Abstract
Carbapenem resistance in Escherichia coli is rare. We report four genetically unrelated carbapenem-resistant E. coli isolates cultured from four patients hospitalized in Tel Aviv Medical Center. PCR, sequencing, and Southern blot analysis identified KPC-2 as the imipenem-hydrolyzing enzyme in all four strains, carried on different plasmids with a possible common origin. This is the first discovery of KPC-2 in E. coli and the first report of this enzyme originating outside the United States.
Carbapenem resistance in Escherichia coli does not occur naturally, and acquired resistance is rare in this species (15). Few cases of carbapenem-resistant E. coli strains have been reported. The first reported case in 1999 described the occurrence of outer-membrane protein deficiency coupled with the plasmid-mediated class C β-lactamase CMY (17). Carbapenem-hydrolyzing, plasmid-mediated metallo-β-lactamases in clinical E. coli strains from various parts of the world have been reported (6, 11, 16, 19), but class A carbapenemases in E. coli have been described in only one case, involving KPC-3 acquisition in a patient in the United States during imipenem therapy (7).
A high prevalence of extended-spectrum-β-lactamase-producing E. coli clinical isolates has been observed and studied in our institution (5), but carbapenem resistance in this species has never been observed. In February 2005, an imipenem-resistant E. coli strain (MIC, 32 μg/ml) was isolated from a urine culture from a 75-year-old woman with multiple chronic illnesses hospitalized at Tel Aviv Medical Center due to syncope. During September and October 2005, three additional carbapenem-resistant E. coli isolates were isolated at our hospital. We examined the epidemiological and genetic relatedness and the clinical scenario as well as the enzymatic mechanism leading to this resistance.
MATERIALS AND METHODS
Setting, bacterial strains, and antibiotic susceptibility testing.
Tel Aviv Medical Center is a 1,200-bed tertiary care teaching hospital, with 70,000 admissions annually. Four clinical isolates of Escherichia coli were isolated from four patients hospitalized during 2005 in our institution. The patients' relevant clinical data were collected from their medical records. Antibiotic susceptibilities were determined by Vitek-2 (BioMerieux Inc., Marcy, France). MICs of all carbapenems (imipenem, meropenem, and ertapenem) were determined by Etest (AB Biodisk, Solna, Sweden).
PFGE.
The genetic relatedness of the four E. coli isolates was analyzed by pulsed-field gel electrophoresis (PFGE). Bacterial DNA was prepared and cleaved with 20U SpeI endonuclease (New England Biolabs, Boston, MA) as previously described (5), and DNA macrorestriction patterns were visually compared and interpreted according to the criteria established by Tenover et al. (18).
Carbapenemase analysis and IEF of β-lactamases.
Visualization of imipenem-hydrolyzing β-lactamase activity by the E. coli clinical isolates was performed by an imipenem inactivation bioassay (21) performed on Mueller-Hinton agar plates (Hy-Labs, Rehovot, Israel), with E. coli ATCC 25922 as the test strain and an imipenem-susceptible E. coli strain as a negative control for carbapenemase production. Screening for production of metallo-β-lactamase was done by disk approximation tests with EDTA and 2-mercaptopropionic acid (1). Analytical isoelectric focusing (IEF) of β-lactamases was performed with crude enzyme preparations from sonicated cell cultures of E. coli isolates and transformants, grown on tryptic soy broth (Biolife Italiana, Milano, Italy). IEF was performed according to the method of Matthew et al. (10), by use of an LKB Multiphor II electrophoresis system apparatus on prepared PAGplates (pH 3.5 to 9.5; Amersham Biosciences, Buckinghamshire, United Kingdom). Beta-lactamase activity was revealed with nitrocefin (0.5 mg/ml; Calbiochem-Novabiochem Corp., San Diego, CA), and pIs were determined by running β-lactamases with known pIs in parallel as controls.
Screening of carbapenemase genes and cloning and sequencing of blaKPC.
Identification of the carbapenemase genes was determined by PCR, with specific primers designed for identification of known class A β-lactamase genes, including blaKPC (4), blaSME (forward, ACTTTGATGGGAGGATTGGCGTCT; reverse, ACCCAATCAGCAGGAACACTAGCA), blaIMI (forward, TCTCACAGGCCAATACAAAGGGCA; reverse, CCGCATAATCATTTGCCGTACCGT), and blaNMC (forward, TAGGTGATATGGCTGCTGCTGCTT; reverse, ACTGCTGCAGGTGTAGATGTGTCA).
The PCR conditions were as follows: 15 min at 95°C and 35 cycles of 1 min at 94°C, 2 min at 68°C, and 3 min at 72°C, followed by an extension step of 10 min at 72°C. PCRs were performed with Hot-StarTaq DNA polymerase (QIAGEN, Hilden, Germany), and the resulting PCR products were analyzed in a 1% agarose gel. Full-length blaKPC PCR products were ligated into pGEM-T easy PCR cloning vector and transformed into competent cells of E. coli JM109 according to the manufacturer's instructions (Promega, WI). Sequencing of cloned genes was performed with SP6 and T7 promoter primers. Sequences were analyzed with an ABI PRISM 3100 genetic analyzer (PE Biosystems), using DNA sequencing analysis software and 3100 data collection software version 1.1. The nucleotide acid and deduced protein sequences were analyzed and compared by use of software available via the Internet at the NCBI website (http://www.ncbi.nlm.nih.gov/).
Transformation experiments, plasmid purification, and Southern analysis.
Plasmid DNA from E. coli strains was isolated by use of a QIAGEN plasmid DNA midi kit (QIAGEN, Hilden, Germany) according to the manufacturer's instructions, and plasmid DNA samples were electrophoresed on 0.8% agarose gels in the presence of 1.0 TAE (Tris-acetate-EDTA) buffer. Transformation experiments with E. coli strain DH5α were carried out by electroporation of plasmid DNA with an Electroporator 2510 (Eppendorf, Hamburg, Germany), and transformants were selected on LB agar plates containing 100 μg/ml AMP. For Southern blot analysis, plasmid DNA from donor E. coli strains and their transformants was digested with BglII, EcoRV, and SmaI endonucleases (New England Biolabs), electrophoresed, transferred to a Hybond N+ membrane (Amersham Biosciences, Buckinghamshire, United Kingdom), and cross-linked with UV light. blaKPC-2 radioactively labeled with random-primer-DNA-labeling mixture (Biological Industries, Beit Haemek, Israel) was used as a probe.
RESULTS
Patients' background, antibiotic susceptibilities, and genetic relatedness.
Carbapenem-resistant E. coli was initially identified at Tel Aviv Medical Center in February 2005 and was subsequently isolated from three additional patients during September and October 2005. This represents an incidence rate of 5.7 cases/100,000 admissions and a proportion of resistance to carbapenem among E. coli isolates of 0.15%. While these isolates were cultured from patients with different clinical backgrounds (Table 1), they were all chronically ill, with extensive exposure to the medical system. Nevertheless, they were hospitalized in different wards and cared for by different staff members, and no common epidemiological link between them was identified. No apparent contact between the patients or travel to the east coast of the United States was documented.
TABLE 1.
Details of patient from whom imipenem-resistant E. coli was isolateda
| Patient | Isolate | Infection site | LOS prior to IpmrE. coli | LOS | Antibiotic treatment
|
Infection/colonization | Outcome | |
|---|---|---|---|---|---|---|---|---|
| 1 mo prior to E. coli isolation | After E. coli isolation | |||||||
| 1 | 157 | Urine | 10 d | 14 d | FQ | No treatment | Colonization | Recovery |
| 2 | 329 | Blood | 1 db | 3 d | No treatment | Empiric i.v. CRO | Infection | Death |
| 3 | 339 | Subphrenic abscess | 30 d | 4 m | BS-CEPH, FQ, MTA, VAN, followed with 14 d IPM until 2 wks before isolation | TZP, AMK | Infection | Recovery |
| 4 | 360 | Urine | 2 db | 18 d | CXMc | VAN, AMK, MTA | Colonization | Recovery |
LOS, length of hospital stay in days (d) or months (m); i.v., intravenous; Ab, antibiotic; BS-CEPH, broad-spectrum cephalosporins; FQ, fluoroquinone; MTA, metronidazole; IPM, imipenem; TZP, piperacillin-tazobactam; AMK, amikacin; CXM, cefuroxime.
Admitted from two different chronic care facilities.
Treated with cefuroxime (CXM) for a urinary tract infection that occurred during hospitalization 1.5 months earlier.
PFGE revealed four different SpeI endonuclease-restricted DNA profiles, with band difference greater than seven, indicating that all four strains were genetically unrelated (18). Three of the patients had recent antibiotic exposure, but only one was treated with carbapenems before E. coli was isolated.
All isolates were multidrug resistant, with different antibiotic susceptibilities (Table 2). All strains were resistant to ertapenem, three were imipenem resistant and one imipenem intermediate, and three were meropenem resistant and one meropenem susceptible.
TABLE 2.
Antibiotic susceptibilities of E. coli clinical strains and their transformants
| E. coli strain or transformantb | MIC (μg/ml)a for:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CRO | CAZ | FEP | TZP | AZT | IPMc | MEMc | ERTc | CIP | SXT | AMK | GEN | |
| 157 | >64 | >64 | >64 | >128 | >64 | 32 | 4 | 32 | >4 | >320 | 8 | <1 |
| T-157 | >64 | 16 | 2 | >128 | >64 | 2 | 0.5 | 1 | <0.25 | >320 | <2 | <1 |
| 329 | >64 | 16 | 8 | >128 | >64 | 32 | >32 | 32 | >4 | >320 | >64 | >16 |
| T-329 | >64 | 8 | 2 | >128 | 32 | 4 | 0.5 | 1.5 | <0.25 | <20 | <2 | <1 |
| 339 | >64 | 16 | 16 | 64 | >64 | 12 | 4 | >32 | >4 | >320 | 4 | >16 |
| T-339 | 4 | 2 | <1 | 16 | 32 | 0.5 | 0.25 | 0.25 | <0.25 | <20 | <2 | <1 |
| 360 | >64 | >64 | >64 | >128 | >64 | 8 | 4 | >32 | >4 | >320 | 32 | 2 |
| T-360 | 8 | 4 | <1 | 64 | 32 | 1 | 0.25 | 1 | <0.25 | <20 | 16 | <1 |
| DH5α | <1 | <1 | <1 | <4 | <1 | 0.19 | 0.012 | 0.006 | <0.25 | <20 | <2 | <1 |
CRO, ceftriaxone; CAZ, ceftazidime; FEP, cefepime; TZP, piperacillin/tazobactam; AZT, aztreonam; IMP, imipenem; MEM, meropenem; ERT, ertapenem; CIP, ciprofloxacin; SXT, trimethoprim-sulfamethoxazole; AMK, amikacin; GEN, gentamicin.
T, transformant.
The MIC for each carbapenem is the average MIC for three measurements obtained from three Etests performed independently.
Detection of a carbapenem-hydrolyzing β-lactamase.
Crude enzyme preparations of all four E. coli strains showed alterations in the shapes of the zones of inhibition around a susceptible E. coli test organism, indicating the production of a β-lactamase with imipenem hydrolysis activity. Phenotypic screening for metallo-β-lactamase was negative in all four strains. IEF showed production of various β-lactamases (Fig. 1A, lanes D), with two distinct bands observed in all four strains migrating as blaTEM (pI 5.4) and a β-lactamase migrating with a pI of 6.7. Isolates 157 and 360 expressed two additional β-lactamases with pIs of 7.4 and above 9.0, and isolate 329 showed a β-lactamase with a pI of 7.9.
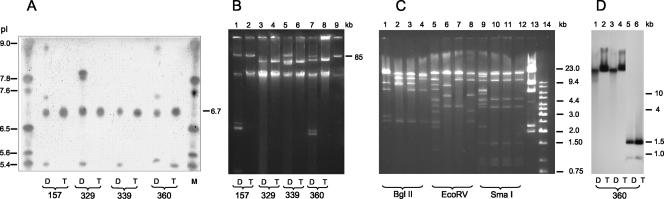
FIG. 1.
IEF, plasmid DNA, and Southern blot analysis of the four KPC-2-producing, carbapenem-resistant E. coli strains (lanes D) and their respective transformants (lanes T). (A) M, pI marker. (B) Lane 9, plasmid AFF2 (85 kb) as a molecular-size marker. (C) Restriction pattern of plasmid DNA from E. coli transformants of strains 157, 329, 339, and 360. Lane 13, lambda DNA digested with HindIII; lane 14, 1-kb DNA ladder (Fermentas). (D) Southern blot analysis of plasmid DNA of strain 360 digested with BglII, EcoRV, or SmaI endonucleases (recognizes position 790 of the OPR of blaKPC-2), respectively, and hybridized with a KPC-2 probe.
Analysis and transfer of KPC-2-encoding plasmid DNA.
Plasmid DNA analysis of the E. coli strains revealed a different band pattern, with a plasmid of >85 kb in three of four strains (Fig. 1B). Transformation of plasmid extracts into E. coli DH5α succeeded, and PCR on plasmid DNA isolated from transformed colonies followed by cloning and sequencing confirmed the presence of blaKPC-2. β-lactamase extracts from all four transformants subjected to IEF showed one distinct band focusing at pI 6.7, corresponding to the pI of KPC-2 (2) (Fig. 1A, lanes T). Transfer of the KPC-2-encoding plasmids raised the MICs of extended-spectrum cephalosporins, aztreonam, and carbapenems compared to what was found for the susceptible E. coli DH5α, but none of the transformants became resistant to imipenem or meropenem (Table 1). Other PCR-screening reactions using specific primers for known class A carbapenemases were all negative.
Plasmid analysis of all four transformants indicated that each had acquired a single plasmid (Fig. 1B, lanes T), with the largest plasmid transferred from parent strain 157 (Fig. 1B, lane 2). Plasmids from strains 329 and 339 appeared similar in size (Fig. 1B, lanes 4 and 6, respectively). Restriction analysis using various endonucleases (Fig. 1C) showed different restriction profiles, but clear similarities (BglII and EcoRV) and even identity (Sma) in the band patterns were noticeable, suggesting the possibility of a common origin. Strain 157 exhibited a unique restriction pattern (Fig. 1C). Southern blot analysis using a blaKPC-2 probe following restriction with BglII, EcoRV, or SmaI, which digests blaKPC-2 at nucleotide 790, proved the presence of blaKPC-2 in the transferred plasmids (Fig. 1D). Although KPC-2-encoding plasmids in the parent strains differed, hybridization was positive at the same position on the blot for each strain (hybridization results are presented only for strain 360 as a representative), supporting the possibility that the plasmids shared a large fragment that contained the gene.
DISCUSSION
Our study is the first to report on KPC-2 in E. coli clinical strains and the first to report KPC-2 outside the United States without an apparent U.S. origin. The enzyme was found in four genetically unrelated carbapenem-resistant isolates of E. coli from four epidemiologically unrelated patients. Three of the patients had been treated recently with antibiotics but only one with a carbapenem. The KPC-2 gene was carried on four different plasmids that contained a common encoding element and probably had a common origin.
Transfer of these plasmids to a susceptible E. coli strain increased the imipenem MIC significantly but did not confer complete resistance, suggesting that an additional mechanism is involved in carbapenem resistance in these strains. Among the possibilities for such a mechanism are porin alterations, which reduce the entry of carbapenems into the cell, as has been shown mainly for imipenem-resistant Klebsiella pneumoniae strains (9, 20) or efflux pumps, a mechanism more common in nonfermenters, such as Pseudomonas aeruginosa and Acinetobacter.
KPC-2 was originally identified in K. pneumoniae strains from Maryland in 2003 (13), and since then, it has been identified in a Salmonella strain from the same area (12), in an Enterobacter strain from Boston (8), and in K. pneumoniae strains from New York (3). KPC class A β-lactamases have been reported to date only in the United States or as originating in the United States, as in the case of the Klebsiella pneumoniae strain in France that was likely acquired by a patient during hospitalization in New York (14). Our study is the first to report KPC-2 outside the United States without an apparent U.S. origin.
PFGE indicated that these E. coli strains were genetically unrelated. Epidemiological investigation revealed that two of the four clinical strains were not acquired in the hospital and were likely imported from two different chronic-health-care facilities, posing the worrying problem of fluctuation of carbapenem resistance in and out of the hospital setting.
Carbapenem resistance in E. coli remains rare, and therefore, at present, there are no clinical outcome data on infections caused by these strains. In our cases, carbapenem treatment did not seem to be a prerequisite for the isolation of carbapenem-resistant E. coli. Nevertheless, the existence of the KPC-2 gene on plasmids that may carry additional antibiotic resistance determinants poses the possibility of dissemination of this carbapenem-encoding gene between and among species.
Acknowledgments
This work was supported in part by a grant from the Israel-United States Binational Science Foundation.
REFERENCES
- 1.Arakawa, Y., N. Shibata, K. Shibayama, H. Kurokawa, T. Yagi, H. Fujiwara, and M. Goto. 2000. Convenient test for screening metallo-β-lactamase-producing gram-negative bacteria using thiol compounds. J. Clin. Microbiol. 38:40-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bratu, S., D. Landman, M. Alam, E. Tolentino, and J. Quale. 2005. Detection of KPC carbapenem-hydrolyzing enzymes in Enterobacter spp. from Brooklyn, New York. Antimicrob. Agents Chemother. 49:776-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bratu, S., D. Landman, R. Haag, R. Rocco, A. Eramo, M. Alam, and J. Quale. 2005. Rapid spread of carbapenem-resistant Klebsiella pneumoniae in New York City. Arch. Intern. Med. 165:1430-1435. [DOI] [PubMed] [Google Scholar]
- 4.Bratu, S., P. Tolaney, U. Karumudi, J. Quale, M. Mooty, S. Nichani, and D. Landman. 2005. Carbapenemase-producing Klebsiella pneumoniae in Brooklyn, N.Y.: molecular epidemiology and in vitro activity of polymyxin B and other agents. J. Antimicrob. Chemother. 56:128-132. [DOI] [PubMed] [Google Scholar]
- 5.Chmelnitsky, I., Y. Carmeli, A. Leavitt, M. J. Schwaber, and S. Navon-Venezia. 2005. CTX-M-2 and a new CTX-M-39 enzyme are the major extended-spectrum beta-lactamases in multiple Escherichia coli clones isolated in Tel Aviv, Israel. Antimicrob. Agents Chemother. 49:4745-4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Galani, I., M. Souli, Z. Chryssouli, D. Katsala, and H. Giamarellou. 2004. First identification of an Escherichia coli clinical isolate producing both metallo-beta-lactamase VIM-2 and extended-spectrum beta-lactamase IBC-1. Clin. Microbiol. Infect. 10:757-760. [DOI] [PubMed] [Google Scholar]
- 7.Hong, T., S. Moland, B. Abdalhamid, N. D. Hanson, J. Wang, C. Sloan, D. Fabian, A. Farajallah, J. Levine, and K. S. Thomson. 2005. Escherichia coli: development of carbapenem resistance during therapy. Clin. Infect. Dis. 40:e84-e86. [DOI] [PubMed] [Google Scholar]
- 8.Hossain, A., M. J. Ferraro, R. M. Pino, R. B. Dew, E. S. Moland, T. J. Lockhart, R. V. Goering, and N. D. Hanson. 2004. Plasmid-mediated carbapenem hydrolyzing enzyme KPC-2 in an Enterobacter sp. Antimicrob. Agents Chemother. 48:4438-4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jacoby, G. A., D. M. Mills, and N. Chow. 2004. Role of β-lactamases and porins in resistance to ertapenem and other β-lactams in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 48:3203-3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matthew, M., M. Harris, M. J. Marshall, and G. W. Rose. 1975. The use of analytical isoelectric focusing for detection and identification of beta-lactamases. J. Gen. Microbiol. 88:169-178. [DOI] [PubMed] [Google Scholar]
- 11.Miriagou, V., E. Tzlepi, D. Gianeneli, and L. S. Tzouvelekis. 2003. Escherichia coli with a self-transferable, multiresistant plasmid coding for metallo-beta-lactamase VIM-1. Antimicrob. Agents Chemother. 47:395-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miriagou, V., L. S. Tzouvelekis, S. Rositter, E. Tzlepi, F. J. Angulo, and J. M. Whichard. 2003. Imipenem resistance in Salmonella clinical strain due to plasmid-mediated class-A carbapenemase KPC-2. Antimicrob. Agents Chemother. 47:1297-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moland, E. S., N. D. Hanson, V. L. Herrera, J. A. Black, T. J. Lockhart, A. Hossain, J. A. Johnson, R. V. Goering, and K. S. Thomson. 2003. Plasmid-mediated, carbapenem-hydrolyzing β-lactamase, KPC-2, in Klebsiella pneumoniae isolates. J. Antimicrob. Chemother. 51:711-714. [DOI] [PubMed] [Google Scholar]
- 14.Naas, T., P. Nordmann, G. Vedel, and C. Poyart. 2005. Plasmid-mediated carbapenem-hydrolyzing β-lactamase KPC in a Klebsiella pneumoniae isolate from France. Antimicrob. Agents Chemother. 49:4423-4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nordmann, P., and L. Poirel. 2002. Emerging carbapenemases in gram-negative aerobes. Clin. Microbiol. Infect. 8:321-331. [DOI] [PubMed] [Google Scholar]
- 16.Poirel, L., J. N. Pham, L. Cabanne, B. J. Gatus, S. M. Bell, and P. Nordmann. 2004. Carbapenem-hydrolysing metallo-beta-lactamases from Klebsiella pneumoniae and Escherichia coli isolated in Australia. Pathology 36:366-367. [DOI] [PubMed] [Google Scholar]
- 17.Stapleton, P. D., K. P. Shannon, and G. L. French. 1999. Carbapenem resistance in Escherichia coli associated with plasmid-determined CMY-4 β-lactamase production and loss of an outer membrane protein. Antimicrob. Agents Chemother. 43:1206-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tórtola, M. T., S. Lavilla, E. Miró, J. J. González, N. Larrosa, M. Sabaté, F. Navarro, and G. Prats. 2005. First detection of a carbapenem-hydrolyzing metalloenzyme in two Enterobacteriaceae isolates in Spain. Antimicrob. Agents Chemother. 49:3492-3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Woodford, N., P. M. Tierno, Jr., K. Young, L. Tysall, M. F. I. Palepou, E. Ward, R. E. Painter, D. F. Suber, D. Shungu, L. L. Silver, K. Inglima, J. Kornblum, and D. Livermore. 2004. Outbreak of Klebsiella pneumonia producing a new carbapenem hydrolyzing class A β-lactamase, KPC-3, in a New York medical center. Antimicrob. Agents Chemother. 48:4793-4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yigit, H., A. M. Queenan, G. J. Andersen, A. Domenech-Sanchez, J. W. Biddle, C. D. Steward, S. Alberti, K. Bush, and F. C. Tenover. 2001. Novel carbapenem-hydrolyzing β-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 45:1151-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]