Abstract
The molecular basis of glycopeptide-intermediate S. aureus (GISA) isolates is not well defined though frequently involves phenotypes such as thickened cell walls and decreased autolysis. We have exploited an isogenic pair of teicoplanin-susceptible (strain MRGR3) and teicoplanin-resistant (strain 14-4) methicillin-resistant S. aureus strains for detailed transcriptomic profiling and analysis of altered autolytic properties. Strain 14-4 displayed markedly deficient Triton X-100-triggered autolysis compared to its teicoplanin-susceptible parent, although microarray analysis paradoxically did not reveal significant reductions in expression levels of major autolytic genes atl, lytM, and lytN, except for sle1, which showed a slight decrease. The most important paradox was a more-than-twofold increase in expression of the cidABC operon in 14-4 compared to MRGR3, which was correlated with decreased expression of autolysis negative regulators lytSR and lrgAB. In contrast, the autolysis-deficient phenotype of 14-4 was correlated with both increased expression of negative autolysis regulators (arlRS, mgrA, and sarA) and decreased expression of positive regulators (agr RNAII and RNAIII). Quantitative bacteriolytic assays and zymographic analysis of concentrated culture supernatants showed a striking reduction in Atl-derived, extracellular bacteriolytic hydrolase activities in 14-4 compared to MRGR3. This observed difference was independent of the source of cell wall substrate (MRGR3 or 14-4) used for analysis. Collectively, our results suggest that altered autolytic properties in 14-4 are apparently not driven by significant changes in the transcription of key autolytic effectors. Instead, our analysis points to alternate regulatory mechanisms that impact autolysis effectors which may include changes in posttranscriptional processing or export.
The intensive use of vancomycin, which has for decades been the only drug uniformly active for treatment of multiresistant nosocomial isolates of Staphylococcus aureus, exerts a high selective pressure for emergence of glycopeptide resistance (52, 53). Since their first discovery in 1997 (32, 33), clinical isolates of glycopeptide-intermediate S. aureus (GISA) have been recovered worldwide (31, 52, 105). A major problem in evaluating the clinical and epidemiological significance of GISA isolates is the limited sensitivity and specificity of glycopeptide resistance phenotypic assays combined with the absence of specific molecular resistance markers (31, 52, 105). This situation is even more problematic when dealing with S. aureus isolates displaying heterogeneous expression of intermediate glycopeptide susceptibility (hGISA) (31, 52, 105). Emergence of GISA seems to result from multifactorial, endogenous changes in contrast to the few vancomycin-resistant S. aureus isolates that have exogenously acquired the vanA gene from enterococci (107).
Numerous investigations were carried out to understand the molecular basis and discover reliable phenotypic markers of GISA isolates (5, 8, 17, 18, 21, 26, 29, 31, 43, 46, 66, 67, 78, 89, 91-93, 105, 108). Several factors have been proposed as contributors to endogenous acquisition of resistance (5, 8), including a thickened cell wall (15, 17, 18, 78, 89), the presence of increased d-Ala-d-Ala residues in cell walls preventing glycopeptides from reaching their true target (31, 91), and other alterations in the composition or regulation of cell wall biosynthesis and turnover (21, 29, 43, 46, 63, 67, 92-94, 102). Some of these studies also mentioned fitness changes characterized by lower in vitro growth rates for GISA compared to non-GISA isolates (63, 66, 71, 89, 90). The functional implication of each of these metabolic changes for decreased glycopeptide susceptibility is not well understood. Furthermore, there is some debate whether these changes occur in all GISA/hGISA isolates or a subset of them (8, 26, 78, 90, 105).
Despite detailed genetic and DNA microarray studies, no comprehensive molecular model that explains the GISA phenotype has emerged. While the list of genes exhibiting altered expression (vraSR, tcaAB, sigB, and msrA2) in GISA compared to non-GISA isolates has been considerably expanded by DNA microarray studies (16, 48, 63, 66), the mutual interactions of these genes and their respective roles in resistance are still elusive (3, 5, 9, 16, 48, 49, 58). A major problem that complicated interpretation of most previous molecular studies has been the difficulty in identifying isogenic non-GISA parents of clinical GISA strains for true comparison. Instead, all reported studies except a recent one (63) were performed with in vitro-selected GISA, derived from multipassaged non-GISA parents (16-18, 43, 67) or isogenic in vitro-derived revertants from clinical GISA, whose fitness properties and virulence could be significantly altered by multiple subcultures (6, 16-18, 66).
Among factors controlling cell wall expansion, remodeling, and daughter cell separation, peptidoglycan hydrolases, referred to as autolysins, that participate in peptidoglycan turnover play an essential role (27, 42, 65). Autolysins that cleave the cell wall in a tightly controlled manner to maintain cell wall integrity during cell division are classified according to their specific cleavage types, e.g., N-acetylmuramidases, N-acetylglucosaminidases, N-acetylmuramyl-l-alanine amidases, endopeptidases, and transglycosylases (27). The major autolysis gene (atl) of S. aureus yields a 63-kDa amidase and a 54-kDa glucosaminidase after processing (2, 22, 69, 97-99, 111). Other autolytic genes include sle1, which encodes an additional N-acetylmuramyl-l-alanine amidase that is distinct from atl (39); lytM, which encodes glycylglycine endopeptidase (75, 76); and lytN, which possibly encodes muramidase activity (96).
Several regulators of autolysis have been described. First, the two-component regulator lytS-lytR is a negative regulator of extracellular peptidoglycan hydrolases via lrgA and lrgB, which in turn control the cidABC operon (10, 11, 25, 28, 81-83). Additional global regulators such as agr, sarA, arlRS, mgrA, and sarV can also influence autolytic activity, thus revealing the complex regulation of autolysis (2, 11, 23, 24, 37, 62, 98, 99).
The decreased autolytic activity of a substantial proportion of clinical or laboratory GISA isolates has been previously reported (7, 35, 43, 70, 71, 85, 91, 102, 108). Two alternative mechanisms were proposed for explaining the autolysis-deficient phenotypes of GISA, one being the reduced expression of atl (43, 108) and the other the occurrence of cell wall alterations that would decrease GISA susceptibility to autolysis (7). While there is a growing consensus that decreased autolysis of GISA strains may, directly or indirectly, contribute to their reduced susceptibility to glycopeptides, the molecular pathways linking defective autolysis to glycopeptide resistance are still undefined (43, 94, 102).
We previously described altered expression of some virulence factors in the teicoplanin-resistant (Teir) strain 14-4 compared to its teicoplanin-susceptible (Teis) parental strain, MRGR3, a methicillin-resistant S. aureus (MRSA) clinical isolate that shows a high degree of virulence in a tissue cage rat model of chronic foreign body MRSA infection (54, 79, 103). In this report, we explored the molecular basis of the markedly reduced autolytic phenotype of the Teir strain compared to its isogenic Teis parent by transcriptomic analysis of a subset of autolysis- and cell wall-associated genes (13).
MATERIALS AND METHODS
Bacterial strains.
MRSA strain MRGR3 is a Teis and vancomycin-susceptible isolate from a patient with catheter-related sepsis in 1979 (54, 103). The average MICs of vancomycin and teicoplanin in cation-adjusted Mueller-Hinton broth (MHB; Difco, Detroit, Mich.) for strain MRGR3 are 1 and 1 to 2 μg/ml, respectively. Strain 14-4 is a stable Teir derivative of MRGR3 that was isolated in a rat model of chronic tissue cage infection followed by two in vitro passages on teicoplanin-containing agar (103). The average MICs of vancomycin and teicoplanin for strain 14-4 are 4 and 16 to 32 μg/ml, respectively (79). The glycopeptide susceptibility or resistance phenotypes of strains MRGR3 and 14-4 were homogeneous, as assessed by population analysis profiles of teicoplanin and vancomycin (data not shown).
Several criteria confirmed the isogenic link of strains MRGR3 and 14-4. First, besides their different susceptibilities to glycopeptides, strains 14-4 and MRGR3 exhibit identical patterns of resistance to penicillin, gentamicin, cholamphenicol, erythromycin, tetracycline and polymyxin B, while remaining susceptible to rifampin, cotrimoxazole, mupirocin, fosfomycin, and all fluoroquinolones. Pulsed-field gel electrophoresis of strains 14-4 and MRGR3 yielded similar patterns except for a single-band difference (57), and their isogenic links were confirmed by a previously described comparative genomic hybridization technique (13), which showed >99.98% identity (data not shown).
In some experiments, parental strain 8325-4 and its atl mutant, SH108, (kindly provided by S. J. Foster) were used as control strains for autolysin production.
Triton X-100-induced autolysis assays.
Autolysis was assayed essentially as described previously (37, 60) on S. aureus cultures grown under three different conditions. In initial experiments (37, 60), overnight cultures of S. aureus were diluted to an optical density at 600 nm (OD600) of 0.05 and grown for 3 h with shaking at 37°C in MHB containing 1 M NaCl until the OD600 reached 0.7 (37). In further experiments, S. aureus cultures were preferably grown in MHB without NaCl supplementation, either with shaking for 3 h or without shaking for 5 h.
Thereafter, cells were harvested, washed twice with ice-cold water, and then resuspended in the same volume of 0.05 M Tris-HCl (pH 7.2) containing 0.05% Triton X-100. Cells were incubated at 30°C with shaking and checked for lysis by measuring the progressive decrease in absorbance (OD600). Autolysis was quantified as a percentage of the initial OD600 remaining at each sampling point.
Zymographic analysis.
Measurement of extracellular bacteriolytic hydrolases was performed essentially as described previously (28, 60, 62). Supernatants from overnight cultures of strains MRGR3 and 14-4 or 8325-4 and its atl mutant, SH108, used as controls, were grown in 10 ml MHB, filter sterilized, and then concentrated 20-fold using Centricon-3 concentrators (Millipore). Various loading amounts (from 1.25 to 20 μg) of extracellular proteins, determined by Bradford assay (Bio-Rad), were separated on sodium dodecyl sulfate-7.5% polyacrylamide gels containing autoclaved and lyophilized cell preparations (1 mg of dry weight per ml) of either strain MRGR3 or 14-4. After electrophoresis, proteins were renatured by overnight incubation in 1% (vol/vol) Triton X-100 in 25 mM Tris-HCl (pH 8.0), and then gels were stained with 1% (wt/vol) methylene blue in 0.05% KOH prior to photography (28, 60, 62).
Quantitative bacteriolytic hydrolase assays.
Bacteriolysis of lyophilized S. aureus by extracellular hydrolases was quantified as previously described (28). Equivalent amounts (100 μg) of concentrated supernatants from either 5-h or overnight cultures of strain MRGR3, 14-4, 8325-4, or its atl mutant, SH108, were added to a suspension of autoclaved and lyophilized S. aureus MRGR3 or 14-4 (1 mg/ml) in 100 mM Tris-HCl (pH 8.0) and incubated at 37°C with shaking. Lytic activity was recorded by measuring the progressive decrease in absorbance (OD600).
Total RNA extraction and labeling.
Cultures of MRGR3 and 14-4 were grown at 37°C for 5 h in MHB without shaking. Then, bacteria were harvested and RNA extraction was performed as previously described (30, 50). Briefly, bacteria were recovered, fixed in acetone-ethanol (1:1), and washed in Tris-sucrose buffer. Samples were treated with ice-cold lysostaphin, and RNA was purified as described previously (50, 79). The absence of contaminating DNA was verified by PCR (50, 79). Purified RNA samples were analyzed using the RNA NanoLab chip on the 2100 Bioanalyser (Agilent, Palo Alto, CA).
DNA microarray preparation, hybridization, and data analysis.
The customized StaphChip oligoarray was designed in our laboratory and manufactured by Agilent Technologies (Palo Alto, CA) as described previously (13). A set of 5,337 S. aureus 40- to 60-mer probes, recognizing 97.5, 93, and 81% of N315, Mu50, and COL open reading frames (ORFs), respectively, were represented in each oligoarray. More than 95% of ORFs evaluated in this study were recognized by two or three nonoverlapping specific probes.
Total RNAs (12 μg) from strains MRGR3 and 14-4 were labeled in parallel with Cy3-dCTP and Cy5-dCTP (New England Nuclear), respectively, using the SuperScript kit (Invitrogen) by following the manufacturer's instructions. Labeled cDNAs were then purified using QIAQuick (QIAGEN) columns.
For competitive hybridization using a dual-label experimental approach, equivalent amounts of Cy3-labeled and Cy5-labeled cDNAs were diluted in 250 μl Agilent hybridization buffer and cohybridized for 17 h at 60°C. Slides were washed, dried under nitrogen flow, and scanned (Agilent) as described previously (13) using 100% photomultiplier power tube power for both wavelengths. All positive and significant-local-background-subtracted signals, obtained using Feature Extraction software (version A6.1.1; Agilent), were corrected for unequal dye incorporation or unequal load of labeled product. The algorithm consisted of a rank consistency filter and a curve fit using the default LOWESS (locally weighted linear regression) method. Irregular or saturated spots were excluded from subsequent analysis. Spots showing a reference signal lower than background plus 2 standard deviations were also excluded from subsequent analysis. Average background values were 328 ± 17 and 363 ± 24 arbitrary units for green and red signals, respectively.
To provide estimates of expression levels of the subset of genes analyzed in our study with respect to the whole transcriptome, all Feature Extraction-processed dye-normalized signals from the oligoarray were subdivided into four categories according to their intensities in the Teis control strain MRGR3: the 25th percentile of probes, yielding the lower-intensity signals (<126 arbitrary units), followed by the 25th to 50th percentile (127 to 924 units), the 50th to 75th percentile (925 to 6,612 units), and the 75th to 100th percentile, yielding the highest-intensity signals (>6,612 units).
Data analysis.
The oligoarray-generated signals across three independent experiments were expressed as mean (± standard error of the mean [SEM]) processed green or red signals separately recorded from each individual probe, or from two or more nonoverlapping probes recognizing common gene transcripts. For all genes whose signal intensities and strain-specific signal ratios originating from multiple probe subsets were equivalent, statistical differences were evaluated by t test on the basis of pooled signals from multiple gene-associated probes (see Table S1 in the supplemental material).
Real-time RT-PCR.
mRNA levels were determined by quantitative reverse transcriptase PCR (RT-PCR) using the one-step reverse transcriptase qPCR Master Mix kit (Eurogentec, Seraing, Belgium), as described previously (79). All primers and probes that were not described previously are listed in Table 1; they were designed using PrimerExpress software (version 1.5; Applied Biosystems) and obtained from Eurogentec or Applied Biosystems. Conditions for reverse transcription, PCR, detection of fluorescence emission, and normalization of the levels of mRNA of the target genes extracted from the different strains on the basis of their 16S rRNA levels were described previously (50, 79, 104). The statistical significance of strain-specific differences in normalized cycle threshold values for each transcript was evaluated by paired t test, and data were considered significant when P was <0.05.
TABLE 1.
TaqMan primers and probes used in this study
| Gene | Forward primer | Reverse primer | Probe (5′-3′)a |
|---|---|---|---|
| arlR | 304F-CCGTTTGATATTGAAGAACTTTTAGCA | 386R-CCGTTGACATCGATAATATCCTTTT | 333T-AATTCGTGCAATTTTACGTCGTCAGCCA |
| atl | 878F-AAACAGCACCAACGGATTACTTATC | 977R-AATGAAGCATAGTCGTGTGTGTGTAC | 911T-TCGGTGCAGTCGGTAACCCTAGATTCA |
| lytR | 475F-GGGATTGGCACACATAATGGT | 576R-AGTGGGATTCAATCGTTTTTCATAA | 500T-CAACCATACATACAACGAATCATAAATACGAAACAACAGA |
| lrgB | 525F-CCGAATTACTAACCCTATTGCCC | 602R-TCTTTGGCTGGTGCTACACCT | 549T-AGGATTAGCACTTGGAACAAGTGGTCACACA |
| cidB | 517F-TTTGAATCTTCTATCGCCAAAGG | 591R-TTCTAGTGCTTTAGCTGTGCCAA | 541T-TTAACGTATGGGAATGCGTCACATGCA |
| mgrA | 142F-TGGGATGAATCTCCTGTAAACGT | 233R-TGTTCCATTCGTTTTAATAATGGTG | 166T-AAGAAAGTCGTAACTGAATTAGCACTCGATACTGGTACA |
| pbp4 | 90F-CCCTGTACAAGCAGCAAATCAA | 159R-AACAGCACTCGTCGGTTCGT | 113T-ATGGTTATGCAGGTTTGTCGGCTGCA |
| pbp2 | 407F-AAAGACGCAGTACTTGCAACTGAAG | 490R-ACCAATTGCACCGAATAAACGT | 437T-CGTTTCTACGAACATGGCGCACTTGATT |
| tcaA | 428F-GTAAAAATGGCACGCGTTATATCTT | 597R-CTTACCACCAGATTTAAACTCATATTTTGT | 532T-CCTACCAAGCAACCAATTGTTAAGCCGAAA |
Probes were labeled with 5′-6-carboxyfluorescein and 3′-6-carboxy tetramethylrhodamine.
Microarray data accession number.
Microarray data analyzed in this study have been deposited in the ArrayExpress database with accession number E-MEXP-378 (http://www.ebi.ac.uk/arrayexpress).
RESULTS
Major differences in Triton X-100-induced autolysis of Teir and Teis strains.
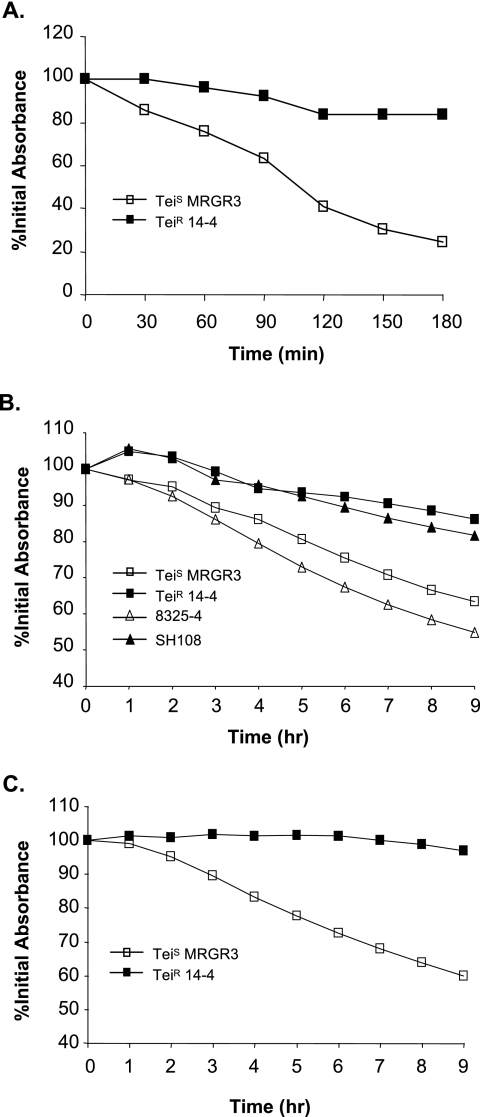
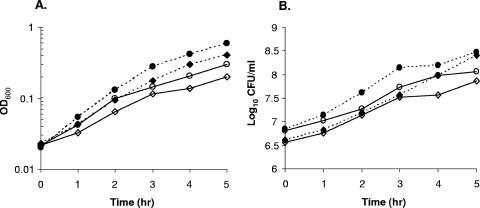
The Teir strain 14-4 exhibited markedly reduced autolysis compared to its isogenic Teis parent, MRGR3 (Fig. 1). Changing the preculture conditions did not lessen the strongly reduced autolytic response of the Teir strain compared to its Teis parent. In initial experiments, we used previously recommended conditions by growing cells incubated with shaking in MHB containing 1 M NaCl (37, 60), which led to autolytic rates of nearly 80% at 3 h for Teis strain MRGR3 compared to ca. 20% for Teir strain 14-4 (Fig. 1A). In further experiments, the autolytic response of Teir and Teis strains was preferably tested on cells that were grown in MHB without NaCl supplementation to be more consistent with conditions used for assaying transcript levels or extracellular bacteriolytic activities. Comparative growth characteristics of Teir strain 14-4 and Teis strain MRGR3, grown in MHB without NaCl supplementation in either shaking or nonshaking conditions, are shown on Fig. 2.
FIG. 1.
Differences in Triton X-100-induced autolysis of Teir strain 14-4 compared to its isogenic Teis parent, MRGR3. Exponential cultures were grown either in shaking conditions for 3 h at 37°C in MHB supplemented with 1 M NaCl (A) or unsupplemented (B) or for 5 h in MHB in nonshaking conditions without NaCl supplement (C). When indicated, strain 8325-4 and its atl mutant, SH108, were used as controls (B). These data are from a single representative experiment and were reproduced several times.
FIG. 2.
Growth curves in MHB of Teir strain 14-4 (⋄) compared to its isogenic Teis parent, MRGR3 (○), in nonshaking (open symbols) or shaking (solid symbols) conditions, recorded as OD600 (A) or viable counts (B).
After growth in shaking conditions, autolysis at 9 h was approximately 40% for Teis MRGR3 and <20% for Teir 14-4 (Fig. 1B). In similar conditions autolytic rates of strain 8325-4 and its atl mutant, SH108, at 9 h were approximately 50% and 20%, respectively, indicating the involvement of Atl-derived products in the Triton X-100-induced autolytic assay.
Finally, after growth in nonshaking conditions, autolytic rates of strains MRGR3 and 14-4 at 9 h were approximately 40% and <10%, respectively (Fig. 1C). Importantly, the autolytic differences recorded on bacterial cultures grown in MHB without shaking and without NaCl supplementation were particularly relevant since identical growth conditions were used for assaying transcript levels of strain 14-4 and MRGR3 by DNA oligoarray or real-time reverse transcription-PCR (RT-PCR).
Reliability of the DNA oligoarray for studying differential gene expression between Teir and Teis strains.
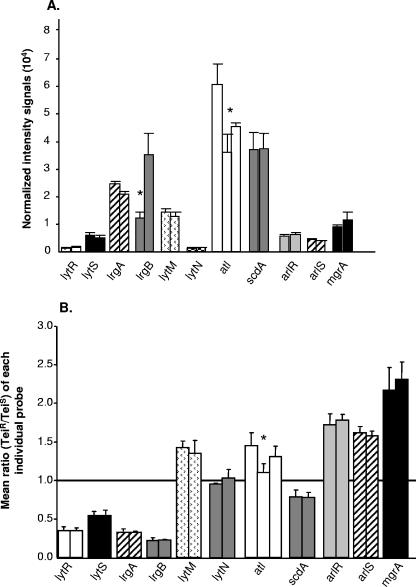
The reliability of the StaphChip oligoarray-generated strain-specific signals was established across three independent experiments by (i) the high interexperiment reproducibility of processed intensity signals recorded on each individual probe for Cy3-labeled or Cy5-labeled hybridized cDNAs from Teis or Teir strains, respectively, and (ii) the equivalent processed intensity signals recorded on multiple, nonoverlapping probes of common gene transcripts (see Table S1 in the supplemental material). As an example, Fig. 3A shows mean ± SEM Cy3-generated signals for a subset of 23 different probes recognizing 23 nonoverlapping regions of 11 different gene transcripts from parental Teis strain MRGR3.
FIG. 3.
(A) Reproducibility of intensity signals generated from nonoverlapping probes covering individual gene transcripts. Shown are normalized intensity signals (mean plus SEM; n = 3) from 23 probes covering 11 different genes from Teis strain MRGR3. *, significantly different intensities. (B) Reproducibility of transcript level changes of Teir strain 14-4 compared to Teis strain MRGR3, expressed as mean (plus SEM; n = 3) ratios (Teir/Teis), for multiple probes recognizing common transcripts. *, significantly different ratios.
For most assayed genes, changes in transcript levels, expressed as ratios of Teir to Teis signal intensities, were highly reproducible, not only on individual probes but also on multiple probes recognizing nonoverlapping regions of each transcript (Fig. 3B and Table 2; see Table S1 in the supplemental material). Collectively, these results provided strong evidence that multiple probes recognizing different regions of each transcript provided reliable data allowing detection of minor, but statistically significant, strain-specific changes in gene expression (Table 2).
TABLE 2.
Ratios of transcript levels of Teir 14-4 to Teis MRGR3
| N315 SA no. | Gene | Description | Probe intensity | Ratio of signal intensity (14-4/MRGR3)
|
P | |
|---|---|---|---|---|---|---|
| Meana | SEM | |||||
| Autolysis genes | ||||||
| SA0905 | atl | Bifunctional precursor autolysin (Atl) | **** | 1.29 | 0.06 | <0.05 |
| SA0423 | sle1 | N-Acetylmuramyl-L-alanine amidase | **** | 0.76 | 0.03 | <0.05 |
| SA0265 | lytM | Peptidoglycan hydrolase | **** | 1.39 | 0.05 | <0.001 |
| SA1090 | lytN | LytN protein | *** | 0.99 | 0.03 | NSc |
| SA2329 | cidA | Hypothetical protein, similar to transcription regulator | ** | 2.24 | 0.14 | <0.001 |
| SA2328 | cidB | Conserved hypothetical protein | **** | 3.25 | 0.05 | <0.001 |
| SA2327 | cidC | Pyruvate oxidase | **** | 2.57 | 0.15 | <0.001 |
| SA0251 | lytR | Two-component response regulator | *** | 0.34 | 0.02 | <0.001 |
| SA0250 | lytS | Two-component sensor histidine kinase | *** | 0.55 | 0.02 | <0.001 |
| SA0252 | lrgA | Murein hydrolase regulator LrgA | **** | 0.33 | 0.01 | <0.001 |
| SA0253 | lrgB | Antiholin-like protein LrgB | **** | 0.22 | 0.01 | <0.001 |
| SA0641 | mgrA | Transcriptional regulator MgrA | **** | 2.24 | 0.10 | <0.001 |
| SA0650 | norA | Quinolone resistance protein | *** | 1.47 | 0.04 | <0.001 |
| SA1248 | arlR | Truncated (putative response regulator ArlR [S | *** | 1.75 | 0.04 | <0.001 |
| SA1246 | arlS | Sensor histidine kinase ArlS | *** | 1.60 | 0.03 | <0.001 |
| SA0573 | sarA | Staphylococcal accessory regulator A | *** | 3.53 | 0.13 | <0.001 |
| SAS065 | RNAIII | Delta hemolysin | **** | 0.02 | 0.00 | <0.001 |
| SA1844 | agrA | Accessory gene regulator A | **** | 0.08 | 0.01 | <0.001 |
| Cell wall-associated genes | ||||||
| SA0702 | llm/tagO | Lipophilic protein affecting bacterial lysis rate and methicillin resistance level | *** | 1.60 | 0.04 | <0.001 |
| SA0909 | fmtA | Autolysis and methicillin resistant-related protein | *** | 1.62 | 0.04 | <0.001 |
| SA2062 | sarV | Staphylococcal accessory regulator A homolog | ** | 1.45 | 0.15 | NS |
| SA1964 | fmtB | FmtB protein | *** | 1.28 | 0.13 | NS |
| SA1028 | ftsA | Cell division protein | **** | 1.36 | 0.04 | <0.001 |
| SA1029 | ftsZ | Cell division protein | **** | 1.10 | 0.04 | <0.05 |
| SA0249 | scdA | Cell division protein and morphogenesis-related protein | **** | 0.78 | 0.03 | <0.001 |
| SA1193 | fmtC/mprF | Oxacillin resistance-related FmtC protein | **** | 0.46 | 0.03 | <0.001 |
| SA1902 | murA | UDP-N-acetylglucosamine 1-carboxyvinyl transferase 1 | **** | 1.35 | 0.08 | <0.01 |
| SA0693 | murB | UDP-N-acetylenolpyruvoylglucosamine reductase | **** | 0.99 | 0.03 | NS |
| SA1562 | murC | DNA translocase stage III sporulation protein homolog | **** | 0.85 | 0.02 | <0.05 |
| SA1026 | murD | UDP-N-acetylmuramoylalanine-d-glutamate ligase | **** | 0.75 | 0.02 | <0.001 |
| SA0876 | murE | UDP-N-acetylmuramoylalanyl-d-glutamate-2,6-diaminopimelate ligase | *** | 0.84 | 0.07 | NS |
| SA1886 | murF | UDP-N-acetylmuramoylalanyl-d-glutamyl-2,6-diaminopimelate-d-alanyl-d-alanyl ligase | **** | 1.23 | 0.04 | <0.01 |
| SA1926 | murZ | UDP-N-acetylglucosamine 1-carboxylvinyl transferase 2 | **** | 2.34 | 0.09 | <0.001 |
| SA1251 | murG | Undecaprenyl-PP-MurNAc-pentapeptide-UDPGlcNAc GlcNAc transferase | **** | 1.03 | 0.03 | NS |
| SA0997 | murI | Glutamate racemase | **** | 0.80 | 0.02 | <0.001 |
| SA1025 | mraY | Phospho-N-muramic acid-pentapeptide translocase | **** | 0.52 | 0.02 | <0.001 |
| SA2057 | femX/fmhB | FmhB protein | **** | 1.29 | 0.05 | NS |
| SA1206 | femA | Essential factor for expression of methicillin resistance | **** | 1.39 | 0.08 | <0.01 |
| SA1207 | femB | FemB protein | **** | 1.03 | 0.05 | NS |
| SA1024 | pbp1 | Penicillin-binding protein 1 | **** | 0.91 | 0.03 | <0.05 |
| SA1283 | pbp2 | Penicillin-binding protein 2 | **** | 1.61 | 0.06 | <0.001 |
| SA1381 | pbp3 | Penicillin-binding protein 3 | **** | 0.91 | 0.04 | <0.05 |
| SA0598 | pbp4 | Penicillin binding protein 4 | *** | 1.44 | 0.05 | <0.001 |
| SA0038 | mecA | Penicillin binding protein 2A | **** | 0.39 | 0.02 | <0.001 |
| SA0592 | tagA | Teichoic acid biosynthesis protein | *** | 0.80 | 0.02 | <0.001 |
| SA0595 | tagB | Teichoic acid biosynthesis protein B | **** | 1.18 | 0.04 | <0.05 |
| SA0597 | tagD | Teichoic acid biosynthesis protein D | *** | 1.03 | 0.02 | NS |
| SA0594 | tagG | Teichoic acid translocation permease protein | **** | 1.06 | 0.03 | NS |
| SA0593 | tagH | Teichoic acid translocation ATP-binding protein | **** | 0.98 | 0.03 | NS |
| SA0596 | tagX | Teichoic acid biosynthesis protein X | **** | 1.01 | 0.05 | NS |
| SA0793 | dltA | d-Alanine-d-alanyl carrier protein ligase | **** | 0.48 | 0.01 | <0.001 |
| SA0794 | dltB | DltB membrane protein | **** | 0.57 | 0.03 | <0.001 |
| SA0795 | dltC | d-Alanine-poly(phosphoribitol) ligase subunit 2 | **** | 0.43 | 0.03 | <0.001 |
| SA0796 | dltD | Poly d-alanine transfer protein | **** | 0.54 | 0.02 | <0.001 |
| Global or antibiotic cell wall regulators | ||||||
| SA2145 | tcaB | TcaB protein | *** | 0.93 | 0.04 | NS |
| SA2480 | drp35 | Drp35 | *** | 1.63 | 0.10 | <0.05 |
| SA1194 | msrA2 | Methionine sulfoxide reductase A | *** | 1.53 | 0.03 | <0.001 |
| SA1195 | msrR | Peptide methionine sulfoxide reductase regulator MsrR | *** | 2.03 | 0.06 | <0.001 |
| SA1257 | msrA1 | Peptide methionine sulfoxide reductase A | **** | 2.05 | 0.06 | <0.001 |
| SA1256 | msrB | Methionine sulfoxide reductase B | **** | 2.13 | 0.05 | <0.001 |
| SA2146 | tcaA | TcaA protein | *** | 2.57 | 0.13 | <0.001 |
| SA2147 | tcaR | TcaR transcription regulator | *** | 2.35 | 0.15 | <0.001 |
| SA1700 | vraR | Two-component response regulator | **** | 3.54 | 0.14 | <0.001 |
| SA1701 | vraS | Two-component sensor histidine kinase | **** | 5.01 | 0.16 | <0.001 |
| SA1691 | sgtB | Monofunctional glycosyltransferase | *** | 8.94 | 0.29 | <0.001 |
| SA1659 | prsA | Peptidyl-prolyl cis/trans isomerase homolog | **** | 6.53 | 0.13 | <0.001 |
| SA1551 | sgtA | Monofunctional glycosyltransferase | *** | 1.21 | 0.06 | <0.05 |
| SA0599 | abcA | ATP-binding cassette transporter A | **** | 0.69 | 0.02 | <0.001 |
| SA1869 | sigB | Sigma factor B | **** | 0.93 | 0.03 | <0.05 |
| SA1872 | rsbU | Sigma B regulation protein RsbU | **** | 0.91 | 0.04 | NS |
| SA1871 | rsbV | Anti-sigma B factor antagonist | **** | 0.98 | 0.05 | NS |
| SA1870 | rsbW | Serine-protein kinase RsbW | **** | 0.93 | 0.05 | NS |
| SA1984 | asp23 | Alkaline shock protein 23, Asp23 | **** | 1.79 | 0.03 | <0.01 |
| SA0108 | sarS | Staphylococcal accessory regulator A homologue | **** | 1.85 | 0.07 | <0.001 |
| SA0018 | yycG | Two-component sensor histidine kinase | **** | 1.12 | 0.03 | <0.05 |
| SA0017 | yycF | Response regulator | **** | 0.99 | 0.04 | NS |
| SA1323 | srrA | Staphylococcal respiratory response protein SrrA | **** | 1.18 | 0.01 | <0.001 |
| SA1322 | srrB | Staphylococcal respiratory response protein SrrB | **** | 1.19 | 0.05 | <0.05 |
| SA0661 | saeR | Response regulator | **** | 1.02 | 0.03 | NS |
| SA0660 | saeS | Histidine protein kinase | **** | 0.99 | 0.01 | NS |
| SA1583 | rot | Repressor of toxins Rot | *** | 1.14 | 0.05 | NS |
| SA2287 | sarU | Staphylococcal accessory regulator A homolog | * | 0.92 | 0.06 | NS |
| SA2286 | sarT | Staphylococcal accessory regulator A homolog | ** | 1.21 | 0.08 | <0.05 |
| Other genes | ||||||
| SA0614 | graR | Hypothetical protein, similar to two-component response regulator | *** | 1.17 | 0.1 | NS |
| SA0615 | graS | Hypothetical protein, similar to two-component sensor histidine kinase | *** | 1.04 | 0.05 | NS |
| SA0639 | graA | Hypothetical protein, similar to ABC transporter for expression of cytochrome bd | **** | 0.97 | 0.04 | NS |
| SA0743 | graB | Hypothetical protein, similar to staphylocoagulase precursor | * | 1.39 | 0.25 | NS |
| SAS044 | graC | 4-Oxalocrotonate tautomerase | ** | 1.08 | 0.04 | NS |
| SA1318 | graD | Hypothetical protein | * | 0.57 | 0.05 | <0.05 |
| SA1337 | graE | Transcription regulator AraC/XylS family homolog | **** | 0.72 | 0.02 | <0.001 |
| SA1339 | malR | Maltose operon transcriptional repressor | *** | 2.14 | 0.12 | <0.001 |
| SAS030 | graF | Hypothetical protein | ** | 2.8 | 0.17 | <0.05 |
| SA0016 | purA | Adenylosuccinate synthase | *** | 0.72 | 0.04 | <0.05 |
| SA1724 | purB | Adenylosuccinate lyase | **** | 1.04 | 0.06 | NS |
| SA0918 | purC | Phosphoribosylaminoimidazolesuccinocarboxamide synthetase homolog | *** | 1.52 | 0.08 | <0.001 |
| SA0926 | purD | Phosphoribosylamine-glycine ligase PurD | **** | 1.35 | 0.06 | <0.05 |
| SA0916 | purE | Hypothetical protein, similar to phosphoribosylaminoimidazole carboxylase PurE | *** | 1.48 | 0.04 | <0.001 |
| SA0922 | purF | Phosphoribosylpyrophosphate amidotransferase PurF | **** | 1.57 | 0.07 | <0.001 |
| SA0925 | purH | Bifunctional purine biosynthesis protein PurH | **** | 1.4 | 0.08 | <0.05 |
| SA0917 | purK | Phosphoribosylaminoimidazole carboxylase carbon dioxide fixation chain | *** | 1.46 | 0.12 | <0.05 |
| SA0921 | purL | Phosphoribosylformylglycinamidine synthetase PurL | **** | 1.42 | 0.06 | <0.05 |
| SA0923 | purM | Phosphoribosylformylglycinamidine cyclo-ligase PurM | **** | 1.44 | 0.04 | <0.001 |
| SA0924 | purN | Phosphoribosylglycinamide formyltransferase | **** | 1.29 | 0.04 | <0.001 |
| SA0920 | purQ | Phosphoribosylformylglycinamidine synthase I PurQ | **** | 1.36 | 0.07 | <0.05 |
| SA0454 | purR | pur operon repressor homologue | **** | 2.23 | 0.18 | <0.05 |
| Proteases | ||||||
| SA1631 | splA | Serine protease SplA | ** | 0.68 | 0.02 | <0.001 |
| SA1630 | splB | Serine protease SplB | ** | 0.74 | 0.06 | <0.05 |
| SA1629 | splC | Serine protease SplC | ** | 0.68 | 0.03 | <0.001 |
| SA1628 | splD | Serine protease SplD | ** | 0.86 | 0.04 | <0.01 |
| SA1627 | splF | Serine protease SplF | ** | 1.68 | 0.19 | <0.01 |
| SA0901 | sspA | Cysteine protease/V8 protease | *** | 1.17 | 0.04 | NS |
| SA0900 | sspB | Cysteine protease precursor | ** | 1.70 | 0.06 | <0.001 |
| SA0899 | sspC | Cysteine protease | * | 0.94 | 0.07 | NS |
| SA2430 | aur | Zinc metalloproteinase aureolysin | ** | 0.92 | 0.06 | NS |
| SA1725 | scpA | Staphopain, cysteine proteinase | ** | 1.36 | 0.15 | NS |
| SA1726 | scpB | Hypothetical protein | ** | 1.50 | 0.03 | <0.001 |
| SA0879 | htrA | Serine protease HtrA | *** | 0.50 | 0.03 | <0.001 |
Recorded in three independent experiments; see text for details.
****, 75th to 100th percentile; ***, 50th to 75th percentile; **, 25th to 50th percentile; *, <25th percentile (see Materials and Methods for details).
NS, not significant.
Comparison of expression levels of autolysis- and cell wall-associated genes between Teir and Teis strains.
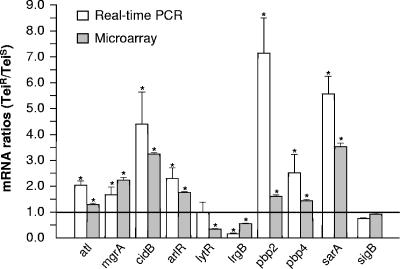
Expression levels of 117 genes involved in autolysis or cell wall biosynthesis or known as related regulators were analyzed by DNA oligoarray (Table 2; Fig. 3B), and a small subset of those transcript levels were also evaluated by real-time RT-PCR (Fig. 4). Overall, 85% of those analyzed genes ranged among the most highly expressed ones of the S. aureus transcriptome, and those with expression levels lower than the 50% percentile were predominantly found within the category of extracellular proteases (Table 2).
FIG. 4.
Comparison of transcript level ratios recorded for 10 different genes in Teir strain 14-4 compared to Teis strain MRGR3 by real-time RT-PCR (white boxes) and DNA microarray (gray boxes). Results are presented as means plus SEM for three experiments. Comparative microarray and RT-PCR values (<5%) for RNAIII are too small to be shown on the graph. In most microarray data, error bars are too small to be shown on the graph. *, significant differences between strains MRGR3 and 14-4.
Surprisingly, transcript levels of most autolysin genes either showed no significant decrease, except for sle1, whose expression slightly decreased, in the autolysis-deficient Teir strain 14-4 compared to its autolysis-proficient Teis parent. Instead, a number of those transcripts were even slightly (atl) or markedly (cidA, cidB, and cidC) increased. Of note, expression of cidA was very weak, compared to the more intense signals of cidB and cidC (Table 2; see Table S1 in the supplemental material). Increased cidABC transcript levels were consistent with the reduced expression of their major autolytic repressors lytSR and lrgAB in the Teir strain compared to the Teis strain, according to previous studies (10, 11, 25, 28, 81, 81, 83), but failed to explain the autolysis-deficient phenotype of the Teir strain. Real-time RT-PCR confirmed increased atl and cidB levels and reduced lrgB levels but showed no significant change in lytR levels (Fig. 4).
In contrast, significantly increased expression levels were recorded for three additional negative regulators of autolysis, namely, mgrA, arlRS, and sarA, which individually or collectively may contribute to the autolysis-deficient phenotype of Teir strain 14-4 (23, 37, 56). An additional explanation for the deficient autolytic phenotype of the Teir strain compared to its Teis parent might be the loss of agr global regulator function, previously reported to promote autolysis, as evidenced by the strong reduction in its RNAII and RNAIII levels (24, 85). The oligoarray-detected changes in mgrA, arlR, sarA, and RNAIII transcript levels in Teir strain 14-4 compared to MRGR3 were all confirmed by real-time RT-PCR (Fig. 4).
The potential influence of additional cell wall-associated factors or global regulators of autolysis was also considered. Whereas transcript levels of llm (59), also referred to as tagO (106), fmtA (47), and the weakly expressed sarV, recently described as a positive regulator of autolysis (62), were slightly increased in strain 14-4 compared to MRGR3, this was not the case for fmtB, ftsA, ftsZ, and scdA (12, 45, 73, 74), which showed marginal (<30%) changes in transcript levels (Table 2). The significance of the twofold decline in fmtC (also called mprF) transcript levels is unclear because of its questionable impact on autolysis (44) and controversial effect on glycopeptide resistance (110).
Differential expression of major cell wall biosynthesis genes by Teir and Teis strains was also studied. No consistent changes were recorded in transcript levels of the cytoplasmic enzymes MurA to MurF, involved in the formation of the monomeric building block N-acetylglucosamine-N-acetylmuramyl pentapeptide or MurI (glutamate racemase) in Teir strain 14-4 compared to MRGR3 (Table 2). The only notable exception was the more-than-twofold increase in murZ transcript levels, which contrasted with the weak increase of its murA functional homolog (19) (Table 2). The molecular basis and biological significance of the differential expression of UDP-N-acetylglucosamine enolpyruvyl transferase isozymes 1 (MurA) and 2 (MurZ) (16) remain unexplained.
Transcript levels of enzymes catalyzing further steps of peptidoglycan synthesis were either decreased by twofold (for MraY) or similar (for MurG, a translocase) in Teir strain 14-4 compared to Teis strain MRGR3 (Table 2). Furthermore, expression levels of FemX, FemA, and FemB factors, catalyzing the assembly of pentaglycine interpeptide bridges (87), remained constant in strain 14-4 compared to MRGR3 (Table 2). Finally, transcript levels of major penicillin-binding proteins (PBPs) involved in the final stages of peptidoglycan synthesis were either similar (PBP1 and PBP3) or significantly increased (PBP2 and PBP4) in the Teir strain compared to the Teis strain (Table 2), as confirmed by real-time RT-PCR (Fig. 4). In line with these findings, MICs for Teir strain 14-4 of oxacillin and imipenem showed a slight twofold increase compared to its Teis parent (data not shown).
These data are compatible with previous observations showing increased production and penicillin-binding activity of PBP2 in some laboratory-derived teicoplanin- and vancomycin-resistant derivatives of methicillin-susceptible S. aureus (67, 88). While the increased expression levels of PBP4 are compatible with previous data recorded on teicoplanin-resistant laboratory isolates (67), an opposite situation was recorded in some clinical isolates displaying intermediate susceptibility to vancomycin (VISA) and exhibiting a lower degree of cell wall cross-linking, which demonstrated decreased expression of PBP4 potentially linked with acquisition of the VISA phenotype (21, 63, 93). As opposed to PBP2 and PBP4, transcript levels for mecA, coding for PBP2A, involved in expression of methicillin resistance, were decreased by more than twofold in strain 14-4 compared to MRGR3 (Table 2).
Transcript levels of the six contiguous genes (tagA, tagH, tagG, tagB, tagX, and tagD) of the cell wall teichoic acid biosynthetic pathway operon were virtually identical in strains 14-4 and MRGR3 (Table 2). In contrast, dltABCD transcript levels were consistently reduced by ca. twofold in the Teir strain compared to the Teis strain (Table 2). While the complete absence of d-alanine esters in teichoic acids was reported to increase simultaneously the autolytic rate and susceptibility to vancomycin of the glycopeptide-susceptible S. aureus strain SA113 (70), a significant contribution of partially decreased dltABCD transcript levels to the autolysis-deficient phenotype of Teir strain 14-4 is uncertain because such a reduction would rather predict an increased susceptibility of S. aureus to teicoplanin (70).
Overall, gene expression levels of most cell wall-building components were not significantly altered in Teir strain 14-4 compared to its Teis isogenic parent, MRGR3. While gene expression levels of the capsular polysaccharide biosynthetic pathway were upregulated in Teir strain 14-4 compared to its Teis parent, this was not the case for the icaABCD operon (data not shown).
Differential expression of additional global or antibiotic-inducible cell wall regulators in Teir and Teis strains.
Expression of several previously identified antibiotic-inducible cell wall regulators was found to be increased in Teir strain 14-4 compared to its Teis parent. With the exception of tcaB transcript levels, which remained constant, those of drp35 and msrA2 were increased by >50%, those of msrR, msrA1, msrB, tcaA, and tcaR by >2-fold, and those of vraR and vraS by 3.5-fold and 5-fold, respectively (Table 2). Two additional genes whose expression levels were highly increased were the putative monofunctional glycosyltransferase gene sgtB (9-fold) (16, 48, 63) and the peptidyl-prolyl cis/trans isomerase homolog prsA (6.5-fold) (48, 63, 101), in contrast to sgtA and abcA, which showed only marginal changes (Table 2).
Differential expression of the SigB regulon, known to influence glycopeptide resistance (3-5) and peptidoglycan hydrolase regulation (83), was also studied. While rsbU, rsbV, rsbW, and sigB transcript levels were similar in Teir and Teis strains, those of their target genes, for instance, asp23 and sarS (4), were increased by ca. 80%. These results were quite similar to those previously recorded by real-time RT-PCR for rsbU, sigB, and asp23 transcript levels of strains 14-4 compared to MRGR3 (79). It should be mentioned that increased expression of tcaR may also contribute to increased sarS transcript levels (64).
In contrast, other strongly expressed global regulators, namely, yycFG, srrAB, and saeRS, remained at nearly identical levels in Teir and Teis strains (Table 2). Among members of the sarA protein family (14), only rot was significantly expressed but at similar levels in both Teir and Teis strains, whereas expression levels of sarU and sarT were too low, as expected from previous studies (61, 86), to allow detection of significant strain-specific differences.
Finally, we also evaluated additional sets of genes reported to be upregulated in two microarray studies comparing vancomycin-intermediate with vancomycin-susceptible isolates (16, 66). In contrast to those studies, we found no significant difference in transcript levels of graR, graS, graA, graB, and graC genes (1) and only minor changes in expression of 13 genes belonging to the purine biosynthetic operon compared to those previously reported apart from a twofold increase of the pur operon repressor purR (66) (Table 2). Interestingly, both previous microarray studies, but not a more recent one (102), demonstrated increased autolytic activity in vancomycin-intermediate isolates (16, 66) in contrast to our study.
Expression of extracellular proteases.
Previous studies indicated a role for extracellular proteases in the processing of cell wall hydrolases (40, 77, 80). Extracellular protease activities are thought to be required for converting the preprotein atl gene product into a bifunctional enzyme (2, 69, 99) and may yield multiple autolytic cleavage products in the extracellular medium (22, 95). Another postulated function of extracellular proteases might be to attenuate or inactivate cell wall hydrolase activities (23, 108).
Transcript levels of 11 protease genes assayed by DNA oligoarray were quite low in both Teir and Teis strains except for sspA, which was more highly expressed (Table 2). The low expression of these proteases in either strain grown for 5 h fits with recent data recorded in our laboratory showing nearly undetectable levels of extracellular proteases in S. aureus grown in similar conditions (50). Previous reports indicate that production of extracellular proteases mainly occurs during later phases of bacterial growth (34, 40, 41). Altogether, the overall low transcript levels of protease-encoding genes, despite minor changes recorded in the Teir strain compared to Teis strain (Table 2), provided marginal evidence for their significant involvement in the autolytic changes of Teir strain 14-4.
Evaluation of extracellular bacteriolytic hydrolase activities.
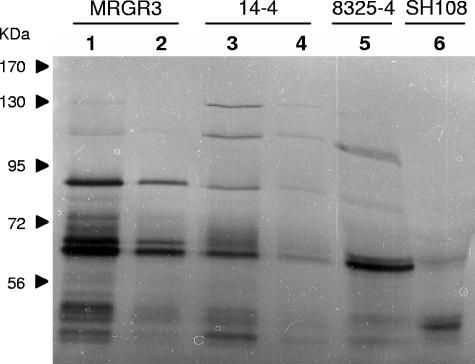
Since Teir strain 14-4 displays an autolysis-deficient phenotype without showing any significant decline in the expression of its autolysis-associated genes compared to its autolysis-proficient Teis parent, MRGR3, we investigated by zymographic analyses and quantitative bacterial hydrolytic assays whether this could result from any potential defect in posttranscriptional expression and/or processing of cell wall hydrolases by the Teir strain compared to its Teis parent. Figure 5 presents bacteriolytic hydrolase profiles of concentrated culture supernatants of strains MRGR3 and 14-4, compared to 8325-4 and its atl mutant, SH108, used as controls, assayed at different protein concentrations against lyophilized MRGR3 cells. The most prominent hydrolytic bands, with estimated molecular sizes of 86 kDa and 62 kDa in concentrated supernatants of strains MRGR3 and 14-4, and the concomitant presence of equivalent hydrolytic bands in strain 8325-4 but not its atl mutant, SH108, led to their tentative identification as Atl-derived products (2). The 86-kDa bacteriolytic band likely represented an intermediate Atl cleavage product, composed of N-acetylmuramyl-l-alanine amidase and two C-terminal (R1 and R2) repeat domains, and the 62-kDa band N-acetylmuramyl-l-alanine amidase (2). When compared at equivalent loading amounts, both 86-kDa and 62-kDa bacteriolytic bands were consistently qualitatively less intense in the Teir strain compared to the Teis parent, MRGR3, especially at the smaller loading amount (1.25 μg) of extracellular proteins (lanes 2 and 4). Besides the 86-kDa and 62-kDa cell wall-hydrolytic bands, there were also two higher-molecular-size hydrolytic bands that likely represented native pro-Atl (134 kDa) and an early 113-kDa Atl-processed product (2, 22). The fact that the 134-kDa and 113-kDa bands were more easily detected in strain 14-4 compared to MRGR3 suggested a less extensive rate of Atl processing by the Teir strain compared to the Teis strain. Nearly identical profiles and strain-specific differences were recorded in zymographic analyses performed in parallel against lyophilized 14-4 cells (not shown).
FIG. 5.
Zymographic analysis of bacteriolytic hydrolase activities of concentrated supernatants of cultures of strains MRGR3 (Teis), 14-4 (Teir), 8325-4, and its atl mutant, SH108, against lyophilized S. aureus MRGR3 cells. Lanes 1 and 3, 5 μg of protein; lanes 2 and 4, 1.25 μg of protein; lanes 5 and 6, 20 μg of protein. Molecular size markers are indicated on the left. The data shown are from a single representative experiment and were reproduced several times.
It is noteworthy that larger amounts (≥10 μg) of extracellular proteins of strain 8325-4 had to be loaded on gels for sodium dodecyl sulfate-polyacrylamide gel electrophoresis-based zymographic analysis, compared to MRGR3 to reveal closely matching Atl-derived bacteriolytic bands. Similar findings were also repeatedly observed with strain RN6390, another member of the NCTC8325 family (data not shown).
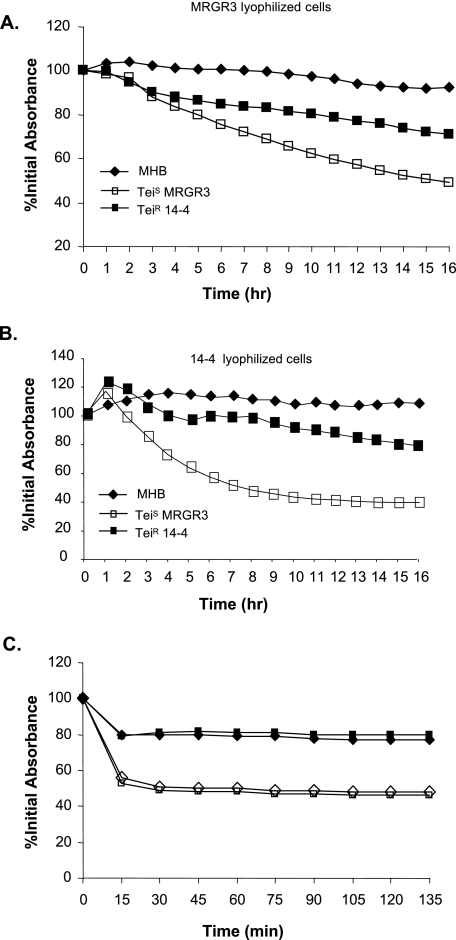
To extend the zymographic analysis, quantitative bacteriolytic assays of extracellular proteins of strains MRGR3 and 14-4 against lyophilized S. aureus suspensions were performed. Nearly identical results were obtained with concentrated supernatants (100 μg/ml) from either 5-h (not shown) or overnight cultures (Fig. 6). Extracellular proteins of strain MRGR3 caused a more significant decrease, compared to 14-4, in the turbidity of lyophilized S. aureus MRGR3 (Fig. 6A). Similar data were recorded with lyophilized S. aureus 14-4 (Fig. 6B). As anticipated from zymographic analyses of strains 8325-4 and SH108 (Fig. 5), the decrease of turbidity of lyophilized S. aureus by 100 μg/ml extracellular proteins from strain 8325-4 was low (<20%) compared to MRGR3, thus preventing detection of any significant difference from its atl mutant (data not shown). The identical susceptibilities of lyophilized substrates of 14-4 and MRGR3 to extracellular bacteriolytic hydrolase fractions from the same strains clearly indicated the absence of any significant change in their respective cell wall compositions that would contribute to the strain-specific autolytic differences. This finding is further supported by recently described observations showing identical muropeptide compositions in strains MRGR3 and 14-4 (57) and by the similar lysostaphin susceptibilities of Teir and Teis strains, whether tested in a nonviable lyophilized or viable state (Fig. 5C). Of note, significantly greater lytic effects were obtained against viable compared to nonviable cell suspensions of strains 14-4 and MRGR3.
FIG. 6.
A and B. Quantitative analysis of extracellular bacteriolytic hydrolase activities in concentrated supernatants of cultures of Teir strain 14-4 and Teis strain MRGR3. Extracellular proteins (100 μg) from either strain or MHB alone (used as a negative control) were added to heat-killed and lyophilized suspensions (1 mg/ml) of either strain MRGR3 (A) or 14-4 (B), and turbidity was monitored for 16 h. C. Lysostaphin susceptibilities of lyophilized (solid symbols) and viable (open symbols) cells of S. aureus MRGR3 (⋄) and 14-4 (□) are shown. The data are from a single representative experiment and were reproduced several times.
DISCUSSION
GISA/hGISA clinical and laboratory isolates frequently exhibit a decreased autolytic activity compared to non-GISA isolates, thus potentially providing a selective advantage to drug-exposed bacterial subclones during emergence of glycopeptide resistance (7, 35, 43, 70, 71, 85, 91, 102, 108). Despite this repeated observation, the molecular links between increased glycopeptide tolerance, glycopeptide resistance, and autolysis are not yet elucidated (7, 108).
A majority of clinical and GISA/hGISA laboratory-derived isolates exhibit cell wall thickening (17, 18, 78, 89), which may significantly contribute to glycopeptide resistance (15-17, 31). Other properties of the cell wall thickening model are increased d-Ala-d-Ala residues in cell walls, forming false targets trapping glycopeptides, and a decrease in the level of muropeptide glutamine amidation (8, 18, 21, 29, 31, 43, 46, 67, 91-93), but these characteristics are not shared by all GISA clinical isolates, thus pointing to the multifactorial origin of GISA (8, 78). Furthermore, it is unclear whether a majority of clinical or laboratory strains of Teir S. aureus exhibit cell wall thickening as extensive as that seen in some VISA isolates.
The molecular basis of cell wall thickening is not well understood (16, 43, 109) and might involve complex regulatory mechanisms (63, 72, 93, 94, 102). Morphological characteristics of clinical and laboratory-derived GISA strains (29, 43, 46, 67, 78, 92, 93) are frequently shared with some autolysin-defective mutants of S. aureus, in particular the presence of cell surface roughness, cell thickening, and sometimes abnormal septum formation (22, 39, 46, 68, 99). Thus, cell wall thickening in GISA might not only result from activated cell wall synthesis as initially suggested (29) but also potentially reflect an imbalance between cell wall synthesis and impaired cell wall remodeling by cell wall hydrolases (8, 43, 93, 94, 102, 108).
Despite intensive investigations, a comprehensive model of cell wall remodeling and autolytic control by peptidoglycan hydrolases, including a complete list of genes and gene pathways controlling cell wall expansion and cell septum and daughter cell separation (27, 42, 65, 100), is not available. A number of genetic loci are reported to influence autolysis (atl, sle1, lytM, lytN, lytSR, lrgAB, cidABC, alrRS, mgrA, agr, sarA, sarV, and RNAIII) (2, 10, 11, 11, 23-25, 28, 37, 62, 81-83, 98, 99), but their mutual interaction and respective contributions to cell wall- and autolysis-controlling molecular processes are still poorly understood and may also be significantly altered by different strain backgrounds.
DNA microarray methods have been shown to be powerful tools for evaluating the impact of individual global regulator (agr, sarA, sigB, rot, vraSR, arlRS, and mgrA) mutations on expression levels of S. aureus genes (4, 20, 48, 51, 55, 84). While recent expression studies explored the molecular mechanisms of glycopeptide resistance (16, 63) and stress responses to cell wall-active antibiotics (48, 101), no extensive functional and transcriptomic study evaluated the regulation and global expression of cell wall hydrolases and autolytic factors in S. aureus.
The development and validation in one of our laboratories of a custom-designed oligoarray for gene expression studies (13) were instrumental for accurate evaluation of gene expression changes in isogenic Teir and Teis strains. Statistical analysis demonstrated significantly different expression levels for all genes whose transcript ratios were either >1.3 or <0.80 in the Teir strain compared to the Teis strain when more than one probe was available for each transcript. Moreover, the choice of appropriate isogenic strains, namely, Teir strain 14-4, which was initially selected in a rat tissue cage infection model and displayed a remarkably stable resistance level after only two in vitro passages, and its Teis MRSA parent, MRGR3, whose virulence has been repeatedly demonstrated in animal studies (103), was essential for recording reliable expression changes even when these strains were grown in antibiotic-free media. We previously demonstrated altered expression of virulence factors including upregulation of fibronectin-binding proteins by Teir strain 14-4 compared to its Teis parent, MRGR3 (79), and ongoing studies indicate that strain 14-4 can also induce experimental infection in a rat tissue cage model (unpublished data).
An interesting and provocative aspect of our gene expression study was the observation of undiminished (atl, lytM, and lytN) or even significantly (more than twofold) increased (cidA, cidB, and cidC) expression of major autolytic genes in the autolysis-deficient strain 14-4 compared to its autolytic parent, MRGR3. These data contrasted with two recent reports that stressed the reduced expression of the atl gene in laboratory or clinical GISA strains displaying autolysis-deficient phenotypes (43, 108). While the increased levels of some autolytic gene transcripts (cidA, cidB, and cidC) recorded in Teir strain 14-4 compared to Teis strain MRGR3 were consistent with the reduced expression of their negative regulators lytSR and lrgAB (10, 11, 25, 28, 81, 81, 83), their mechanisms of autolytic regulation are currently unknown (82). At least, our transcriptomic and RT-PCR data seem to rule out a significant role of the cidABC operon in the autolysis-deficient phenotype of Teir strain 14-4.
In contrast to the above-mentioned results, other global regulators such as arlRS, mgrA, and sarA, known as negative regulators of autolysis, may individually or in concert contribute to the autolysis-deficient phenotype of strain 14-4 compared to MRGR3. Furthermore, the strongly reduced expression of the positive autolytic regulator agr, known to be frequently downregulated in GISA (85) as well in strain 14-4 (79), may also play a significant role in the deficient autolysis of our Teir strain. Further work is required to understand the mutual interactions and respective contribution of those global regulators to the cell wall- and autolysis-controlling pathways (23, 24, 36, 37).
The potential contribution of the sigB operon was also carefully examined in our study since this regulon was previously described as one of the preferred mutation sites associated with first-step teicoplanin resistance in SigB-deficient S. aureus strains (3, 5). In view of the modest increase found in expression of the SigB-dependent asp23 gene in Teir strain 14-4 compared to Teis strain MRGR3, and in line with a previous report (79), our data do not support a major role for this regulon in the expression of teicoplanin resistance and deficient autolysis by strain 14-4.
In contrast to the similar transcript levels of several global regulators (yycFG, srrAB, saeRS, rot, sarU, and sarT) recorded in Teir and Teis strains, other antibiotic-inducible regulators reported in previous microarray studies of antibiotic-exposed S. aureus cells (16, 48, 101), such as drp35, msrA2, msrR, msrA1, msrB, tcaA, tcaR, and especially vraR and vraS, were found upregulated in Teir 14-4 compared to Teis MRGR3. It should be stressed that overexpression of those antibiotic-inducible regulators, recently identified as a cell wall stress stimulon (101), occurred constitutively in Teir strain 14-4 compared to its Teis parent, especially in the absence of any antibiotic exposure. A recent microarray study comparing isogenic vancomycin-susceptible and VISA MRSA clinical isolates also described the constitutive overexpression of the cell wall stress stimulon in a VISA derivative that was selected after a 2-month period of extensive chemotherapy and whose vancomycin MIC reached 8 μg/ml (63). Nevertheless, the specific contribution of the cell wall stress stimulon to decreased glycopeptide susceptibility is difficult to evaluate due to the fact that this stimulon is also triggered in glycopeptide-susceptible strains exposed to vancomycin or other cell wall antimicrobial agents (16, 48, 63, 101). Site-specific mutagenesis combined with transcriptomic studies may help to more precisely define the individual as well as combined contributions of major stress cell wall stimulon components to the emergence of the GISA phenotype, as done for vraRS (49).
It should be mentioned that, despite some similarities in gene expression changes, the phenotypic properties of our isogenic Teir strain 14-4 differ in several aspects from the above-mentioned, late-stage VISA isolate (63, 89, 93), which was shown to display striking alterations in cell wall composition and morphology, including abnormally thick cell walls, markedly decreased growth rate, and formation of multicellular aggregates during growth in antibiotic-free liquid medium (93). In contrast, the vancomycin-susceptible (MIC: 4 μg/ml) but Teir strain 14-4 used for our study showed minor differences in growth rate and no evidence of multicellular aggregates, which fit with its identical peptidoglycan cell wall composition compared to its Teis parent (57). Furthermore, ongoing transmission electron microscopy studies failed to detect any major alteration in either cell wall thickness or septum formation for strain 14-4 compared to MRGR3 (unpublished data).
Our study also revealed the lack of significant differences in expression of major genes (ftsA, ftsZ, and scdA) coding for regulators of cell wall septum and daughter cell formation (12, 45, 73, 74). Thus, despite the likely occurrence of common molecular pathways contributing to decreased susceptibility to both teicoplanin and vancomycin, some glycopeptide-specific resistance phenotypes should also be considered. One argument for glycopeptide-specific resistance mechanisms is provided by the relatively easy in vitro selection of teicoplanin-resistant derivatives by one or two passages (38), while selection of stable VISA subclones seem to require a larger number of in vitro passages (38) and a potentially higher fitness cost (63, 89, 93). This may indicate that a larger number of mutational and regulatory changes are required for emergence of isolates displaying decreased susceptibility to vancomycin compared to teicoplanin.
The mechanisms of the autolysis-deficient phenotype of strain 14-4 were found to be linked with a decreased content of extracellular bacteriolytic hydrolase activities, as shown by zymographic analyses and quantitative bacteriolytic activities of concentrated supernatants from the Teir strain compared to its Teis parent against lyophilized suspensions of S. aureus. Further studies are required to elucidate the molecular components of the decreased autolytic bacteriolytic hydrolases and the altered rate of Atl processing by the Teir strain compared to the Teis strain. The relatively low extracellular bacteriolytic activity of strain 8325-4 and other members of the 8325 family such as RN6390 (not shown) compared to Teis strain MRGR3 remains unexplained and would deserve further studies.
In conclusion, our results suggest that altered autolytic properties in 14-4 are apparently not driven by significant changes in the transcription of key autolytic effectors. Instead, our analysis points to alternate regulatory mechanisms that impact autolysis effectors, which may include changes in posttranscriptional processing or export. Microarray studies offer a unique opportunity not only to evaluate global gene expression changes associated with resistance phenotype but also to compare such changes in different backgrounds or lineages of susceptible and resistant S. aureus isogenic pairs. Detailed comparison of global gene regulation in isolates from different clinically and epidemiologically relevant clonotypes of susceptible and resistant S. aureus strains may also bring key information for a deeper understanding of the molecular basis of virulence expression in this highly important nosocomial and community-acquired pathogen.
Supplementary Material
Acknowledgments
This work was supported by grants 4049-63250 from National Research Program NRP49 (“Antibiotic Resistance”), 3200B0-108401 (to D.L.), 3200B0-103951 (to P.V.), PP00B-103002/1 (to J.S.), 404940-106296/1 (to P.F.), 3100A0-100425 (to W.K.), and 3200B0-103793 (to P.M.) from the Swiss National Science Foundation.
We thank S. J. Foster (University of Sheffield, United Kingdom) for sending strain SH108 and B. Fleury for helpful comments.
Footnotes
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Aritaka, N., H. Hanaki, L. Z. Cui, and K. Hiramatsu. 2001. Combination effect of vancomycin and β-lactams against a Staphylococcus aureus strain, Mu3, with heterogeneous resistance to vancomycin. Antimicrob. Agents Chemother. 45:1292-1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baba, T., and O. Schneewind. 1998. Targeting of muralytic enzymes to the cell division site of gram-positive bacteria: repeat domains direct autolysin to the equatorial surface ring of Staphylococcus aureus. EMBO J. 17:4639-4646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bischoff, M., and B. Berger-Bächi. 2001. Teicoplanin stress-selected mutations increasing σB activity in Staphylococcus aureus. Antimicrob. Agents Chemother. 45:1714-1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bischoff, M., P. Dunman, J. Kormanec, D. Macapagal, E. Murphy, W. Mounts, B. Berger-Bachi, and S. Projan. 2004. Microarray-based analysis of the Staphylococcus aureus σB regulon. J. Bacteriol. 186:4085-4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bischoff, M., M. Roos, J. Putnik, A. Wada, P. Glanzmann, P. Giachino, P. Vaudaux, and B. Berger-Bächi. 2001. Involvement of multiple genetic loci in Staphylococcus aureus teicoplanin resistance. FEMS Microbiol. Lett. 194:77-82. [DOI] [PubMed] [Google Scholar]
- 6.Boyle-Vavra, S., S. K. Berke, J. C. Lee, and R. S. Daum. 2000. Reversion of the glycopeptide resistance phenotype in Staphylococcus aureus clinical isolates. Antimicrob. Agents Chemother. 44:272-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyle-Vavra, S., M. Challapalli, and R. S. Daum. 2003. Resistance to autolysis in vancomycin-selected Staphylococcus aureus isolates precedes vancomycin-intermediate resistance. Antimicrob. Agents Chemother. 47:2036-2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boyle-Vavra, S., H. Labischinski, C. C. Ebert, K. Ehlert, and R. S. Daum. 2001. A spectrum of changes occurs in peptidoglycan composition of glycopeptide-intermediate clinical Staphylococcus aureus isolates. Antimicrob. Agents Chemother. 45:280-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brandenberger, M., M. Tschierske, P. Giachino, A. Wada, and B. Berger-Bächi. 2000. Inactivation of a novel three-cistronic operon tcaR-tcaA-tcaB increases teicoplanin resistance in Staphylococcus aureus. Biochim. Biophys. Acta Gen. Subj. 1523:135-139. [DOI] [PubMed] [Google Scholar]
- 10.Brunskill, E. W., and K. W. Bayles. 1996. Identification and molecular characterization of a putative regulatory locus that affects autolysis in Staphylococcus aureus. J. Bacteriol. 178:611-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brunskill, E. W., and K. W. Bayles. 1996. Identification of LytSR-regulated genes from Staphylococcus aureus. J. Bacteriol. 178:5810-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brunskill, E. W., B. L. M. De Jonge, and K. W. Bayles. 1997. The Staphylococcus aureus scdA gene: a novel locus that affects cell division and morphogenesis. Microbiology 143:2877-2882. [DOI] [PubMed] [Google Scholar]
- 13.Charbonnier, Y., B. Gettler, P. Francois, M. Bento, A. Renzoni, P. Vaudaux, W. Schlegel, and J. Schrenzel. 17. June 2005, posting date. A generic approach for the design of whole-genome oligoarrays, validated for genomotyping, deletion mapping and gene expression analysis on Staphylococcus aureus. BMC Genomics 6:95. [Online.] http://www.biomedcentral.com/1471-2164/6/95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheung, A. L., A. S. Bayer, G. Zhang, H. Gresham, and Y. Q. Xiong. 2004. Regulation of virulence determinants in vitro and in vivo in Staphylococcus aureus. FEMS Immunol. Med. Microbiol. 40:1-9. [DOI] [PubMed] [Google Scholar]
- 15.Cui, L., A. Iwamoto, J. Q. Lian, H. M. Neoh, T. Maruyama, Y. Horikawa, and K. Hiramatsu. 2006. Novel mechanism of antibiotic resistance originating in vancomycin-intermediate Staphylococcus aureus. Antimicrob. Agents Chemother. 50:428-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cui, L., J. Q. Lian, H. M. Neoh, E. Reyes, and K. Hiramatsu. 2005. DNA microarray-based identification of genes associated with glycopeptide resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 49:3404-3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cui, L., X. Ma, K. Sato, K. Okuma, F. C. Tenover, E. M. Mamizuka, C. G. Gemmell, M. N. Kim, M. C. Ploy, N. El Solh, V. Ferraz, and K. Hiramatsu. 2003. Cell wall thickening is a common feature of vancomycin resistance in Staphylococcus aureus. J. Clin. Microbiol. 41:5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cui, L. Z., H. Murakami, K. Kuwahara-Arai, H. Hanaki, and K. Hiramatsu. 2000. Contribution of a thickened cell wall and its glutamine nonamidated component to the vancomycin resistance expressed by Staphylococcus aureus Mu50. Antimicrob. Agents Chemother. 44:2276-2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Du, W., J. R. Brown, D. R. Sylvester, J. Huang, A. F. Chalker, C. Y. So, D. J. Holmes, D. J. Payne, and N. G. Wallis. 2000. Two active forms of UDP-N-acetylglucosamine enolpyruvyl transferase in gram-positive bacteria. J. Bacteriol. 182:4146-4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dunman, P. M., E. Murphy, S. Haney, D. Palacios, G. Tucker-Kellogg, S. Wu, E. L. Brown, R. J. Zagursky, D. Shlaes, and S. J. Projan. 2001. Transcription profiling-based identification of Staphylococcus aureus genes regulated by the agr and/or sarA loci. J. Bacteriol. 183:7341-7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Finan, J. E., G. L. Archer, M. J. Pucci, and M. W. Climo. 2001. Role of penicillin-binding protein 4 in expression of vancomycin resistance among clinical isolates of oxacillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 45:3070-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Foster, S. J. 1995. Molecular characterization and functional analysis of the major autolysin of Staphylococcus aureus 8325/4. J. Bacteriol. 177:5723-5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fournier, B., and D. C. Hooper. 2000. A new two-component regulatory system involved in adhesion autolysis and extracellular proteolytic activity of Staphylococcus aureus. J. Bacteriol. 182:3955-3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fujimoto, D. F., and K. W. Bayles. 1998. Opposing roles of the Staphylococcus aureus virulence regulators, Agr and Sar, in Triton X-100- and penicillin-induced autolysis. J. Bacteriol. 180:3724-3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fujimoto, D. F., E. W. Brunskill, and K. W. Bayles. 2000. Analysis of genetic elements controlling Staphylococcus aureus lrgAB expression: Potential role of DNA topology in SarA regulation. J. Bacteriol. 182:4822-4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Geisel, R., F. J. Schmitz, A. C. Fluit, and H. Labischinski. 2001. Emergence, mechanism, and clinical implications of reduced glycopeptide susceptibility in Staphylococcus aureus. Eur. J. Clin. Microbiol. Infect. Dis. 20:685-697. [DOI] [PubMed] [Google Scholar]
- 27.Giesbrecht, P., T. Kersten, H. Maidhof, and J. Wecke. 1998. Staphylococcal cell wall: morphogenesis and fatal variations in the presence of penicillin. Microbiol. Mol. Biol. Rev. 62:1371-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Groicher, K. H., B. A. Firek, D. F. Fujimoto, and K. W. Bayles. 2000. The Staphylococcus aureus lrgAB operon modulates murein hydrolase activity and penicillin tolerance. J. Bacteriol. 182:1794-1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hanaki, H., K. Kuwahara-Arai, S. Boyle-Vavra, R. S. Daum, H. Labischinski, and K. Hiramatsu. 1998. Activated cell-wall synthesis is associated with vancomycin resistance in methicillin-resistant Staphylococcus aureus clinical strains Mu3 and Mu50. J. Antimicrob. Chemother. 42:199-209. [DOI] [PubMed] [Google Scholar]
- 30.Herbert, S., P. Barry, and R. P. Novick. 2001. Subinhibitory clindamycin differentially inhibits transcription of exoprotein genes in Staphylococcus aureus. Infect. Immun. 69:2996-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hiramatsu, K. 2001. Vancomycin-resistant Staphylococcus aureus: a new model of antibiotic resistance. Lancet Infect. Dis. 1:147-155. [DOI] [PubMed] [Google Scholar]
- 32.Hiramatsu, K., N. Aritaka, H. Hanaki, S. Kawasaki, Y. Hosoda, S. Hori, Y. Fukuchi, and I. Kobayashi. 1997. Dissemination in Japanese hospitals of strains of Staphylococcus aureus heterogeneously resistant to vancomycin. Lancet 350:1670-1673. [DOI] [PubMed] [Google Scholar]
- 33.Hiramatsu, K., H. Hanaki, T. Ino, K. Yabuta, T. Oguri, and F. C. Tenover. 1997. Methicillin-resistant Staphylococcus aureus clinical strain with reduced vancomycin susceptibility. J. Antimicrob. Chemother. 40:135-136. [DOI] [PubMed] [Google Scholar]
- 34.Horsburgh, M. J., J. L. Aish, I. J. White, L. Shaw, J. K. Lithgow, and S. J. Foster. 2002. σB modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4. J. Bacteriol. 184:5457-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hussain, F. M., S. Boyle-Vavra, P. B. Shete, and R. S. Daum. 2002. Evidence for a continuum of decreased vancomycin susceptibility in unselected Staphylococcus aureus clinical isolates. J. Infect. Dis. 186:661-667. [DOI] [PubMed] [Google Scholar]
- 36.Ingavale, S., W. Van Wamel, T. T. Luong, C. Y. Lee, and A. L. Cheung. 2005. Rat/MgrA, a regulator of autolysis, is a regulator of virulence genes in Staphylococcus aureus. Infect. Immun. 73:1423-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ingavale, S. S., W. Van Wamel, and A. L. Cheung. 2003. Characterization of RAT, an autolysis regulator in Staphylococcus aureus. Mol. Microbiol. 48:1451-1466. [DOI] [PubMed] [Google Scholar]
- 38.Kaatz, G. W., S. M. Seo, N. J. Dorman, and S. A. Lerner. 1990. Emergence of teicoplanin resistance during therapy of Staphylococcus aureus endocarditis. J. Infect. Dis. 162:103-108. [DOI] [PubMed] [Google Scholar]
- 39.Kajimura, J., T. Fujiwara, S. Yamada, Y. Suzawa, T. Nishida, Y. Oyamada, I. Hayashi, J. Yamagishi, H. Komatsuzawa, and M. Sugai. 2005. Identification and molecular characterization of an N-acetylmuramyl-l-alanine amidase Sle1 involved in cell separation of Staphylococcus aureus. Mol. Microbiol. 58:1087-1101. [DOI] [PubMed] [Google Scholar]
- 40.Karlsson, A., and S. Arvidson. 2002. Variation in extracellular protease production among clinical isolates of Staphylococcus aureus due to different levels of expression of the protease repressor SarA. Infect. Immun. 70:4239-4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karlsson, A., P. Saravia-Otten, K. Tegmark, E. Morfeldt, and S. Arvidson. 2001. Decreased amounts of cell wall-associated protein A and fibronectin-binding proteins in Staphylococcus aureus sarA mutants due to up-regulation of extracellular proteases. Infect. Immun. 69:4742-4748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koch, A. L. 2001. Autolysis control hypotheses for tolerance to wall antibiotics. Antimicrob. Agents Chemother. 45:2671-2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koehl, J. L., A. Muthaiyan, R. K. Jayaswal, K. Ehlert, H. Labischinski, and B. J. Wilkinson. 2004. Cell wall composition and decreased autolytic activity and lysostaphin susceptibility of glycopeptide-intermediate Staphylococcus aureus. Antimicrob. Agents Chemother. 48:3749-3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Komatsuzawa, H., K. Ohta, T. Fujiwara, G. H. Choi, H. Labischinski, and M. Sugai. 2001. Cloning and sequencing of the gene, fmtC, which affects oxacillin resistance in methicillin-resistant Staphylococcus aureus. FEMS Microbiol. Lett. 203:49-54. [DOI] [PubMed] [Google Scholar]
- 45.Komatsuzawa, H., K. Ohta, M. Sugai, T. Fujiwara, P. Glanzmann, B. Berger-Bächi, and H. Suginaka. 2000. Tn551-mediated insertional inactivation of the fmtB gene encoding a cell wall-associated protein abolishes methicillin resistance in Staphylococcus aureus. J. Antimicrob. Chemother. 45:421-431. [DOI] [PubMed] [Google Scholar]
- 46.Komatsuzawa, H., K. Ohta, S. Yamada, K. Ehlert, H. Labischinski, J. Kajimura, T. Fujiwara, and M. Sugai. 2002. Increased glycan chain length distribution and decreased susceptibility to moenomycin in a vancomycin-resistant Staphylococcus aureus mutant. Antimicrob. Agents Chemother. 46:75-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Komatsuzawa, H., M. Sugai, K. Ohta, T. Fujiwara, S. Nakashima, J. Suzuki, C. Y. Lee, and H. Suginaka. 1997. Cloning and characterization of the fmt gene which affects the methicillin resistance level and autolysis in the presence of Triton X-100 in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 41:2355-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kuroda, M., H. Kuroda, T. Oshima, F. Takeuchi, H. Mori, and K. Hiramatsu. 2003. Two-component system VraSR positively modulates the regulation of cell-wall biosynthesis pathway in Staphylococcus aureus. Mol. Microbiol. 49:807-821. [DOI] [PubMed] [Google Scholar]
- 49.Kuroda, M., K. Kuwahara-Arai, and K. Hiramatsu. 2000. Identification of the up- and down-regulated genes in vancomycin-resistant Staphylococcus aureus strains Mu3 and Mu50 by cDNA differential hybridization method. Biochem. Biophys. Res. Commun. 269:485-490. [DOI] [PubMed] [Google Scholar]
- 50.Li, D., A. Renzoni, T. Estoppey, C. Bisognano, P. Francois, W. L. Kelley, D. P. Lew, J. Schrenzel, and P. Vaudaux. 2005. Induction of fibronectin adhesins in quinolone-resistant Staphylococcus aureus by subinhibitory levels of ciprofloxacin or by sigma B transcription factor activity is mediated by two separate pathways. Antimicrob. Agents Chemother. 49:916-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liang, X., L. Zheng, C. Landwehr, D. Lunsford, D. Holmes, and Y. Ji. 2005. Global regulation of gene expression by ArlRS, a two-component signal transduction regulatory system of Staphylococcus aureus. J. Bacteriol. 187:5486-5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu, C., and H. F. Chambers. 2003. Staphylococcus aureus with heterogeneous resistance to vancomycin: epidemiology, clinical significance, and critical assessment of diagnostic methods. Antimicrob. Agents Chemother. 47:3040-3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lowy, F. D. 2003. Antimicrobial resistance: the example of Staphylococcus aureus. J. Clin. Investig. 111:1265-1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lucet, J. C., M. Herrmann, P. Rohner, R. Auckenthaler, F. A. Waldvogel, and D. P. Lew. 1990. Treatment of experimental foreign body infection caused by methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 34:2312-2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Luong, T. T., P. M. Dunman, E. Murphy, S. J. Projan, and C. Y. Lee. 2006. Transcription profiling of the mgrA regulon in Staphylococcus aureus. J. Bacteriol. 188:1899-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Luong, T. T., S. W. Newell, and C. Y. Lee. 2003. mgr, a novel global regulator in Staphylococcus aureus. J. Bacteriol. 185:3703-3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Majcherczyk, P. A., T. McKenna, P. Moreillon, and P. Vaudaux. 2006. The discriminatory power of MALDI-TOF mass spectrometry to differentiate between isogenic teicoplanin-susceptible and teicoplanin-resistant strains of methicillin-resistant Staphylococcus aureus. FEMS Microbiol. Lett. 255:233-239. [DOI] [PubMed] [Google Scholar]
- 58.Maki, H., N. McCallum, M. Bischoff, A. Wada, and B. Berger-Bachi. 2004. tcaA inactivation increases glycopeptide resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 48:1953-1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Maki, H., T. Yamaguchi, and K. Murakami. 1994. Cloning and characterization of a gene affecting the methicillin resistance level and the autolysis rate in Staphylococcus aureus. J. Bacteriol. 176:4993-5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mani, N., P. Tobin, and R. K. Jayaswal. 1993. Isolation and characterization of autolysis-defective mutants of Staphylococcus aureus created by Tn917-lacZ mutagenesis. J. Bacteriol. 175:1493-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Manna, A. C., and A. L. Cheung. 2003. sarU, a sarA homolog, is repressed by SarT and regulates virulence genes in Staphylococcus aureus. Infect. Immun. 71:343-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Manna, A. C., S. S. Ingavale, M. Maloney, W. Van Wamel, and A. L. Cheung. 2004. Identification of sarV (SA2062), a new transcriptional regulator, is repressed by SarA and MgrA (SA0641) and involved in the regulation of autolysis in Staphylococcus aureus. J. Bacteriol. 186:5267-5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McAleese, F., S. W. Wu, K. Sieradzki, P. Dunman, E. Murphy, S. Projan, and A. Tomasz. 2006. Overexpression of genes of the cell wall stimulon in clinical isolates of Staphylococcus aureus exhibiting vancomycin-intermediate-S. aureus-type resistance to vancomycin. J. Bacteriol. 188:1120-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McCallum, N., M. Bischoff, H. Maki, A. Wada, and B. Berger-Bachi. 2004. TcaR, a putative MarR-like regulator of sarS expression. J. Bacteriol. 186:2966-2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Milohanic, E., R. Jonquieres, P. Cossart, P. Berche, and J. L. Gaillard. 2001. The autolysin Ami contributes to the adhesion of Listeria monocytogenes to eukaryotic cells via its cell wall anchor. Mol. Microbiol. 39:1212-1224. [DOI] [PubMed] [Google Scholar]
- 66.Mongodin, E., J. Finan, M. W. Climo, A. Rosato, S. Gill, and G. L. Archer. 2003. Microarray transcription analysis of clinical Staphylococcus aureus isolates resistant to vancomycin. J. Bacteriol. 185:4638-4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Moreira, B., S. Boyle-Vavra, B. L. M. DeJonge, and R. S. Daum. 1997. Increased production of penicillin-binding protein 2, increased detection of other penicillin-binding proteins, and decreased coagulase activity associated with glycopeptide resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 41:1788-1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nishi, H., H. Komatsuzawa, S. Yamada, T. Fujiwara, M. Ohara, K. Ohta, M. Sugiyama, T. Ishikawa, and M. Sugai. 2003. Moenomycin resistance is associated with vancomycin-intermediate susceptibility in Staphylococcus aureus. Microbiol. Immunol. 47:927-935. [DOI] [PubMed] [Google Scholar]
- 69.Oshida, T., M. Sugai, H. Komatsuzawa, Y.-M. Hong, H. Suginaka, and A. Tomasz. 1995. A Staphylococcus aureus autolysin that has an N-acetylmuramoyl-l-alanine amidase domain and an endo-β-N-acetylglucosaminidase domain: cloning, sequence analysis, and characterization. Proc. Natl. Acad. Sci. USA 92:285-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Peschel, A., C. Vuong, M. Otto, and F. Götz. 2000. The d-alanine residues of Staphylococcus aureus teichoic acids alter the susceptibility to vancomycin and the activity of autolytic enzymes. Antimicrob. Agents Chemother. 44:2845-2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pfeltz, R. F., V. K. Singh, J. L. Schmidt, M. A. Batten, C. S. Baranyk, M. J. Nadakavukaren, R. K. Jayaswal, and B. J. Wilkinson. 2000. Characterization of passage-selected vancomycin-resistant Staphylococcus aureus strains of diverse parental backgrounds. Antimicrob. Agents Chemother. 44:294-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pfeltz, R. F., and B. J. Wilkinson. 2004. The escalating challenge of vancomycin resistance in Staphylococcus aureus. Curr. Drug Targets Infect. Disord. 4:273-294. [DOI] [PubMed] [Google Scholar]
- 73.Pinho, M. G., and J. Errington. 2003. Dispersed mode of Staphylococcus aureus cell wall synthesis in the absence of the division machinery. Mol. Microbiol. 50:871-881. [DOI] [PubMed] [Google Scholar]
- 74.Pinho, M. G., and J. Errington. 2005. Recruitment of penicillin-binding protein PBP2 to the division site of Staphylococcus aureus is dependent on its transpeptidation substrates. Mol. Microbiol. 55:799-807. [DOI] [PubMed] [Google Scholar]
- 75.Ramadurai, L., and R. K. Jayaswal. 1997. Molecular cloning, sequencing, and expression of lytM a unique autolytic gene of Staphylococcus aureus. J. Bacteriol. 179:3625-3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ramadurai, L., K. J. Lockwood, M. J. Nadakavukaren, and R. K. Jayaswal. 1999. Characterization of a chromosomally encoded glycylglycine endopeptidase of Staphylococcus aureus. Microbiology 145(Pt. 4):801-808. [DOI] [PubMed] [Google Scholar]
- 77.Reed, S. B., C. A. Wesson, L. E. Liou, W. R. Trumble, P. M. Schlievert, G. A. Bohach, and K. W. Bayles. 2001. Molecular characterization of a novel Staphylococcus aureus serine protease operon. Infect. Immun. 69:1521-1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Reipert, A., K. Ehlert, T. Kast, and G. Bierbaum. 2003. Morphological and genetic differences in two isogenic Staphylococcus aureus strains with decreased susceptibilities to vancomycin. Antimicrob. Agents Chemother. 47:568-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Renzoni, A., P. Francois, D. Li, W. L. Kelley, D. P. Lew, P. Vaudaux, and J. Schrenzel. 2004. Modulation of fibronectin adhesins and other virulence factors in a teicoplanin-resistant derivative of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 48:2958-2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rice, K., R. Peralta, D. Bast, J. De Azavedo, and M. J. McGavin. 2001. Description of staphylococcus serine protease (ssp) operon in Staphylococcus aureus and nonpolar inactivation of sspA-encoded serine protease. Infect. Immun. 69:159-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rice, K. C., B. A. Firek, J. B. Nelson, S. J. Yang, T. G. Patton, and K. W. Bayles. 2003. The Staphylococcus aureus cidAB operon: evaluation of its role in regulation of murein hydrolase activity and penicillin tolerance. J. Bacteriol. 185:2635-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rice, K. C., J. B. Nelson, T. G. Patton, S. J. Yang, and K. W. Bayles. 2005. Acetic acid induces expression of the Staphylococcus aureus cidABC and lrgAB murein hydrolase regulator operons. J. Bacteriol. 187:813-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rice, K. C., T. Patton, S. J. Yang, A. Dumoulin, M. Bischoff, and K. W. Bayles. 2004. Transcription of the Staphylococcus aureus cid and lrg murein hydrolase regulators is affected by sigma factor B. J. Bacteriol. 186:3029-3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Said-Salim, B., P. M. Dunman, F. M. McAleese, D. Macapagal, E. Murphy, P. J. McNamara, S. Arvidson, T. J. Foster, S. J. Projan, and B. N. Kreiswirth. 2003. Global regulation of Staphylococcus aureus genes by Rot. J. Bacteriol. 185:610-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sakoulas, G., G. M. Eliopoulos, V. G. Fowler, Jr., R. C. Moellering, Jr., R. P. Novick, N. Lucindo, M. R. Yeaman, and A. S. Bayer. 2005. Reduced susceptibility of Staphylococcus aureus to vancomycin and platelet microbicidal protein correlates with defective autolysis and loss of accessory gene regulator (agr) function. Antimicrob. Agents Chemother. 49:2687-2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schmidt, K. A., A. C. Manna, S. Gill, and A. L. Cheung. 2001. SarT, a repressor of α-hemolysin in Staphylococcus aureus. Infect. Immun. 69:4749-4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Schneider, T., M. M. Senn, B. Berger-Bachi, A. Tossi, H. G. Sahl, and I. Wiedemann. 2004. In vitro assembly of a complete, pentaglycine interpeptide bridge containing cell wall precursor (lipid II-Gly5) of Staphylococcus aureus. Mol. Microbiol. 53:675-685. [DOI] [PubMed] [Google Scholar]
- 88.Shlaes, D. M., J. H. Shlaes, S. Vincent, L. Etter, P. D. Fey, and R. V. Goering. 1993. Teicoplanin-resistant Staphylococcus aureus expresses a novel membrane protein and increases expression of penicillin-binding protein 2 complex. Antimicrob. Agents Chemother. 37:2432-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sieradzki, K., T. Leski, J. Dick, L. Borio, and A. Tomasz. 2003. Evolution of a vancomycin-intermediate Staphylococcus aureus strain in vivo: multiple changes in the antibiotic resistance phenotypes of a single lineage of methicillin-resistant S. aureus under the impact of antibiotics administered for chemotherapy. J. Clin. Microbiol. 41:1687-1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sieradzki, K., R. B. Roberts, S. W. Haber, and A. Tomasz. 1999. The development of vancomycin resistance in a patient with methicillin-resistant Staphylococcus aureus infection. N. Engl. J. Med. 340:517-523. [DOI] [PubMed] [Google Scholar]
- 91.Sieradzki, K., and A. Tomasz. 1997. Inhibition of cell wall turnover and autolysis by vancomycin in a highly vancomycin-resistant mutant of Staphylococcus aureus. J. Bacteriol. 179:2557-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sieradzki, K., and A. Tomasz. 1999. Gradual alterations in cell wall structure and metabolism in vancomycin-resistant mutants of Staphylococcus aureus. J. Bacteriol. 181:7566-7570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sieradzki, K., and A. Tomasz. 2003. Alterations of cell wall structure and metabolism accompany reduced susceptibility to vancomycin in an isogenic series of clinical isolates of Staphylococcus aureus. J. Bacteriol. 185:7103-7110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sieradzki, K., and A. Tomasz. 2006. Inhibition of the autolytic system by vancomycin causes mimicry of vancomycin-intermediate Staphylococcus aureus-type resistance, cell concentration dependence of the MIC, and antibiotic tolerance in vancomycin-susceptible S. aureus. Antimicrob. Agents Chemother. 50:527-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sugai, M., T. Akiyama, H. Komatsuzawa, Y. Miyake, and H. Suginaka. 1990. Characterization of sodium dodecyl sulfate-stable Staphylococcus aureus bacteriolytic enzymes by polyacrylamide gel electrophoresis. J. Bacteriol. 172:6494-6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sugai, M., T. Fujiwara, H. Komatsuzawa, and H. Suginaka. 1998. Identification and molecular characterization of a gene homologous to epr (endopeptidase resistance gene) in Staphylococcus aureus. Gene 224:67-75. [DOI] [PubMed] [Google Scholar]
- 97.Sugai, M., H. Komatsuzawa, T. Akiyama, Y. M. Hong, T. Oshida, Y. Miyake, T. Yamaguchi, and H. Suginaka. 1995. Identification of endo-β-N-acetylglucosaminidase and N-acetylmuramyl-l-alanine amidase as cluster-dispersing enzymes in Staphylococcus aureus. J. Bacteriol. 177:1491-1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sugai, M., S. Yamada, S. Nakashima, H. Komatsuzawa, A. Matsumoto, T. Oshida, and H. Suginaka. 1997. Localized perforation of the cell wall by a major autolysin: atl gene products and the onset of penicillin-induced lysis of Staphylococcus aureus. J. Bacteriol. 179:2958-2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Takahashi, J., H. Komatsuzawa, S. Yamada, T. Nishida, H. Labischinski, T. Fujiwara, M. Ohara, J. Yamagishi, and M. Sugai. 2002. Molecular characterization of an atl null mutant of Staphylococcus aureus. Microbiol. Immunol. 46:601-612. [DOI] [PubMed] [Google Scholar]
- 100.Tomasz, A. 2000. The staphylococcal cell wall, p. 351-360. In: V. A. Fischetti, R. P. Novick, P. Ferrieri, D. A. Portnoy, and J. I. Rood (ed.), gram-positive pathogens. American Society for Microbiogy, Washington, D.C.
- 101.Utaida, S., P. M. Dunman, D. Macapagal, E. Murphy, S. J. Projan, V. K. Singh, R. K. Jayaswal, and B. J. Wilkinson. 2003. Genome-wide transcriptional profiling of the response of Staphylococcus aureus to cell-wall-active antibiotics reveals a cell-wall-stress stimulon. Microbiology 149:2719-2732. [DOI] [PubMed] [Google Scholar]
- 102.Utaida, S., R. F. Pfeltz, R. K. Jayaswal, and B. J. Wilkinson. 2006. Autolytic properties of glycopeptide-intermediate Staphylococcus aureus Mu50. Antimicrob. Agents Chemother. 50:1541-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Vaudaux, P., P. Francois, B. Berger-Bächi, and D. Lew. 2001. In vivo emergence of subpopulations expressing teicoplanin or vancomycin-resistant phenotypes in a glycopeptide-susceptible, methicillin-resistant strain of Staphylococcus aureus. J. Antimicrob. Chemother. 47:163-170. [DOI] [PubMed] [Google Scholar]
- 104.Vaudaux, P., P. Francois, C. Bisognano, W. L. Kelley, D. P. Lew, J. Schrenzel, R. A. Proctor, P. J. McNamara, G. Peters, and C. Von Eiff. 2002. Increased expression of clumping factor and fibronectin-binding proteins by hemB mutants of Staphylococcus aureus expressing small colony variant phenotypes. Infect. Immun. 70:5428-5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Walsh, T. R., and R. A. Howe. 2002. The prevalence and mechanisms of vancomycin resistance in Staphylococcus aureus. Annu. Rev. Microbiol. 56:657-675. [DOI] [PubMed] [Google Scholar]
- 106.Weidenmaier, C., J. F. Kokai-Kun, S. A. Kristian, T. Chanturiya, H. Kalbacher, M. Gross, G. Nicholson, B. Neumeister, J. J. Mond, and A. Peschel. 2004. Role of teichoic acids in Staphylococcus aureus nasal colonization, a major risk factor in nosocomial infections. Nat. Med. 10:243-245. [DOI] [PubMed] [Google Scholar]
- 107.Weigel, L. M., D. B. Clewell, S. R. Gill, N. C. Clark, L. K. McDougal, S. E. Flannagan, J. F. Kolonay, J. Shetty, G. E. Killgore, and F. C. Tenover. 2003. Genetic analysis of a high-level vancomycin-resistant isolate of Staphylococcus aureus. Science 302:1569-1571. [DOI] [PubMed] [Google Scholar]
- 108.Wootton, M., P. M. Bennett, A. P. MacGowan, and T. R. Walsh. 2005. Reduced expression of the atl autolysin gene and susceptibility to autolysis in clinical heterogeneous glycopeptide-intermediate Staphylococcus aureus (hGISA) and GISA strains. J. Antimicrob. Chemother. 56:944-947. [DOI] [PubMed] [Google Scholar]
- 109.Wootton, M., R. A. Howe, R. Hillman, T. R. Walsh, P. M. Bennett, and A. P. MacGowan. 2001. A modified population analysis profile (PAP) method to detect hetero-resistance to vancomycin in Staphylococcus aureus in a UK hospital. J. Antimicrob. Chemother. 47:399-403. [DOI] [PubMed] [Google Scholar]
- 110.Wootton, M., A. P. MacGowan, and T. R. Walsh. 2005. Expression of tcaA and mprF and glycopeptide resistance in clinical glycopeptide-intermediate Staphylococcus aureus (GISA) and heteroGISA strains. Biochim. Biophys. Acta 1726:326-327. [DOI] [PubMed] [Google Scholar]
- 111.Yamada, S., M. Sugai, H. Komatsuzawa, S. Nakashima, T. Oshida, A. Matsumoto, and H. Suginaka. 1996. An autolysin ring associated with cell separation of Staphylococcus aureus. J. Bacteriol. 178:1565-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.