Abstract
The possibility that unexpressed antibiotic resistance genes are carried by bacterial genomes is seldom investigated. Potential silencing of the resistance genes blaOXA-2, aadA1, sul1, and tetA carried on the plasmid pVE46 in a recent porcine isolate of Escherichia coli was investigated following oral inoculation of the strain into organic piglets. A small proportion of isolates recovered from feces did not express one or more resistance genes, despite retaining the pVE46 plasmid. Different combinations of unexpressed resistance genes were observed, and 12 representative isolates were selected for further study. Surprisingly, in most cases the resistance genes and their promoters, although not expressed, were intact, with fully wild-type sequences. Apart from four isolates exhibiting intermediate-level tetracycline resistance, no mRNA for the unexpressed genes was detected. Silencing of resistance genes was reversible at low frequencies between 10−6 and 10−10. Introduction of the plasmid from silenced isolates to another strain restored expression, indicating that gene silencing was a property of the host chromosome rather than the plasmid itself. When the same recent porcine E. coli strain carrying the unrelated plasmid RP1 was inoculated into piglets, three isolates (of 9,492) that no longer expressed RP1-encoded resistance genes were recovered. As with pVE46, in most cases the coding sequences and promoter regions of these genes were found to be intact, but they were not transcribed. Such gene silencing indicates a previously unrecognized form of transcriptional control that overrides standard expression signals to shut down gene expression. These findings suggest that unexpressed resistance genes may occur in the wild and hence may have clinical implications.
The emergence and spread of antibiotic resistance constitute a contemporary problem that threatens the efficacy of treatment of many bacterial infections (20). One suggestion to curb the rising incidence of antibiotic resistance and possibly reverse the trend is to reduce the use of the agent(s) concerned. This idea arises from the belief that the acquisition and expression of antibiotic resistance genes reduce competitive fitness of a bacterial strain in the absence of antibiotic selection, because expression of antibiotic resistance requires extra resources and/or compromises the cell's normal metabolic functions (1, 4). While it is true that acquisition of antibiotic resistance can frequently impose an initial fitness cost, bacteria can often minimize the burden by acquiring compensatory mutations that restore fitness, wholly or in part, both in vivo and in vitro (5, 6, 10, 19, 28). The prevalence of resistance may also fail to decrease because the gene(s) conferring resistance to the antibiotic being restricted may be linked to resistance genes for drugs that are still in use (3, 12). In principle, bacteria could also reduce the fitness cost of resistance by silencing the genes when not required, but this possibility has not been extensively studied. Whereas gene silencing in eukaryotic cells is well established (32), there is little documented evidence for gene silencing in bacteria in general or for silencing of resistance genes in particular (34). However, rare instances of gene silencing in prokaryotes, mediated by the H-NS protein (15, 18) and by proteins involved in plasmid partitioning (22, 26), have been reported.
In clinical settings, carriage of antibiotic resistance genes is generally assumed on the basis of phenotype, and in most genotypic investigations only resistant isolates are screened for the presence of particular genes conferring antibiotic resistance. Accordingly, if silent resistance genes were present, most surveys of resistant bacteria would fail to detect them. Here we present evidence for in vivo silencing of antibiotic resistance genes carried on two resistance plasmids, pVE46 and RP1.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Escherichia coli 345-2 was isolated from pig feces in 2001 and chosen as a progenitor strain for development due to its excellent growth characteristics and total antibiotic susceptibility (13). Rifampin-resistant derivatives were isolated, and the fittest mutant (E. coli 345-2RifC) was chosen as a study strain after assessment by in vitro and in vivo competition (13). Resistance plasmids pVE46 and RP1 (11, 23) were introduced into E. coli 345-2RifC by conjugation (21). pVE46 is an IncN conjugative plasmid, related to R46 (7), and accommodates the antibiotic resistance genes blaOXA-2 and sul1 as part of a class 1 integron (14, 17) and aadA1, dhfr1, and sat1 as part of a class 2 integron (17) plus tetA and its regulator, tetR. RP1 is a conjugative plasmid, belonging to the IncP-1 incompatibility group, which accommodates the antibiotic resistance genes aphA, blaTEM-2, and tetA and its regulator, tetR. The tetA genes of pVE46 and RP1 are not identical but share 75% identity at the nucleotide level (V. I. Enne, unpublished data).
In an attempt to obtain an E. coli 345-2RifC variant better adapted to pVE46, E. coli 345-2RifC/pVE46 was propagated for 200 generations in laboratory culture media. One colony from nutrient agar was inoculated into 100 ml nutrient broth, and the culture was incubated at 37°C with shaking (170 rpm) for 24 h. This culture was diluted 1/100 in fresh broth, and growth continued for 24 h. The transfer process was repeated until 200 generations had passed. At this point it was assumed that the bacterial host would have adapted to carriage of pVE46 if adaptation improved the competitive fitness. The culture is hereafter referred to as postpassage E. coli 345-2RifC/pVE46. This procedure was not carried out with E. coli 345-2RifC/RP1.
In vitro fitness assays.
Paired growth competition experiments were performed with Davis minimal medium with glucose (25 mg/ml) to determine the fitness impact of pVE46 upon E. coli 345-2RifC, as described previously (13). Plasmid-carrying strains were identified on Iso-Sensitest agar (Oxoid, Basingstoke, United Kingdom) containing ampicillin (25 μg/ml). A nalidixic acid-resistant derivative of E. coli 345-2RifC was used in a parallel experiment to assess the frequency of pVE46 transfer to the plasmid-free competitor.
Animal experiments to investigate bacterial fitness and retention of antibiotic resistance in vivo.
Animal experiments were conducted as described previously (13). Briefly, for each experiment, six 5-week-old organic piglets, i.e., animals that had not been fed drugs routinely, were housed as a single group for 2 weeks to acclimatize the animals. The animals in each group were then divided equally into two enclosures, except in the case of the animals inoculated with postpassage E. coli 345-2RifC/pVE46, where the enclosures contained four and two animals. All animals were then screened for carriage of rifampin-resistant coliforms, to establish the detection limit of E. coli 345-2RifC.
Each strain (E. coli 345-2RifC, prepassage E. coli 345-2RifC/pVE46, postpassage E. coli 345-2RifC/pVE46, and E. coli 345-2RifC/RP1) was grown separately to stationary phase in nutrient broth, harvested, resuspended in an antacid solution, and inoculated into the piglets orally, in a single dose of 1010 CFU per animal. Fecal samples were collected by digital manipulation on days 3, 5, 7, 10, 12, 14, 17, 19, and 21 postinoculation, suspended in saline, and spread at appropriate dilutions onto MacConkey agar containing rifampin (50 μg/ml). Plates were incubated overnight at 37°C, and all colonies obtained were replica plated onto MacConkey agar containing rifampin and the appropriate antibiotic(s) for the plasmid under study (for pVE46, this consisted of one plate containing 25 μg/ml tetracycline and 25 μg/ml ampicillin and one plate containing 500 μg/ml sulfamethoxazole and 25 μg/ml streptomycin; for RP1, this consisted of one plate containing 25 μg/ml ampicillin and 30 μg/ml kanamycin and one plate containing 25 μg/ml tetracycline), followed by replica plating onto MacConkey agar containing rifampin only. All procedures complied with the Animals (Scientific Procedures) Act 1986 of Great Britain and were performed under Home Office license.
Characterization of isolates exhibiting loss of antibiotic resistance.
Colonies failing to grow on medium containing antibiotics other than rifampin were first typed biochemically using the API 20E bacterial identification system (bioMérieux, Marcy l'Etoile, France), and then the profiles obtained were compared with that of E. coli 345-2RifC. Any colony demonstrating a different profile was presumed to be an unrelated isolate and was ignored. The antibiotic susceptibility pattern of any isolate that failed to grow in the presence of one or more test antibiotics but that showed the same API profile as E. coli 345-2RifC was then confirmed by Etest (AB Biodisk, Solna, Sweden). The relatedness of selected isolates was investigated by randomly amplified polymorphic DNA (RAPD) PCR, as described previously (2). The presence of pVE46 was initially confirmed by PCR for the plasmid-specific origin of transfer, oriT (see below), followed by plasmid extraction from selected isolates using the PureLink HiPure plasmid DNA purification kit (Invitrogen, Paisley, United Kingdom) according to the manufacturer's instructions. The presence of RP1 was confirmed by plasmid extraction from all isolates. Plasmids were visualized with UV light following agarose gel electrophoresis on 0.8% (wt/vol) agarose gels containing ethidium bromide (1 μg/ml).
PCR and DNA sequencing.
The presence in isolates retrieved from pig feces of the resistance genes blaOXA-2, sul1, aadA1, and tetA of pVE46 and aphA, blaTEM-2, and tetA of RP1, as well as their respective promoters and regulators, was determined by PCR. Each PCR mixture consisted of 12.5 μl Extensor Hi-Fidelity PCR master mix (ABgene, Epsom, United Kingdom), 16 pmol of each primer in a 0.5-μl volume (Operon, Cologne, Germany), 10.5 μl Eppendorf molecular-biology-grade water (Fisher Scientific, Loughborough, United Kingdom), and 1 μl of template, prepared by suspending one bacterial colony in 100 μl molecular-biology-grade water and boiling it for 5 min. The primers used are described in Table 1. PCR amplification consisted of 1 cycle at 95°C for 5 min, followed by 35 cycles of 95°C for 1 min, annealing between 50 and 60°C as appropriate for the primer pair for 1 min and at 68°C for 1 min, and 1 cycle at 68°C for 10 min. A positive control containing template prepared from postpassage E. coli 345-2RifC/pVE46 or E. coli 345-2RifC/RP1 and a negative control where no template was added were included in each batch of PCR mixture. Following amplification, PCR products were mixed with a sixfold concentration of loading dye (Bioline, London, United Kingdom), separated on 1% (wt/vol) agarose gels containing ethidium bromide, and visualized by UV light. PCR products were purified using a QIAquick PCR purification kit (QIAGEN, Crawley, United Kingdom) according to the manufacturer's instructions and were sent for sequencing to the Advanced Biotechnology Centre, Imperial College, London, United Kingdom. Sequence analysis was carried out using the DNASTAR Lasergene software package.
TABLE 1.
Descriptions of oligonucleotide primers
| Primer | Sequence (5′-3′) | Positions (gene)a | Purpose |
|---|---|---|---|
| AADA1F | ATGAGGGAAGCGGTGATCGC | 1 to 20 (pVE46 aadA1) | Amplification of aadA1, aadA1 RT-PCR |
| AADA1R | CGGCTTGAACGAATTGTTAG | 814 to 795 (pVE46 aadA1) | Amplification of aadA1 |
| AADA1RTR | TTTGCCGACTACCTTGGTGA | 789 to 770 (pVE46 aadA1) | aadA1 RT-PCR |
| AADAPROF | GACATAAAACGCTCCTTGTC | −1486 to −1467 (pVE46 aadA1) | Amplification of aadA1 promoter |
| AADAPROR | CACAATTCTTCCTCAGAGGT | −1148 to −1167 (pVE46 aadA1) | Amplification of aadA1 promoter |
| APHAPROF | GAGTGAGGCGATTGGAAACC | −236 to −217 (RP1 aphA) | Amplification of aphA 5′ end and aphA promoter |
| APHAPROR | GAGCATTCGCTCACTGGGAT | 419 to 400 (RP1 aphA) | Amplification of aphA 5′ end and aphA promoter |
| APHAF | AACGGCATATCAAGTGCTGA | 312 to 331 (RP1 aphA) | Amplification of aphA 3′ end, aphA RT-PCR |
| APHAR | GAAAAGTTCGTCCAGCAGGA | 813 to 794 (RP1 aphA) | Amplification of aphA 3′ end, aphA RT-PCR |
| ORITF | CCCTGATACTTTTGGGCTTC | 1527 to 1546 (pVE46 oriT) | Amplification of oriT |
| ORITR | ACCGATTACCTCCTGAAACC | 114 to 94 (pVE46 oriT) | Amplification of oriT |
| OXA2F | TTGGGCATTAAGGAAAAGTT | −21 to −2 (pVE46 blaOXA-2) | Amplification of blaOXA-2 |
| OXA2R | GTTGAAGTAACCGGCGCTGC | 875 to 859 (pVE46 blaOXA-2) | Amplification of blaOXA-2 |
| OXA2PROF | ATGCCTCGACTTCGCTGCTG | −347 to −328 (pVE46 blaOXA-2) | Amplification of blaOXA-2 promoter |
| OXA2PROR | TGGATGCCCGAGGCATAGAC | −111 to −130 (pVE46 blaOXA-2) | Amplification of blaOXA-2 promoter |
| OXA2RTF | TTCAAGCCAAAGGCACGATAG | 113 to 133 (pVE46 blaOXA-2) | blaOXA-2 RT-PCR |
| OXA2RTR | TCCGAGTTGACTGCCGGGTTG | 815 to 795 (pVE46 blaOXA-2) | blaOXA-2 RT-PCR |
| RPSLF | CTCGCAAAGTTGCGAAAAGC | 38 to 57 (E. coli rpsL) | rpsL RT-PCR |
| RPSLR | TTCACGCCATACTTGGAACG | 359 to 340 (E. coli rpsL) | rpsL RT-PCR |
| SUL1F | CTTTGTAGGTATGGGGCTCA | −81 to −62 (pVE46 sul1) | Amplification of sul1 |
| SUL1R | TGACGAGCCCAGCATGTCTG | 916 to 897 (pVE46 sul1) | Amplification of sul1 |
| SUL1PROF | CGCTGGGTTTGCCGTTTCTC | −694 to −675 (pVE46 sul1) | Amplification of sul1 promoter |
| SUL1PROR | TCTAGCCGCCGGCTCTCATC | 67 to 48 (pVE46 sul1) | Amplification of sul1 promoter |
| SUL1RTF | CCGATATTGCTGAGGCGGACT | 338 to 358 (pVE46 sul1) | sul1 RT-PCR |
| SUL1RTR | CCAACGCCGCTTCAGCTT | 604 to 586 (pVE46 sul1) | sul1 RT-PCR |
| TEMF | ATGAGTATTCAACATTTCCG | 1 to 20 (RP1 blaTEM-2) | Amplification of blaTEM-2, blaTEM-2 RT-PCR |
| TEMR | GACGCTCAGTGGAACGAAAA | 1000 to 981 (RP1 blaTEM-2) | Amplification of blaTEM-2 |
| TEMPROF | TTAGACGTCAGGTGGCACTT | −139 to −120 (RP1 blaTEM-2) | Amplification of blaTEM-2 promoter |
| TEMPROR | GAGCAAAAACAGGAAGGCAA | 79 to 60 (RP1 blaTEM-2) | Amplification of blaTEM-2 promoter |
| TEMRTR | CCAATGCTTAATCAGTGACG | 858 to 839 (RP1 blaTEM-2) | blaTEM-2 RT-PCR |
| TETA1F | AGTAGAGCGCTGGCTGTTGC | −218 to −199 (pVE46 tetA) | Amplification of tetA 5′ end and tetA promoter |
| TETA1R | CATACCCACGCCGAAACAAG | 432 to 413 (pVE46 tetA) | Amplification of tetA 5′ end and tetA promoter |
| TETA2F | GTTGCTGGCGCCTATATCGC | 343 to 362 (pVE46 tetA) | Amplification of tetA 3′ end |
| TETA2R | TCAGGTCGAGGTGGCCCGAC | 1176 to 1157 (pVE46 tetA) | Amplification of tetA 3′ end |
| TETRF | TTCAGCTAGGTGACTTTGCT | −64 to −45 (pVE46 tetR) | Amplification of tetR |
| TETRR | GTGCCTGACTGCGTTAGCAA | 746 to 727 (pVE46 tetR) | Amplification of tetR |
| TETARTF | ACAATGCGCTCATCGTCATC | 11 to 30 (pVE46 tetA) | tetA RT-PCR |
| TETARTR | GGCGCCTACAATCCATGC | 1125 to 1108 (pVE46 tetA) | tetA RT-PCR |
| RP1TET1AF | CGGCTGCAACTTTGTCATGC | −123 to −104 (RP1 tetA) | Amplification of tetA 5′ end |
| RP1TETA1R | CGCCGAAAATGACCCAAAGC | 712 to 693 (RP1 tetA) | Amplification of tetA 5′ end |
| RP1TETA2F | CCGCTCAGCTTCGTTCGGTG | 595 to 614 (RP1 tetA) | Amplification of tetA 3′ end |
| RP1TETA2R | CGATCAGCGATCGGCTCGTT | 1202 to 1183 (RP1 tetA) | Amplification of tetA 3′ end |
| RP1TETRTETPROF | GCAGCGGTCCTGATCAATCG | 663 to 644 (RP1 tetR) | Amplification of tetA promoter and tetR |
| RP1TETRTETPROR | CTCCGCTGGTCCGATTGAAC | −20 to −39 (RP1 tetA) | Amplification of tetA promoter and tetR |
| RP1TETARTF | CGGCCAATCTTGCTCGTCTC | 217 to 236 (RP1 tetA) | tetA RT-PCR |
| RP1TETARTR | CCTGCCTGGACAACATTGCT | 982 to 936 (RP1 tetA) | tetA RT-PCR |
Positions are relative to the start codon of the gene.
RNA extraction and RT-PCR.
RNA was isolated from bacteria growing exponentially in nutrient broth using an RNeasy kit (QIAGEN). Since tetA expression is inducible, RNA was isolated from bacteria grown in nutrient broth containing a subinhibitory concentration of tetracycline (250 ng/ml). Isolated RNA was treated with RNase-free DNase (Promega, Southampton, United Kingdom), and then RNA samples were adjusted to a concentration of 100 ng/μl by adding molecular-biology-grade water following measurement of the optical density at 260 nm. mRNA was detected by reverse transcription (RT)-PCR using a OneStep RT-PCR kit (QIAGEN); for each reaction, primers to detect resistance gene mRNA were multiplexed with primers to detect the rpsL gene, to act as an internal control for the quality and quantity of mRNA. Each reaction mixture contained 1 μl RT-PCR enzyme mix, 1 μl 10 mM deoxynucleoside triphosphate mix, 5 μl 5× RT-PCR buffer, 1 μl RNA, 16 μl RNase-free water, and 16 pmol of each resistance gene primer, as well as 16 pmol of each primer to detect rpsL (each in a 0.5-μl volume) (Table 1). For the RT-PCRs, a negative control that contained no template was included in each batch. A control experiment was also performed to detect the residual presence of DNA by use of a standard PCR with the digested RNA preparations. RT-PCR amplification comprised 1 cycle of reverse transcription at 50°C for 30 min, directly followed by the PCR which consisted of 1 cycle at 95°C for 15 min, 35 cycles at 94°C for 1 min, appropriate annealing for each resistance gene primer pair between 50 and 58°C for 1 min and at 72°C for 1 min, and a final cycle at 72°C for 10 min. RT-PCR products were separated on agarose gels and visualized as described for PCRs.
Investigation of reversion to resistance.
The recovery of resistance by isolates with intact but silent pVE46 resistance genes was assessed by plating undiluted (up to 1 ml) and serially diluted stationary-phase nutrient broth cultures onto Iso-Sensitest agar containing the appropriate antibiotic (ampicillin, 25 μg/ml; streptomycin, 25 μg/ml; sulfamethoxazole, 500 μg/ml; or tetracycline, 25 μg/ml). Total cell counts for calculation of reversion frequencies were determined by plating serial dilutions of the same culture onto antibiotic-free medium. A representative proportion of revertants was then chosen, and the susceptibilities of these chosen revertants to ampicillin, streptomycin, sulfamethoxazole, and tetracycline were investigated by disc diffusion assays (8).
Conjugal transfer to investigate isolates with silent resistance genes.
Conjugation experiments were carried out by the agar mating method (21). Briefly, bacteria were grown to midlogarithmic phase in nutrient broth. Equal numbers of recipient and donor bacterial cells (based on the optical density at 600 nm, approximately 107 cells) were harvested by centrifugation, and each cell type was resuspended in 100 μl of fresh broth. The donor and recipient cells were then immediately mixed, the mixture was plated onto nutrient agar, and the plates were incubated at 37°C overnight. Approximately one-eighth of the resulting lawn of bacteria was harvested with a sterile swab and suspended in 1 ml of sterile water. Dilutions of the suspension in 100-μl volumes were then spread onto Iso-Sensitest agar containing two antibiotics (to select for transconjugants) and Iso-Sensitest agar containing one antibiotic (to measure donor numbers). The broth cultures of donors and recipients were also spread separately, undiluted, onto medium containing the antibiotics used to select for transconjugants, to ensure that spontaneous mutation to resistance did not confound the results.
To investigate the transfer of pVE46 plasmids with silent resistance genes, strains L6, L8, and L10 were mated with a high-level streptomycin-resistant mutant of E. coli 345-2RifC, E. coli 345-2RifC RpsL(K42R) (13), and E. coli K-12 JM109. For comparison, experiments were also performed with the wild-type postpassage E. coli 345-2RifC/pVE46 strain acting as a donor. Conjugation mixtures were spread onto Iso-Sensitest agar plates containing 500 μg/ml streptomycin [345-2RifC RpsL(K42R)] or 50 μg/ml nalidixic acid (JM109) and 25 μg/ml ampicillin or ampicillin only to measure donor frequency. Each cross was repeated three times to determine conjugation frequency, and from each cross, five colonies were chosen at random and their susceptibilities to ampicillin, nalidixic acid, spectinomycin, streptomycin, sulfamethoxazole, tetracycline, and rifampin were determined by disc diffusion assays (8). For experiments where E. coli JM109 was used as a recipient, transconjugants were also streaked onto MacConkey agar, to ensure that they possessed the Lac− phenotype of E. coli JM109, rather than the Lac+ phenotype of E. coli 345-2RifC.
To investigate the specificity of gene silencing, the plasmids RP1 (11, 23), R100 (29), and the integrative and conjugative element R391 (9), all carried by E. coli K-12, were introduced into isolates L1 and L2 (Table 2) and post-laboratory passage E. coli 345-2RifC/pVE46. Transconjugants were selected by plating dilutions of the conjugation mixture onto Iso-Sensitest agar containing rifampin (50 μg/ml) and chloramphenicol (25 μg/ml) for R100, rifampin and kanamycin (30 μg/ml) for R391 and RP1, or chloramphenicol or kanamycin only to determine donor numbers. Each cross was performed in triplicate. Five transconjugants per cross were chosen at random and subjected to susceptibility testing for ampicillin, chloramphenicol, kanamycin, rifampin, streptomycin, sulfamethoxazole, and tetracycline by disc diffusion assays (8).
TABLE 2.
Characteristics of 12 selected isolates of E. coli 345-2RifC/pVE46 in which phenotypic loss of antibiotic resistance was detected, as well as those of the parent strain with and without pVE46
| Isolate | MIC (μg/ml)a
|
blaOXA-2
|
aadA1
|
sul1
|
tetA
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMP | STR | SUL | TET | PCRb | RT-PCRc | PCRb | RT-PCRc | PCRb | RT-PCRc | PCRb | RT-PCRc | |
| L1 | 0.5 | 4 | 24 | 0.38 | + | − | + | − | − | − | − | − |
| L2 | 1 | 2 | 6 | 0.5 | + | − | + | − | + | − | + | − |
| L3 | 0.75 | 4 | 24 | 0.5 | − | − | + | − | − | − | − | − |
| L4 | 1 | 2 | 24 | 4 | + | − | + | − | + | − | + | ++ |
| L5 | 1.5 | 2 | 32 | 0.75 | + | − | + | − | + | − | + | − |
| L6 | 48 | 4 | 16 | 8 | + | +++ | + | − | + | − | + | ++ |
| L7 | 48 | 4 | >1,024 | 1.5 | + | +++ | + | − | + | +++ | − | − |
| L8 | 64 | 64 | >1,024 | 0.5 | + | +++ | + | +++ | + | +++ | + | − |
| L9 | 48 | 64 | >1,024 | 12 | + | +++ | + | +++ | + | +++ | + | ++ |
| L10 | 96 | 4 | 32 | 0.5 | + | +++ | + | − | + | − | + | − |
| L11 | 96 | 32 | >1,024 | 0.75 | + | +++ | + | +++ | + | +++ | − | − |
| L12 | 64 | 2 | >1,024 | 12 | + | +++ | + | − | + | +++ | + | + |
| 345-2RifC | 1 | 2 | 24 | 2 | − | − | − | − | − | − | − | − |
| 345-2RifC/pVE46 | 64 | 64 | >1,024 | 48 | + | +++ | + | +++ | + | +++ | + | +++ |
AMP, ampicillin; STR, streptomycin; SUL, sulfamethoxazole; TET, tetracycline.
+, gene could be amplified by PCR and had wild-type sequence; −, gene could not be amplified by PCR.
Strength of RT-PCR signal (arbitrary units): −, no signal; +, weak signal; ++, intermediate signal; +++, strong signal.
RESULTS
Stability of plasmid resistance genes during passage of E. coli 345-2RifC in animals.
In this study we set out to investigate the stability of the expression of resistance genes on pVE46, an IncN plasmid related to R46 (7), while the E. coli host was established in a pig gastrointestinal tract. The pVE46 resistance genes investigated were aadA1, blaOXA-2, sul1, and tetA, which confer resistance to streptomycin/spectinomycin, β-lactams, sulfonamides, and tetracycline, respectively, and are expressed from four separate promoters. The first resistance genes are expressed constitutively, while expression of tetA is inducible by tetracycline. pVE46 was introduced by conjugation into E. coli 345-2RifC (13), a rifampin-resistant derivative of a recent porcine isolate, to create E. coli 345-2RifC/pVE46, referred to as the prepassage isolate. A rifampin-resistant derivative of E. coli 345-2 was used in order to facilitate recovery of the strain from the porcine gut, because rifampin resistance is rare in that environment (V. I. Enne and A. A. Delsol, unpublished results). Acquisition of pVE46 imposed a fitness cost per generation of 2.6% ± 1.2% on E. coli 345-2RifC when grown in Davis minimal medium containing glucose. This slight disadvantage was not ameliorated (2.8% ± 0.9%) after 200 generations of growth in antibiotic-free nutrient broth (the strain in this case being referred to as the postpassage isolate).
E. coli 345-2RifC and pre- and post-laboratory passage E. coli 345-2RifC/pVE46 cell lines were each inoculated orally into six piglets which had not demonstrated the presence of rifampin-resistant coliforms prior to the start of the experiment. Recovery of fed bacteria from the animals' feces was then monitored (Fig. 1) by plating the bacteria onto rifampin-containing medium. There were no significant differences in the rates of recovery of the different isolates over a period of 3 weeks, indicating that bacterial fitness in vivo was not significantly affected by plasmid carriage. Maintenance of the resistance profile conferred by pVE46 while the strains were in the pig gut was investigated by replica plating. With prepassage E. coli 345-2RifC/pVE46, 10,088 colonies were screened over the 3-week period; all retained resistance to the four antibiotics tested. In contrast, of 10,269 postpassage E. coli 345-2RifC/pVE46 isolates screened, 52 were susceptible to one or more of the antibiotics to which pVE46 confers resistance. These 52 isolates were recovered over 14 days, starting 5 days after inoculation, from four animals housed in the same enclosure. Bacteria recovered from two animals housed in a second enclosure and treated identically but independently all had the resistance profile expected from carriage of pVE46.
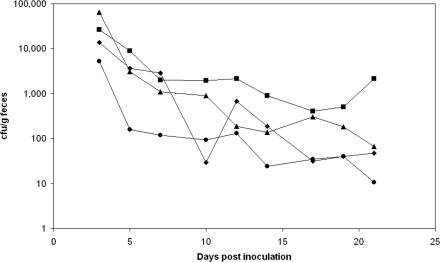
FIG. 1.
Recovery of E. coli 345-2RifC (diamonds), prepassage E. coli 345-2RifC/pVE46 (squares), postpassage E. coli 345-2RifC/pVE46 (triangles), and E. coli 345-2RifC/RP1 (circles) from pig feces. Variations of up to a factor of 1,000 were seen between individual animals infected with the same strain. Hence, standard deviations are large, and so, for clarity, error bars have not been included; however, they overlapped at all time points. Analysis of variance indicates no statistically significant difference in the recovery rates of the strains (for comparison of pre- and postpassage E. coli 345-2RifC/pVE46 with E. coli 345-2RifC, F = 1.87, P = 0.157; for comparison of E. coli 345-2RifC/RP1 with E. coli 345-2RifC, F = 3.031, P = 0.085).
Characterization of E. coli 345-2RifC/pVE46 isolates that failed to express plasmid-encoded antibiotic resistance.
Of the 52 isolates that did not display the antibiotic resistance profile of the strain fed to the pigs, all retained the plasmid pVE46 (confirmed by PCR) and the rifampin-resistant property of the fed strain. Biochemical testing using API 20E strips confirmed that all 52 isolates had the same biochemical profile as the fed strain, postpassage E. coli 345-2RifC/pVE46. All 52 isolates showed losses of tetracycline resistance. Twelve exhibited partial losses (MICs, 4 to 12 μg/ml); the rest were fully susceptible to tetracycline (MICs, 0.25 to 1 μg/ml). Of the four resistance combinations recovered, 27 isolates had lost the full resistance profile conferred by pVE46; 2 had lost resistance to sulfamethoxazole, streptomycin, and tetracycline; 12 had lost resistance to streptomycin and tetracycline; and 11 had lost only tetracycline resistance. As a control and to determine if the resistance profile conferred by pVE46 is stable during prolonged laboratory growth in a medium lacking antibiotics, 10,239 colonies obtained from a postpassage E. coli 345-2RifC/pVE46 laboratory culture were tested. All displayed the pVE46 resistance profile, indicating that the resistance genes and their expression levels are stable during laboratory culture, a finding consistent with that obtained using prepassage E. coli 345-2RifC/pVE46 in the pig experiment.
Twelve of the 52 variants, representing the different resistance profiles observed, were selected for further study (Table 2). They were genetically typed by RAPD PCR, and the same banding profile was demonstrated by the 12 variants and by E. coli 345-2RifC/pVE46. Plasmid extraction from the 12 isolates confirmed that pVE46 was still present in each isolate. The complete open reading frames of the pVE46 resistance genes, aadA1, blaOXA-2, sul1, and tetA, from all 12 isolates were amplified by PCR, and the products sequenced. The sequences were compared to those obtained from the postpassage E. coli 345-2RifC/pVE46 isolate (Table 2). Unexpectedly, in most cases where resistance was not expressed, intact resistance genes with wild-type sequences were found. In addition, the sequences of the promoter regions for aadA1, blaOXA-2, sul1, and tetA, as well as that of the tetA repressor gene, tetR (which shares its promoter region with tetA), were found to be unchanged in those isolates that retained resistance genes, with the exception of one isolate. In that isolate, the tetA promoter and tetR repressor could not be amplified, despite the presence of an intact tetA gene. In the small remainder of cases, the resistance genes and the corresponding promoters could not be amplified by PCR (Table 2). Gene transcription from the intact but silent resistance genes was then assessed by RT-PCR analysis (Table 2, Fig. 2). Transcripts of fully silent resistance genes were not detected in any of the 12 isolates; in the four variants that displayed intermediate levels of tetracycline resistance, some tetA mRNA was found but less than in the parent strain (Table 2), consistent with the lower levels of resistance observed.
FIG. 2.
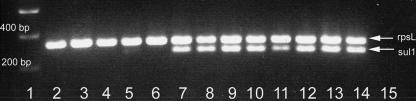
Multiplex RT-PCR analysis of isolates carrying pVE46 for the sul1 gene, incorporating RT-PCR for the rpsL gene as a control for mRNA quality and quantity. Lane 1, Hyperladder I DNA marker (Bioline, London, United Kingdom); lanes 2 to 13, isolates L1 to L12, respectively; lane 14, postpassage 345-2RifC/pVE46; lane 15, negative control (no template).
Silencing of RP1-encoded antibiotic resistance by E. coli 345-2RifC/RP1 after animal passage.
Due to the unexpected nature of the results obtained with postpassage E. coli 345-2RifC/pVE46, the animal experiment was repeated with another, unrelated plasmid, the IncP-1 plasmid RP1 (11, 23). RP1 encodes β-lactam resistance through the blaTEM-2 β-lactamase, tetracycline resistance through the tetA gene, and kanamycin resistance through the aphA gene. RP1 was introduced into E. coli 345-2RifC by conjugation. E. coli 345-2RifC/RP1 was then fed to six organic piglets, and its recovery was monitored over a 3-week period (Fig. 1). No significant differences were detected between the rates of recovery of E. coli 345-2RifC and E. coli 345-2RifC/RP1. Maintenance of the E. coli 345-2RifC/RP1 resistance profile in vivo was monitored by replica plating. Of 9,492 colonies recovered, 3 had lost the RP1 resistance profile (Table 3). Biochemical testing using API 20E strips and RAPD analysis showed that the three isolates had biochemical and RAPD profiles indistinguishable from that of E. coli 345-2RifC/RP1 (Table 3). All three isolates retained the RP1 plasmid. They were recovered from two different animals, housed in separate enclosures, over a period of 6 days.
TABLE 3.
Characteristics of three isolates of E. coli 345-2RifC/RP1 in which phenotypic loss of antibiotic resistance was detected, as well as those of the parent strain with and without RP1
| Isolate | MIC (μg/ml)a
|
blaTEM-2
|
tetA
|
aphA
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| AMP | TET | KAN | PCRb | RT-PCRc | PCRb | RT-PCRc | PCRb | RT-PCRc | |
| P1 | 4 | 1.5 | >256 | + | − | − | − | − | − |
| P2 | 3 | 2 | 2 | + | − | + | − | + | − |
| P3 | 4 | 3 | 2 | + | − | + | − | + | − |
| 345-2 RifC/RP1 | >256 | >256 | 64 | + | +++ | + | +++ | + | +++ |
| 345-2RifC | 1 | 2 | 4 | − | − | − | − | − | − |
AMP, ampicillin; TET, tetracycline; KAN, kanamycin.
+, gene could be amplified by PCR and had wild-type sequence; −, gene could not be amplified by PCR.
Strength of RT-PCR signal (arbitrary units): −, no signal; +, weak signal; ++, intermediate signal; +++, strong signal.
Closer examination of the three isolates that had lost the RP1 resistance profile revealed that one isolate, P1, had lost ampicillin and tetracycline resistance, and that the other two, isolates P2 and P3, had lost resistance to all three antibiotics conferred by RP1 (Table 3). The three isolates were screened for the presence of the intact open reading frames of all RP1 resistance genes, as well the corresponding promoter regions and the tetR repressor (Table 3). Amplification products were obtained in most cases, except for the aphA, tetA, and tetR genes of isolate P1 and their corresponding promoters. The sequences of all genes that were amplified matched precisely those of the parent strain, indicating that the isolates that failed to demonstrate RP1-encoded antibiotic resistance nonetheless retained wild-type resistance genes and their promoters. RT-PCR was performed to detect transcription of the resistance genes, but no mRNA was detected despite retention of intact genes and promoters (Table 3).
Analysis of silencing of pVE46-encoded antibiotic resistance.
Recovery of expression of the silenced genes, and hence resistance, was investigated in the 12 selected isolates carrying silent pVE46 genes. The ability of each isolate to recover resistance was investigated separately for all antibiotics to which resistance had been lost. Depending on the particular resistance gene and isolate in question, reversion was detected at frequencies between 10−6 and 10−10 in all cases where isolates had retained intact resistance genes and promoters but not in cases where resistance genes had been lost. The full antibiotic susceptibility profiles of 15 revertants, chosen to represent a cross section of those obtained, were investigated. In all cases where the cell line possessed intact but silent resistance genes, the lost resistance profile was fully recovered when just a single antibiotic was used for selection. However, selection of resistance to one antibiotic did not lead to recovery of resistance in those cases where the presence of resistance genes could no longer be demonstrated. For example, a revertant of isolate L5 (Table 2) selected with sulfamethoxazole also recovered resistance to ampicillin, streptomycin, and tetracycline, while a revertant of isolate L3 (Table 2) selected with streptomycin recovered only streptomycin resistance.
The pVE46 plasmids of isolates L6, L8, and L10 were then transferred, by conjugation, into E. coli 345-2RifC RpsL(K42R), which had no recent history of growth in animals, and the laboratory strain E. coli K-12 JM109. In each case, ampicillin was used to select transconjugants because all three isolates retained expression of blaOXA-2. In every case where pVE46 resistance genes had been retained but silenced, expression of the pVE46 resistance profile was fully restored in the new host. The experimental design prevents verification of expression of aadA1-mediated streptomycin resistance in the E. coli 345-2RifC RpsL(K42R) host; therefore expression of the aadA1 gene was verified by investigating spectinomycin resistance. Further, transfer frequencies [approximately 10−4 per donor cell for E. coli 345-2RifC RpsL(K42R) and approximately 10−3 per donor cell for E. coli JM109] were comparable to those obtained for the pVE46 plasmid from the progenitor strain.
A selection of other antibiotic resistance-encoding elements was transferred into the pVE46-carrying isolates L1 and L2 (Table 2) by conjugation: the plasmids R100 (29), encoding resistance to chloramphenicol, streptomycin, sulfonamide, and tetracycline, and RP1 (11, 23) and the integrative and conjugative element R391 (9), encoding kanamycin resistance. As a control, these elements were also transferred into the progenitor post-laboratory passage E. coli 345-2RifC/pVE46 strain. Antibiotics to which pVE46 does not encode resistance (kanamycin and chloramphenicol) were used to select transconjugants, to avoid possible confusion between transconjugants and revertants. The frequencies of plasmid transfer into the silenced isolates L1 and L2 were the same as those obtained using wild-type post-laboratory passage E. coli 345-2RifC/pVE46 as the recipient, i.e., approximately 10−4 per donor cell for RP1 and R100 and 10−6 per donor cell for R391. The susceptibility profiles of 15 transconjugants per cross were determined. In all cases the transconjugants exhibited resistance to the antibiotics to which the newly introduced element conferred resistance but remained susceptible to those antibiotics to which resistance encoded by pVE46 had been silenced. For example, transconjugants of L1 that received RP1 were ampicillin, kanamycin, and tetracycline resistant but remained susceptible to streptomycin and sulfonamide, while transconjugants of L2 that received R100 were resistant to chloramphenicol, streptomycin, sulfonamide, and tetracycline but remained susceptible to ampicillin.
DISCUSSION
The findings reported in this paper indicate that the expression of multiple unrelated resistance genes on R plasmids, which are normally expressed either constitutively or inducibly from their own promoters, can be coordinately overridden or prevented by a change in the host. Carriage of unexpressed genes by bacteria is a well-established phenomenon (16, 24), including carriage of unexpressed resistance genes, e.g., the cryptic cfi gene of Bacteroides fragilis encoding a metallo-β-lactamase (25), but in these cases the absence of expression has been attributed to the absence of effective promoter sequences. It is also known that in integron gene cassette arrays, the further a cassette is located from the integron promoter, the lower the level of expression, due to polarity effects (14). However, in the cases of gene silencing reported here, neither of these explanations pertains. The silenced genes each possess what are normally effective promoters, and several of them are not components of integrons. Those that are have not changed position within the integron and neither has the integron promoter changed.
Our data indicate that expression of intact antibiotic resistance gene systems can be switched off in bacteria, i.e., resistance genes can be silenced. Furthermore, the process is reversible. The mechanism by which this occurs is unknown, but the fact that recovery of gene expression was detected in a small proportion of the cell populations, at frequencies akin to single-site mutation frequencies, suggests that it may be the result of mutation, or some other low-frequency genetic change. It is likely that gene silencing has resulted from a chromosomal alteration in E. coli 345-2RifC, because resistance gene expression is recovered when pVE46 is transferred to a new host, provided that the resistance gene is intact. The fact that after prolonged laboratory growth in antibiotic-free medium the postpassage E. coli 345-2RifC/pVE46 culture yielded no evidence of resistance gene silencing and the fact that isolates carrying silenced pVE46 resistance genes were recovered from animals held in one enclosure but not the other suggest that the variants with silenced resistance genes probably arose while the bacterium was in the pig. Given that isolates carrying silenced resistance genes were recovered from all animals in one enclosure but from none in the other, it is likely that a variant(s) in which expression of pVE46 resistance genes had been silenced arose in one animal and that animal-to-animal spread occurred subsequently. It is interesting to note that among the 52 isolates with silenced pVE46 resistance genes, individual isolates exhibit different patterns of gene silencing. This suggests an initial genetic change that predisposes a cell to gene silencing, followed by a quantitatively variable imposition in progeny cells. The observation that resistance gene silencing apparently arose while the bacterium was in the animal gut but was not seen after laboratory culture when similar numbers of isolates were tested may explain why this phenomenon has not been reported previously, as the use of an animal model to investigate the fate of antibiotic resistance is rare. It is possible that the silent strains have a selective advantage in the animal gut and that once they have arisen through a form of mutation, they then have a selective advantage over other strains, which enables them to outgrow competitors to the point where they are detectable. This would also explain why silent isolates were not detected in all enclosures, as a relatively rare mutational event, presumably random, would be required first. Another explanation for this phenomenon not being reported earlier is that most routine studies screen only resistant isolates for the presence of resistance genes, while susceptible isolates are not usually investigated, often due to constraints on resources.
Remarkably, similar results were obtained with another, unrelated plasmid, RP1. In this case, isolates with silent resistance genes were recovered from animals in both enclosures used. However, isolate P1, which was from an animal in a different enclosure than those from which P2 and P3 were isolated, appears to have undergone a complex genetic change. Isolate P1 remained kanamycin resistant, despite testing negative by PCR for the aphA gene of RP1. The kanamycin MIC of the isolate was also more than threefold higher than that resulting from the expression of RP1 aphA. The reason for the apparent anomaly was not investigated. It seems unlikely that this isolate gave rise to the silenced isolates from the second enclosure but rather that P1 arose independently from P2 and P3.
Although silencing was observed with two different plasmids, and hence does not seem an isolated phenomenon, it does appear to be specific to the plasmid originally carried by the host. This is demonstrated by the fact that other plasmids could be introduced into isolates carrying silent resistance genes by conjugation at normal frequencies and that resistance genes on the newly introduced plasmids were expressed, although pVE46 resistance genes remained silent. The specificity of gene silencing in the affected isolates is not known; however, the fact that pVE46 transferred from isolates with silenced resistance genes to other strains by conjugation indicates that not all nonessential plasmid genes are affected, i.e., transfer functions are clearly still expressed. At present, we cannot offer a comprehensive explanation for the reported phenomenon; however, it is possible to rule out several possibilities. Silencing is not due to mutational changes in the genes themselves or in their promoters, nor is it due to integration of the plasmid into the chromosome (demonstrated by plasmid extraction and by the fact that plasmids encoding silenced resistance genes conjugated at normal frequency). As the change that is responsible appears to be chromosomally encoded, it is unlikely to be mediated by plasmid-partioning proteins, as reported in other instances of bacterial gene silencing (22, 26). The changes observed are also unlikely to result from a reduction in plasmid copy number, as both study plasmids are thought to be naturally low in copy number. In the case of RP1, which has an estimated copy number of 2.6 per cell (30), halving the plasmid copy number would result in just one copy of the plasmid per cell. However, the MICs of antibiotics to which RP1 confers resistance are decreased by factors of 32 or more in the isolates with silent resistance genes (Table 3). In the case of pVE46, the exact copy number of the plasmid is not known, but the related plasmid R46 is thought to have a copy number of 10 (31). Accordingly, an eightfold reduction in plasmid copy number would reduce the copy number of pVE46 per cell to approximately one; yet, with the expectation of isolates exhibiting intermediate-level resistance to tetracycline, the MICs of all antimicrobials have been lowered by factors of 16 or more in isolates with silent resistance genes (Table 2). Also, the fact that in the case of pVE46 many isolates retained resistance to some antibiotics but not to others argues strongly against the suggestion that a reduction in plasmid copy number is responsible for the silencing of antibiotic resistance genes.
Our finding has practical implications; it suggests that there may be reservoirs of antibiotic resistance genes in bacteria that we are unaware of and that cannot be detected by phenotype. Until the extent of gene silencing is known, the scale of the problem, if any, will remain unknown. Reports of unexpressed bacterial resistance genes are rare; they include the mecA gene in Staphylococcus sciuri (33), the cfiA gene in Bacteroides spp. (25), and the catB9 gene on a Vibrio cholerae superintegron (27). However, in general, genes such as these are silent because they lack promoters that express the gene sufficiently to lower the antibiotic susceptibility of the host. This is not the case with the derivatives described in this report. They retain their original promoter sequences, which function perfectly well in the parent and other strains; the resistance genes are silent, not because promoters have been lost or changed, but because their activities have been suppressed. The phenomenon of silencing has potentially important implications for therapy because it may explain some therapy failures where the strain responsible for the infection was initially susceptible to the drug(s) used in treatment but subsequently became resistant. It has also not escaped our attention that, once the mechanism is understood, it might be possible to harness the phenomenon to give new leases of life to antibiotics whose use is currently compromised by resistance.
Acknowledgments
This work was supported by the Department for the Environment, Food and Rural Affairs (DEFRA), United Kingdom (project code OD 2007).
REFERENCES
- 1.Andersson, D. I., and B. R. Levin. 1999. The biological cost of antibiotic resistance. Curr. Opin. Microbiol. 2:489-493. [DOI] [PubMed] [Google Scholar]
- 2.Avison, M. B., S. Underwood, A. Okazaki, T. R. Walsh, and P. M. Bennett. 2004. Analysis of AmpC β-lactamase expression and sequence in biochemically atypical ceftazidime-resistant Enterobacteriaceae from paediatric patients. J. Antimicrob. Chemother. 53:584-591. [DOI] [PubMed] [Google Scholar]
- 3.Bischoff, K. M., D. G. White, M. E. Hume, T. L. Poole, and D. J. Nisbet. 2005. The chloramphenicol resistance gene cmlA is disseminated on transferable plasmids that confer multiple-drug resistance in swine Escherichia coli. FEMS Microbiol. Lett. 243:285-291. [DOI] [PubMed] [Google Scholar]
- 4.Bjorkman, J., and D. I. Andersson. 2000. The cost of antibiotic resistance from a bacterial perspective. Drug Resist. Updates 3:237-245. [DOI] [PubMed] [Google Scholar]
- 5.Bjorkman, J., I. Nagaev, O. G. Berg, D. Hughes, and D. I. Andersson. 2000. Effects of environment on compensatory mutations to ameliorate costs of antibiotic resistance. Science 287:1479-1482. [DOI] [PubMed] [Google Scholar]
- 6.Bouma, J. E., and R. E. Lenski. 1988. Evolution of a bacteria/plasmid association. Nature 335:351-352. [DOI] [PubMed] [Google Scholar]
- 7.Brown, A. M. C., and N. S. Willetts. 1981. A physical and genetic map of the IncN plasmid R46. Plasmid 5:188-201. [DOI] [PubMed] [Google Scholar]
- 8.BSAC Working Party on Susceptibility Testing. 2001. BSAC standardized disc susceptibility testing method. J. Antimicrob. Chemother. 48(Suppl. S1):S43-S57. [Google Scholar]
- 9.Coetzee, J. N., N. Datta, and R. W. Hedges. 1972. R factors from Proteus rettgeri. J. Gen. Microbiol. 72:543-552. [DOI] [PubMed] [Google Scholar]
- 10.Dahlberg, C., and L. Chao. 2003. Amelioration of the cost of conjugative plasmid carriage in Escherichia coli K12. Genetics 165:1641-1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Datta, N., R. W. Hedges, E. J. Shaw, R. B. Sykes, and M. H. Richmond. 1971. Properties of an R factor from Pseudomonas aeruginosa. J. Bacteriol. 108:1244-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Enne, V. I., D. M. Livermore, P. Stephens, and L. M. C. Hall. 2001. Persistence of sulphonamide resistance in Escherichia coli in the UK despite national prescribing restriction. Lancet 357:1325-1328. [DOI] [PubMed] [Google Scholar]
- 13.Enne, V. I., A. A. Delsol, G. R. Davis, S. L. Hayward, J. M. Roe, and P. M. Bennett. 2005. Assessment of the fitness impacts on Escherichia coli of acquisition of antibiotic resistance genes encoded by different types of genetic element. J. Antimicrob. Chemother. 56:544-551. [DOI] [PubMed] [Google Scholar]
- 14.Fluit, A. C., and F. J. Schmitz. 1999. Class 1 integrons, gene cassettes, mobility, and epidemiology. Eur. J. Clin. Microbiol. Infect. Dis. 18:761-770. [DOI] [PubMed] [Google Scholar]
- 15.Goransson, M., B. Sonden, P. Nilsson, B. Dagberg, K. Forsman, K. Emanuelsson, and B. E. Uhlin. 1990. Transcriptional silencing and thermoregulation of gene expression in Escherichia coli. Nature 344:682-685. [DOI] [PubMed] [Google Scholar]
- 16.Hall, B. G. 1999. Transposable elements as activators of cryptic genes in E. coli. Genetica. 107:181-187. [PubMed] [Google Scholar]
- 17.Hall, R. M., and C. M. Collis. 1998. Antibiotic resistance in Gram-negative bacteria: the role of gene cassettes and integrons. Drug Resist. Updates 1:109-119. [DOI] [PubMed] [Google Scholar]
- 18.Hommais, F., E. Krin, C. Laurent-Winter, O. Soutorina, A. Malpertuy, J. Le Caer, A. Danchin, and P. Bertin. 2001. Large-scale monitoring of pleiotropic regulation of gene expression by the prokaryotic nucleoid-associated protein H-NS. Mol. Microbiol. 40:20-36. [DOI] [PubMed] [Google Scholar]
- 19.Johnsen, P. J., G. S. Simonsen, O. Olsvik, T. Midtvedt, and A. Sundsfjord. 2002. Stability, persistence, and evolution of plasmid-encoded VanA glycopeptide resistance in enterococci in the absence of antibiotic selection in vitro and in gnotobiotic mice. Microb. Drug Resist. 8:161-170. [DOI] [PubMed] [Google Scholar]
- 20.Levy, S. B. 2001. Antibiotic resistance: consequences of inaction. Clin. Infect. Dis. 33:S124-S129. [DOI] [PubMed] [Google Scholar]
- 21.Livermore, D. M., and J. D. Williams. 1996. β-lactams: mode of action and mechanisms of bacterial resistance, p. 56. In V. Lorian (ed.), Antibiotics in laboratory medicine, 4th ed. Williams & Wilkins, Baltimore, Md.
- 22.Lynch, A. S., and J. C. Wang. 1995. SopB protein-mediated silencing of genes linked to the sop locus of Escherichia coli F-plasmid. Proc. Natl. Acad. Sci. USA 92:1896-1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pansegrau, W., E. Lanka, P. T. Barth, D. H. Figurski, D. G. Guiney, D. Haas, D. R. Helinski, H. Schwab, V. A. Stanisich, and C. M. Thomas. 1994. Complete nucleotide sequence of Birmingham IncP alpha plasmids. Compilation and comparative analysis. J. Mol. Biol. 239:623-663. [DOI] [PubMed] [Google Scholar]
- 24.Parker, L. L., and B. G. Hall. 1990. Characterization and nucleotide sequence of the cryptic cel operon of Escherichia coli K12. Genetics 124:455-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Podglajen, I., J. Breuil, and E. Collatz. 1994. Insertion of a novel DNA sequence IS1186, upstream of the silent carbapenemase gene cfiA, promotes expression of carbapenem resistance in clinical isolates of Bacteroides fragilis. Mol. Microbiol. 12:105-114. [DOI] [PubMed] [Google Scholar]
- 26.Rodionov, O., M. Lobocka, and M. Yarmolinski. 1999. Silencing of genes flanking the P1 plasmid centromere. Science 283:546-549. [DOI] [PubMed] [Google Scholar]
- 27.Rowe-Magnus, D. A., A. Guerout, and D. Mazel. 2002. Bacterial resistance evolution by recruitment of super-integron gene cassettes. Mol. Microbiol. 43:1657-1669. [DOI] [PubMed] [Google Scholar]
- 28.Schrag, S. J., and V. Perrot. 1996. Reducing antibiotic resistance. Nature 381:120-121. [DOI] [PubMed] [Google Scholar]
- 29.Tanaka, N., J. H. Cramer, and R. H. Rownd. 1976. EcoRI restriction endonuclease map of the composite R plasmid NR1. J. Bacteriol. 127:619-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taylor, D. E., and E. C. Brose. 1988. Modified Birboim-Doly method for rapid detection of plasmid copy number. Nucleic Acids Res. 16:9056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Winans, S. C., and G. C. Walker. 1985. Entry exclusion determinant(s) of IncN plasmid pKM101. J. Bacteriol. 161:411-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wolffe, A. P., and M. A. Matzke. 1999. Epigenetics: regulation through repression. Science 286:481-486. [DOI] [PubMed] [Google Scholar]
- 33.Wu, S. W., H. de Lancastre, and A. Tomasz. 2001. Recruitment of the mecA gene homologue of Staphylococcus sciuri into a resistance determinant and expression of the resistant phenotype in Staphylococcus aureus. J. Bacteriol. 183:2417-2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yarmolinski, M. 2000. Transcriptional silencing in bacteria. Curr. Opin. Microbiol. 3:138-143. [DOI] [PubMed] [Google Scholar]