ-secretase to release an intracellular domain [111]. Binding of ApoER2 ligands, ApoE and α2M, as well as F-spondin to ApoE influences ApoER2 proteolysis. Currently, the function of these processed fragments is unknown. Subsequently, it will be essential to determine if the proteolytic processing of ApoER2 could alter its interactions with APP and NMDA receptors and influence Aβ levels and/or synaptic activity.
Conclusion
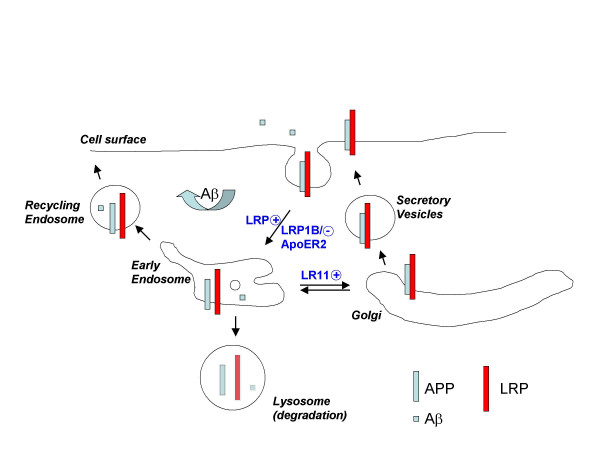
Alterations in APP processing to favor Aβ production and the accumulation of Aβ in the brain are key pathogenic events in AD. A number of proteins, including several members of the LDL receptor family, have been found to interact with APP and regulate its trafficking and processing. In this review, we have discussed several possible mechanisms by which APP trafficking and processing are regulated by LRP, LRP1B, SorLA/LR11 and ApoER2 (Fig. 3). For LRP and LRP1B, the expression and endocytosis of these receptors may have opposing roles in their ability to influence APP endocytosis and thus result in increased Aβ levels with LRP and decreased Aβ levels with LRP1B expression. Expression of SorLA/LR11 alters trafficking of APP to discrete intracellular compartments that result in a decrease in Aβ levels. The uncovering of an interaction between ApoER2, APP, and F-spondin reveals a complex between the extracellular matrix and ApoER2 at the cell surface that can decrease APP processing to Aβ. Although we have focused primarily on the roles of these LDL receptor family members in APP, these receptors are also regulated by alternative splicing and subject to proteolysis that can influence intricate intracellular signaling pathways. Future studies are needed to determine if interactions between these receptors, APP and other ligands or co-receptors can activate downstream signaling cascades that may have ultimately effect the pathogenesis of AD. The regulation of APP processing to Aβ is inherently complex; nonetheless, the discovery that these LDL receptor family members are able to affect its processing is an important step to uncovering new therapies to reduce Aβ and its associated dementia.
Figure 3.
Model of APP processing pathways mediated by LDL receptor family members. LRP endocytosis enhances APP endocytosis and processing to Aβ. Due to its slow rate of endocytosis LRP1B retains APP at the cell surface and decreases its processing to Aβ. ApoER2 enhances interactions between APP and F-spondin at the cell surface and also decreases its processing to Aβ. SorLA/LR11 may shuttle APP to the Golgi and prevent its processing by β-secretase in the early endosome, thus decreasing processing to Aβ.
Abbreviations
LDL, low-density lipoprotein; AD, Alzheimer's disease; Aβ, amyloid-β peptide; APP, β-amyloid precursor protein; APLP1, amyloid precursor like proteins-1; APLP2, amyloid precursor like proteins-2; sAPP, soluble APP fragment; sAPPβ, soluble β-cleaved APP fragment; BACE, β-site APP cleaving enzyme; CTF, C-terminal fragment; PNII, protease nexin II; ER, endoplasmic reticulum; CHO, Chinese hamster ovary; LRP, low-density lipoprotein receptor-related protein; VLDLR, very low density lipoprotein receptor; apoER2, apoE receptor 2; sorLA, sortingprotein-related receptor containing LDLR class A repeats; RAP, receptor-associated protein; α2M, alpha-2-macroglobulin; tPA, tissue-type plasminogen activator; FE65L1, FE65-like protein; EGFBP, epidermal growth factor binding protein; mLRP4, LRP minireceptor; mLRP1B4, LRP1B minireceptor; KPI, Kunitz proteinase inhibitor; vps10p, vacuolar protein sorting 10 protein; LTP, long-term potentiation.
Acknowledgments
Acknowledgements
Work in the authors' laboratory is supported by grants from the National Institutes of Health, the Alzheimer's Association and the American Health Assistance Foundation. We wish to thank Dr. Amy Caruano-Yzermans for critical reading of this manuscript.
Contributor Information
Judy A Cam, Email: judy.cam@med.nyu.edu.
Guojun Bu, Email: bu@wustl.edu.
References
- Selkoe DJ. Deciphering the genesis and fate of amyloid beta-protein yields novel therapies for Alzheimer disease. J Clin Invest. 2002;110:1375–1381. doi: 10.1172/JCI200216783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glenner GG, Wong CW. Alzheimer's disease: initial report of the purification and characterization of a novel cerebrovascular amyloid protein. Biochem Biophys Res Commun. 1984;120:885–890. doi: 10.1016/S0006-291X(84)80190-4. [DOI] [PubMed] [Google Scholar]
- Kang J, Lemaire HG, Unterbeck A, Salbaum JM, Masters CL, Grzeschik KH, Multhaup G, Beyreuther K, Muller-Hill B. The precursor of Alzheimer's disease amyloid A4 protein resembles a cell-surface receptor. Nature. 1987;325:733–736. doi: 10.1038/325733a0. [DOI] [PubMed] [Google Scholar]
- Iwatsubo T, Odaka A, Suzuki N, Mizusawa H, Nukina N, Ihara Y. Visualization of A beta 42(43) and A beta 40 in senile plaques with end-specific A beta monoclonals: evidence that an initially deposited species is A beta 42(43) Neuron. 1994;13:45–53. doi: 10.1016/0896-6273(94)90458-8. [DOI] [PubMed] [Google Scholar]
- Citron M. Strategies for disease modification in Alzheimer's disease. Nat Rev Neurosci. 2004;5:677–685. doi: 10.1038/nrn1495. [DOI] [PubMed] [Google Scholar]
- Burdick D, Soreghan B, Kwon M, Kosmoski J, Knauer M, Henschen A, Yates J, Cotman C, Glabe C. Assembly and aggregation properties of synthetic Alzheimer's A4/beta amyloid peptide analogs. J Biol Chem. 1992;267:546–554. [PubMed] [Google Scholar]
- Jarrett JT, Berger EP, Lansbury PTJ. The carboxy terminus of the beta amyloid protein is critical for the seeding of amyloid formation: implications for the pathogenesis of Alzheimer's disease. Biochemistry. 1993;32:4693–4697. doi: 10.1021/bi00069a001. [DOI] [PubMed] [Google Scholar]
- Snyder SW, Ladror US, Wade WS, Wang GT, Barrett LW, Matayoshi ED, Huffaker HJ, Krafft GA, Holzman TF. Amyloid-beta aggregation: selective inhibition of aggregation in mixtures of amyloid with different chain lengths. Biophys J. 1994;67:1216–1228. doi: 10.1016/S0006-3495(94)80591-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowan E, Pickford F, Kim J, Onstead L, Eriksen J, Yu C, Skipper L, Murphy MP, Beard J, Das P, Jansen K, Delucia M, Lin WL, Dolios G, Wang R, Eckman CB, Dickson DW, Hutton M, Hardy J, Golde T. Abeta42 is essential for parenchymal and vascular amyloid deposition in mice. Neuron. 2005;47:191–199. doi: 10.1016/j.neuron.2005.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkitadze MD, Bitan G, Teplow DB. Paradigm shifts in Alzheimer's disease and other neurodegenerative disorders: the emerging role of oligomeric assemblies. J Neurosci Res. 2002;69:567–577. doi: 10.1002/jnr.10328. [DOI] [PubMed] [Google Scholar]
- Goate A, Chartier-Harlin MC, Mullan M, Brown J, Crawford F, Fidani L, Giuffra L, Haynes A, Irving N, James L, et al. Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer's disease. Nature. 1991;349:704–706. doi: 10.1038/349704a0. [DOI] [PubMed] [Google Scholar]
- Cruts M, Rademakers R. Alzheimer Disease & Frontotemporal Dementia Mutation Database http://www.molgen.ua.ac.be/ADMutations/
- Prasher VP, Farrer MJ, Kessling AM, Fisher EM, West RJ, Barber PC, Butler AC. Molecular mapping of Alzheimer-type dementia in Down's syndrome. Ann Neurol. 1998;43:380–383. doi: 10.1002/ana.410430316. [DOI] [PubMed] [Google Scholar]
- Kang J, Muller-Hill B. Differential splicing of Alzheimer's disease amyloid A4 precursor RNA in rat tissues: PreA4(695) mRNA is predominantly produced in rat and human brain. Biochem Biophys Res Commun. 1990;166:1192–1200. doi: 10.1016/0006-291X(90)90992-V. [DOI] [PubMed] [Google Scholar]
- Rohan de Silva HA, Jen A, Wickenden C, Jen LS, Wilkinson SL, Patel AJ. Cell-specific expression of beta-amyloid precursor protein isoform mRNAs and proteins in neurons and astrocytes. Brain Res Mol Brain Res. 1997;47:147–156. doi: 10.1016/S0169-328X(97)00045-4. [DOI] [PubMed] [Google Scholar]
- Mattson MP. Cellular actions of beta-amyloid precursor protein and its soluble and fibrillogenic derivatives. Physiol Rev. 1997;77:1081–1132. doi: 10.1152/physrev.1997.77.4.1081. [DOI] [PubMed] [Google Scholar]
- Zheng H, Jiang M, Trumbauer ME, Hopkins R, Sirinathsinghji DJ, Stevens KA, Conner MW, Slunt HH, Sisodia SS, Chen HY, Van der Ploeg LH. Mice deficient for the amyloid precursor protein gene. Ann N Y Acad Sci. 1996;777:421–426. doi: 10.1111/j.1749-6632.1996.tb34456.x. [DOI] [PubMed] [Google Scholar]
- Herms J, Anliker B, Heber S, Ring S, Fuhrmann M, Kretzschmar H, Sisodia S, Muller U. Cortical dysplasia resembling human type 2 lissencephaly in mice lacking all three APP family members. Embo J. 2004;23:4106–4115. doi: 10.1038/sj.emboj.7600390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heber S, Herms J, Gajic V, Hainfellner J, Aguzzi A, Rulicke T, von Kretzschmar H, von Koch C, Sisodia S, Tremml P, Lipp HP, Wolfer DP, Muller U. Mice with combined gene knock-outs reveal essential and partially redundant functions of amyloid precursor protein family members. J Neurosci. 2000;20:7951–7963. doi: 10.1523/JNEUROSCI.20-21-07951.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yankner BA, Duffy LK, Kirschner DA. Neurotrophic and neurotoxic effects of amyloid beta protein: reversal by tachykinin neuropeptides. Science. 1990;250:279–282. doi: 10.1126/science.2218531. [DOI] [PubMed] [Google Scholar]
- Turner PR, O'Connor K, Tate WP, Abraham WC. Roles of amyloid precursor protein and its fragments in regulating neural activity, plasticity and memory. Prog Neurobiol. 2003;70:1–32. doi: 10.1016/S0301-0082(03)00089-3. [DOI] [PubMed] [Google Scholar]
- Oltersdorf T, Fritz LC, Schenk DB, Lieberburg I, Johnson-Wood KL, Beattie EC, Ward PJ, Blacher RW, Dovey HF, Sinha S. The secreted form of the Alzheimer's amyloid precursor protein with the Kunitz domain is protease nexin-II. Nature. 1989;341:144–147. doi: 10.1038/341144a0. [DOI] [PubMed] [Google Scholar]
- Smith RP, Higuchi DA, Broze GJJ. Platelet coagulation factor XIa-inhibitor, a form of Alzheimer amyloid precursor protein. Science. 1990;248:1126–1128. doi: 10.1126/science.2111585. [DOI] [PubMed] [Google Scholar]
- Cao X, Sudhof TC. A transcriptionally active complex of APP with Fe65 and histone acetyltransferase Tip60. Science. 2001;293:115–120. doi: 10.1126/science.1058783. [DOI] [PubMed] [Google Scholar]
- Nordstedt C, Caporaso GL, Thyberg J, Gandy SE, Greengard P. Identification of the Alzheimer beta/A4 amyloid precursor protein in clathrin-coated vesicles purified from PC12 cells. J Biol Chem. 1993;268:608–612. [PubMed] [Google Scholar]
- Koo EH, Squazzo SL. Evidence that production and release of amyloid beta-protein involves the endocytic pathway. J Biol Chem. 1994;269:17386–17389. [PubMed] [Google Scholar]
- Chen WJ, Goldstein JL, Brown MS. NPXY, a sequence often found in cytoplasmic tails, is required for coated pit-mediated internalization of the low density lipoprotein receptor. J Biol Chem. 1990;265:3116–3123. [PubMed] [Google Scholar]
- Lai A, Sisodia SS, Trowbridge IS. Characterization of sorting signals in the beta-amyloid precursor protein cytoplasmic domain. J Biol Chem. 1995;270:3565–3573. doi: 10.1074/jbc.270.38.22176. [DOI] [PubMed] [Google Scholar]
- Cescato R, Dumermuth E, Spiess M, Paganetti PA. Increased generation of alternatively cleaved beta-amyloid peptides in cells expressing mutants of the amyloid precursor protein defective in endocytosis. J Neurochem. 2000;74:1131–1139. doi: 10.1046/j.1471-4159.2000.741131.x. [DOI] [PubMed] [Google Scholar]
- Koo EH, Squazzo SL, Selkoe DJ, Koo CH. Trafficking of cell-surface amyloid beta-protein precursor. I. Secretion, endocytosis and recycling as detected by labeled monoclonal antibody. J Cell Sci. 1996;109:991–998. doi: 10.1242/jcs.109.5.991. [DOI] [PubMed] [Google Scholar]
- Perez RG, Soriano S, Hayes JD, Ostaszewski B, Xia W, Selkoe DJ, Chen X, Stokin GB, Koo EH. Mutagenesis identifies new signals for beta-amyloid precursor protein endocytosis, turnover, and the generation of secreted fragments, including Abeta42. J Biol Chem. 1999;274:18851–18856. doi: 10.1074/jbc.274.27.18851. [DOI] [PubMed] [Google Scholar]
- Vassar R, Bennett BD, Babu-Khan S, Kahn S, Mendiaz EA, Denis P, Teplow DB, Ross S, Amarante P, Loeloff R, Luo Y, Fisher S, Fuller J, Edenson S, Lile J, Jarosinski MA, Biere AL, Curran E, Burgess T, Louis JC, Collins F, Treanor J, Rogers G, Citron M. Beta-secretase cleavage of Alzheimer's amyloid precursor protein by the transmembrane aspartic protease BACE. Science. 1999;286:735–741. doi: 10.1126/science.286.5440.735. [DOI] [PubMed] [Google Scholar]
- Ray WJ, Yao M, Mumm J, Schroeter EH, Saftig P, Wolfe M, Selkoe DJ, Kopan R, Goate AM. Cell surface presenilin-1 participates in the gamma-secretase-like proteolysis of Notch. J Biol Chem. 1999;274:36801–36807. doi: 10.1074/jbc.274.51.36801. [DOI] [PubMed] [Google Scholar]
- Pasternak SH, Bagshaw RD, Guiral M, Zhang S, Ackerley CA, Pak BJ, Callahan JW, Mahuran DJ. Presenilin-1, nicastrin, amyloid precursor protein, and gamma-secretase activity are co-localized in the lysosomal membrane. J Biol Chem. 2003;278:26687–26694. doi: 10.1074/jbc.M304009200. [DOI] [PubMed] [Google Scholar]
- Parvathy S, Hussain I, Karran EH, Turner AJ, Hooper NM. Cleavage of Alzheimer's amyloid precursor protein by alpha-secretase occurs at the surface of neuronal cells. Biochemistry. 1999;38:9728–9734. doi: 10.1021/bi9906827. [DOI] [PubMed] [Google Scholar]
- Hussain MM, Kancha RK, Zhou Z, Luchoomun J, Zu H, Bakillah A. Chylomicron assembly and catabolism: role of apolipoproteins and receptors. Biochim Biophys Acta. 1996;1300:151–170. doi: 10.1016/0005-2760(96)00041-0. [DOI] [PubMed] [Google Scholar]
- Strittmatter WJ, Saunders AM, Schmechel D, Pericak-Vance M, Enghild J, Salvesen GS, Roses AD. Apolipoprotein E: high-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc Natl Acad Sci U S A. 1993;90:1977–1981. doi: 10.1073/pnas.90.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herz J, Hamann U, Rogne S, Myklebost O, Gausepohl H, Stanley KK. Surface location and high affinity for calcium of a 500-kd liver membrane protein closely related to the LDL-receptor suggest a physiological role as lipoprotein receptor. Embo J. 1988;7:4119–4127. doi: 10.1002/j.1460-2075.1988.tb03306.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willnow TE, Orth K, Herz J. Molecular dissection of ligand binding sites on the low density lipoprotein receptor-related protein. J Biol Chem. 1994;269:15827–15832. [PubMed] [Google Scholar]
- Obermoeller-McCormick LM, Li Y, Osaka H, FitzGerald DJ, Schwartz AL, Bu G. Dissection of receptor folding and ligand-binding property with functional minireceptors of LDL receptor-related protein. J Cell Sci. 2001;114:899–908. doi: 10.1242/jcs.114.5.899. [DOI] [PubMed] [Google Scholar]
- Neels JG, van Den Berg BM, Lookene A, Olivecrona G, Pannekoek H, van Zonneveld AJ. The second and fourth cluster of class A cysteine-rich repeats of the low density lipoprotein receptor-related protein share ligand-binding properties. J Biol Chem. 1999;274:31305–31311. doi: 10.1074/jbc.274.44.31305. [DOI] [PubMed] [Google Scholar]
- Bu G. The roles of receptor-associated protein (RAP) as a molecular chaperone for members of the LDL receptor family. Int Rev Cytol. 2001;209:79–116. doi: 10.1016/s0074-7696(01)09011-8. [DOI] [PubMed] [Google Scholar]
- Croy JE, Brandon T, Komives EA. Two apolipoprotein E mimetic peptides, ApoE(130-149) and ApoE(141-155)2, bind to LRP1. Biochemistry. 2004;43:7328–7335. doi: 10.1021/bi036208p. [DOI] [PubMed] [Google Scholar]
- Croy JE, Shin WD, Knauer MF, Knauer DJ, Komives EA. All three LDL receptor homology regions of the LDL receptor-related protein bind multiple ligands. Biochemistry. 2003;42:13049–13057. doi: 10.1021/bi034752s. [DOI] [PubMed] [Google Scholar]
- Kristensen T, Moestrup SK, Gliemann J, Bendtsen L, Sand O, Sottrup-Jensen L. Evidence that the newly cloned low-density-lipoprotein receptor related protein (LRP) is the alpha 2-macroglobulin receptor. FEBS Lett. 1990;276:151–155. doi: 10.1016/0014-5793(90)80530-V. [DOI] [PubMed] [Google Scholar]
- Mikhailenko I, Battey FD, Migliorini M, Ruiz JF, Argraves K, Moayeri M, Strickland DK. Recognition of alpha 2-macroglobulin by the low density lipoprotein receptor-related protein requires the cooperation of two ligand binding cluster regions. J Biol Chem. 2001;276:39484–39491. doi: 10.1074/jbc.M104382200. [DOI] [PubMed] [Google Scholar]
- Li Y, Marzolo MP, van Kerkhof P, Strous GJ, Bu G. The YXXL motif, but not the two NPXY motifs, serves as the dominant endocytosis signal for low density lipoprotein receptor-related protein. J Biol Chem. 2000;275:17187–17194. doi: 10.1074/jbc.M000490200. [DOI] [PubMed] [Google Scholar]
- Li Y, Lu W, Marzolo MP, Bu G. Differential functions of members of the low density lipoprotein receptor family suggested by their distinct endocytosis rates. J Biol Chem. 2001;276:18000–18006. doi: 10.1074/jbc.M101589200. [DOI] [PubMed] [Google Scholar]
- Gotthardt M, Trommsdorff M, Nevitt MF, Shelton J, Richardson JA, Stockinger W, Nimpf J, Herz J. Interactions of the low density lipoprotein receptor gene family with cytosolic adaptor and scaffold proteins suggest diverse biological functions in cellular communication and signal transduction. J Biol Chem. 2000;275:25616–25624. doi: 10.1074/jbc.M000955200. [DOI] [PubMed] [Google Scholar]
- Trommsdorff M, Borg JP, Margolis B, Herz J. Interaction of cytosolic adaptor proteins with neuronal apolipoprotein E receptors and the amyloid precursor protein. J Biol Chem. 1998;273:33556–33560. doi: 10.1074/jbc.273.50.33556. [DOI] [PubMed] [Google Scholar]
- Zhuo M, Holtzman DM, Li Y, Osaka H, DeMaro J, Jacquin M, Bu G. Role of tissue plasminogen activator receptor LRP in hippocampal long-term potentiation. J Neurosci. 2000;20:542–549. doi: 10.1523/JNEUROSCI.20-02-00542.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacskai BJ, Xia MQ, Strickland DK, Rebeck GW, Hyman BT. The endocytic receptor protein LRP also mediates neuronal calcium signaling via N-methyl-D-aspartate receptors. Proc Natl Acad Sci U S A. 2000;97:11551–11556. doi: 10.1073/pnas.200238297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conboy L, Murphy KJ, Regan CM. Amyloid precursor protein expression in the rat hippocampal dentate gyrus modulates during memory consolidation. J Neurochem. 2005;95:1677–1688. doi: 10.1111/j.1471-4159.2005.03484.x. [DOI] [PubMed] [Google Scholar]
- Deane R, Wu Z, Sagare A, Davis J, Du Yan S, Hamm K, Xu F, Parisi M, LaRue B, Hu HW, Spijkers P, Guo H, Song X, Lenting PJ, Van Nostrand WE, Zlokovic BV. LRP/amyloid beta-peptide interaction mediates differential brain efflux of Abeta isoforms. Neuron. 2004;43:333–344. doi: 10.1016/j.neuron.2004.07.017. [DOI] [PubMed] [Google Scholar]
- Gylys KH, Fein JA, Tan AM, Cole GM. Apolipoprotein E enhances uptake of soluble but not aggregated amyloid-beta protein into synaptic terminals. J Neurochem. 2003;84:1442–1451. doi: 10.1046/j.1471-4159.2003.01643.x. [DOI] [PubMed] [Google Scholar]
- Narita M, Holtzman DM, Schwartz AL, Bu G. Alpha2-macroglobulin complexes with and mediates the endocytosis of beta-amyloid peptide via cell surface low-density lipoprotein receptor-related protein. J Neurochem. 1997;69:1904–1911. doi: 10.1046/j.1471-4159.1997.69051904.x. [DOI] [PubMed] [Google Scholar]
- Qiu Z, Strickland DK, Hyman BT, Rebeck GW. Alpha2-macroglobulin enhances the clearance of endogenous soluble beta-amyloid peptide via low-density lipoprotein receptor-related protein in cortical neurons. J Neurochem. 1999;73:1393–1398. doi: 10.1046/j.1471-4159.1999.0731393.x. [DOI] [PubMed] [Google Scholar]
- Arelin K, Kinoshita A, Whelan CM, Irizarry MC, Rebeck GW, Strickland DK, Hyman BT. LRP and senile plaques in Alzheimer's disease: colocalization with apolipoprotein E and with activated astrocytes. Brain Res Mol Brain Res. 2002;104:38–46. doi: 10.1016/S0169-328X(02)00203-6. [DOI] [PubMed] [Google Scholar]
- Qiu Z, Strickland DK, Hyman BT, Rebeck GW. Elevation of LDL receptor-related protein levels via ligand interactions in Alzheimer disease and in vitro. J Neuropathol Exp Neurol. 2001;60:430–440. doi: 10.1093/jnen/60.5.430. [DOI] [PubMed] [Google Scholar]
- Rebeck GW, Harr SD, Strickland DK, Hyman BT. Multiple, diverse senile plaque-associated proteins are ligands of an apolipoprotein E receptor, the alpha 2-macroglobulin receptor/low-density-lipoprotein receptor-related protein. Ann Neurol. 1995;37:211–217. doi: 10.1002/ana.410370212. [DOI] [PubMed] [Google Scholar]
- Wavrant-DeVrieze F, Perez-Tur J, Lambert JC, Frigard B, Pasquier F, Delacourte A, Amouyel P, Hardy J, Chartier-Harlin MC. Association between the low density lipoprotein receptor-related protein (LRP) and Alzheimer's disease. Neurosci Lett. 1997;227:68–70. doi: 10.1016/S0304-3940(97)00304-2. [DOI] [PubMed] [Google Scholar]
- Kang DE, Saitoh T, Chen X, Xia Y, Masliah E, Hansen LA, Thomas RG, Thal LJ, Katzman R. Genetic association of the low-density lipoprotein receptor-related protein gene (LRP), an apolipoprotein E receptor, with late-onset Alzheimer's disease. Neurology. 1997;49:56–61. doi: 10.1212/wnl.49.1.56. [DOI] [PubMed] [Google Scholar]
- Van Leuven F, Stas L, Thiry E, Nelissen B, Miyake Y. Strategy to sequence the 89 exons of the human LRP1 gene coding for the lipoprotein receptor related protein: identification of one expressed mutation among 48 polymorphisms. Genomics. 1998;52:138–144. doi: 10.1006/geno.1998.5408. [DOI] [PubMed] [Google Scholar]
- Scott WK, Yamaoka LH, Bass MP, Gaskell PC, Conneally PM, Small GW, Farrer LA, Auerbach SA, Saunders AM, Roses AD, Haines JL, Pericak-Vance MA. No genetic association between the LRP receptor and sporadic or late-onset familial Alzheimer disease. Neurogenetics. 1998;1:179–183. doi: 10.1007/s100480050026. [DOI] [PubMed] [Google Scholar]
- Kamboh MI, Ferrell RE, DeKosky ST. Genetic association studies between Alzheimer's disease and two polymorphisms in the low density lipoprotein receptor-related protein gene. Neurosci Lett. 1998;244:65–68. doi: 10.1016/S0304-3940(98)00141-4. [DOI] [PubMed] [Google Scholar]
- Bullido MJ, Guallar-Castillon P, Artiga MJ, Ramos MC, Sastre I, Aldudo J, Frank A, Coria F, Rodriguez-Artalejo F, Valdivieso F. Alzheimer's risk associated with human apolipoprotein E, alpha-2 macroglobulin and lipoprotein receptor related protein polymorphisms: absence of genetic interactions, and modulation by gender. Neurosci Lett. 2000;289:213–216. doi: 10.1016/S0304-3940(00)01304-5. [DOI] [PubMed] [Google Scholar]
- Baum L, Chen L, Ng HK, Chan YS, Mak YT, Woo J, Chiu HF, Pang CP. Low density lipoprotein receptor related protein gene exon 3 polymorphism association with Alzheimer's disease in Chinese. Neurosci Lett. 1998;247:33–36. doi: 10.1016/S0304-3940(98)00294-8. [DOI] [PubMed] [Google Scholar]
- Hollenbach E, Ackermann S, Hyman BT, Rebeck GW. Confirmation of an association between a polymorphism in exon 3 of the low-density lipoprotein receptor-related protein gene and Alzheimer's disease. Neurology. 1998;50:1905–1907. doi: 10.1212/wnl.50.6.1905. [DOI] [PubMed] [Google Scholar]
- Lambert JC, Wavrant-De Vrieze F, Amouyel P, Chartier-Harlin MC. Association at LRP gene locus with sporadic late-onset Alzheimer's disease. Lancet. 1998;351:1787–1788. doi: 10.1016/S0140-6736(05)78749-3. [DOI] [PubMed] [Google Scholar]
- Kang DE, Pietrzik CU, Baum L, Chevallier N, Merriam DE, Kounnas MZ, Wagner SL, Troncoso JC, Kawas CH, Katzman R, Koo EH. Modulation of amyloid beta-protein clearance and Alzheimer's disease susceptibility by the LDL receptor-related protein pathway. J Clin Invest. 2000;106:1159–1166. doi: 10.1172/JCI11013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beffert U, Arguin C, Poirier J. The polymorphism in exon 3 of the low density lipoprotein receptor-related protein gene is weakly associated with Alzheimer's disease. Neurosci Lett. 1999;259:29–32. doi: 10.1016/S0304-3940(98)00888-X. [DOI] [PubMed] [Google Scholar]
- Knauer MF, Orlando RA, Glabe CG. Cell surface APP751 forms complexes with protease nexin 2 ligands and is internalized via the low density lipoprotein receptor-related protein (LRP) Brain Res. 1996;740:6–14. doi: 10.1016/S0006-8993(96)00711-1. [DOI] [PubMed] [Google Scholar]
- Fiore F, Zambrano N, Minopoli G, Donini V, Duilio A, Russo T. The regions of the Fe65 protein homologous to the phosphotyrosine interaction/phosphotyrosine binding domain of Shc bind the intracellular domain of the Alzheimer's amyloid precursor protein. J Biol Chem. 1995;270:30853–30856. doi: 10.1074/jbc.270.52.30853. [DOI] [PubMed] [Google Scholar]
- Borg JP, Ooi J, Levy E, Margolis B. The phosphotyrosine interaction domains of X11 and FE65 bind to distinct sites on the YENPTY motif of amyloid precursor protein. Mol Cell Biol. 1996;16:6229–6241. doi: 10.1128/mcb.16.11.6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebeck GW, Moir RD, Mui S, Strickland DK, Tanzi RE, Hyman BT. Association of membrane-bound amyloid precursor protein APP with the apolipoprotein E receptor LRP. Brain Res Mol Brain Res. 2001;87:238–245. doi: 10.1016/S0169-328X(01)00006-7. [DOI] [PubMed] [Google Scholar]
- Kinoshita A, Whelan CM, Smith CJ, Mikhailenko I, Rebeck GW, Strickland DK, Hyman BT. Demonstration by fluorescence resonance energy transfer of two sites of interaction between the low-density lipoprotein receptor-related protein and the amyloid precursor protein: role of the intracellular adapter protein Fe65. J Neurosci. 2001;21:8354–8361. doi: 10.1523/JNEUROSCI.21-21-08354.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cam JA, Zerbinatti CV, Li Y, Bu G. Rapid endocytosis of the low density lipoprotein receptor-related protein modulates cell surface distribution and processing of the beta-amyloid precursor protein. J Biol Chem. 2005;280:15464–15470. doi: 10.1074/jbc.M500613200. [DOI] [PubMed] [Google Scholar]
- Ye S, Huang Y, Mullendorff K, Dong L, Giedt G, Meng EC, Cohen FE, Kuntz ID, Weisgraber KH, Mahley RW. Apolipoprotein (apo) E4 enhances amyloid beta peptide production in cultured neuronal cells: apoE structure as a potential therapeutic target. Proc Natl Acad Sci U S A. 2005;102:18700–18705. doi: 10.1073/pnas.0508693102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbinatti CV, Wozniak DF, Cirrito J, Cam JA, Osaka H, Bales KR, Zhuo M, Paul SM, Holtzman DM, Bu G. Increased soluble amyloid-beta peptide and memory deficits in amyloid model mice overexpressing the low-density lipoprotein receptor-related protein. Proc Natl Acad Sci U S A. 2004;101:1075–1080. doi: 10.1073/pnas.0305803101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Arnim CA, Kinoshita A, Peltan ID, Tangredi MM, Herl L, Lee BM, Spoelgen R, Hshieh TT, Ranganathan S, Battey FD, Liu CX, Bacskai BJ, Sever S, Irizarry MC, Strickland DK, Hyman BT. The low density lipoprotein receptor-related protein (LRP) is a novel beta-secretase (BACE1) substrate. J Biol Chem. 2005;280:17777–17785. doi: 10.1074/jbc.M414248200. [DOI] [PubMed] [Google Scholar]
- May P, Reddy YK, Herz J. Proteolytic processing of low density lipoprotein receptor-related protein mediates regulated release of its intracellular domain. J Biol Chem. 2002;277:18736–18743. doi: 10.1074/jbc.M201979200. [DOI] [PubMed] [Google Scholar]
- Lleo A, Waldron E, von Arnim CA, Herl L, Tangredi MM, Peltan ID, Strickland DK, Koo EH, Hyman BT, Pietrzik CU, Berezovska O. Low density lipoprotein receptor-related protein (LRP) interacts with presenilin 1 and is a competitive substrate of the amyloid precursor protein (APP) for gamma-secretase. J Biol Chem. 2005;280:27303–27309. doi: 10.1074/jbc.M413969200. [DOI] [PubMed] [Google Scholar]
- Ehehalt R, Keller P, Haass C, Thiele C, Simons K. Amyloidogenic processing of the Alzheimer beta-amyloid precursor protein depends on lipid rafts. J Cell Biol. 2003;160:113–123. doi: 10.1083/jcb.200207113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May P, Rohlmann A, Bock HH, Zurhove K, Marth JD, Schomburg ED, Noebels JL, Beffert U, Sweatt JD, Weeber EJ, Herz J. Neuronal LRP1 functionally associates with postsynaptic proteins and is required for normal motor function in mice. Mol Cell Biol. 2004;24:8872–8883. doi: 10.1128/MCB.24.20.8872-8883.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CX, Musco S, Lisitsina NM, Forgacs E, Minna JD, Lisitsyn NA. LRP-DIT, a putative endocytic receptor gene, is frequently inactivated in non-small cell lung cancer cell lines. Cancer Res. 2000;60:1961–1967. [PubMed] [Google Scholar]
- Liu CX, Li Y, Obermoeller-McCormick LM, Schwartz AL, Bu G. The putative tumor suppressor LRP1B, a novel member of the low density lipoprotein (LDL) receptor family, exhibits both overlapping and distinct properties with the LDL receptor-related protein. J Biol Chem. 2001;276:28889–28896. doi: 10.1074/jbc.M102727200. [DOI] [PubMed] [Google Scholar]
- Langbein S, Szakacs O, Wilhelm M, Sukosd F, Weber S, Jauch A, Lopez Beltran A, Alken P, Kalble T, Kovacs G. Alteration of the LRP1B gene region is associated with high grade of urothelial cancer. Lab Invest. 2002;82:639–643. doi: 10.1038/labinvest.3780458. [DOI] [PubMed] [Google Scholar]
- Sonoda I, Imoto I, Inoue J, Shibata T, Shimada Y, Chin K, Imamura M, Amagasa T, Gray JW, Hirohashi S, Inazawa J. Frequent silencing of low density lipoprotein receptor-related protein 1B (LRP1B) expression by genetic and epigenetic mechanisms in esophageal squamous cell carcinoma. Cancer Res. 2004;64:3741–3747. doi: 10.1158/0008-5472.CAN-04-0172. [DOI] [PubMed] [Google Scholar]
- Marschang P, Brich J, Weeber EJ, Sweatt JD, Shelton JM, Richardson JA, Hammer RE, Herz J. Normal development and fertility of knockout mice lacking the tumor suppressor gene LRP1b suggest functional compensation by LRP1. Mol Cell Biol. 2004;24:3782–3793. doi: 10.1128/MCB.24.9.3782-3793.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Knisely JM, Lu W, McCormick LM, Wang J, Henkin J, Schwartz AL, Bu G. Low density lipoprotein (LDL) receptor-related protein 1B impairs urokinase receptor regeneration on the cell surface and inhibits cell migration. J Biol Chem. 2002;277:42366–42371. doi: 10.1074/jbc.M207705200. [DOI] [PubMed] [Google Scholar]
- Ulery PG, Beers J, Mikhailenko I, Tanzi RE, Rebeck GW, Hyman BT, Strickland DK. Modulation of beta-amyloid precursor protein processing by the low density lipoprotein receptor-related protein (LRP). Evidence that LRP contributes to the pathogenesis of Alzheimer's disease. J Biol Chem. 2000;275:7410–7415. doi: 10.1074/jbc.275.10.7410. [DOI] [PubMed] [Google Scholar]
- Pietrzik CU, Busse T, Merriam DE, Weggen S, Koo EH. The cytoplasmic domain of the LDL receptor-related protein regulates multiple steps in APP processing. Embo J. 2002;21:5691–5700. doi: 10.1093/emboj/cdf568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cam JA, Zerbinatti CV, Knisely JM, Hecimovic S, Li Y, Bu G. The low density lipoprotein receptor-related protein 1B retains beta-amyloid precursor protein at the cell surface and reduces amyloid-beta peptide production. J Biol Chem. 2004;279:29639–29646. doi: 10.1074/jbc.M313893200. [DOI] [PubMed] [Google Scholar]
- Yamazaki H, Bujo H, Kusunoki J, Seimiya K, Kanaki T, Morisaki N, Schneider WJ, Saito Y. Elements of neural adhesion molecules and a yeast vacuolar protein sorting receptor are present in a novel mammalian low density lipoprotein receptor family member. J Biol Chem. 1996;271:24761–24768. doi: 10.1074/jbc.271.40.24761. [DOI] [PubMed] [Google Scholar]
- Jacobsen L, Madsen P, Moestrup SK, Lund AH, Tommerup N, Nykjaer A, Sottrup-Jensen L, Gliemann J, Petersen CM. Molecular characterization of a novel human hybrid-type receptor that binds the alpha2-macroglobulin receptor-associated protein. J Biol Chem. 1996;271:31379–31383. doi: 10.1074/jbc.271.49.31379. [DOI] [PubMed] [Google Scholar]
- Jacobsen L, Madsen P, Jacobsen C, Nielsen MS, Gliemann J, Petersen CM. Activation and functional characterization of the mosaic receptor SorLA/LR11. J Biol Chem. 2001;276:22788–22796. doi: 10.1074/jbc.M100857200. [DOI] [PubMed] [Google Scholar]
- Andersen OM, Reiche J, Schmidt V, Gotthardt M, Spoelgen R, Behlke J, von Arnim CA, Breiderhoff T, Jansen P, Wu X, Bales KR, Cappai R, Masters CL, Gliemann J, Mufson EJ, Hyman BT, Paul SM, Nykjaer A, Willnow TE. Neuronal sorting protein-related receptor sorLA/LR11 regulates processing of the amyloid precursor protein. Proc Natl Acad Sci U S A. 2005;102:13461–13466. doi: 10.1073/pnas.0503689102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herz J, Clouthier DE, Hammer RE. LDL receptor-related protein internalizes and degrades uPA-PAI-1 complexes and is essential for embryo implantation. Cell. 1992;71:411–421. doi: 10.1016/0092-8674(92)90511-A. [DOI] [PubMed] [Google Scholar]
- Scherzer CR, Offe K, Gearing M, Rees HD, Fang G, Heilman CJ, Schaller C, Bujo H, Levey AI, Lah JJ. Loss of apolipoprotein E receptor LR11 in Alzheimer disease. Arch Neurol. 2004;61:1200–1205. doi: 10.1001/archneur.61.8.1200. [DOI] [PubMed] [Google Scholar]
- Andersen OM, Schmidt V, Spoelgen R, Gliemann J, Behlke J, Galatis D, McKinstry WJ, Parker MW, Masters CL, Hyman BT, Cappai R, Willnow TE. Molecular Dissection of the Interaction between Amyloid Precursor Protein and Its Neuronal Trafficking Receptor SorLA/LR11. Biochemistry. 2006;45:2618–2628. doi: 10.1021/bi052120v. [DOI] [PubMed] [Google Scholar]
- Offe K, Dodson SE, Shoemaker JT, Fritz JJ, Gearing M, Levey AI, Lah JJ. The lipoprotein receptor LR11 regulates amyloid beta production and amyloid precursor protein traffic in endosomal compartments. J Neurosci. 2006;26:1596–1603. doi: 10.1523/JNEUROSCI.4946-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoelgen R, von Arnim CA, Thomas AV, Peltan ID, Koker M, Deng A, Irizarry MC, Andersen OM, Willnow TE, Hyman BT. Interaction of the cytosolic domains of sorLA/LR11 with the amyloid precursor protein (APP) and beta-secretase beta-site APP-cleaving enzyme. J Neurosci. 2006;26:418–428. doi: 10.1523/JNEUROSCI.3882-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohm C, Seibel N, Henkel B, Steiner H, Haass C, Hampe W. SorLA signaling by regulated intramembrane proteolysis. J Biol Chem. 2006. [DOI] [PubMed]
- Hampe W, Riedel IB, Lintzel J, Bader CO, Franke I, Schaller HC. Ectodomain shedding, translocation and synthesis of SorLA are stimulated by its ligand head activator. J Cell Sci. 2000;113 Pt 24:4475–4485. doi: 10.1242/jcs.113.24.4475. [DOI] [PubMed] [Google Scholar]
- Trommsdorff M, Gotthardt M, Hiesberger T, Shelton J, Stockinger W, Nimpf J, Hammer RE, Richardson JA, Herz J. Reeler/Disabled-like disruption of neuronal migration in knockout mice lacking the VLDL receptor and ApoE receptor 2. Cell. 1999;97:689–701. doi: 10.1016/S0092-8674(00)80782-5. [DOI] [PubMed] [Google Scholar]
- Weeber EJ, Beffert U, Jones C, Christian JM, Forster E, Sweatt JD, Herz J. Reelin and ApoE receptors cooperate to enhance hippocampal synaptic plasticity and learning. J Biol Chem. 2002;277:39944–39952. doi: 10.1074/jbc.M205147200. [DOI] [PubMed] [Google Scholar]
- Beffert U, Weeber EJ, Durudas A, Qiu S, Masiulis I, Sweatt JD, Li WP, Adelmann G, Frotscher M, Hammer RE, Herz J. Modulation of synaptic plasticity and memory by Reelin involves differential splicing of the lipoprotein receptor Apoer2. Neuron. 2005;47:567–579. doi: 10.1016/j.neuron.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Hoe HS, Pocivavsek A, Chakraborty G, Fu Z, Vicini S, Ehlers MD, Rebeck GW. Apolipoprotein E receptor 2 interactions with the N-methyl-D-aspartate receptor. J Biol Chem. 2006;281:3425–3431. doi: 10.1074/jbc.M509380200. [DOI] [PubMed] [Google Scholar]
- Hoe HS, Wessner D, Beffert U, Becker AG, Matsuoka Y, Rebeck GW. F-spondin interaction with the apolipoprotein E receptor ApoEr2 affects processing of amyloid precursor protein. Mol Cell Biol. 2005;25:9259–9268. doi: 10.1128/MCB.25.21.9259-9268.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho A, Sudhof TC. Binding of F-spondin to amyloid-beta precursor protein: a candidate amyloid-beta precursor protein ligand that modulates amyloid-beta precursor protein cleavage. Proc Natl Acad Sci U S A. 2004;101:2548–2553. doi: 10.1073/pnas.0308655100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoe HS, Rebeck GW. Regulation of ApoE receptor proteolysis by ligand binding. Brain Res Mol Brain Res. 2005;137:31–39. doi: 10.1016/j.molbrainres.2005.02.013. [DOI] [PubMed] [Google Scholar]