Abstract
Proctolin is a bioactive neuropeptide that modulates interneuronal and neuromuscular synaptic transmission in a wide variety of arthropods. We present several lines of evidence to propose that the orphan G protein-coupled receptor CG6986 of Drosophila is a proctolin receptor. When expressed in mammalian cells, CG6986 confers second messenger activation after proctolin application, with an EC50 of 0.3 nM. In competition-based studies, the CG6986 receptor binds proctolin with high affinity (IC50 = 4 nM). By microarray analysis, CG6986 transcript is consistently detected in head mRNA of different genotypes, and under different environmental conditions. By blot analysis, anti-CG6986 antibodies detect a band in tissue homogenates similar to the predicted size of the protein. Proctolin receptor immunosignals are found in the hindgut, heart, and in distinct neuronal populations of the CNS; such patterns correlate with previous demonstrations of proctolin biological activity, and in several instances, with areas of proctolin peptide immunosignals. The identification of a bona fide proctolin receptor provides the basis for a mechanistic analysis of this critical synaptic modulator.
The pentapeptide proctolin (RYLPT) was the first neuropeptide to be isolated and sequenced from insects (1). It was purified based on its myotropic actions on the insect hindgut, and was subsequently found to have wide distribution throughout the arthropods (2–4). In the crab Cancer, proctolin has potent effects on defined neural circuits within the stomatogastric system (5) and at the neuromuscular junction (6). In insects, proctolin is a major peptide cotransmitter and it modulates contractions of both somatic and visceral muscles (7, 8). Proctolin-like immunosignals are found within a small number of neurons in the larval CNS of Drosophila and these include efferents to body-wall muscles, heart, and viscera (9).
The mechanisms of actions of proctolin have also been investigated. Proctolin elevates levels of inositol trisphosphate (IP3) and cAMP, and increases calcium entry through voltage-gated ion channels (10–13). Several studies have suggested the existence of multiple proctolin receptor subtypes based on binding studies and activities of proctolin (14, 15) or its structural analogues (16–19). However, more recent studies (15, 20, 21) of proctolin binding support a model featuring a single proctolin receptor. The identification of a specific proctolin receptor will help address issues regarding possibilities of multiple receptors, the precise nature of proctolin-dependent signaling pathways, and phylogenetic differences in proctolin signaling. The significance of such studies is substantial because proctolin exerts such a widespread role as a synaptic modulator in arthropod physiology.
Analysis of the completed Drosophila genomic sequences (22, 23) identified >100 genes encoding G protein-coupled receptors (GPCRs). Further analysis indicated that 44 such receptors are likely to have peptide ligands and that the majority of these are derived from ancestors for mammalian peptide GPCR genes (24). The CG6986 gene encodes a predicted peptide GPCR that is distantly related to the thyrotropin-releasing factor receptor family (24). Because proctolin was previously shown to signal through heterotrimeric G proteins in the cockroach (20), we included this peptide among other potential ligands with which to test functional activation of the orphan peptide GPCR potentially encoded by CG6986. We found that CG6986 expression confers selective proctolin sensitivity to a heterologous cell type. We then extended our studies to further implicate CG6986 as a strong candidate for a Drosophila proctolin receptor.
Materials and Methods
Molecular Cloning.
We constructed a full-length CG6986 coding sequence appropriate for expression (25) in HEK cells by stepwise PCR amplification using RACE-generated cDNAs and a CG6986 EST as templates. Details of the cloning steps and reagents are found in Supporting Materials and Methods, which is published as supporting information on the PNAS web site, www.pnas.org.
Cell Culture, Transfections, and Functional Assays.
HEK-293 cells that contain a FLP recombination target (FRT) site integrated within the genome (Invitrogen) were cultured on DMEM supplemented with 10% FBS and 100 units per μg per ml of a penicillin/streptomycin solution. Cells were transfected with a total of 10 μg of plasmid DNA [a 9:1 ratio of pOG44 to CG6986 in pcDNA5/FRT] by using Lipofectamine (Invitrogen) according to the manufacturer's recommendations. After 48 h, the cells were selected on DMEM containing 100 μg/ml hygromycin B. Cells were split at a 1:5 ratio every 3 days and maintained under an atmosphere of 5% CO2 at 37°C in a humidified incubator. After selection for resistance to antibiotics, cells were assayed for ligand-dependent receptor activation by monitoring changes in intracellular calcium after incubation with the dye, Fluo3-AM (Molecular Probes). Details of dye incubation and fluorescence measurements are provided in Supporting Materials and Methods.
Binding Assays.
Membranes were prepared by differential centrifugation through sucrose. Proctolin was iodinated by a chloramine-T method (26). Conditions for determination of specific binding of 125I-labeled proctolin to membranes were as follows: membranes were added to a 1.5-ml polypropylene microfuge tube containing 100 pM 125I-labeled proctolin in 50 mM Tris⋅HCl, pH 7.5, 1× Hanks' balanced salt solution, 1.5% BSA, and protease inhibitor in the presence of various amounts of unlabeled proctolin or other peptides. For HEK cells transfected with CG6986, the ability of unlabeled proctolin to displace binding of the label to membranes was compared with six other neuropeptides. Binding reactions were conducted at room temperature for 2 h and were terminated by centrifugation and subsequent washing. Pellets were collected for determination of radioactivity on a Packard Gamma II counter. Raw counts obtained from the binding assays were converted to percent total binding, and resulting data were analyzed by nonlinear regression analysis with graphpad prism software (San Diego). More complete details are provided in Supporting Materials and Methods.
Immunocytochemistry and Western Blots.
A synthetic peptide corresponding to the C-terminal 20 aa of the predicted CG6986 product was used to generate rabbit antisera (Multiple Peptide Systems, San Diego). An antiserum raised in rabbit to proctolin conjugated to thyroglobulin with glutaraldehyde was produced commercially (Euro Diagnostica, Malmö, Sweden). HEK cells were fixed for 60 min at RT in 4% paraformaldehyde in PBS, containing 7% picric acid (vol/vol). The cells were washed and stained for 1 h at room temperature in anti-CG6986; cells were washed, then stained with Cy3-conjugated anti-rabbit antibodies (Jackson Laboratories, West Grove, PA). For Western blots, SDS gels were electrotransferred to poly(vinylidene difluoride) membranes, then incubated with primary antibody at a 1:750 dilution for 1 h, and signals were detected after incubation with an alkaline phosphatase-conjugated anti-rabbit antibody.
Immunocytochemistry was performed on tissues of adults and third-instar larvae of the Oregon R strain. Fresh-frozen, 15-μm cryostat sections were collected from adult heads, thoraces, and abdomens and fixed in 4% paraformaldehyde in sodium phosphate buffer for 1 h. Sections were incubated for 24 h with diluted antireceptor (1:1,000) and antipeptide (1:2,000) antisera. We used either Cy3-tagged secondary antiserum or the biotin-streptavidine method (with peroxidase detection) using a DAKO ABC kit following manufacturer's recommendations. Whole-mount staining was performed on adult and larval CNS and intestine by using Cy3 detection. We tested preimmune sera at a 1:1000 dilution, and antisera preadsorbed with synthetic peptide immunogen (100 nmol/ml). Images were obtained on a Zeiss Axioplan microscope equipped with a Hamamatsu charge-coupled device camera (Ichinocho, Japan) using OPENLAB 3.1.2 software (Improvision, Coventry, U.K.) and PHOTOSHOP 6.0 software (Adobe Systems, Mountain View, CA).
Results
Functional Assay.
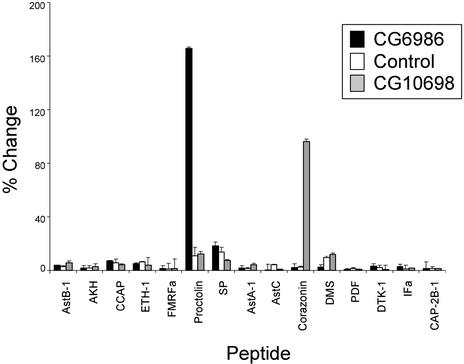
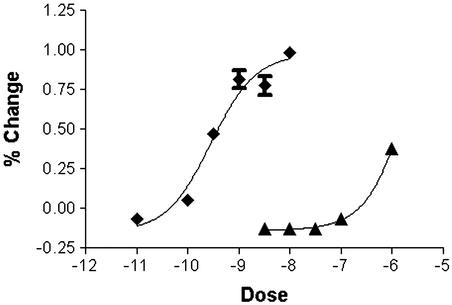
We generated six independent HEK 293 lines that were stably transfected with a cDNA corresponding to CG6986 and one line stably transfected with a cDNA corresponding to CG10698. We tested these lines with a library of insect bioactive peptides for receptor activation by monitoring changes in calcium-dependent fluorescence. The addition of individual peptides to untransfected cells resulted in small responses (Fig. 1). CG6986-transfected cells responded to proctolin at 10 nM (P < 10−12), but not to any of 14 other peptides tested (Fig. 1). At 1 μM, both proctolin and sex peptide (SP) produced responses (P < 10−8 and P < 10−4, respectively), although the magnitude of the response to proctolin was much larger. Further, CG6986 was sensitive to proctolin at subnanomolar concentrations, whereas it did not respond to SP at concentrations <1 μM (Fig. 2). CG10698-transfected cells displayed specific responses to corazonin with an EC50 comparable to values reported (27, 28) and did not respond to proctolin at doses as high as 1 μM. From the dose-response curve (Fig. 2), we calculated an EC50 value for proctolin on CG6986 cells of 0.3 nM (R2 = 0.9662). That value suggested that proctolin is a good candidate to be an endogenous ligand of the receptor encoded by CG6986.
Figure 1.
Effects of peptides on HEK cells stably expressing CG6986 or CG10698. Fluorescence was measured before and immediately after peptide addition. The concentration of all peptides shown was 10−8 M. Compared with control cells, only proctolin induced significant changes in cells expressing CG6986 and only corazonin induced significant changes in cells expressing CG10698. AstB, allatostatin B; AKH, adipokinetic hormone; CCAP, crustacean cardioacceleratory peptide; ETH, ecdysis-triggering hormone; FMRF, DPKQDFMRFamide; Ast, allatostatin A; AstC, allatostatin C; DMS, dromyosuppressin; PDF, pigment dispersing factor; IFa, IF amide; dTK, Drosophila tachykinin; and CAP-2B1, cardioacceleratory peptide 2B1.
Figure 2.
Dose responsiveness of HEK cells expressing CG6986 to proctolin and SP application. The percentage changes due to proctolin (■) and SP (▴) addition were computed as means ± SE from eight wells tested per dose per peptide.
Binding Properties.
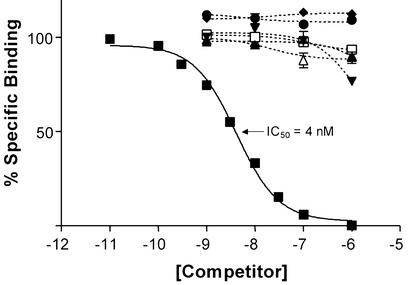
To test the hypothesis that proctolin might be an endogenous ligand of CG6986, we characterized CG6986 binding properties with a RRA. Membranes prepared from HEK cells that were stably transfected with the CG6986 receptor bound 125I-proctolin specifically. For label at 0.1 nM, total specific binding was composed of ≈8% of the added 125I-proctolin. Nonspecific binding (addition of 1 μM unlabeled proctolin) constituted only 1% of the label bound. The binding of 125I-proctolin was displaced by addition of unlabeled proctolin in a concentration-dependent manner (Fig. 3), with an IC50 of 4 nM (R2 = 0.9941). In contrast, label was not displaced to any similar degree by corazonin, [Leu-5]enkephalin, [Met-5]enkephalin, FMRFamide, leucomyosupressin (LMS), or leucokinin. Lack of such competition by other peptides further supports the specificity of binding exhibited by synthetic proctolin. Membranes prepared from untransfected control cells exhibited no specific binding for 125I-proctolin (data not shown).
Figure 3.
Competition for 125I-proctolin binding to membranes prepared from HEK cells stably transfected with CG6986. Membranes were incubated with 100 pM 125I-proctolin and various concentrations of unlabeled proctolin (■) [Leu-5]enkephalin (▴), [Met-5]enkephalin (▾), leucomyosupressin (♦), FMRFamide (●), leucokinin VIII (□), or corazonin (▵) for 2 h at 30°C. Values indicate means ± SE (n = 3).
Expression.
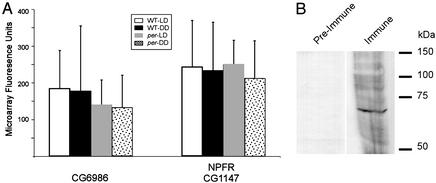
We evaluated potential expression of both transcript and protein levels in Drosophila tissues. Measures of CG6986 RNA levels were mined from data collected in previously described microarray experiments that studied adult head RNA (ref. 29; raw data are available at http://circadian.wustl.edu). That data set includes measurements from two genotypes (y w and period w). Each genotype was also measured in each of two environmental conditions (12 h light, 12 h dark, and constant darkness). Finally, the data includes measurements made at 4-h intervals to monitor possible diurnal and/or circadian variation. CG6986 is expressed in adult head at low, but sustained levels (Fig. 4A) that match those of a previously identified peptide GPCR [CG1147, neuropeptide F receptor (NFPR); ref. 30]. This consistent level of expression is notable given that roughly half of the ≈14,000 genes represented on the microarray were not reliably detected with adult head RNA (29). The average expression levels for these peptide GPCRs did not change significantly as a function of genotype, of environmental condition (Fig. 4A), or of time of day (data not shown). CG6986 protein levels were detected with an antibody directed to the C terminus of the predicted protein. HEK cells that were stably transfected with CG6986, but not with untransfected cells, were specifically stained (data not shown). By Western blot analysis (n = 5), we observed a band of ≈60 kDa (Fig. 4B), which is close to the predicted value of 62 kDa for the CG6986 protein (Berkeley Drosophila Genome Project).
Figure 4.
In vivo expression of CG6986. (A) Histograms indicating means (±SE) for average fluorescence of CG6986 and CG1147 on each of several genome-wide microarrays (ref. 29; http://circadian.wustl.edu). For the different conditions listed: WT LD, n = 18; WT DD, n = 12; per LD, n = 12; and for per DD, n = 12. (B) Western blot analysis of whole adult bodies stained with antisera to CG6986-predicted protein. MW standards are indicated in kDa.
Distribution of Immunosignals.
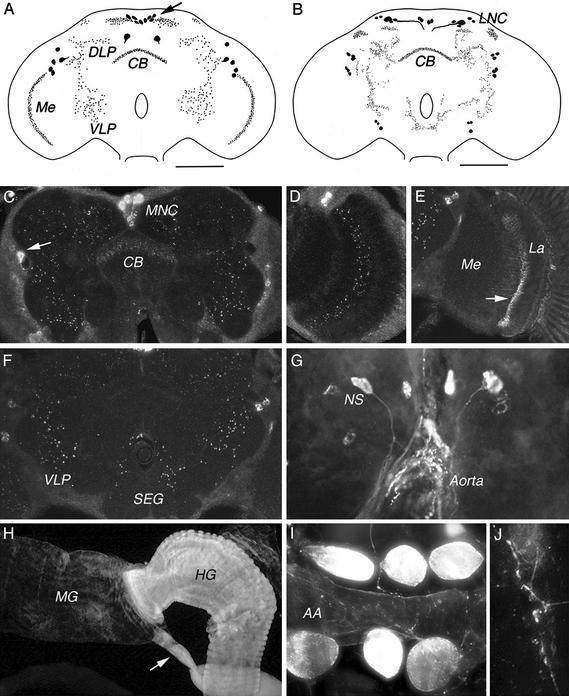
In the adult brain, punctate labeling was observed in the neuropil of the superior median and lateral protocerebrum, of the ventral lateral protocerebrum, of the upper division of central body, and of the subesophageal ganglion (Fig. 5A and C–F). In the optic lobe, labeling was detected in the serpentine layer of the medulla (Fig. 5D) and in a basal layer of the lamina (Fig. 5E). Labeling was also seen in a number of neuronal cell bodies in the brain and optic lobes (Fig. 5A). Two prominently labeled cell bodies are located posteriorly in the superior median protocerebrum and eight others are located in the median neurosecretory cell group of protocerebrum. There are two pairs of labeled cell bodies in the lateral dorsal protocerebrum and a pair in the medulla.
Figure 5.
Immunolabeling in Drosophila tissues with antiserum to proctolin receptor (A and C–J) and to proctolin peptide (B). (A) Tracing of receptor distribution in adult Drosophila brain (compressed frontal view). A few clusters of cell bodies are seen dorsal to the medulla (Me). Posterior to the central body (CB) and in the median neurosecretory cell group (arrow). Punctate labeling is seen in the Me, dorsally in the CB, adjacent to the median neurosecretory cells and in dorsal (DLP) and ventral (VLP) lateral protocerebrum. (B) Tracing of antiproctolin labeling. Cell bodies are seen in lateral neurosecretory cell group (LNC), dorsal to the medulla and in vertrolateral protocerebrum. Faint punctate processes were also seen in the medulla, but were not drawn. (C–F) Proctolin receptor labeling in adult brain. (C) Anterior brain section with punctate labeling in the CB and protocerebral neuropil. Cell bodies are seen in the MNC group and lateral protocerebrum (arrow). (D) Labeling in the serpentine layer of medulla (optic lobe) and in a pair of dorsal cell bodies. (E) Labeling of neural processes in basal neuropil of lamina (optic lobe). (F) Labeling in the VLP and subesophageal ganglion (SEG). (G) Labeled neurons in larval brain; some of these are neurosecretory cells (NS) with processes arborizing on the wall of the anterior aorta adjacent to the ring gland. (H) Labeling of the adult intestine, in the muscle layer of the hindgut (HG) and basal portion of Malpighian tubules (arrow), but not in the midgut (MG). (I) In the abdominal aorta (AA) of the adult, motor nerve terminals on the heart muscle and adjacent pericardial cells are labeled. (J) Detail of labeled neuronal terminals on heart.
Fig. 5B shows the proctolin immunoreactive cell bodies that are present in adult brains. Mostly, there appears good correspondence between the distributions of peptide and receptor immunosignals. For example, peptide immunoreactive processes were seen in the upper division of the central body, in the lateral and superior median protocerebrum, and in the medulla (not shown).
The adult thoracic abdominal ganglia displayed very little proctolin receptor immunolabeling. In the larval brain, at least two pairs of median neurosecretory cells were immunolabeled together with three more pairs of protocerebral neurons (Fig. 5G). In the adult and larval hindguts, intense immunolabeling could be seen in the muscle layers (Fig. 5H) and Malpighian tubules (especially their basal portions). In adult heart, we observed distinct labeling of neuronal terminals (Fig. 5 I and J), as well as strong labeling of the adjacent pericardial cells (Fig. 5I). Preimmune serum and preabsorbed receptor antiserum produced no labeling in the nervous system or muscle.
Discussion
We demonstrated that the GPCR encoded by CG6986 is likely an integral component of proctolin signaling in Drosophila. Our evidence stems from results of three independent experimental approaches. We showed (i) a functional and highly specific response to proctolin, (ii) high-affinity binding of the pentapeptide to the receptor, and (iii) anatomical distribution of the receptor consistent with previously described proctolin sensitivity.
We showed that the expression of CG6986 in the heterologous HEK system specifically conferred sensitivity for the neuropeptide proctolin. The high sensitivity of this functional assay to proctolin is consistent with EC50 values previously estimated from in vivo physiological studies of proctolin action (20). However, expression in a heterologous cell line is not strictly comparable to normal receptor expression in vivo; although a similarity in EC50 values is suggestive, we do not consider that evidence alone to constitute compelling proof. Therefore, to establish independent evidence, we evaluated the binding properties of this receptor for proctolin. Our estimation of the IC50 value of 4 nM for proctolin competition is in accord with IC50 values for other peptide GPCRs (30), and suggests a high-affinity interaction. Significantly, that value is within an order of magnitude of the estimated proctolin EC50 value. We also note that the IC50 estimation represents 10–100 times greater sensitivity than was seen for proctolin binding to tissue membrane preparations from larger insects (20, 21). That discrepancy may derive from several cellular differences between the mammalian cell line and Drosophila tissues. Alternatively, variation in IC50 estimations may derive from inherent differences in proctolin receptors among different arthropod species, or differences because of receptor interactions with other signaling molecules.
The “promiscuous” G protein Gα16 (31) has been used in several recent studies of Drosophila peptide GPCRs to improve the chances of generating a measurable response (e.g., refs. 28, 32–33). Addition of this subunit proved unnecessary for study of the CG6986 receptor: Coexpression of Gα16 did not alter proctolin sensitivity in HEK cells (data not shown), but did increase responses to other peptides (E.C.J., unpublished work). We assume that the increase in calcium-dependent fluorescence after receptor activation in HEK cells is the result of Gq coupling and concomitant activation of the phospholipase C pathway (34). There is substantial evidence implicating phosphoinositol metabolism as a component of proctolin signaling (11, 12, 18, 20), but other pathways have also been proposed (35–37). Proctolin signaling by means of CG6986 could be responsible for all of these divergent observations (see refs. 38 and 39 for examples of GPCRs coupling to multiple signaling pathways). Alternatively, it is possible that additional proctolin receptors may be necessary to explain all of the proctolin actions so far demonstrated.
Microarray analysis of the CG6986 transcript confirmed that it is expressed consistently in the head of adult flies. Although absolute levels of CG6986 mRNA appeared low, they were comparable to those of the neuropeptide F GPCR, CG1147 (30). Proctolin binding sites from locust hindgut and oviducal muscles display an estimated size ≈50 kDa after partial purification (40). We estimated the size of the CG6986 protein at ≈60 kDa by Western Blot analysis, a size consistent with the computational prediction. There are several possible reasons for the apparent size difference, including species variation, and/or the possibility of additional genes that encode alternative functional proctolin receptors.
By immunolocalization, this receptor is highly expressed in tissues that were previously shown to be physiologically responsive to proctolin. Such data represent still another line of evidence to implicate CG6986 as a proctolin receptor. Specifically, marked CG6986 receptor expression in the hindgut is consistent with the potent myostimulatory effects of proctolin on that tissue in cockroaches (1). A similar physiological action of proctolin was confirmed for the case of the Drosophila hindgut, which is also innervated by proctolin-immunoreactive neurons originating in the abdominal ganglia (9). Expression of the CG6986 receptor in Drosophila Malpighian tubules is consistent with reports that, in the locust, proctolin induces tubule secretion and stimulates tubule writhing (41). Finally, there appears an excellent correspondence in several brain regions between the distribution of proctolin receptor immunosignals and those of proctolin-expressing neurites. There are no previous descriptions of the spatial distribution of proctolin in the adult brain of Drosophila. However, studies in the larger fly Calliphora (42) revealed a number of neurons similar to the ones shown herein for Drosophila.
Information regarding locations and times of proctolin receptor expression can be of value in the interpretation of proctolin signaling. For example, several sites where proctolin was assumed to act directly, such as hindgut and heart, were immunolabeled here. Several groups have postulated a role for proctolin in regulating heart rate of various insects (16, 43). We did not observe staining in the heart proper, but did observe minor staining in the anterior aorta and in some nerve terminals apposed to cardiac tissue. There are conflicting reports on proctolin action on heart rate in Drosophila (9, 44). Given the mechanism of cardiac pacemaking and its modulation (45), any direct action of proctolin on heart muscle is likely to be excitatory, whereas indirect action onto neurons that innervate the heart could have a net excitatory or inhibitory action (46). Proctolin receptor immunosignals on neurosecretory cells suggest the peptide may act as a releasing (or inhibiting) factor of other, specific peptide hormones. Identifying those cells, and relating their secretory products to proctolin actions, may be a useful step in providing detail to neural circuits that control discrete physiological functions. An unexpected role for proctolin in visual processing was indicated by the presence of both peptide and receptor immunostaining in the medulla and lamina. Likewise, the expression of receptor immunosignals in pericardial cells suggests that proctolin may have unpredicted roles in cardiac physiology.
In its transmembrane domains, CG6986 shares only limited identity with other peptide GPCRs (24). It is distantly related to the candidate dFMRFamide receptor CG2114 (32, 33) and to the orphan CG16726 receptor. The primary significance of the present work lies in its potential to use a genetic analysis in Drosophila to explore the functions and mechanisms of proctolin signaling in vivo. However, the discovery of a candidate proctolin receptor will also promote further studies of proctolin-signaling mechanisms underlying synaptic modulation in other arthropods. For example, three different proctolin-expressing neurons produce robust modifications of the pyloric circuit in Cancer borealis (5). However, the precise details of the regulatory effects depend on which proctolin neuron is activated, and the mechanisms that pattern such fundamental synaptic modulation are unknown. Clearly, information about sites of proctolin receptor expression will present a useful means with which to pursue such issues. Comparative studies may also reveal evolutionary divergences, as proctolin signaling may not be a uniform feature of arthropod phylogeny. blastp searches of the Anopheles (mosquito) genome reveals an absence of candidate CG6986 orthologues (47). The mosquito genome contains a candidate proctolin gene (48), but its organization is dissimilar to the best candidate found in the Drosophila genome (CG7105). Notably, CG7105 has an expression pattern similar to that of proctolin and proctolin-precursor immunolabeling (D.R.N., A. M. E. Winther, R. J. Siviter, M. Dushay, A. D. Shirras, and R. E. Isaac, unpublished work). The mosquito hindgut lacks proctolin sensitivity (49). Likewise, the moth Manduca displays proctolin-immunoreactive neurons, but there is no reported evidence of functional responsiveness to the peptide (50). Given these suggestions of evolutionary variation, the ability to assay for proctolin receptor expression will strengthen efforts to interpret the occurrence and significance of proctolin modulation in arthropods.
Supplementary Material
Acknowledgments
We thank Anne Karlsson for technical assistance, Cornelis Grimmelikhuijzen and Michael Adams for sharing unpublished information, Erik Kubli and Jan Veenstra for samples of synthetic peptides, and Randy Hewes for comments on the manuscript. We also thank the Berkeley Drosophila Genome Project for genomic and EST sequence information. The work was supported by a Keck Fellowship (to E.C.J.), a grant from the Human Frontier Science Program (to D.R.N. and P.H.T.), grants from the McDonnell Center (to P.H.T.), and National Institutes of Health Grant NS21749 (to P.H.T.).
Abbreviations
- GPCR
G protein-coupled receptor
- SP
sex peptide
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Brown B, Starratt A. J Insect Physiol. 1975;21:1879–1881. [Google Scholar]
- 2.Osborne R H. Pharmacol Ther. 1996;69:117–142. doi: 10.1016/0163-7258(95)02054-3. [DOI] [PubMed] [Google Scholar]
- 3.Konopinska D, Rosinski G. J Pept Sci. 1999;5:533–546. doi: 10.1002/(SICI)1099-1387(199912)5:12<533::AID-PSC225>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 4.Christie A E, Baldwin D H, Marder E, Graubard K. Cell Tissue Res. 1997;288:135–148. doi: 10.1007/s004410050801. [DOI] [PubMed] [Google Scholar]
- 5.Nusbaum M P, Blitz D M, Swensen A M, Wood D, Marder E. Trends Neurosci. 2001;24:146–154. doi: 10.1016/s0166-2236(00)01723-9. [DOI] [PubMed] [Google Scholar]
- 6.Rathmayer W, Djokaj S, Gaydukov A, Kreissl S. J Neurosci. 2002;22:708–717. doi: 10.1523/JNEUROSCI.22-03-00708.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O'Shea M, Adams M. Adv Insect Physiol. 1986;19:1–28. [Google Scholar]
- 8.Orchard I, Belanger J H, Lange A B. J Neurobiol. 1989;20:470–496. doi: 10.1002/neu.480200515. [DOI] [PubMed] [Google Scholar]
- 9.Anderson M S, Halpern M E, Keshishian H. J Neurosci. 1988;8:242–255. doi: 10.1523/JNEUROSCI.08-01-00242.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hiripi L, Rozsa K S, Miller T A. Experientia. 1979;35:1287–1288. doi: 10.1007/BF01963960. [DOI] [PubMed] [Google Scholar]
- 11.Lange A B. Arch Insect Biochem Physiol. 1988;10:201–209. [Google Scholar]
- 12.Baines R A, Lange A B, Downer R G. J Comp Neurol. 1990;297:479–486. doi: 10.1002/cne.902970402. [DOI] [PubMed] [Google Scholar]
- 13.Hinton M, Osborne R H. Insect Biochem Mol Biol. 1996;26:111–117. [Google Scholar]
- 14.Puiroux J, Pedelaborde A, Loughton B G. Insect Biochem Mol Biol. 1992;22:547–551. [Google Scholar]
- 15.Puiroux J, Pedelaborde A, Loughton B G. Insect Biochem Mol Biol. 1992;22:859–865. [Google Scholar]
- 16.Bartosz-Bechowski H, Rosinski G, Konopinska D, Sujak P, Sobotka W. Int J Pept Protein Res. 1990;36:450–456. doi: 10.1111/j.1399-3011.1990.tb01305.x. [DOI] [PubMed] [Google Scholar]
- 17.Konopinska D. J Pept Res. 1997;49:457–466. [PubMed] [Google Scholar]
- 18.Lange A B, Orchard I, Konopinska I. J Insect Physiol. 1993;39:347–351. [Google Scholar]
- 19.Baines R A, Walther C, Hinton J M, Osborne R H, Konopinska D. J Neurophysiol. 1996;75:2647–2650. doi: 10.1152/jn.1996.75.6.2647. [DOI] [PubMed] [Google Scholar]
- 20.Mazzocco-Manneval C, Kuczer M, Konopinska D, Fournier B, Loughton B G, Puiroux J. Peptides (Tarrytown, NY) 1998;19:1641–1651. doi: 10.1016/s0196-9781(98)00120-x. [DOI] [PubMed] [Google Scholar]
- 21.Gray A S, Hancock J T, Osborne R H. Peptides (Tarrytown, NY) 2000;21:189–196. doi: 10.1016/s0196-9781(99)00199-0. [DOI] [PubMed] [Google Scholar]
- 22.Adams M D, Celniker S E, Holt R A, Evans C A, Gocayne J D, Amanatides P G, Scherer S E, Li P W, Hoskins R A, Galle R F, et al. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 23.Brody T, Cravchik A. J Cell Biol. 2000;150:F83–F88. doi: 10.1083/jcb.150.2.f83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hewes R, Taghert P. Genome Res. 2001;11:1126–1142. doi: 10.1101/gr.169901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kozak M. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crim J W, Garczynski S F, Brown M R. Peptides (Tarrytown, NY) 2002;23:2045–2051. doi: 10.1016/s0196-9781(02)00192-4. [DOI] [PubMed] [Google Scholar]
- 27.Park Y, Kim Y, J, Adams M E. Proc Natl Acad Sci USA. 2002;99:11423–11428. doi: 10.1073/pnas.162276199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cazzamali G, Grimmelikhuijzen C J. Proc Natl Acad Sci USA. 2002;99:12073–12078. doi: 10.1073/pnas.192442799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin Y, Han M, Shimada B, Wang L, Gibler T M, Amarakone A, Awad T A, Stormo G D, Van Gelder R N, Taghert P H. Proc Natl Acad Sci USA. 2002;99:9562–9567. doi: 10.1073/pnas.132269699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garczynski S F, Brown M R, Shen P, Murray T F, Crim J W. Peptides (Tarrytown, NY) 2002;23:773–780. doi: 10.1016/s0196-9781(01)00647-7. [DOI] [PubMed] [Google Scholar]
- 31.Offermanns S, Simon M I. J Biol Chem. 1995;270:15175–15180. doi: 10.1074/jbc.270.25.15175. [DOI] [PubMed] [Google Scholar]
- 32.Cazzamali G, Saxild N, Grimmelikhuijzen C. Biochem Biophys Res Commun. 2002;298:31–36. doi: 10.1016/s0006-291x(02)02398-7. [DOI] [PubMed] [Google Scholar]
- 33.Meeusen T, Mertens I, Clynen E, Baggerman G, Nichols R, Nachman R J, Huybrechts R, De Loof A, Schoofs L. Proc Natl Acad Sci USA. 2002;99:15363–15368. doi: 10.1073/pnas.252339599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Singer A U, Waldo G L, Harden T K, Sondek J. Nat Struct Biol. 2002;9:32–36. doi: 10.1038/nsb731. [DOI] [PubMed] [Google Scholar]
- 35.Swales L S, Evans P D. Histochemistry. 1988;90:233–239. doi: 10.1007/BF00492512. [DOI] [PubMed] [Google Scholar]
- 36.Baines R A, Downer R G. Arch Insect Biochem Physiol. 1992;20:215–229. doi: 10.1002/arch.940200306. [DOI] [PubMed] [Google Scholar]
- 37.Wegener C, Nässel D R. J Neurophysiol. 2000;84:3056–3066. doi: 10.1152/jn.2000.84.6.3056. [DOI] [PubMed] [Google Scholar]
- 38.Cheng J, Baldassare J J, Raben D M. Biochem J. 1999;337:97–104. [PMC free article] [PubMed] [Google Scholar]
- 39.Real V, Hannan F, Hall L M, Evans P D. J Neurosci. 1997;17:6545–6553. doi: 10.1523/JNEUROSCI.17-17-06545.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Puiroux J, Pédelaborde A, Loughton B G. Insect Biochem Mol Biol. 1998;28:887–893. [Google Scholar]
- 41.Coast G M. Peptides (Tarrytown, NY) 1998;19:469–480. doi: 10.1016/s0196-9781(97)00461-0. [DOI] [PubMed] [Google Scholar]
- 42.Nässel D R, O'Shea M. J Comp Neurol. 1987;265:437–454. doi: 10.1002/cne.902650311. [DOI] [PubMed] [Google Scholar]
- 43.Sliwowska J, Rosinski G, Nässel D R. Peptides (Tarrytown, NY) 2001;22:209–217. doi: 10.1016/s0196-9781(00)00384-3. [DOI] [PubMed] [Google Scholar]
- 44.Zornik E, Paisley K, Nichols R. Peptides (Tarrytown, NY) 1999;20:45–51. doi: 10.1016/s0196-9781(98)00151-x. [DOI] [PubMed] [Google Scholar]
- 45.Johnson E, Sherry T, Ringo J, Dowse H. J Comp Physiol B. 2002;172:227–236. doi: 10.1007/s00360-001-0246-8. [DOI] [PubMed] [Google Scholar]
- 46.McGaw I J, Wilkens J L, McMahon BR, Airriess C N. J Exp Biol. 1995;198:2547–2550. doi: 10.1242/jeb.198.12.2547. . . [DOI] [PubMed] [Google Scholar]
- 47.Hill C A, Fox A N, Pitts R J, Kent L B, Tan P L, Chrystal M A, Cravchik A, Collins F H, Robertson H M, Zwiebel L J. Science. 2002;298:176–178. doi: 10.1126/science.1076196. [DOI] [PubMed] [Google Scholar]
- 48.Riehle M A, Garczynski S F, Crim J W, Hill C A, Brown M R. Science. 2002;298:172–175. doi: 10.1126/science.1076827. [DOI] [PubMed] [Google Scholar]
- 49.Messer A C, Brown M R. J Exp Biol. 1995;198:2325–2336. doi: 10.1242/jeb.198.11.2325. [DOI] [PubMed] [Google Scholar]
- 50.Davis N T, Velleman S G, Kingan T G, Keshishian H. J Comp Neurol. 1989;283:71–85. doi: 10.1002/cne.902830107. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.