Abstract
The methicillin-resistant Staphylococcus aureus (MRSA) population in the Hospital Universitario Nuestra Señora de Candelaria over a 5-year period (1998 to 2002) was marked by shifts in the circulation of pandemic clones. Here, we investigated the emergence of high-level mupirocin resistance (Hi-Mupr). In addition to clonal spread, transfer of ileS2-carrying plasmids played a significant role in the dissemination of Hi-Mupr among pandemic MRSA lineages.
Most hospital-acquired methicillin-resistant Staphylococcus aureus (MRSA) isolates are members of five MRSA pandemic lineages or clonal complexes (CCs), namely, CC5, CC8, CC22, CC30, and CC45 (6, 14). Mupirocin constitutes the cornerstone of avoidance of MRSA carriage and ulterior infection, but resistance has emerged, and its spreading is worrisome, with transferable high-level mupirocin resistance (Hi-Mupr) being of clinical significance (3, 4, 5, 11, 23, 25). Hi-Mupr is associated with an additional isoleucyl-tRNA synthetase that is encoded by the ileS2 gene (8, 10). The ileS2 gene was commonly reported on plasmids that differed in size, restriction patterns, and ability to be transferred in conjugation experiments (12, 13, 16, 21, 24, 26).
We reported that 95.5% of the 375 MRSA isolates obtained from patients at Hospital Universitario Nuestra Señora de Candelaria (located in Tenerife, Canary Islands, Spain) between 1998 and 2002 belonged to six clones fitting in pandemic lineages (i.e., CC5, CC8, CC22, and CC30) (20). In the present work, we investigated Hi-Mupr in such a staphylococcal population.
(Parts of this work were presented during the 1st International Workshop for Origin and Evolution of Bacterial Pathogens, Baeza, Spain, October 2004, and during the XX Congreso Nacional de Microbiología, Cáceres, Spain, September 2005.)
Isolates were screened for mupirocin resistance by the diskdiffusion and Etest methods (7, 15). The ileS2 gene was detected by a multiplex PCR (17, 18). Plasmid DNA was extracted with the QIAprep spin plasmid kit (QIAGEN, Hilden, Germany) with the addition of lysostaphin (Sigma Chemical Co., St. Louis, Mo.) and digested with EcoRI and HindIII. Curing of plasmids and conjugation experiments were performed as previously described (24). The primers used are listed in Table 1.
TABLE 1.
Amplification and sequencing primers used in this study
| Target gene | Primer namea | Sequence (5′-3′) | Reference or source | GenBank accession no. | Predicted PCR product size (bp) |
|---|---|---|---|---|---|
| Amplification | |||||
| ileS2 | MupA | TATATTATGCGATGGAAGGTTGG | 1 | 458 | |
| MupB | AATAAAATCAGCTGGAAGTGTTG | ||||
| traD-traE | TraD-E.F | GTAGCACAATTAGATCATGCAGTG | This study | NC_005024 | 212 |
| TraD-E.R | TGCCATAATCATAAACTCCCTTC | ||||
| traK | TraK-F | TTGCCGAAGATAGCGAATTG | This study | NC_005024 | 1,888 |
| TraK-R | CTGCAATACCTTCGGTCAGTTC | ||||
| Sequencing | |||||
| traK | TraK-R1 | CGCCACTAGGGTCAGTTACAAC | This study | NC_005024 | |
| TraK-F2 | GTCAGCTGGGTATGTTATTCCTAATG | This study | NC_005024 | ||
| TraK-R2 | GGGTGGTCTTTAGATAATTCATTAAAGC | This study | NC_005024 | ||
| TraK-F3 | AACCTAATCAAGATGAAGAAGGATCAG | This study | NC_005024 | ||
| TraK-R3 | ATATTTGGCCATTCGTCTAG | This study | NC_005024 | ||
| TraK-F4 | CTTCTCTCAAATGTTCCAACAGC | This study | NC_005024 | ||
| TraK-R4 | GCTTTATCCTGTTGCATATTAC | This study | NC_005024 | ||
| TraK-F5 | GCTACACTGGAAGGGCATTACTAAATG | This study | NC_005024 | ||
| TraK-R5 | TTCGTAGAATGCTTTCTTAG | This study | NC_005024 |
TraK-F and TraK-R primers were used in amplification and sequencing.
The ileS2 locus was associated with conjugative plasmids in each Hi-Mupr isolate.
Hi-Mupr was manifested by 48 MRSA isolates (12.8%) (Table 2) and increased significantly from 0% in 1998 to 15.6% in 2002 (P < 0.001). Each Hi-Mupr isolate belonged to one of the six major MRSA clones and amplified the ileS2 gene fragment. Non-Hi-Mupr isolates did not show the ileS2 amplicon. Pulsed-field gel electrophoresis (PFGE) subtypes shown by Hi-Mupr isolates, excepting PFGE subtype B4, also included mupirocin-susceptible isolates, suggesting intrahospital acquisition of Hi-Mupr by circulating clones.
TABLE 2.
Phenotypic and genotypic characteristics of high-level mupirocin-resistant MRSA isolates
| MRSA clone (lineage)b and isolate no. | Isolation date (day/mo/yr) | Hospital wardc | Sample origind | PFGE subtypeb | HindIII-REAP patterne | ileS2 locus polymorph | ileS2-carrying plasmid type | Resistance transferred to standard recipientf |
|---|---|---|---|---|---|---|---|---|
| ST125-IVA (CC5) | ||||||||
| HUNSC-356 | 06/11/2001 | Internal medicine | Wound | C1 | XI | E | pMUP2 | + |
| HUNSC-364 | 29/11/2001 | Internal medicine | Sputum | C1 | XI | E | pMUP2 | ND |
| HUNSC-414 | 22/03/2002 | Internal medicine | Nasal exudate | C1 | XI | E | pMUP2 | ND |
| HUNSC-453 | 22/05/2002 | Internal medicine | Wound | C1 | XII | E | pMUP2 | + |
| HUNSC-464 | 20/06/2002 | Internal medicine | Sputum | C1 | XI | E | pMUP2 | ND |
| HUNSC-485 | 28/08/2002 | Vascular surgery | Wound | C1 | XII | E | pMUP2 | ND |
| HUNSC-488 | 03/09/2002 | ICU | BAS | C1 | XIII | E | pMUP2 | + |
| HUNSC-492 | 13/09/2002 | Internal medicine | Nasal exudate | C1 | XI | E | pMUP2 | ND |
| HUNSC-505 | 07/10/2002 | Vascular surgery | Wound | C1 | XIII | E | pMUP2 | ND |
| HUNSC-510 | 23/10/2002 | Vascular surgery | Wound | C1 | XIII | E | pMUP2 | ND |
| HUNSC-523 | 13/11/2002 | Vascular surgery | Wound | C1 | XIV | E | pMUP2 | + |
| HUNSC-525 | 19/11/2002 | Vascular surgery | Wound | C1 | XIV | E | pMUP2 | ND |
| HUNSC-534 | 12/12/2002 | Internal medicine | Nasal exudate | C1 | XV | E | pMUP2 | + |
| HUNSC-535 | 12/12/2002 | Internal medicine | Nasal exudate | C1 | XV | E | pMUP2 | ND |
| HUNSC-537 | 17/12/2002 | Internal medicine | Nasal exudate | C1 | XV | E | pMUP2 | ND |
| HUNSC-538 | 17/12/2002 | Internal medicine | Nasal exudate | C1 | XV | E | pMUP2 | ND |
| HUNSC-540 | 18/12/2002 | Internal medicine | Nasal exudate | C1 | XV | E | pMUP2 | ND |
| HUNSC-541 | 31/12/2002 | Internal medicine | Nasal exudate | C1 | XV | E | pMUP2 | ND |
| ST146-IVA (CC5) HUNSC-521 | 09/11/2002 | Hematology | Wound | D1 | V | A | pMUP4 | ND |
| ST247-IA (CC8) | ||||||||
| HUNSC-229 | 04/05/2000 | Vascular surgery | Sputum | A1 | I | D | pMUP1 | + |
| HUNSC-246 | 08/04/2000 | ICU | Blood | A1 | I | D | pMUP1 | ND |
| HUNSC-267 | 07/11/2000 | Neurology | BAS | A1 | I | D | pMUP1 | ND |
| HUNSC-268 | 14/08/2000 | Neurology | Wound | A1 | I | D | pMUP1 | ND |
| HUNSC-182 | 19/01/1999 | ICU | BAS | A2 | II | D | pMUP1 | + |
| HUNSC-183 | 09/02/1999 | ICU | BAS | A2 | II | D | pMUP1 | ND |
| HUNSC-242 | 29/06/2000 | General surgery | Wound | A6 | I | D | pMUP1 | ND |
| HUNSC-243 | 04/07/2000 | General surgery | Blood | A6 | I | D | pMUP1 | ND |
| HUNSC-244 | 07/07/2000 | General surgery | Catheter | A6 | I | D | pMUP1 | ND |
| HUNSC-264 | 16/10/2000 | Digestive surgery | Wound | A8 | III | E | pMUP2 | + |
| ST22-IV (CC22) | ||||||||
| HUNSC-684 | 21/10/2002 | Dermatology | Wound | E1 | XVI | E | pMUP2 | + |
| HUNSC-799 | 06/04/2002 | Vascular surgery | Wound | E1 | XVI | E | pMUP2 | ND |
| ST36-II (CC30) | ||||||||
| HUNSC-234 | 06/06/2000 | Vascular surgery | Wound | B1 | IV | B | pMUP3 | + |
| HUNSC-238 | 14/06/2000 | Vascular surgery | Wound | B1 | IV | B | pMUP3 | ND |
| HUNSC-256 | 11/08/2000 | Internal medicine | Wound | B1 | V | A | pMUP4 | ND |
| HUNSC-277 | 14/02/2001 | Internal medicine | Nasal exudate | B1 | V | A | pMUP4 | + |
| HUNSC-298 | 04/04/2001 | Traumatology | Wound | B1 | VI | E | pMUP5 | + |
| HUNSC-319 | 14/05/2001 | Internal medicine | Wound | B1 | VI | E | pMUP5 | ND |
| HUNSC-326 | 25/06/2001 | Internal medicine | Nasal exudate | B1 | V | A | pMUP4 | ND |
| HUNSC-339 | 30/07/2001 | Traumatology | Wound | B1 | VII | E | pMUP6 | ND |
| HUNSC-363 | 01/12/2001 | Vascular surgery | Blood | B1 | VIII | C | pMUP7 | + |
| HUNSC-393 | 11/02/2002 | Thoracic surgery | Sputum | B1 | IX | A | pMUP8 | + |
| HUNSC-396 | 19/02/2002 | Internal medicine | Wound | B1 | IX | A | pMUP8 | ND |
| HUNSC-401 | 02/03/2002 | Traumatology | Wound | B1 | VI | E | pMUP5 | ND |
| HUNSC-491 | 10/09/2002 | General surgery | Wound | B1 | X | D | pMUP9 | + |
| HUNSC-499 | 30/09/2002 | Traumatology | Catheter | B1 | I | D | pMUP1 | ND |
| HUNSC-503 | 07/10/2002 | Traumatology | Wound | B1 | I | D | pMUP1 | ND |
| HUNSC-459 | 06/06/2002 | U. medicine | Wound | B4 | VII | E | pMUP6 | + |
| HUNSC-489 | 03/09/2002 | Cardiology | Wound | B4 | VII | E | pMUP6 | ND |
Each Hi-Mupr isolate showed a MIC of ≥1,024 μg/ml.
Results of PFGE, multilocus sequence, and staphylococcal chromosome cassette typing were already included in the analysis of MRSA clones (20).
ICU, intensive care unit; U. medicine, urgency medicine.
BAS, bronchial aspirate.
Patterns obtained from total plasmid DNA.
ND, not determined; +, isolate produced transconjugants.
HindIII restriction endonuclease analysis of plasmid DNA (REAP) revealed 16 patterns (i.e., I to XVI) (Table 2), each hybridizing with the ileS2-specific probe. Thus, the ileS2 gene was harbored by plasmids; this result was consistent with data from previous studies (12, 21). The different sizes of ileS2-hybridizing plasmid fragments permitted five polymorphs of the ileS2 locus (i.e., A to E) to be distinguished. Polymorphs A, B, D, and E were characterized by a single HindIII-hybridizing fragment, while polymorph C was defined by two hybridizing fragments (Tables 2 and 3).
TABLE 3.
Structural group, ileS2 locus polymorph, and clonal complex distribution of Hi-Mupr plasmid types
| Structural group | ileS2 locus polymorph | Size(s) (kb) of restriction fragments hybridizing with the ileS2 probea | ileS2-carrying plasmid type (kb)b | No. of isolates (%) | MRSA clone(s) | Clonal complex(es) |
|---|---|---|---|---|---|---|
| S1 | A | 4.3 | pMUP4 (26) | 4 (8.3) | ST146-IVA, ST36-II | CC5, CC30 |
| pMUP8 (41) | 2 (4.2) | ST36-II | CC30 | |||
| S2 | B | 5.0 | pMUP3 (34) | 2 (4.2) | ST36-II | CC30 |
| C | 4.7, 5.0 | pMUP7 (39) | 1 (2.1) | ST36-II | CC30 | |
| S3 | D | 5.2 | pMUP1 (33) | 11 (22.9) | ST247-IA, ST36-II | CC8, CC30 |
| pMUP9 (35) | 1 (2.1) | ST36-II | CC30 | |||
| S4 | E | 5.5 | pMUP2 (40) | 21 (43.8) | ST125-IVA, ST247-IA, ST22-IV | CC5, CC8, CC22 |
| pMUP5 (47) | 3 (6.2) | ST36-II | CC30 | |||
| pMUP6 (46) | 3 (6.2) | ST36-II | CC30 |
Aproximated sizes by comparison with a lambda HindIII marker.
Plasmid sizes were determined by summation of restriction fragments using agarose gel electrophoresis following separate digestions with EcoRI and HindIII.
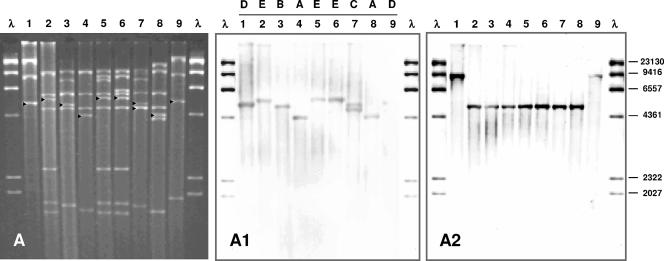
One Hi-Mupr isolate of each restriction endonuclease analysis pattern (n = 16) was randomly selected for conjugation assays. The Hi-Mupr was transferred from each isolate (Table 2). Overall, nine different ileS2-carrying plasmids (i.e., pMUP1 to pMUP9) were identified in transconjugants (Fig. 1). Susceptibility tests and curing experiments support that idea that these plasmids mediate Hi-Mupr. Plasmids differed from each other by at least one HindIII restriction fragment, but some bands in common permitted four structural groups (i.e., S1 to S4) to be distinguished (Table 3). EcoRI digestions corroborated these results (data not shown). Plasmids included in each group displayed the same ileS2 locus polymorph (Fig. 1A1). The exception was group S2, given that plasmids pMUP3 and pMUP7 showed different polymorphs due to an additional ileS2-hybridizing fragment in pMUP7, which appeared as a double-intensity ca. 4.7-kb HindIII band, suggesting the existence of two copies of the ileS2 gene in pMUP7 (Fig. 1A, lane 7). Plasmids belonging to distinct groups differed from each other by at least five bands and showed different polymorphs.
FIG. 1.
Analysis of ileS2-carrying plasmid types by restriction endonuclease analysis and Southern hybridization. (A) HindIII restriction patterns of plasmid types extracted from S. aureus transconjugants of Hi-Mupr MRSA isolates. ▸, ileS2-hybridizing fragments. (A1) Southern hybridization with the ileS2-containing DNA as a probe, showing the different ileS2 polymorphs (A to E). (A2) Southern hybridization using the traK DNA fragment as probe. Lanes 1 through 9, plasmid types pMUP1 to pMUP9. λ, lambda phage DNA digested with HindIII and digoxigenin labeled as a molecular size standard. Numbers on the right correspond to the molecular sizes (in base pairs) of λ DNA HindIII restriction fragments.
ileS2-carrying plasmids are related to the pSK41 family of staphylococcal multiresistance plasmids.
It has been suggested that mupirocin resistance plasmids belong to the pSK41 family of staphylococcal conjugative plasmids (12) (the pSK41 sequence was obtained from GenBank, accession no. NC_005024). The putative relationship of pSK41 to ileS2-carrying plasmids was examined at the traK gene level. The traK gene of pSK41 is involved in the tra gene system for conjugation (2, 9). The traK gene was detected in each ileS2-carrying plasmid (Fig. 1A2). The nucleotide sequence of the open reading frame (1,641 bp) in the ca. 1.8-kb traK PCR products from plasmids included in groups S1 and S2 was identical to the pSK41 traK gene sequence. Group S3 plasmids showed 41 nucleotide differences, plus the insertion of 6 nucleotides, compared to the pSK41 traK sequence (>97% identity). The three plasmid types included in group S4 showed one nucleotide difference (>99% identity). These results indicated that the traK gene of pSK41 was conserved in ileS2-carrying plasmids. The plasmids included in groups S1, S2, and S4 had a single 4.7-kb traK-hybridizing fragment. The plasmids in group S3 presented a ca. 9-kb hybridization band. Additionally, the traD-traE region was detected in each Hi-Mupr isolate and transconjugants. Our results are consistent with the concept of ileS2-carrying plasmids being members of the pSK41 family of conjugative plasmids.
Distribution of Hi-Mupr plasmids among pandemic lineages (CCs).
Some of the ileS2-carrying plasmids were detected in the background of different MRSA clones belonging to different CCs (Tables 2 and 3), indicating plasmid transfer. Furthermore, we found plasmids of the four structural groups in the collection of Hi-Mupr ST36-II epidemiologically related isolates. The emergence of Hi-Mupr in ST36-II is of health concern because it is the predominant clone, completely replacing the Iberian clone (ST247-IA) (19, 20, 22). Unlike ST36-II isolates, Hi-Mupr isolates belonging to clones ST125-IVA and ST22-IV shared a single, unique ileS2-carrying plasmid, which indicates clonal dissemination in the spread of Hi-Mupr.
Conclusions.
All major MRSA pandemic clones already circulating in the hospital acquired ileS2-carrying plasmids, resulting in a multiclonal population structure. In addition to clonal spread and plasmid transfer, the finding of the ileS2 gene in plasmids differing in size and restriction patterns show that gene transposition constitutes an important mode of dissemination of Hi-Mupr. Intervention strategies in the current scenario should identify dissemination of MRSA clones and/or ileS2-carrying mobile elements.
Nucleotide sequence accession numbers.
The nucleotide sequences of the entire traK homolog genes from mupirocin resistance plasmids pMUP1, pMUP2, pMUP3, pMUP4, pMUP5, pMUP6, pMUP7, pMUP8, and pMUP9 identified in this study have been deposited in GenBank under accession numbers DQ232628, DQ232629, DQ232630, DQ232631, DQ232632, DQ232633, DQ232634, DQ232635, and DQ232636, respectively.
Acknowledgments
We are grateful to Mark Enright and Manuel Espinosa for critical reading of the manuscript, Warren B. Grubb for providing the S. aureus WBG541 strain, and Ninive Batista and Francisco Javier González-Paredes for their technical assistance in susceptibility tests and traK gene sequencing, respectively. We thank SmithKline Beecham Pharmaceuticals (United Kingdom) for the gift of mupirocin powder.
This study was supported by grants 2001/020 from the Consejería de Educación, Cultura y Deportes, FUNCIS 2002/038 and 2005/50 from the Canary Islands Autonomous Government, 2001/3150 from the Fondo de Investigación Sanitaria, and BIO2002/00953 from the Ministerio de Ciencia y Tecnología, Government of Spain, to S.M.-A. E.P.-R. was supported by a grant from the Consejería de Educación, Cultura y Deportes, and C.L.-A. was supported by a grant from FUNCIS, Canary Islands Autonomous Government. S.M.-A. was partially supported by Fondo de Investigación Sanitaria contract 99/3060.
REFERENCES
- 1.Anthony, R. M., A. M. Connor, E. G. M. Power, and G. L. French. 1999. Use of the polymerase chain reaction for rapid detection of high-level mupirocin resistance in staphylococci. Eur. J. Clin. Microbiol. 18:30-34. [DOI] [PubMed] [Google Scholar]
- 2.Berg, T., N. Firth, S. Apisiridej, A. Hettiaratchi, A. Leelaporn, and R. A. Skurray. 1998. Complete nucleotide sequence of pSK41: evolution of staphylococcal conjugative multiresistance plasmids. J. Bacteriol. 180:4350-4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradley, S. F., M. A. Ramsey, T. M. Morton, and C. A. Kauffman. 1995. Mupirocin resistance: clinical and molecular epidemiology. Infect. Control Hosp. Epidemiol. 16:354-358. [DOI] [PubMed] [Google Scholar]
- 4.Cookson, B. D. 1998. The emergence of mupirocin resistance: a challenge to infection control and antibiotic prescribing practice. J. Antimicrob. Chemother. 41:11-18. [DOI] [PubMed] [Google Scholar]
- 5.Eltringham, I. 1997. Mupirocin resistance and methicillin-resistant Staphylococcus aureus (MRSA). J. Hosp. Infect. 35:1-8. [DOI] [PubMed] [Google Scholar]
- 6.Enright, M. C., D. A. Robinson, G. Randle, E. J. Feil, H. Grundmann, and B. Spratt. 2002. The evolutionary history of methicillin resistant Staphylococcus aureus (MRSA). Proc. Natl. Acad. Sci. USA 99:7687-7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Finlay, J. E., L. A. Miller, and J. A. Poupard. 1997. Interpretative criteria for testing susceptibility of staphylococci to mupirocin. Antimicrob. Agents Chemother. 41:1137-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gilbart, J., C. R. Perry, and B. Slocombe. 1993. High-level mupirocin resistance in Staphylococcus aureus: evidence for two distinct isoleucyl-tRNA synthetases. Antimicrob. Agents Chemother. 37:32-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grohmann, E., G. Muth, and M. Espinosa. 2003. Conjugative plasmid transfer in gram-positive bacteria. Microbiol. Mol. Biol. Rev. 67:277-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hodgson, J. E., S. P. Curnock, K. G. H. Dyke, R. Morris, D. R. Sylvester, and M. S. Gross. 1994. Molecular characterization of the gene encoding high-level mupirocin resistance in Staphylococcus aureus J2870. Antimicrob. Agents Chemother. 38:1205-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hurdle, J. G., A. J. O'Neill, L. Mody, I. Chopra, and S. F. Bradley. 2005. In vivo transfer of high-level mupirocin resistance from Staphylococcus epidermidis to methicillin-resistant Staphylococcus aureus associated with failure of mupirocin prophylaxis. J. Antimicrob. Chemother. 56:1166-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morton, T. M., J. L. Johnston, J. Patterson, and G. L. Archer. 1995. Characterization of a conjugative staphylococcal mupirocin resistance plasmid. Antimicrob. Agents Chemother. 39:1272-1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Needham, C., M. Rahman, K. G. H. Dyke, and W. C. Noble. 1994. An investigation of plasmids from Staphylococcus aureus that mediate resistance to mupirocin and tetracycline. Microbiology 140:2577-2583. [DOI] [PubMed] [Google Scholar]
- 14.Oliveira, D. C., A. Tomasz, and H. de Lencastre. 2002. Secrets of success of a human pathogen: molecular evolution of pandemic clones of methicillin-resistant Staphylococcus aureus. Lancet Infect. Dis. 2:180-189. [DOI] [PubMed] [Google Scholar]
- 15.Palepou, M. F. I., A. P. Johnson, B. D. Cookson, H. Beattie, A. Charlett, and N. Woodford. 1998. Evaluation of disc diffusion and Etest for determining the susceptibility of Staphylococcus aureus to mupirocin. J. Antimicrob. Chemother. 41:577-583. [DOI] [PubMed] [Google Scholar]
- 16.Pawa, A., W. C. Noble, and S. A. Howell. 2000. Co-transfer of plasmids in association with conjugative transfer of mupirocin or mupirocin and penicillin resistance in methicillin-resistant Staphylococcus aureus. J. Med. Microbiol. 49:1103-1107. [DOI] [PubMed] [Google Scholar]
- 17.Pérez-Roth, E., F. Claverie-Martín, J. Villar, and S. Méndez-Álvarez. 2001. Multiplex PCR for simultaneous identification of Staphylococcus aureus and detection of methicillin and mupirocin resistance. J. Clin. Microbiol. 39:4037-4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pérez-Roth, E., F. Claverie-Martín, N. Batista, A. Moreno, and S. Méndez-Álvarez. 2002. Mupirocin resistance in methicillin-resistant Staphylococcus aureus clinical isolates in a Spanish hospital. Co-application of multiplex PCR assay and conventional microbiology methods. Diagn. Microbiol. Infect. Dis. 43:123-128. [DOI] [PubMed] [Google Scholar]
- 19.Pérez-Roth, E., F. Lorenzo-Díaz, and S. Méndez-Álvarez. 2003. Establishment and clonal dissemination of the methicillin-resistant Staphylococcus aureus UK-16 epidemic strain in a Spanish hospital. J. Clin. Microbiol. 41:5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pérez-Roth, E., F. Lorenzo-Díaz, N. Batista, A. Moreno, and S. Méndez-Álvarez. 2004. Tracking methicillin-resistant Staphylococcus aureus clones during a 5-year period (1998 to 2002) in a Spanish hospital. J. Clin. Microbiol. 42:4649-4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rahman, M., S. Connolly, W. C. Noble, B. Cookson, and I. Phillips. 1990. Diversity of staphylococci exhibiting high-level resistance to mupirocin. J. Med. Microbiol. 33:97-100. [DOI] [PubMed] [Google Scholar]
- 22.Santos-Sanches, I., M. Ramirez, H. Troni, M. Abecassis, M. Papua, A. Tomasz, and H. de Lencastre. 1995. Evidence for the geographic spread of methicillin-resistant Staphylococcus aureus clone between Portugal and Spain. J. Clin. Microbiol. 33:1243-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmitz, F. J., E. Lindenlauf, B. Hofmann, A. C. Fluit, J. Verhoef, H. P. Heinz, and M. E. Jones. 1998. The prevalence of low- and high-level mupirocin resistance in staphylococci from 19 European hospitals. J. Antimicrob. Chemother. 42:489-495. [DOI] [PubMed] [Google Scholar]
- 24.Udo, E. E., and L. E. Jacob. 1998. Conjugative transfer of high-level mupirocin resistance and the mobilization of non-conjugative plasmids in Staphylococcus aureus. Microb. Drug Resist. 4:185-193. [DOI] [PubMed] [Google Scholar]
- 25.Upton, A., S. Lang, and H. Heffernan. 2003. Mupirocin and Staphylococcus aureus: a recent paradigm of emerging antibiotic resistance. J. Antimicrob. Chemother. 51:613-617. [DOI] [PubMed] [Google Scholar]
- 26.Woodford, N., A. P. Watson, S. Patel, M. Jevon, D. J. Waghorn, and B. D. Cookson. 1998. Heterogeneous location of the mupA high-level mupirocin resistance gene in Staphylococcus aureus. J. Med. Microbiol. 47:829-835. [DOI] [PubMed] [Google Scholar]