Abstract
The purpose of the present study was to assess the in vitro activity of daptomycin against a well-defined collection of methicillin-resistant Staphylococcus aureus (MRSA) isolates (n = 98), including heterogeneously glycopeptide-resistant MRSA (hGISA) strains. Susceptibility testing was performed by using the Etest system. Daptomycin was potent against both glycopeptide-susceptible and hGISA strains.
Methicillin-resistant Staphylococcus aureus (MRSA) is an important nosocomial pathogen (1). According to a report from the National Nosocomial Infection Surveillance System, ca. 60% of all S. aureus isolated from patients in intensive care units in U.S. hospitals were methicillin resistant in 2003 (12). Infections caused by MRSA strains are associated with longer hospital stay, more days of antibiotic administration, and higher costs than infections caused by methicillin-susceptible Staphylococcus aureus (MSSA) strains (1, 5). Reports of vancomycin intermediately susceptible S. aureus (VISA), first isolated in Japan in 1997 (9), and vancomycin-resistant S. aureus in the United States (4) caused widespread alarm among physicians since it was feared that we are entering an era of untreatable S. aureus infections. Vancomycin remains the standard for treating most MRSA infections; however, concerns over increases in the rates of heteroresistance and tolerance to this agent, combined with its pharmacodynamic and clinical shortcomings and the increasingly important role of gram-positive bacterial infections in clinical medicine, have motivated the development of newer agents. Daptomycin is a cyclic lipopeptide antibiotic that is rapidly bactericidal in vitro against a broad spectrum of gram-positive bacteria. Its unique mechanism of action involves calcium-dependent binding to the bacterial plasma membrane and disruption of membrane function (3).
The purpose of the present study was to assess the in vitro activity of daptomycin against MRSA isolates collected in The Netherlands using a well-defined collection of strains that included heterogeneously glycopeptide-resistant MRSA strains (hGISA).
The MRSA isolates were collected in The Netherlands between 1989 and 1998 and are part of the MRSA strain collection of the National Institute of Public Health and Environmental Protection, Bilthoven, The Netherlands. Identification as S. aureus and methicillin resistance were determined by duplex PCR for the mecA gene and coagulase gene as described previously (10, 19). In the present study a total of 98 strains were tested, including 19 hGISA strains, which were determined by using modified population analysis profiles (20, 21). The MIC of daptomycin was determined by using the Etest system (AB Biodisk, Solna, Sweden) with a concentration range of 0.016 to 256 μg/ml. The daptomycin Etest contained a concentration gradient of daptomycin with a standard amount of calcium throughout the strip. E-test strips were applied to the surface of 150-mm Mueller-Hinton agar plates. Plates were incubated at 35°C in ambient air for 18 to 24 h prior to reading the MIC results. The MICs of the following antimicrobial agents were determined simultaneously: vancomycin, teicoplanin, gentamicin, cotrimoxazole, ciprofloxacin, erythromycin, clindamycin, and rifampin. All MICs were determined with the Etest system. For vancomycin and teicoplanin, the Etest strips were placed on brain heart infusion agar, and a large inoculum (no. 2 McFarland standard) and an extended incubation time (48 h) were used to be able to detect hGISA isolates. Isolates were categorized as susceptible or resistant to an antimicrobial agent according to the breakpoints published by the Clinical and Laboratory Standards Institute (CLSI; formerly the National Committee for Clinical Laboratory Standards) (11). The breakpoint for daptomycin is 1 mg/liter for S. aureus (both methicillin-resistant and methicillin-susceptible strains).
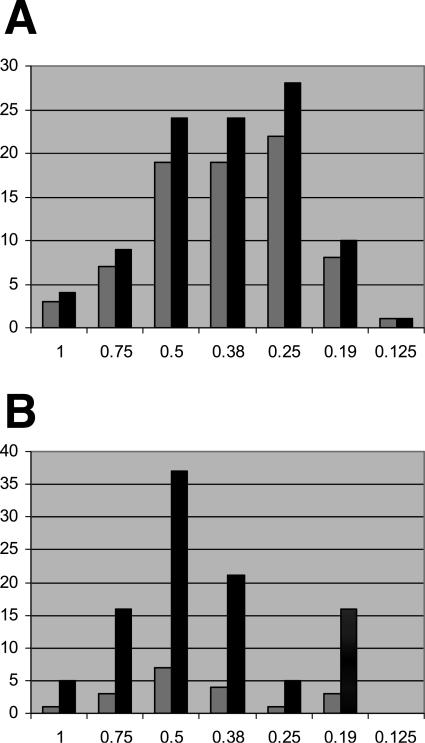
For 98 MRSA isolates, the MIC range for daptomycin was 0.125 to 1.0 μg/ml (Table 1 and Fig. 1), with MICs at which 50 and 90% of the isolates tested are inhibited (MIC50 and MIC90, respectively) of 0.38 and 0.75 μg/ml, respectively. The MIC range for daptomycin against hGISA isolates was 0.19 to 1.0 μg/ml. The MICs of MRSA isolates for other antibiotics are outlined in Table 1. The MICs of daptomycin correlated weakly, but statistically significantly, with those of vancomycin (r = 0.318; P = 0.001) and teicoplanin (r = 0.334; P = 0.001) by using log-linear regression analysis.
TABLE 1.
In vitro activities of daptomycin and other antimicrobial agents against 98 MRSA strains isolated in The Netherlands
| Antibiotic | MIC (mg/liter)
|
% Susceptibilitya
|
|||
|---|---|---|---|---|---|
| Range | MIC50 | MIC90 | S | R | |
| Daptomycin | 0.125-1 | 0.38 | 0.75 | 100 | 0 |
| Oxacillin | 0.19-≥256 | 256 | 256 | 0 | 100b |
| Vancomycinc | 2-6 | 3.0 | 4.0 | ||
| Teicoplaninc | 1-16 | 3.0 | 12.0 | ||
| Gentamicin | 0.38-≥256 | 1.0 | 256 | 54.1 | 43.9 |
| Cotrimoxazole | 0.032-≥32 | 0.094 | 32.0 | 87.8 | 11.2 |
| Ciprofloxacin | 0.25-≥32 | 32.0 | 32.0 | 24.5 | 72.4 |
| Erythromycin | 0.19-≥256 | 256 | 256 | 26.5 | 72.4 |
| Clindamycin | 0.047-≥256 | 0.19 | 256 | 58.2 | 41.8 |
| Rifampin | 0.004-≥32 | 0.012 | 32.0 | 69.4 | 21.3 |
S, sensitive; R, resistant.
Identification as S. aureus and methicillin resistance were determined by duplex PCR for the mecA gene and coagulase gene (19).
The Etest system with a large inoculum and 48 h of incubation was used.
FIG. 1.
MICs for daptomycin for 79 glycopeptide-susceptible MRSA strains (A) and 19 heterogeneously glycopeptide-resistant MRSA strains (hGISA) (B) collected in The Netherlands as determined by using the Etest system. Gray bars, total number of isolates tested; black bars, percentage of the total tested.
The results are generally comparable with other data on European and North American antibiotic-resistant clinical isolates that were phenotypically characterized (2, 8, 14, 18). In a recent study, the in vitro activity of daptomycin against 337 gram-positive European clinical isolates with known resistance genes was determined (7). MICs were determined by CLSI methodology using microtiter plates containing antibiotic solutions and physiologic concentrations of Ca2+ (50 μg/ml). For 38 MRSA isolates, the MIC range for daptomycin was 0.03 to 0.5 μg/ml, with MIC50 and MIC90 values for the isolates tested of 0.25 and 0.5 μg/ml, respectively. The daptomycin MICs determined in our study appear to be slightly higher. One explanation for these results may be the limited number of isolates tested in the study by Fluit et al. (7) and because we selected for hGISA in our study.
It appears that S. aureus with reduced susceptibility to vancomycin also exhibits reduced susceptibility to daptomycin (13, 16). In concordance with findings by Cui et al. (6), we found a positive correlation between reduced daptomycin susceptibility and reduced susceptibility to glycopeptides. Although the correlation coefficient is relatively weak, it is highly significant. In the study by Cui et al., the reduction of daptomycin susceptibility showed a strong correlation with the increment of cell wall thickness. The level of daptomycin susceptibility correlated strongly and positively with that of vancomycin susceptibility (r = 0.814; P < 0.0001) and with cell wall thickness (r = 0.883; P < 0.0001). It is conceivable that the changes mediating reduced susceptibility to vancomycin in S. aureus, including a thickened cell wall, changes in cellular metabolism, and enhanced cell wall turnover, interfere with the antimicrobial action of daptomycin. In a recent study, the bactericidal activity of vancomycin and daptomycin against 105 well-characterized S. aureus strains with decreased susceptibility against vancomycin (88 hGISA and 17 VISA), as well as 105 vancomycin-suseptible MRSA strains (VSSA) were tested by using a CLSI broth microdilution method (15). All VSSA and hGISA strains were inhibited by ≤1 mg of daptomycin/liter. However, a slight skewing toward higher daptomycin MIC results was noted when the hGISA (MIC50 = 0.5 mg/liter, and MIC90 = 1 mg/liter) and VISA (MIC50 = 1 mg/liter, and MIC90 = 2 mg/liter) were compared to VSSA strains (MIC50 and MIC90 = 0.5 mg/liter). Among the 17 VISA strains evaluated, 7 (41.2%) showed a daptomycin MIC of 2 mg/liter and 1 (5.9%) had a daptomycin MIC of 4 mg/liter. However, all daptomycin minimum bactericidal concentration (MBC) results were at the MIC or twofold higher than the MIC, and the MBC/MIC ratios were not significantly affected by the susceptibility to vancomycin. Conversely, 69.3% of hGISA strains and all VISA strains showed a vancomycin MBC/MIC ratio consistent with tolerance (tolerance was defined as an MBC/MIC ratio of ≥32 or ≥16 with an associated MBC at ≥32 mg/liter for vancomycin). The increased MIC of daptomycin associated with hGISA is therefore most likely not caused by an intrinsic resistance mechanism but by the thickening of the cell wall. Further studies are needed before final conclusions can be made.
In conclusion, daptomycin exhibits broad in vitro activity against a collection of MRSA strains collected in The Netherlands. The in vitro data presented in this and other studies, together with promising clinical results (17), show that daptomycin is a valuable new addition to the arsenal of antimicrobial agents for the treatment of infections caused by gram-positive microorganisms.
REFERENCES
- 1.Archer, G. L. 1998. Staphylococcus aureus: a well-armed pathogen. Clin. Infect. Dis. 26:1179-1181. [DOI] [PubMed] [Google Scholar]
- 2.Barry, A. L., P. C. Fuchs, and S. D. Brown. 2001. In vitro activities of daptomycin against 2,789 clinical isolates from 11 North American medical centers. Antimicrob. Agents Chemother. 45:1919-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carpenter, C. F., and H. F. Chambers. 2004. Daptomycin: another novel agent for treating infections due to drug-resistant gram-positive pathogens. Clin. Infect. Dis. 38:994-1000. [DOI] [PubMed] [Google Scholar]
- 4.Chang, S., D. M. Sievert, J. C. Hageman, M. L. Boulton, F. C. Tenover, F. P. Downes, S. Shah, J. T. Rudrik, G. R. Pupp, W. J. Brown, D. Cardo, S. K. Fridkin, et al. 2003. Infection with vancomycin-resistant Staphylococcus aureus containing the vanA resistance gene. N. Engl. J. Med. 348:1342-1347. [DOI] [PubMed] [Google Scholar]
- 5.Cosgrove, S. E., Y. Qi, K. S. Kaye, S. Harbath, A. W. Karchmer, and Y. Carmeli. 2005. The impact of methicillin resistance in Staphylococcus aureus bacteremia on patient outcomes: mortality, length of stay, and hospital charges. Infect. Control Hosp. Epidemiol. 26:166-174. [DOI] [PubMed] [Google Scholar]
- 6.Cui, L., E. Tominaga, H. Neoh, and K. Hiramatsu. 2006. Correlation between reduced daptomycin susceptibility and vancomycin resistance in vancomycin-intermediate Staphylococcus aureus. Antimicrob. Agents Chemother. 50:1079-1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fluit, A. C., F. J. Schmitz, J. Verhoef, and D. Milatovic. 2004. In vitro activity of daptomycin against gram-positive European clinical isolates with defined resistance determinants. Antimicrob. Agents Chemother. 48:1007-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fuchs, P. C., A. L. Barry, and S. D. Brown. 2002. In vitro bactericidal activity of daptomycin against staphylococci. J. Antimicrob. Chemother. 49:467-470. [DOI] [PubMed] [Google Scholar]
- 9.Hiramatsu, K., H. Hanaki, T. Ino, K. Yabuta, T. Oguri, and F. C. Tenover. 1997. Methicillin-resistant Staphylococcus aureus clinical strain with reduced vancomycin susceptibility. J. Antimicrob. Chemother. 40:135-136. [DOI] [PubMed] [Google Scholar]
- 10.Kluytmans, J., A. Van Griethuysen, P. Willemse, and P. Van Keulen. 2002. Performance of CHROMagar selective medium and oxacillin resistance screening agar base for identifying Staphylococcus aureus and detecting methicillin resistance. J. Clin. Microbiol. 40:2480-2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.National Committee for Clinical Laboratory Standards. 2006. Performance standards for antimicrobial susceptibility testing. Approved standard M100-S16. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 12.National Nosocomial Infections Surveillance System. 2004. National Nosocomial Infections Surveillance (NNIS) System Report, data summary from January 1992 through June 2004, issued October 2004. Am. J. Infect. Control 32:470-485. [DOI] [PubMed] [Google Scholar]
- 13.Patel, J. B., L. A. Jevitt, J. Hageman, L. C. McDonald, and F. C. Tenover. 2006. An association between reduced susceptibility to daptomycin and reduced susceptibility to vancomycin in Staphylococcus aureus. Clin. Infect. Dis. 42:1652-1653. [DOI] [PubMed] [Google Scholar]
- 14.Rybak, M. J., E. Hershberger, T. Moldovan, and R. G. Grucz. 2000. In vitro activities of daptomycin, vancomycin, linezolid, and quinupristin-dalfopristin against staphylococci and enterococci, including vancomycin-intermediate and -resistant strains. Antimicrob. Agents Chemother. 44:1062-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sader, H. S., P. R. Rhomberg, and R. N. Jones. 2005. Bactericidal activity of vancomycin and daptomycin tested against heterogeneous and homogeneous vancomycin-intermediate Staphylococcus aureus (hVISA and VISA) strains, p. 978. In Program and abstracts book, 15th European Congress of Clinical Microbiology and Infectious Diseases. European Congress of Clinical Microbiology and Infectious Diseases, Copenhagen, Denmark.
- 16.Sakoulas, G., J. Alder, G. M. Eliopoulos, R. C. Moellering, C. Thauvin-Eliopoulos, and G. M. Eliopoulos. 2005. Induction of daptomycin heterogeneous susceptibility in Staphylococcus aureus by exposure to vancomycin. Antimicrob. Agents Chemother. 50:1581-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schriever, C., C. Fernandez, K. Rodvold, and L. Danziger. 2005. Daptomycin: a novel cyclic lipopeptide antimicrobial. Am. J. Health Syst. Pharm. 62:1145-1158. [DOI] [PubMed] [Google Scholar]
- 18.Silverman, J. A., N. Oliver, O. Andrew, and T. Li. 2001. Resistance studies with daptomycin. Antimicrob. Agents Chemother. 45:1799-1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Griethuysen, A. J., M. Pouw, N. van Leeuwen, M. Heck, P. Willemse, A. Buiting, and J. Kluytmans. 1999. Rapid slide latex agglutination test for detection of methicillin resistance in Staphylococcus aureus. J. Clin. Microbiol. 37:2789-2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Griethuysen, A. J., A. van′t Veen, A. Buiting, T. Walsh, and J. Kluytmans. 2003. High percentage of methicillin-resistant Staphylococcus aureus isolates with reduced susceptibility to glycopeptides in The Netherlands. J. Clin. Microbiol. 41:2487-2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walsh, T. R., A. Bolmström, A. Qwärnström, P. Ho, M. Wootton, R. A. Howe, A. P. MacGowan, and D. Diekema. 2001. Evaluation of current methods for detection of staphylococci with reduced susceptibility to glycopeptides. J. Clin. Microbiol. 39:2439-2444. [DOI] [PMC free article] [PubMed] [Google Scholar]