Abstract
A newly identified 16S rRNA methyltransferase gene, rmtC, was accompanied by an ISEcp1 element at its 5′ end. This ISEcp1 element, which contained a transposase gene, tnpA, provided a promoter activity for expression of the adjacent rmtC; and this structure enabled the rmtC gene to be transposed onto another plasmid in Escherichia coli.
Four types of plasmid-mediated 16S rRNA methyltransferase genes, rmtA, rmtB, rmtC, and armA, which confer high levels of resistance to various aminoglycosides, have been found worldwide among a number of pathogenic gram-negative rods (3, 4, 8, 14, 17-19). The distribution of these plasmid-mediated 16S rRNA methyltransferase genes among pathogenic bacteria seems attributable to the fact that these genes are associated with some bacterium-specific DNA recombination systems, such as a transposon (3, 5, 7, 16). In fact, it was recently reported that transposition of armA was mediated by a composite transposon, Tn1548 (5). However, little is known about the transposition system of the other three plasmid-mediated 16S rRNA methyltransferase genes, rmtA, rmtB, and rmtC. Therefore, in the present study we characterized in detail the transposition system of rmtC, which was located on a plasmid (pARS68) found in a clinical Proteus mirabilis strain, ARS68 (14).
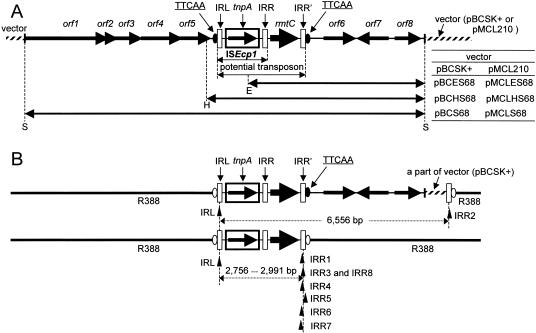
A SacI-digested 11-kb fragment carrying rmtC was cloned from pARS68, and both strands were entirely sequenced. By using the bacterial genetic code, ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html) was used to search for open reading frames (ORFs). A schematic representation of the cloned fragment is shown in Fig. 1A. ISEcp1, which contained a transposase gene, tnpA, was located just upstream of rmtC. Although there were several ORFs (orf1 to orf8) around ISEcp1 and rmtC, their functions remained unknown, even though their sequences were compared with the sequences in the public databases of GenBank and EMBL by using the BLAST and FASTA search tools.
FIG. 1.
(A) Schematic organization of genes in the 11-kb SacI fragment of pBCS68. ORFs and genes are shown as arrows indicating transcription orientation. The IRL, IRR, and IRR′ motifs are indicated by open rectangles. Possible 5-bp target sites (TTCAA) are shown as a closed ellipse. DNA fragments carried by recombinant plasmids used in the transposition experiments are indicated by double-headed arrows with the corresponding recombinant plasmid names. Fragments on pBCHS68 and pMCLHS68 were generated by ligating the fragment on pBCES68 or pMCLES68, and the PCR fragments were amplified by using primers supplemented with restriction sites (HindIII or EcoRI). Restriction sites: S, SacI; H, HindIII; E, EcoRI. (B) Transposition of the ISEcp1-rmtC element onto R388 from pBCS68. The positions of the IRRs (IRR1 to IRR8) were confirmed by transposition experiments with the ISEcp1-rmtC element in E. coli cells by using R388 as the recipient plasmid. Positions of the 5-bp duplicated target sites are shown as open ellipses. Upward-pointing arrowheads indicate the positions of the IRL and the IRR′ extremities of each probable transposed fragment (horizontal broken lines). This result reveals that the IRR′ elements of ISEcp1 are not so rigorous but are comparatively multifarious or flexible and that the 5-bp target sites or recombination junctions at the IRR′ side are also diverse.
It is well known that ISEcp1 is often located at the 5′ ends of several β-lactamase genes, such as blaCTX-M and blaCMY (1, 2, 6, 9, 11, 12, 15), and enables these genes to be transposed to other DNA target sites (2, 10, 13). Moreover, ISEcp1 provides promoter activity for expression of a downstream CTX-M-type β-lactamase gene (2, 12). These findings strongly suggested that the transposition and expression of rmtC, as seen in pARS68, were also regulated by ISEcp1. ISEcp1 was bracketed with two imperfect 14-bp inverted repeat (IR) sequences (the left IR [IRL] and the right IR [IRR]) (Fig. 1A). A putative 5-bp target site (TTCAA) was located in the immediate vicinity of IRL (Fig. 1A). This 5-bp target site might be duplicated, most likely after the insertion of a DNA fragment mediated by a transposon, and is subsequently located on both faces of a transposed DNA fragment. The presence of duplicated 5-bp target sites can be a trace that the insertion of a DNA fragment by transposon occurred at that position. Considering the flanking genetic organizations of rmtC, it is speculated that a DNA fragment containing ISEcp1 and rmtC bracketed on one side by IRL and on the other by a putative second IRR, IRR′, constituting a potential transposon on pARS68. To determine if rmtC could transpose with ISEcp1 and to determine the structural limits of the transposable unit, we tried to identify a potential transposon carrying ISEcp1 and rmtC bracketed with IRL and putative IRR′ by an in vitro transposition experiment. For this purpose, the transposition of rmtC carried by several donor recombinant plasmids based on two different backbones (Fig. 1A) to the recipient plasmid R388 (TMPr) was investigated with a standard mating assay. Escherichia coli DH5α harboring R388 together with various recombinant plasmids and E. coli HB101 (STRr) were used as the donor and recipient strains, respectively. Transconjugants were selected on Luria-Bertani agar plates supplemented with gentamicin (10 μg/ml), trimethoprim (50 μg/ml), and streptomycin (100 μg/ml). Transconjugants were obtained when E. coli DH5α carrying recombinant plasmids (pBCS68, pBCHS68, pMCLS68, and pMCLHS68) (Fig. 1A) was used as a donor strain at a frequency of ca. 10−7 to 10−6 per recipient. On the other hand, transconjugants could not be obtained (frequency, < 10−9 per recipient) when E. coli DH5α carrying plasmids (pBCES68 and pMCLES68) lacking a part of ISEcp1 was used (FIG. 1A). These findings strongly suggested that ISEcp1 plays an essential role in the transposition of rmtC.
Twenty recombinant plasmids carrying rmtC with the backbone of plasmid R388 obtained when pBCS68 was used as a donor plasmid were extracted from the transconjugants. The sequences of both terminal ends of the transposed fragments were determined in detail by direct sequencing of these recombinant plasmids with customized primers. As a result, eight types of transposed fragments, which were structurally different from each other, were obtained. All transposed fragments analyzed contained both ISEcp1 and rmtC, and each left end (IRL) of those fragments was perfectly identical (Fig. 1B). However, the right end of each fragment (IRR1 to IRR8) varied (Fig. 1B and Table 1). IRR was within 459 bases of the end of rmtC in seven of eight cases, although only IRR2 belonged to the cloning vector region (Fig. 1B). The locations of IRR3 and IRR8 were adjacent to the typical 5-bp nucleotide sequence, TTCAA, which seemed to be an innate target site on pARS68. Therefore, it is probable that the 2,973-bp fragment bracketed on the left side by IRL and on the right end by IRR3 or IRR8 constituted a potential transposon on plasmid pARS68.
TABLE 1.
Characteristics of inverted repeats and target sites for ISEcp1-mediated transposition
| Description of sequence | Nucleotide sequence of IRL and IRRs (14 bp)a | 5-bp duplicated target site sequence | Size of transposed fragment (bp) |
|---|---|---|---|
| IRL of ISEcp1 | 5′-CCTAGATTCTACGT-3′ | TTCAA | |
| Expected perfect IRR of ISEcp1 (complementary sequence of IRL) | 5′-ACGTAGAATCTAGG-3′ | ||
| IRR of ISEcp1 | 5′-ACGTGGAATTTAGG-3′ | ||
| IRR′ | 5′-CCTAGGAACTCGGC-3′ | TTCAA | 2,973 |
| IRR1 | 5′-GCCTGGGATTTCGA-3′ | TTCTT | 2,943 |
| IRR2 | 5′-GAACAGTATTTGGT-3′ | TATGT | 6,556 |
| IRR3 | 5′-CCTAGGAACTCGGC-3′ | ACGCA | 2,973 |
| IRR4 | 5′-TCCTAGGAACTCGG-3′ | GCCAA | 2,972 |
| IRR5 | 5′-ATATGGTGTTTCCT-3′ | ATGAA | 2,991 |
| IRR6 | 5′-AATCTTTTTTCGGA-3′ | AATTT | 2,774 |
| IRR7 | 5′-ACGCCGTAACTCGG-3′ | GTGAC | 2,756 |
| IRR8 | 5′-CCTAGGAACTCGGC-3′ | GAAAT | 2,973 |
Underlining of IRRs indicates nucleotide residues identical to the corresponding complementary nucleotide residues of the IRL sequence. IRR′, IRR3, and IRR8 showed identical nucleotide sequences.
The numbers of base pairs in IRR, which is identical to those in IRL, ranged from three to nine (Table 1). The 3′ ends of the IRRs identified varied (GA, GT, GC, GG, or CT), although it was reported that ISEcp1B needs a guanosine (G) residue at the 3′ ends of the IRRs when it transposed the adjacent genes blaKLU-A and blaCTX-M-19 (10, 13). In any event, it was commonly observed that ISEcp1 and ISEcp1B were able to transpose adjacent antibiotic resistance genes by using IRRs composed of a wide variety of nucleotide sequences.
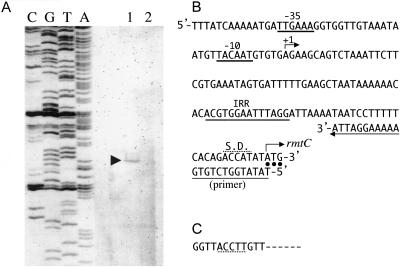
Primer extension analysis of RNA from RmtC-producing E. coli transformant, E. coli DH5α(pBC-KB1) (14), revealed the start residue of the mRNA transcription of rmtC (Fig. 2A and 2B). Transcription was initiated at an A (adenine) residue, located 99 nucleotides upstream of an AUG translation initiation codon of rmtC. This position was located within ISEcp1 near its IRR. Although diversity in the start residue of transcription was observed among ISEcp1-bearing antimicrobial resistance genes, including rmtC, blaCTX-M, and blaCMY, ISEcp1 commonly provides promoter sequences within the right-end region near its IRR for expression of downstream antibiotic resistance genes (2, 6, 12).
FIG. 2.
Identification of transcriptional start site for rmtC in E. coli DH5α(pBC-KB1) by primer extension analysis (14). (A) Lanes C, G, T, and A, results of sequencing reactions performed with DNA prepared by PCR as a template and the primer; lane 1, primer elongation product obtained with total RNA of E. coli DH5α(pBC-KB1); lane 2, primer elongation product obtained with total RNA of E. coli DH5α (pBCSK+). (B) Promoter region of rmtC. +1, transcriptional start site for rmtC; putative −35 and −10 promoter sequences upstream of transcriptional start site are underlined with a thin line; dots indicate the ATG start codon of rmtC; a probable Shine-Dalgarno (S.D.) sequence is marked with a dotted line; the IRR of ISEcp1 is underlined with double thin lines; the primer used in primer extension analysis shown with an arrow at bottom. (C) Complementary sequence of the 3′ terminus of the 16S rRNA of Proteus mirabilis. The portion of the nucleotide residues that coincide with the upstream region of ATG start codon of rmtC is marked with a dashed line. The sequence was referred to the EMBL/GenBank database and can be found under accession no. AJ301682.
Recently, it was experimentally confirmed that ISEcp1B could transpose upstream of chromosomally located blaKLU-A of Kluyvera ascorbata, which is thought to be a progenitor of CTX-M type β-lactamases, and, consequently, could also transpose blaKLU-A to other target sites in E. coli (10). This hybrid structure of ISEcp1B and blaKLU-A seems to be the origin of that of ISEcp1 and blaCTX-M, which is widely distributed among members of the family Enterobacteriaceae worldwide. Although the overall schemes for the development of the hybrid structure of ISEcp1 and rmtC have not been elucidated, it is probable that ISEcp1 first transposed into the 5′ end of chromosome-carrying rmtC in unknown aminoglycoside-producing bacteria and that subsequently the ISEcp1-rmtC element transposed to other DNA target sites on a residential plasmid of the Enterobacteriaceae. To understand the development of the hybrid structure of ISEcp1 and rmtC, it would be necessary to identify natural reservoirs of rmtC. In conclusion, we report here that ISEcp1 plays an essential role in the transposition and expression of a 16S rRNA methyltransferase gene, rmtC.
Nucleotide sequence accession number.
The nucleotide sequence of the 11-kbp SacI fragment shown in Fig. 1A was submitted to the EMBL/GenBank database through the DNA Data Bank of Japan (DDBJ) and can be found under accession no. AB194779.
Acknowledgments
We are grateful to Chihiro Sasakawa for providing plasmid R388 and Kumiko Kai and Fusako Yokokawa for technical assistance.
The bacterial strains and plasmids used in this study were collected by studies supported by the Ministry of Health, Labor and Welfare of Japan (grants H15-Shinkou-9 and H18-Shinkou-11). Genetic analyses were supported by a grant from the Ministry of Education, Culture, Sports, Science and Technology of Japan (grant 16017313). The research activity of J. Wachino was supported by a Scholarship for Young Scientists provided by the Japan Society for the Promotion of Science.
REFERENCES
- 1.Baraniak, A., J. Fiett, W. Hryniewicz, P. Nordmann, and M. Gniadkowski. 2002. Ceftazidime-hydrolysing CTX-M-15 extended-spectrum β-lactamase (ESBL) in Poland. J. Antimicrob. Chemother. 50:393-396. [DOI] [PubMed] [Google Scholar]
- 2.Cao, V., T. Lambert, and P. Courvalin. 2002. ColE1-like plasmid pIP843 of Klebsiella pneumoniae encoding extended-spectrum β-lactamase CTX-M-17. Antimicrob. Agents Chemother. 46:1212-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Doi, Y., K. Yokoyama, K. Yamane, J. Wachino, N. Shibata, T. Yagi, K. Shibayama, H. Kato, and Y. Arakawa. 2004. Plasmid-mediated 16S rRNA methylase in Serratia marcescens conferring high-level resistance to aminoglycosides. Antimicrob. Agents Chemother. 48:491-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Galimand, M., P. Courvalin, and T. Lambert. 2003. Plasmid-mediated high-level resistance to aminoglycosides in Enterobacteriaceae due to 16S rRNA methylation. Antimicrob. Agents Chemother. 47:2565-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Galimand, M., S. Sabtcheva, P. Courvalin, and T. Lambert. 2005. Worldwide disseminated armA aminoglycoside resistance methylase gene is borne by composite transposon Tn1548. Antimicrob. Agents Chemother. 49:2949-2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Giles, W. P., A. K. Benson, M. E. Olson, R. W. Hutkins, J. M. Whichard, P. L. Winokur, and P. D. Fey. 2004. DNA sequence analysis of regions surrounding blaCMY-2 from multiple Salmonella plasmid backbones. Antimicrob. Agents Chemother. 48:2845-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gonzalez-Zorn, B., A. Catalan, J. A. Escudero, L. Dominguez, T. Teshager, C. Porrero, and M. A. Moreno. 2005. Genetic basis for dissemination of armA. J. Antimicrob. Chemother. 56:583-585. [DOI] [PubMed] [Google Scholar]
- 8.Gonzalez-Zorn, B., T. Teshager, M. Casas, M. C. Porrero, M. A. Moreno, P. Courvalin, and L. Dominguez. 2005. armA and aminoglycoside resistance in Escherichia coli. Emerg. Infect. Dis. 11:954-956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karim, A., L. Poirel, S. Nagarajan, and P. Nordmann. 2001. Plasmid-mediated extended-spectrum β-lactamase (CTX-M-3 like) from India and gene association with insertion sequence ISEcp1. FEMS Microbiol. Lett. 201:237-241. [DOI] [PubMed] [Google Scholar]
- 10.Lartigue, M., L. Poirel, D. Aubert, and P. Nordmann. 2006. In vitro analysis of ISEcp1B-mediated mobilization of naturally occurring β-lactamase gene blaCTX-M of Kluyvera ascorbata. Antimicrob. Agents Chemother. 50:1282-1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Literacka, E., J. Empel, A. Baraniak, E. Sadowy, W. Hryniewicz, and M. Gniadkowski. 2004. Four variants of the Citrobacter freundii AmpC-type cephalosporinases, including novel enzymes CMY-14 and CMY-15, in a Proteus mirabilis clone widespread in Poland. Antimicrob. Agents Chemother. 48:4136-4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poirel, L., J. W. Decousser, and P. Nordmann. 2003. Insertion sequence ISEcp1B is involved in expression and mobilization of a blaCTX-M β-lactamase gene. Antimicrob. Agents Chemother. 47:2938-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poirel, L., M. F. Lartigue, J. W. Decousser, and P. Nordmann. 2005. ISEcp1B-mediated transposition of blaCTX-M in Escherichia coli. Antimicrob. Agents Chemother. 49:447-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wachino, J., K. Yamane, K. Shibayama, H. Kurokawa, N. Shibata, S. Suzuki, Y. Doi, K. Kimura, Y. Ike, and Y. Arakawa. 2006. Novel plasmid-mediated 16S rRNA methylase, RmtC, found in a Proteus mirabilis isolate demonstrating extraordinary high-level resistance against various aminoglycosides. Antimicrob. Agents Chemother. 50:178-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu, S. W., K. Dornbusch, G. Kronvall, and M. Norgren. 1999. Characterization and nucleotide sequence of a Klebsiella oxytoca cryptic plasmid encoding a CMY-type β-lactamase: confirmation that the plasmid-mediated cephamycinase originated from the Citrobacter freundii AmpC β-lactamase. Antimicrob. Agents Chemother. 43:1350-1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamane, K., Y. Doi, K. Yokoyama, T. Yagi, H. Kurokawa, N. Shibata, K. Shibayama, H. Kato, and Y. Arakawa. 2004. Genetic environments of the rmtA gene in Pseudomonas aeruginosa clinical isolates. Antimicrob. Agents Chemother. 48:2069-2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamane, K., J. Wachino, Y. Doi, H. Kurokawa, and Y. Arakawa. 2005. Global spread of multiple aminoglycoside resistance genes. Emerg. Infect. Dis. 11:951-953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yan, J. J., J. J. Wu, W. C. Ko, S. H. Tsai, C. L. Chuang, H. M. Wu, Y. J. Lu, and J. D. Li. 2004. Plasmid-mediated 16S rRNA methylases conferring high-level aminoglycoside resistance in Escherichia coli and Klebsiella pneumoniae isolates from two Taiwanese hospitals. J. Antimicrob. Chemother. 54:1007-1012. [DOI] [PubMed] [Google Scholar]
- 19.Yokoyama, K., Y. Doi, K. Yamane, H. Kurokawa, N. Shibata, K. Shibayama, T. Yagi, H. Kato, and Y. Arakawa. 2003. Acquisition of 16S rRNA methylase gene in Pseudomonas aeruginosa. Lancet 362:1888-1893. [DOI] [PubMed] [Google Scholar]