Abstract
A qnrB2 determinant was described for a new complex sul1-type integron from Salmonella enterica serovar Keurmassar. The genetic structure contained two class 1 integrons surrounding two common regions (CRs) separated by a partial 3′ conserved segment. The qnrB2 gene is adjacent to the first CR.
Quinolone resistance in gram-negative bacteria is mainly due to chromosomal mutations in genes encoding quinolone targets (DNA gyrase and topoisomerase IV) and/or in regulatory genes of outer membrane proteins or efflux pumps (7). The first plasmid-mediated quinolone resistance determinant, qnrA, was recently characterized (8), and several Qnr proteins (QnrA-like, QnrB, and QnrS) have since been described (6, 9; G. A. Jacoby, K. Walsh, D. Mills, V. Walker, A. Robicsek, H. Oh, and D. C. Hooper, Abstr. 44th Intersci. Conf. Antimicrob. Agents Chemother., abstr. C2-1898a, 2004). Epidemiological studies of the distribution of qnr determinants show that qnr-positive strains frequently express extended-spectrum β-lactamases (ESBLs) (9). The genetic environment of qnr genes has been characterized for qnrA variants and qnrS (6) but not, to our knowledge, for qnrB determinants. The qnrA variants are located on plasmids and are often embedded in complex sul1-type integrons (9). These structures, first described for In6 and In7 (17), contain two partial copies of the 3′ conserved segment (3′-CS) of class 1 integrons, surrounding a common region (CR) which contains the orf513 that encodes a putative recombinase (11, 18). Several antibiotic resistance genes have been described downstream of the CR, including blaDHA-1 (20), dfrA10 (10, 11), cat (16, 17), blaCTX-M genes (4), dfrA19 (21), and blaCMY-9 (3).
We have previously characterized two class-1 integrons in eight clonal strains of the newly described Keurmassar serovar of Salmonella enterica subsp. enterica (5). One integron contained the aadA2 cassette and the other the aac(6′)-IIc and ereA2 cassettes. The strain was resistant to amikacin, chloramphenicol, gentamicin, netilmicin, spectinomycin, streptomycin, sulfamethoxazole, tetracycline, tobramycin, and trimethoprim, and expressed the ESBL SHV-12; all the resistances were transferred en bloc to Escherichia coli by conjugation.
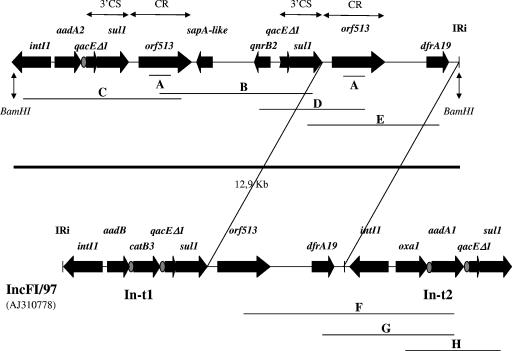
Here, we detected orf513 by means of specific PCR method A (Fig. 1, Table 1). PCR A was positive with both the original strain and its transconjugants. Thus, to further investigate the genetic organization of the complex sul1-type integron, plasmid DNA from the transconjugants was extracted and amplified by PCR with primers located in known sequences (PCRs B and C) (Fig. 1, Table 1). Sequence analysis of fragment B showed, downstream of the CR, a 2,505-bp fragment containing (i) a 726-bp sequence with 90% identity to a fragment of the Klebsiella pneumoniae plasmid pRBDHA, including the first 535 bp of the sapA gene (AJ971343), and (ii) a 645-bp sequence with 100% identity to the qnrB2 gene (DQ351242) (Fig. 1). In the plasmid pRBDHA, the sapA and sapB genes of the sap operon (ABC transporter family) were also adjacent to the CR and may have originated from S. enterica serovar Typhimurium (20). Sequence analysis of fragment C showed, upstream of the CR, the aadA2-containing class 1 integron (Fig. 1).
FIG. 1.
Genetic organization of the qnrB2-containing complex sul1-type integron. Comparison with the organization of the plasmid IncFI/97 showed that the 12.9-kb BamHI fragment harbors the same orf513-dfrA19-containing fragment. Analysis by PCR mapping and sequencing of PCR products F, G, and H showed that integron In-t2 of IncFI/97 is located at the 3′ end of the 12.9-kb BamHI fragment. The horizontal bars correspond to PCR products, the gray ovals correspond to attC sites, and the thick arrows show the genes or ORFs and their direction of transcription.
TABLE 1.
Primer sequences used for PCR mapping
| Amplicon (target) | Primer | Sequence (5′-3′) | Size of amplicon (bp) | Reference |
|---|---|---|---|---|
| A (orf513) | 341A | CGCCCACTCAAACAAACG | 468 | 15 |
| 341B | GAGGCTTTGGTGTAACCG | 15 | ||
| B (orf513-sul1) | 341STOP | ACATTAGTCGGCCAGCGG | 4,262 | 15 |
| SulR | GATTGCGCTTCGCAGATCTCCAGG | 12 | ||
| C (intI1-orf513) | intI1L | ACATGTGATGGCGACGCACGA | 3,943 | 12 |
| 341B | GAGGCTTTGGTGTAACCG | 15 | ||
| D (qnrB2-orf513) | qnrL | TGAACCACTGAACGTCGC | 2,696 | This study |
| 341B | GAGGCTTTGGTGTAACCG | 15 | ||
| E (sul1-dfrA19) | SulL | CCTGGAGATCTGCGAAGCGCAATC | 3,298 | This study |
| dfrR | CTCGCTGGCACTGGAAT | This study | ||
| F (orf513-aadA1) | 341STOP | ACATTAGTCGGCCAGCGG | 5,333 | 15 |
| aadAR | AAGTATGACGGGCTGATACTG | This study | ||
| G (dfrA19-aadA1) | dfrL | ATTCCAGTGCCAGCGAG | 3,531 | This study |
| aadAR | AAGTATGACGGGCTGATACTG | This study | ||
| H (oxa1-qacEΔ1) | oxaL | CTGAAATTGCTCAATTCAATAAAGC | 2,431 | This study |
| SulR2 | GTCGTTATAGCCCTATCTCGCGTC | This study |
To determine the outer boundaries of this complex sul1-type integron, plasmid DNA from the transconjugants was digested with BamHI. The fragments were then cloned into pUC18, and transformants were selected on brain heart infusion agar containing spectinomycin (25 μg/ml). A recombinant plasmid with a 12.9-kb BamHI insert was selected. PCRs A, B, and C were positive for this fragment, and sequence analysis of the boundaries with primers in pUC18 showed that the 5′-CS of class 1 integron was present at the left-hand boundary. The right-hand 700 bp showed 100% identity to the 3′ end of the complex sul1-type integron In-t1 containing the dfrA19 gene, downstream of the CR. This integron has been described for plasmid IncFI/97 of S. enterica serovar Typhimurium (AJ310778) (21) (Fig. 1). We postulated that a second copy of the CR might be present on the 12.9-kb fragment and thus performed PCRs D and E (Fig. 1, Table 1). The PCR products were sequenced, confirming the presence (downstream of the sequence containing the first CR, the sapA-like, and qnrB2) of a fragment 100% identical to the CR and also the dfrA19-associated gene of plasmid IncFI/97 (Fig. 1). In the plasmid, this sequence was followed by the class 1 integron In-t2 containing the oxa1 and aadA1 cassettes (Fig. 1). By a combination of PCRs with primers located in the orf513, oxa1, aadA1, dfrA19, and sul1 genes, we successfully amplified products F, G, and H from the transconjugant plasmid DNA (Fig. 1, Table 1). Sequencing of these fragments confirmed that the In-t2 integron was present downstream of the dfrA19-containing sequence. This structure may have resulted from RecA-dependent homologous recombination between two copies of sul1 or orf513.
To our knowledge, this is the first characterization of the genetic organization of a qnrB determinant. The variant qnrB2 gene has previously been detected on plasmid pMG301 in Citrobacter koseri (DQ351242) (Jacoby et al., Abstr. 44th Intersci. Conf. Antimicrob. Agents Chemother.), but its genetic environment has not yet been characterized. In the complex sul1-type integron described here, the qnrB2 gene is in the opposite orientation (Fig. 1). To our knowledge, the only resistance gene so far found to be located downstream of the CR, in the opposite orientation, is blaDHA-1 (20). The qnrA determinants previously described for complex sul1-type integrons were all inserted in the same orientation as orf513 (9). Some antibiotic resistance genes located downstream of the CR have been shown to originate from bacterial chromosome segments (1, 2, 3, 15, 19). The reservoir of qnrA-like genes was recently shown to be Shewanella algae (14). The origin of qnrB is unknown, although QnrB-like proteins were recently found in members of the Vibrionaceae family (13).
The S. enterica serovar Keurmassar strain expressed the ESBL SHV-12. Previous studies have shown an association between qnr-positive isolates and ESBLs, the two determinants sometimes being located on the same plasmid (9). In our S. enterica serovar Keurmassar strain, all the resistances, including the ESBL phenotype, were transferred en bloc. Thus, the SHV-12 determinant may be located on the same plasmid as the complex sul1-type integron described here.
We describe a new complex sul1-type integron in an S. enterica serovar Keurmassar strain containing two complete class-1 integrons surrounding two CRs separated by a partial 3′CS (Fig. 1). The resistance genes qnrB2 and dfrA19 were found adjacent to the two CRs.
Nucleotide sequence accession number.
The nucleotide sequence of the 12.9-kb BamHI fragment has been deposited in the EMBL-GenBank databases under accession number AM234698.
Acknowledgments
This work was supported by grants from the French Ministère de la Recherche and from Conseil Régional du Limousin. A. Gassama was supported by a grant from EGIDE, and N. Raked was supported by a grant from Fondation pour la Recherche Médicale.
We thank Emmanuelle Charpentier for critical reading and Bertrand Luzier and Pierre Rivet for technical assistance and sequencing.
REFERENCES
- 1.Arduino, S. M., P. H. Roy, G. A. Jacoby, B. E. Orman, S. A. Pineiro, and D. Centron. 2002. blaCTX-M-2 is located in an unusual class 1 integron (In35) which includes Orf513. Antimicrob. Agents Chemother. 46:2303-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Di Conza, J., J. A. Ayala, P. Power, M. Mollerach, and G. Gutkind. 2002. Novel class 1 integron (InS21) carrying blaCTX-M-2 in Salmonella enterica serovar Infantis. Antimicrob. Agents Chemother. 46:2257-2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Doi, Y., N. Shibata, K. Shibayama, K. Kamachi, H. Kurokawa, K. Yokoyama, T. Yagi, and Y. Arakawa. 2002. Characterization of a novel plasmid-mediated cephalosporinase (CMY-9) and its genetic environment in an Escherichia coli clinical isolate. Antimicrob. Agents Chemother. 46:2427-2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eckert, C., V. Gautier, and G. Arlet. 2006. DNA sequence analysis of the genetic environment of various blaCTX-M genes. J. Antimicrob. Chemother. 57:14-23. [DOI] [PubMed] [Google Scholar]
- 5.Gassama-Sow, A., A. Aïdara-Kane, N. Raked, F. Denis, and M. C. Ploy. 2004. Integron-associated antibiotic resistance in the newly described Salmonella enterica serovar Keurmassar emerging in Senegal. Emerg. Infect. Dis. 10:1339-1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hata, M., M. Suzuki, M. Matsumoto, M. Takahashi, K. Sato, S. Ibe, and K. Sakae. 2005. Cloning of a novel gene for quinolone resistance from a transferable plasmid in Shigella flexneri 2b. Antimicrob. Agents Chemother. 49:801-803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jacoby, G. A. 2005. Mechanisms of resistance to quinolones. Clin. Infect. Dis. 15(Suppl. 2):S120-S126. [DOI] [PubMed] [Google Scholar]
- 8.Martinez-Martinez, L., A. Pascual, and G. A. Jacoby. 1998. Quinolone resistance from a transferable plasmid. Lancet 351:797-799. [DOI] [PubMed] [Google Scholar]
- 9.Nordmann, P., and L. Poirel. 2005. Emergence of plasmid-mediated resistance to quinolones in Enterobacteriaceae. J. Antimicrob. Chemother. 56:463-469. [DOI] [PubMed] [Google Scholar]
- 10.Parsons, Y., R. M. Hall, and H. W. Stokes. 1991. A new trimethoprim resistance gene, dhfrX, in the In7 integron of plasmid pDGO100. Antimicrob. Agents Chemother. 35:2436-2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Partridge, S. R., and R. M. Hall. 2003. In34, a complex In5 family class 1 integron containing orf513 and dfrA10. Antimicrob. Agents Chemother. 47:342-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ploy, M. C., F. Denis, P. Courvalin, and T. Lambert. 2000. Molecular characterization of integrons in Acinetobacter baumannii. Description of a hybrid class 2 integron. Antimicrob. Agents Chemother. 44:2684-2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poirel, L., A. Liard, J. M. Rodriguez-Martinez, and P. Nordmann. 2005. Vibrionaceae as a possible source of Qnr-like quinolone resistance determinants. J. Antimicrob. Chemother. 56:1118-1121. [DOI] [PubMed] [Google Scholar]
- 14.Poirel, L., J. M. Rodriguez-Martinez, H. Mammeri, A. Liard, and P. Nordmann. 2005. Origin of plasmid-mediated quinolone resistance determinant QnrA. Antimicrob. Agents Chemother. 49:3523-3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sabaté, M., F. Navarro, E. Miró, S. Campoy, B. Mirelis, J. Barbé, and G. Prats. 2002. Novel complex sul1-type integron in Escherichia coli carrying blaCTX-M-9. Antimicrob. Agents Chemother. 46:2656-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sørum, H., T. M. L'Abée-Lund, A. Solberg, and A. Wold. 2003. Integron-containing IncU R plasmid Pras1 and pAr-32 from the fish pathogen Aeromonas salmonicida. Antimicrob. Agents Chemother. 47:1285-1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stokes, H. W., C. Tomaras, Y. Parsons, and R. M. Hall. 1993. The partial 3′-conserved segment duplications in the integrons In6 from pSa and In7 from pDGO100 have a common origin. Plasmid 30:39-50. [DOI] [PubMed] [Google Scholar]
- 18.Valentine, C. R., R. M. Heinrich, S. L. Chissoe, and A. Roe. 1994. DNA sequence of direct repeats of the sulI gene of plasmid pSa. Plasmid 32:222-227. [DOI] [PubMed] [Google Scholar]
- 19.Verdet, C., G. Arlet, G. Barnaud, P. H. Lagrange, and A. Phillipon. 2000. A novel integron in Salmonella enterica serovar Enteritidis, carrying the blaDHA-1 gene and its regulator ampR, originated from Morganella morganii. Antimicrob. Agents Chemother. 44:222-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Verdet, C., Y. Benzerara, V. Gautier, O. Adam, Z. Ould-Hocine, and G. Arlet. 2006. Emergence of DHA-1-producing Klebsiella spp. in the Parisian region: genetic organization of the ampC and ampR genes originating from Morganella morganii. Antimicrob. Agents Chemother. 50:607-617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Villa, L., P. Visca, F. Tosini, C. Pezzella, and A. Carattoli. 2002. Composite integron array generated by insertion of an ORF341-type integron within a Tn21-like element. Microb. Drug Resist. 8:1-8. [DOI] [PubMed] [Google Scholar]