Abstract
PBP 10, an antibacterial, cell membrane-permeant rhodamine B-conjugated peptide derived from the polyphosphoinositide binding site of gelsolin, interacts selectively with both lipopolysaccharides (LPS) and lipoteichoic acid (LTA), the distinct components of gram-negative and gram-positive bacteria, respectively. Isolated LPS and LTA decrease the antimicrobial activities of PBP 10, as well as other antimicrobial peptides, such as cathelicidin-LL37 (LL37) and mellitin. In an effort to elucidate the mechanism of bacterial killing by PBP 10, we compared its effects on artificial lipid bilayers and eukaryotic cell membranes with the actions of the mellitin, magainin II, and LL37 peptides. This study reveals that pore formation is unlikely to be involved in PBP 10-mediated membrane destabilization. We also investigated the effects of these peptides on platelets and red blood cells (RBCs). Comparison of these antimicrobial peptides shows that only mellitin has a toxic effect on platelets and RBCs in a concentration range concomitant with its bactericidal activity. The hemolytic activities of the PBP 10 and LL37 peptides significantly increase when RBCs are osmotically swollen in hypotonic solution, indicating that these antibacterial peptides may take advantage of the more extended form of bacterial membranes in exerting their killing activities. Additionally, we found that LL37 hemolytic activity was much higher when RBCs were induced to expose phosphatidylserine to the external leaflet of their plasma membranes. This finding suggests that asymmetrical distribution of phospholipids in the external membranes of eukaryotic cells may represent an important factor in determining the specificity of antibacterial peptides for targeting bacteria rather than eukaryotic cells.
Antimicrobial peptides are gene-encoded host-derived molecules that provide a first line of defense against gram-negative and gram-positive bacteria, fungi, mycobacteria, and some enveloped viruses (44, 45). They are found in all species, ranging from protozoa to vertebrates. Generally, they contain 15 to 45 amino acid residues with a net positive charge (lysine or arginine rich) and are amphipathic (46). After targeting bacterial membranes, their killing action is faster than the bacterial growth rate, which accounts for the large antibacterial effect (4). However, our understanding of the peptide-mediated bacterium-killing mechanism is still incomplete. Many of these peptides are believed to specifically kill bacteria by directly interacting with and disrupting the negatively charged bacterial membrane (36, 46). Two general mechanisms, “barrel stave” and “carpet” (31), have been proposed to describe the process of phospholipid membrane permeation by membrane-active peptides. According to the “barrel stave model,” as few as three molecules are required to induce a membrane pore. To allow pore formation, the inserted molecules should have distinct structures, like amphipathic α-helices. On the other hand, according to the “carpet model,” peptides bind to the phospholipid membrane surface until a threshold concentration is reached and only then permeate it in a detergent-like manner (17, 31). Antimicrobial peptides might also act through mechanisms other than bacterial-membrane destabilization. In addition to its bactericidal activity, LL37 activates airway epithelial cells, followed by release of interleukin-8 (41). LL37 also affects the toxicity of bacterium-derived factors, neutralizing lipopolysaccharide (LPS) bioactivity (20). Cecropin P1 and PR-39 peptides from the pig small intestine have been shown to prevent bacterial protein and DNA synthesis (5). The antimicrobial peptides magainin II and temporins (B and L) were found to modulate the hydrolytic activity of secretory phospholipase A2 (47).
Recently, we reported that a cell membrane-permeant rhodamine B-conjugated peptide based on the phosphatidylinositol-4,5-bisphosphate (PIP2) binding site of gelsolin (GS 160-169 [rhodamine B-QRLFQVKGRR], named PBP 10) can kill the gram-negative Escherichia coli and Pseudomonas aeruginosa and the gram-positive Streptococcus pneumoniae (10). A net positive charge, short sequence, and ability to cross cell membranes represent features of PBP 10 that bear a major resemblance to those of the antibiotic peptides. Circular dichroism and nuclear magnetic resonance evaluations have shown a coil-to-helix transition when gelsolin residues 150 to 169 bind PIP2 (43). The helical structure produces an amphipathic molecule with positive charges arranged on one face and hydrophobic residues on the other (10). Unique properties of PBP 10 include polyphosphoinositide binding due to its cationic charge and hydrophobicity, which are due in part to the linked rhodamine B. PBP 10 enters cells passively and can affect many eukaryotic cellular functions that are dependent on phosphoinositide signaling, most significantly, actin organization and vesicle trafficking (3, 9, 12, 16).
In exploring mechanisms to explain the bacterium-killing activity of PBP 10, we compared the effect of PBP 10 on artificial membranes with the action of the pore-forming peptides mellitin and magainin II, as well as with the “carpet-like” membrane-covering LL37 peptide (28). This study shows that PBP 10 does not form pores during membrane destabilization. The abilities of LPS and lipoteichoic acid (LTA) to inhibit the antibacterial activities of all tested peptides, including PBP 10, suggests that these molecules represent specific bacterial targets recognized by antibacterial peptides. We have also investigated the effects of PBP 10, LL37, magainin II, and mellitin on platelet function and red blood cell (RBC) membranes. Among these peptides, only mellitin has a toxic effect on platelets and RBCs at a concentration similar to that at which it kills bacteria.
MATERIALS AND METHODS
Materials.
Magainin II (M-7402), synthetic mellitin (M-4171), l-α-phosphatidylcholine (PC) (P-6638), l-α-phosphatidyl-l-serine (P-6641), sodium dithionite (S-256), thrombin (T-6884), calcium ionophore A23187 (C-7522), fatty-acid-free bovine serum albumin (A6003), apyrase (A-6535), Sepharose CL-2B (CL-2B-300), Sephadex G-75 (G-75-120), and Fura 2-AM (A-9210) were obtained from Sigma. Brain phosphatidylinositol-4,5-bisphosphate, triammonium salt (840046), and Escherichia coli total lipid extract (100500) were obtained from Avanti Polar Lipids. p-Xylene-bis-pyridinium bromide (DPX) (X-1525) and 8-aminonaphthalene-1,3,6-trisulfonic acid (ANTS), disodium salt (A-350), were obtained from Molecular Probes. LL37 was synthesized using an automatic peptide synthesizer at the Louisiana State University Medical Center Core Laboratories. The quality of the obtained peptide was monitored by mass spectrometry and capillary electrophoresis. The set of gelsolin-related peptides, including QRL and QRLFQVKGRR, were prepared either as free peptides or conjugated to rhodamine B with an amide link at the N terminus of the peptide (denoted with RhB- as a prefix to the peptide sequence) as previously described (12).
Interaction of LTA with fluorescent peptides derived from the gelsolin PIP2 binding site.
The fluorescence of rhodamine B-QRLFQVKGRR (Iem, 590 nm; Iex, 560 nm) was measured 15 minutes after the addition of various concentrations of LTA (from Staphylococcus aureus [Sigma]), LPS (Escherichia coli serotype O26:B6 [Sigma]), PIP2, or phosphatidylethanolamine (PE) to 2 μM peptide solutions (PBP 10 or RhB-QRL) in buffer A (10 mM TRIS, 10 mM MES [morpholineethanesulfonic acid], pH 7.0). Because the quantum yield of rhodamine B is higher in a lipophilic environment, binding of the peptide to a membrane changes rhodamine B fluorescence.
Bactericidal assay.
A single colony each of gram-negative Escherichia coli (kanamycin-resistant SG 13009 or MG1655) and gram-positive Bacillus subtilis (ATCC 6051) was selected from a plate and grown to mid-log phase (optical density at 600 nm [OD600], ∼0.3) in 5 ml of LB (Becton-Dickinson, Cockeysville, MD) in a shaking incubator (300 rpm; 37°C). One milliliter of the bacterial suspension was centrifuged at 4,000 rpm for 5 min at room temperature, and the bacterial pellet was resuspended in phosphate-buffered saline. This suspension was diluted 1:1,000. Serial dilutions of antibacterial peptides were mixed with the diluted bacterial suspension, with or without different lipids (LPS, LTA, PIP2, PC, phosphatidylserine [PS], or bacterial-lipid [BL] extract from Escherichia coli). The tubes were incubated at 37°C for 1 hour and transferred to ice. Duplicate 10-μl aliquots of 10-fold dilutions (undiluted, 1:10, 1:100, and 1:1,000) of these mixtures were plated on sectors of LB agar plates, and the plates were incubated overnight at 37°C. The number of colonies in the duplicate samples at each dilution was determined the following morning, and the numbers of CFU in the individual mixtures were determined from the dilution factor. The MIC of PBP 10 was determined by a broth microdilution method with Mueller-Hinton (MH) broth according to the procedures outlined by the CLSI (Formerly NCCLS) (18). A series of twofold dilutions of PBP 10 in MH broth was prepared from a stock solution and placed in 96-well plates, and then a dilution of bacteria (grown in half-concentrated MH broth) was added. After incubation for 18 h at 37°C, the MIC was read (OD600) as the lowest concentration of PBP 10 (μg/ml) resulting in inhibition of visible bacterial growth.
LUV preparation.
Lipids dissolved in chloroform were transformed into dry lipid film by solvent evaporation under a stream of nitrogen. In the next step, they were kept under vacuum for 3 h, and then the dry lipid film was hydrated and vortexed. The suspension was freeze-thawed 5 times and then extruded 11 times through polycarbonate filters (100 nm for large unilammelar vesicles [LUVs] used in phospholipid redistribution assays and 200 nm for ANTS leakage assays). The LUV size (hydrodynamic diameter) was determined by dynamic light scattering (DLS). The LUV phospholipid composition was determined by thin-layer chromatography analysis (8, 11), and the concentration of the resulting preparation was determined by phosphorus analysis (1).
Redistribution of NBD phospholipids.
LUVs were prepared as described above. 1-Oleoyl-2-[6(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino] caproyl (NBD)-PC was incorporated in the outer leaflet by adding NBD-PC dissolved in ethanol, at a ratio of 1 to 100 NBD-PC-unlabeled PC, to LUVs in a stirred suspension. Vesicles (100 nmol lipid/ml) were incubated for 60 min at room temperature for equilibration. To evaluate the flip-flop rate of PC-NBD in PC LUVs in the presence of the magainin II, LL37, and PBP 10 peptides, we used a technique described by Matsuzaki et al. (22). The LUVs were incubated with peptides for 30 min at 37°C and placed in a cuvette of an LS-5B spectrofluorometer (Perkin-Elmer). When required, trypsin was added for the last 5 min of the incubation period. The NBD fluorescence was recorded (Iex = 470 nm; Iem = 540 nm) until a constant baseline (F0) was obtained. Detection of the NBD-PC present in the LUV's inner leaflet was monitored after reduction of the NBD groups of the analogs present in the outer leaflet by dithionite ion, resulting in a loss of fluorescence intensity due to the conversion of the NBD group to its seven-amino derivative. Once the decrease in fluorescence emission caused by the addition of dithionite (10 mM) to the intact LUVs reached a plateau value (F1), Triton X-100 (1%) was added to expose any NBD-PC protected in the inner leaflet and to provide the basal fluorescence signal (F2). The fraction of NBD-PC redistributed into the inner leaflets of LUVs was calculated from the expression (F1 − F2)/(F0 − F2).
ANTS leakage assay.
For the ANTS/DPX leakage assay, LUVs were prepared as described above. The dry phospholipid film was hydrated with buffer B (12.5 mM ANTS, 45 mM DPX, 22.5 mM NaCl, 10 mM glycine, 10 mM HEPES, pH 7.4). Following the extrusion process, liposomes were separated from unencapsulated material on a Sephadex G-75 column. DPX is highly water soluble, and when coencapsulated with ANTS, it efficiently quenches ANTS fluorescence by collisional transfer. The liposomes initially containing both ANTS and DPX emit about 4% of the fluorescence of the liposomes treated with Triton X-100, which is assumed to produce 100% leakage (14). Changes in the fluorescence of 1.5 ml ANTS/DPX LUV samples (2 μM phospholipid concentrations) were monitored with an SLM-Aminco MC200 fluorometer set at an Iex wavelength of 380 nm and an Iem wavelength of 530 nm for 10 min after the addition of the peptides.
Measurement of vesicle diameter.
The LUV sizes (hydrodynamic diameters) were determined by DLS using a DynaPro 99 instrument. The method measures the diffusion constant of the vesicles using the autocorrelation function of scattered-light intensity, and the diameter of the vesicle is calculated from the relation D = kT/6πη Rh, where D is the translational diffusion constant, k is Boltzmann’s constant, T is temperature, η is the solvent viscosity, and Rh is the hydrodynamic radius. To determine if the LUVs' phospholipid flip-flop or ANTS leakage is related to liposome size changes (indicating vesicle fusion or disruption), LUVs treated with peptides were measured by DLS before and after lipid redistribution or ANTS leakage assays.
Isolation of human platelets, platelet aggregation, and membrane integrity.
Freshly donated blood from healthy volunteers was collected in acid-citrate dextrose. Platelet-rich plasma was obtained after centrifugation (15 min; 110 × g; room temperature) of blood supplemented with apyrase (0.5 U/ml). The platelets were sedimented by centrifugation (10 min; 1,000 × g), suspended in buffer C (139 mM NaCl, 2.8 mM KCl, 0.8 mM MgCl2, 0.8 mM KH2PO4, 8.9 mM NaHCO3, 10 mM HEPES, 5.6 mM glucose, 0.3% albumin, pH 7.35), and filtered on a 50-ml column of Sepharose 2B equilibrated with the same buffer. The cell count was determined using a Z2 Coulter particle counter and size analyzer. To conduct the aggregation assay, 1-ml samples of 2 × 108 platelets per ml in buffer C containing 2 mM CaCl2 were introduced into a prewarmed cuvette (37°C) of a Chronolog Lumi-Aggregometer (Chrono-Log Corp., Havertown, PA). Aggregation was initiated by adding human thrombin and stirring the mixture at 1,000 rpm. Changes in light transmission were recorded using a PowerLab/200 instrument and MacLab Chart program version 3.2. When required, the platelet suspension was preincubated with antibacterial peptides before platelet aggregation was induced. Platelets labeled with the fluorescent indicator dye Fura-2 according to the method of Pollock et al. (34) and resuspended in buffer C containing 2 mM CaCl2 were used to measure platelet membrane integrity in the presence of different antibacterial peptides. Changes in fluorescence were monitored with an SLM-Aminco 8100 fluorometer set at an Iex of 340 nm and an Iem of 510 nm.
Red blood cell osmotic fragility and hemolysis.
A turbidity shift (cloudy to clear) occurs in a suspension of RBCs when the integrity of the plasma membrane is compromised. This change was detected by optical density at 625 nm after RBCs, obtained by centrifugation of heparinized blood (1,300 × g; 10 min; 4°C), washed three times in buffer D (140 mM NaCl, 10 mM HEPES, 5 mM glucose, 0.1 mM EGTA, pH 7.4), and pretreated with antibacterial peptides, were added to hypotonic Ringer solution (21). A percent hemolytic index (HI) was calculated using the following formula: HI (%) = 100 × (OD of tested compound − OD of negative control)/(OD of positive control − OD of negative control). RBCs suspended in deionized water were used as a positive control. The hemolytic activities of the peptides against RBCs were tested as described previously (27). Peptides dissolved in phosphate-buffered saline were added to 100 μl of RBCs (hematocryt, ∼5%), and incubation was continued for 1 h at 37°C. The samples were then centrifuged at 1,300 × g for 10 min. Release of hemoglobin as a measure of hemolysis was monitored by measuring the absorbance of the supernatant at 540 nm. One hundred percent hemolysis (positive control) was taken from samples in which 1% Triton X-100 was added. When required, RBCs were activated with 4 μM calcium ionophore (A23187) in the presence of 2 mM CaCl2 or 0.1 mM EGTA (control sample) for 20 min at 37°C. Phosphatidylserine exposure in A23187-activated RBCs was evaluated using an annexin V-fluorescein isothiocyanate binding test (data not shown).
RESULTS
LTA interaction with gelsolin polyphosphoinositide binding sequence.
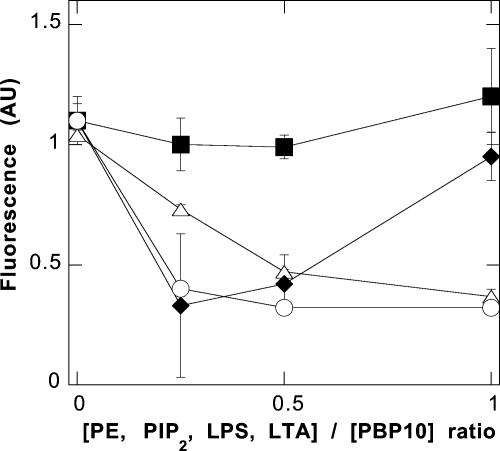
In addition to the previously described interaction of the PBP 10 peptide with PIP2 and LPS (7, 9), fluorescence measurements showed an interaction between LTA and PBP 10. The effect of LTA on the fluorescence of PBP 10 is shown in Fig. 1. As previously reported, upon interaction of LPS with PBP 10, there was an initial decrease in fluorescence at low LPS/peptide ratios; the peptide fluorescence increased strongly with increasing LPS, suggestive of insertion of the peptide-bound rhodamine B into a more hydrophobic environment (7). At the molar ratios tested, only the first stage of lowered fluorescence was seen with LTA or PIP2, and no change was seen after the addition of PE. There was also no significant fluorescence change after the addition of LTA to a control peptide with the sequence rhodamine B-QRL (RhB-QRL) (data not shown).
FIG. 1.
LTA and LPS bind to PBP 10. Changes in PBP 10 rhodamine B fluorescence in the presence of different lipids (PE, squares; PIP2, triangles; LPS, diamonds; LTA, circles) indicate its specific interaction with PIP2, LPS, and LTA. The data shown are means or means ± standard deviations of two to four experiments. AU, arbitrary units.
Bactericidal activity.
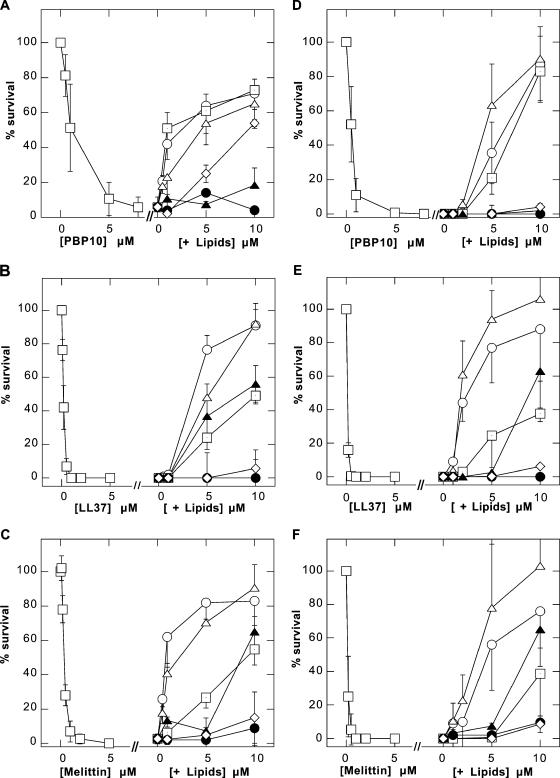
By using a standard bacterium-killing assay, we compared the antimicrobial activities of the naturally occurring antibacterial agents mellitin and LL37 peptide and the synthetic rhodamine B-labeled derivative of the PIP2 binding site of gelsolin by themselves and after their preincubation with LPS, LTA, PIP2, PC, PS, and an extract of BL from Escherichia coli (Fig. 2). Previous reports indicated that LPS could interfere with the antimicrobial activities of different cathelicidin-LL37 derivates (2). Similarly, we found that LPS and LTA are powerful inhibitors of the antibacterial activities of the LL37, mellitin, and PBP 10 peptides against Escherichia coli SG 13009, Bacillus subtilis ATCC 6051 (Fig. 2), and Escherichia coli MG1655 (data not shown). In the case of PBP 10, the effectiveness of PIP2 was similar to that of LPS (Fig. 2A). Under the same conditions PC, PS, and BL did not significantly affect the number of growing colonies. The MICs of the PBP 10 peptide, considered to be the lowest PBP 10 concentrations at which observable growth of Escherichia coli SG13009 and Bacillus subtilis ATCC 6051 was inhibited, were 12.5 and 3.125 μg/ml, respectively. These values are in the range previously determined for other membrane-active peptides (18). Similar MICs and killing values for the PBP 10 peptide against the bacteria tested indicate that a threshold concentration of PBP 10 is necessary for antibacterial activity and that once this concentration is reached, PBP 10 causes bacterial death. This behavior is consistent with the mechanisms of action of other membrane-active antibacterial molecules (37).
FIG. 2.
Bactericidal activities of (A and D) PBP 10, (B and E) LL37, and (C and F) mellitin (all squares) against gram-negative Escherichia coli SG13009 (A to C) and gram-positive Bacillus subtilis ATCC 6051 (D to F) by themselves and their inhibition in the presence of increasing concentrations of LPS (circles), LTA (open triangles), or PIP2 (squares with dots). Other membrane phospholipids have limited or no effect (PS, filled triangles; PC, filled circles; BL, diamonds). The data shown are means ± standard deviations of three to five experiments.
Phospholipid flip-flop in asymmetrically labeled PC/PC-NBD vesicles.
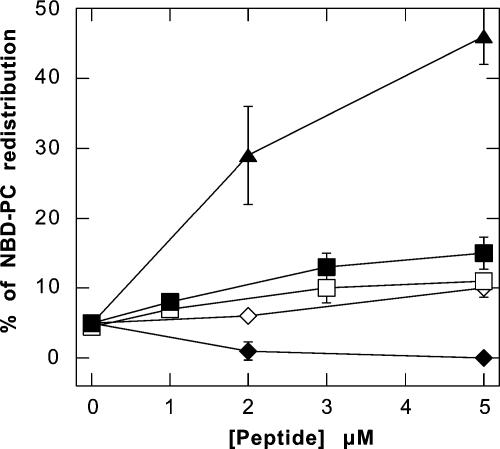
We evaluated the flip-flop rates of PC-NBD in PC LUVs after the addition of magainin II, LL37, and PBP 10. Figure 3 shows that only magainin II (a pore-forming peptide digestible by trypsin) (25) induced LUV membrane phospholipid redistribution to a significant extent (45%). The interaction of antibacterial peptides with LUVs prepared from BL results in vesicle aggregation (Fig. 4B), which interferes with the NBD-analog redistribution assay, preventing the investigation of peptide-mediated phospholipid redistribution in BL vesicles.
FIG. 3.
Flip-Flop of LUV phospholipid after addition of PBP 10 (open squares), LL37 (filled squares), LL37 plus trypsin (open diamonds), magainin II (filled diamonds), and magainin II plus trypsin (filled triangles) evaluated by monitoring NBD-PC redistribution. PC LUVs were labeled with 1% NBD-PC in the outer monolayer. After the addition of peptides, the LUVs were incubated for 30 min at 37°C. The amount of NBD-PC flipped to the inner monolayer was determined by dithionite assay. The data shown are means ± standard deviations of three or four experiments.
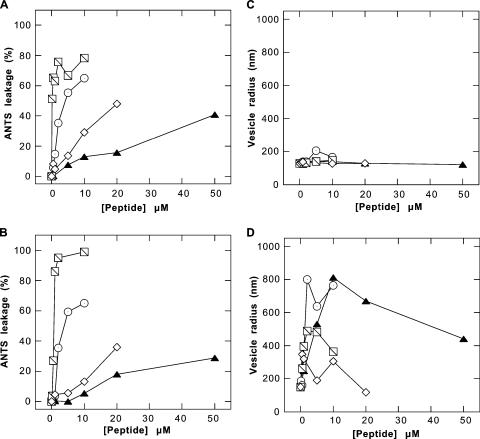
FIG. 4.
Comparison of leakage of ANTS/DPX containing PC (A) and BL (B) liposomes induced with PBP 10 (filled triangles), LL37 (circles), mellitin (slashed squares), and magainin II (open diamonds). LUVs were prepared by lipid extrusion using 200-nm filters. Changes in their sizes during the leakage assay were monitored by DLS and are presented as average hydrodynamic radii for PC (C) and BL (D) LUVs. An increase in the hydrodynamic radius indicates vesicle aggregation. Each data point represents the mean of two experiments.
Vesicle leakage assay.
Pore formation and membrane disruption would be expected to allow the leakage of small molecules from lipid vesicles. By using LUVs containing a mixture of ANTS and DPX, we were able to evaluate the leakage of ANTS fluorophore, which indicates membrane damage caused by antibacterial peptides. Figure 4 shows that the PBP 10 peptide can damage vesicles composed of either PC or BL, but only at concentrations 5 to 10 times higher than those required to ensure its Escherichia coli-killing property (Fig. 2). Also, the percentage of LUVs damaged by PBP 10 was much lower than the damage caused by mellitin, LL37, or magainin II. The percentages of ANTS leakage from PC and BL were similar. The efficiencies of equimolar concentrations of antibacterial peptides decreased in the sequence mellitin > LL37 > magainin II > PBP 10 and were not significantly different in PC and BL vesicles, supporting the concept that the membrane-destabilizing effects of these antibacterial agents are not specifically related to the membrane phospholipid composition.
Effects of antibacterial agents on platelets and red blood cells.
The resting state and thrombin-mediated activation of platelets are both altered by perturbations of the plasma membrane and provide a sensitive system by which to detect the effects of amphipathic peptides. The leakage of hemoglobin from RBCs provides an independent quantitative assay of the extent of membrane permeabilization. Neither the resting state of platelets nor platelet aggregation induced by thrombin was affected by preincubation of platelet suspensions with magainin II or LL37 (Fig. 5A). The possibility that these peptides may disrupt eukaryotic cell membranes was also tested in experiments in which calcium influx across the plasma membrane was detected after Fura-2-labeled platelets were mixed with the LL37 peptide (Fig. 5B) and by evaluating hemoglobin release from RBCs (Fig. 6). Both assays showed no significant effect of either LL37 or magainin II at a concentration sufficient to kill bacteria with high efficiency.
FIG. 5.
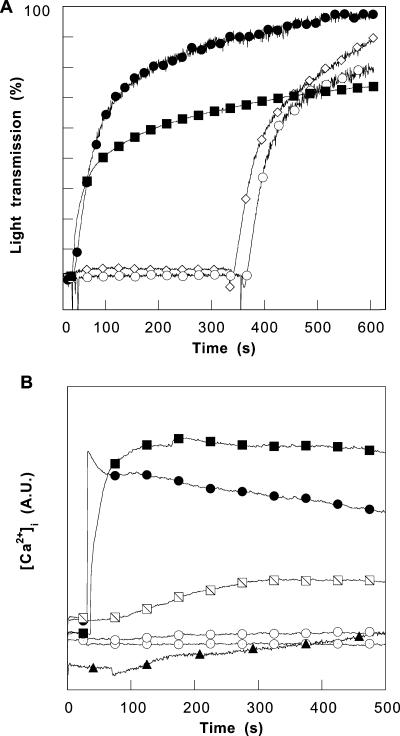
Human platelet aggregation induced by 1 U/ml of thrombin in the presence of PBP 10, LL37, magainin II, and mellitin. (A) Changes in light transmission in platelet suspensions after adding 1 U/ml thrombin (filled circles), 5 μM mellitin (filled squares), 5 μM magainin II (followed by thrombin addition; open diamonds), and 5 μM LL37 peptide (followed by thrombin addition; open circles). (B) Changes in fluorescence in Fura-2-labeled platelets indicate Ca2+ influx after 1-U/ml thrombin stimulation (filled circles), addition of 0.25 μM or 1 μM mellitin (slashed and filled squares, respectively), addition of 1 μM or 5 μM LL37 (circles with dots and empty circles, respectively), and addition of 20 μM RhB-QRLFQVKGRR peptide (filled triangles). The data shown in panels A and B were obtained in a single experiment. Two other experiments produced similar data. A.U., arbitrary units.
FIG. 6.
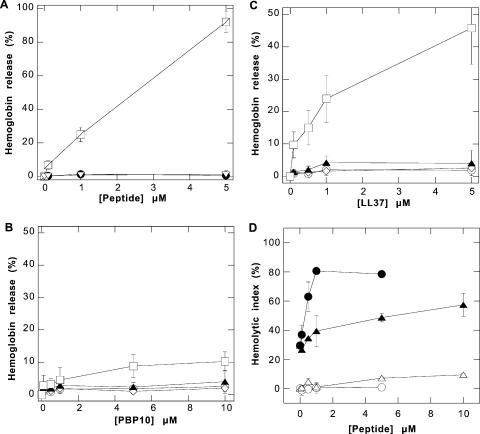
Effects of PBP 10, LL37, magainin II, and mellitin peptides on RBCs. (A) Hemoglobin release from human red blood cells after the addition of mellitin (squares), magainin II (open diamonds), or calcium ionophore (triangles). (B and C) Hemoglobin release from RBCs after the addition of PBP 10 and LL37 peptides, respectively. In this experiment, RBCs were preincubated in the presence of EGTA (circles), calcium (diamonds), A23187 plus EGTA (triangles), or A23187 plus calcium (squares) for 20 min at 37°C before the peptides were added. (D) HI indicating osmotic fragility of red blood cells in the presence of LL37 (circles) and PBP 10 (triangles), measured as a change in the optical density at 625 nm in isotonic (open circles and open triangles) and hypotonic (filled circles and filled triangles) buffers. The data shown are means ± standard deviations of four or five experiments.
In contrast to their tolerance of LL37 and magainin II, RBCs and platelets were significantly damaged by mellitin. The increased light transmission of platelet suspensions following the addition of mellitin in the absence of thrombin suggests platelet lysis (Fig. 5A), the change in the fluorescence of Fura-2-labeled platelets (Fig. 5B) shows the entry of extracellular Ca2+, and the nearly total hemoglobin release from RBCs (Fig. 6A) results from red blood cell lysis. PBP 10 induced negligible increases of Fura-2-labeled platelet fluorescence and hemoglobin release from RBCs, similar to the effects of the LL37 peptide and magainin II (Fig. 5B and 6). A disruptive effect of LL37 and PBP 10 on red blood cells was observed only in RBCs activated with calcium ionophore in the presence of sufficient calcium to induce plasma membrane PS exposure (6) in isotonic buffers. Increased hemoglobin release (∼40%), observed after adding LL37 peptide to calcium-ionophore-activated RBCs (Fig. 6C), suggests that the presence of negatively charged phospholipids in the external leaflets of plasma membranes increases the bioactivity of LL37. This observation also suggests that phospholipid asymmetry of the eukaryotic plasma membrane is one of the factors that determines the low toxicity of antimicrobial peptides against host cells compared to that against bacteria. When RBCs are moderately swollen by decreasing the osmolarity of the medium, PBP 10 and, to a greater extent, LL37 begin to exhibit a significant ability to destroy membrane integrity. As shown in Fig. 6D, LL37 significantly increases hemolysis at concentrations above 0.5 μM in hypotonic solution, and PBP 10 is significantly lytic at 5 μM. This observation suggests that these antibacterial peptides may take advantage of the more extended form of bacterial bilayer membranes in exerting their antibiotic function.
DISCUSSION
In order to better understand the principles governing the interactions of antibacterial peptides with target microbes and host cells, we conducted several experiments to compare the effects of the synthetic peptide derivative PBP 10 on biological membranes to those of the natural bioactive peptides LL37, magainin II, and mellitin. All of the peptides were found to have significant bactericidal activities, but their abilities to damage artificial and eukaryotic membranes differed greatly from each other. Membrane permeabilization and cell lysis occurring as a result of peptide interaction with membrane phospholipids may represent a mechanism by which antibacterial peptides exert their killing actions; however, the coexistence of additional mechanisms, such as effects on other bacterial-cell components or metabolic pathways (32), cannot be ruled out.
Based on data indicating specific interactions of the PBP 10 peptide with LPS and LTA and the inhibitory effects of LPS and LTA on bacterium-killing activity by PBP 10, LL37, and mellitin, two hypotheses can be proposed. First, the presence of LPS and LTA in external bacterial membranes can specifically attract these peptides, increasing peptide adsorption to the bacterial surface (24, 31, 38). In this context, LPS and LTA are the bacterial targets, mediating the peptide's entry into bacteria, the initial step of the bacterium-killing cascade (30). It is also possible that the specific membrane structures formed by LPS or LTA in association with other bacterial-wall molecules form binding sites for antibacterial peptides (24). The second hypothesis suggests an opposite role for bacterial-membrane components, i.e., that LPS and LTA represent the first line of bacterial defense against host antibacterial peptides.
Inhibition of antibacterial peptides by molecules released from growing or dead bacteria may have a clinical significance, especially when bacterial infection takes place in the airways or oral cavity or on the skin surface, the areas in which antibacterial peptides are normally active (4, 40). On a molar basis, LPS as an inhibitor of LL37 and mellitin is much stronger than any other lipid bilayer constituent from gram-negative bacteria or eukaryotic cells, including PS, PIP2, or a mixture of bacterial phospholipids that is rich in PE but devoid of LPS. Only LTA, the acid glycolipid from the outer membranes of gram-positive bacteria, has similar inhibitory effects under the conditions assayed (Fig. 2). The partial inhibition by PS and PIP2 confirms the primarily electrostatic nature of the interaction that inactivates LL37 and mellitin, but the higher affinities of LPS and LTA suggest some specificity. The reported resistance to antibacterial peptides in Salmonella and Staphylococcus strains has been related to variations in the chemical compositions of LPS and teichoic acids present in gram-negative and gram-positive bacterial cell walls, respectively (19, 33).
The observed increase in hemolytic activity of the LL37 peptide against RBCs that expose PS (Fig. 6) confirms the importance of electrostatic interactions between antibacterial peptides and phospholipids. In resting cells, phospholipids are distributed asymmetrically over both halves of the cell plasma membrane (13, 26), with PS and PE essentially located in the inner leaflet. This membrane asymmetry may be one factor allowing the lower lytic activities of antibacterial peptides against eukaryotic cells than against bacteria. The observed hemolytic effect of LL37 against RBCs that expose PS supports the hypothesis that the interaction of cationic antibiotics with the cell surface is regulated by exposed charges rather than the electronegative transmembrane gradient pointing toward the cell interior. In this context, cancer cells and vascular endothelial cells in tumors that expose PS (42, 48) may represent specific targets for host antibacterial peptides.
Rapid flip-flop of membrane lipids triggered by pore-forming peptides is a well-characterized phenomenon, and lateral diffusion along the wall of the pores composed of the peptides and the lipids has been proposed as a model to explain this process (15, 22, 23). The effects of the PBP 10 and LL37 peptides on phospholipid redistribution (5 to 10% of PC-NBD redistribution) are unlikely to be induced by pore formation because of their low rates, and they probably result from peptide penetration into the bilayer rather than from pore formation. Our previous data show that PBP 10 can induce significant PC-NBD redistribution in LUVs containing PIP2 (9) but not in vesicles lacking phosphoinositides. Additionally, comparison of the results from experiments to evaluate phospholipid flip-flop with the results of peptide-mediated ANTS leakage revealed that magainin II-induced flip-flop of membrane phospholipids occurred at a peptide concentration threefold lower than that required for significant ANTS leakage, in agreement with previous reports (32). Similarly, PBP 10 lacks the ability to form pores, since the fluorescence of Fura-2 (a Ca2+ indicator) in platelet cytosol did not change in the presence of extracellular calcium after the addition of PBP 10.
With regard to hemolytic activity, previous research has shown that many antibacterial peptides are able to lyse RBCs, but usually their lytic concentrations are high compared to their bacterium-killing concentrations (29, 35). We found that antimicrobial peptides that do not damage human cells in isotonic solutions could be rendered hemolytic under hypo-osmotic conditions that by themselves stress but do not permeabilize the cell membrane. Red blood cell ghosts prepared in hypotonic solution preserve their membrane skeletons (39), suggesting that mechanical stretching of the membrane, which induces an increase in RBC membrane tension under hypo-osmotic conditions, facilitates membrane distortion in the presence of antibacterial peptides.
In conclusion, we have shown that a synthetic peptide, PBP 10, derived from the phosphoinositide-binding site of gelsolin, like natural antibacterial peptides, targets the bacterial-cell wall due to interactions with LPS and LTA. The osmotic conditions under which antibacterial peptides function can modulate their efficiencies. Host cells that expose negatively charged phospholipids can represent additional targets for antibacterial peptides, indicating their involvement in tissue elimination of apoptotic or necrotic cells.
Acknowledgments
This work was supported by NIH grants R01 HL67286 and AR38910.
We thank Mark Goulian, Department of Physics, University of Pennsylvania, for providing us with different bacterial strains and for helpful discussions.
REFERENCES
- 1.Bartlett, G. R. 1959. Colorimetric assay methods for free and phosphorylated glyceric acids. J. Biol. Chem. 234:469-471. [PubMed] [Google Scholar]
- 2.Bartlett, K. H., P. B. McCray, Jr., and P. S. Thorne. 2004. Reduction in the bactericidal activity of selected cathelicidin peptides by bovine calf serum or exogenous endotoxin. Int. J. Antimicrob. Agents 23:606-612. [DOI] [PubMed] [Google Scholar]
- 3.Beaulieu, V., N. Da Silva, N. Pastor-Soler, C. R. Brown, P. J. Smith, D. Brown, and S. Breton. 2005. Modulation of the actin cytoskeleton via gelsolin regulates vacuolar H+-ATPase recycling. J. Biol. Chem. 280:8452-8463. [DOI] [PubMed] [Google Scholar]
- 4.Boman, H. G. 2003. Antibacterial peptides: basic facts and emerging concepts. J. Intern. Med. 254:197-215. [DOI] [PubMed] [Google Scholar]
- 5.Boman, H. G., B. Agerberth, and A. Boman. 1993. Mechanisms of action on Escherichia coli of cecropin P1 and PR-39, two antibacterial peptides from pig intestine. Infect. Immun. 61:2978-2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bucki, R., C. Bachelot-Loza, A. Zachowski, F. Giraud, and J. C. Sulpice. 1998. Calcium induces phospholipid redistribution and microvesicle release in human erythrocyte membranes by independent pathways. Biochemistry 37:15383-15391. [DOI] [PubMed] [Google Scholar]
- 7.Bucki, R., P. C. Georges, Q. Espinassous, M. Funaki, J. J. Pastore, R. Chaby, and P. A. Janmey. 2005. Inactivation of endotoxin by human plasma gelsolin. Biochemistry 44:9590-9597. [DOI] [PubMed] [Google Scholar]
- 8.Bucki, R., M. Gorska, M. Zendzian-Piotrowska, and J. Gorski. 2000. Effect of triiodothyronine on the content of phospholipids in the rat liver nuclei. J. Physiol. Pharmacol. 51:535-540. [PubMed] [Google Scholar]
- 9.Bucki, R., P. A. Janmey, R. Vegners, F. Giraud, and J. C. Sulpice. 2001. Involvement of phosphatidylinositol 4,5-bisphosphate in phosphatidylserine exposure in platelets: use of a permeant phosphoinositide-binding peptide. Biochemistry 40:15752-15761. [DOI] [PubMed] [Google Scholar]
- 10.Bucki, R., J. J. Pastore, P. Randhawa, R. Vegners, D. J. Weiner, and P. A. Janmey. 2004. Antibacterial activities of rhodamine B-conjugated gelsolin-derived peptides compared to those of the antimicrobial peptides cathelicidin LL37, magainin II, and melittin. Antimicrob. Agents Chemother. 48:1526-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bucki, R., M. Zendzian-Piotrowska, A. Nawrocki, and J. Gorski. 1997. Effect of increased uptake of plasma fatty acids by the liver on lipid metabolism in the hepatocellular nuclei. Prostaglandins Leukot. Essent. Fatty Acids 57:27-31. [DOI] [PubMed] [Google Scholar]
- 12.Cunningham, C. C., R. Vegners, R. Bucki, M. Funaki, N. Korde, J. H. Hartwig, T. P. Stossel, and P. A. Janmey. 2001. Cell permeant polyphosphoinositide-binding peptides that block cell motility and actin assembly. J. Biol. Chem. 276:43390-43399. [DOI] [PubMed] [Google Scholar]
- 13.Devaux, P. F. 1991. Static and dynamic lipid asymmetry in cell membranes. Biochemistry 30:1163-1173. [DOI] [PubMed] [Google Scholar]
- 14.Ellens, H., J. Bentz, and F. C. Szoka. 1985. H+- and Ca2+-induced fusion and destabilization of liposomes. Biochemistry 24:3099-3106. [DOI] [PubMed] [Google Scholar]
- 15.Fattal, E., S. Nir, R. A. Parente, and F. C. Szoka, Jr. 1994. Pore-forming peptides induce rapid phospholipid flip-flop in membranes. Biochemistry 33:6721-6731. [DOI] [PubMed] [Google Scholar]
- 16.Fu, H., L. Bjorkman, P. Janmey, A. Karlsson, J. Karlsson, C. Movitz, and C. Dahlgren. 2004. The two neutrophil members of the formylpeptide receptor family activate the NADPH-oxidase through signals that differ in sensitivity to a gelsolin derived phosphoinositide-binding peptide. BMC Cell Biol. 5:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gazit, E., A. Boman, H. G. Boman, and Y. Shai. 1995. Interaction of the mammalian antibacterial peptide cecropin P1 with phospholipid vesicles. Biochemistry 34:11479-11488. [DOI] [PubMed] [Google Scholar]
- 18.Giacometti, A., O. Cirioni, G. Greganti, M. Quarta, and G. Scalise. 1998. In vitro activities of membrane-active peptides against gram-positive and gram-negative aerobic bacteria. Antimicrob. Agents Chemother. 42:3320-3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo, L., K. B. Lim, C. M. Poduje, M. Daniel, J. S. Gunn, M. Hackett, and S. I. Miller. 1998. Lipid A acylation and bacterial resistance against vertebrate antimicrobial peptides. Cell 95:189-198. [DOI] [PubMed] [Google Scholar]
- 20.Larrick, J. W., M. Hirata, R. F. Balint, J. Lee, J. Zhong, and S. C. Wright. 1995. Human CAP18: a novel antimicrobial lipopolysaccharide-binding protein. Infect. Immun. 63:1291-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Light, D. B., P. K. Dahlstrom, R. T. Gronau, and N. L. Baumann. 2001. Extracellular ATP activates a P2 receptor in necturus erythrocytes during hypotonic swelling. J. Membr. Biol. 182:193-202. [DOI] [PubMed] [Google Scholar]
- 22.Matsuzaki, K., O. Murase, N. Fujii, and K. Miyajima. 1996. An antimicrobial peptide, magainin 2, induced rapid flip-flop of phospholipids coupled with pore formation and peptide translocation. Biochemistry 35:11361-11368. [DOI] [PubMed] [Google Scholar]
- 23.Matsuzaki, K., O. Murase, N. Fujii, and K. Miyajima. 1995. Translocation of a channel-forming antimicrobial peptide, magainin 2, across lipid bilayers by forming a pore. Biochemistry 34:6521-6526. [DOI] [PubMed] [Google Scholar]
- 24.Matsuzaki, K., K. Sugishita, and K. Miyajima. 1999. Interactions of an antimicrobial peptide, magainin 2, with lipopolysaccharide-containing liposomes as a model for outer membranes of gram-negative bacteria. FEBS Lett. 449:221-224. [DOI] [PubMed] [Google Scholar]
- 25.Matsuzaki, K., S. Yoneyama, and K. Miyajima. 1997. Pore formation and translocation of melittin. Biophys. J. 73:831-838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Op den Kamp, J. A. 1979. Lipid asymmetry in membranes. Annu. Rev. Biochem. 48:47-71. [DOI] [PubMed] [Google Scholar]
- 27.Oren, Z., J. Hong, and Y. Shai. 1999. A comparative study on the structure and function of a cytolytic alpha-helical peptide and its antimicrobial beta-sheet diastereomer. Eur. J. Biochem. 259:360-369. [DOI] [PubMed] [Google Scholar]
- 28.Oren, Z., J. C. Lerman, G. H. Gudmundsson, B. Agerberth, and Y. Shai. 1999. Structure and organization of the human antimicrobial peptide LL-37 in phospholipid membranes: relevance to the molecular basis for its non-cell-selective activity. Biochem. J. 341:501-513. [PMC free article] [PubMed] [Google Scholar]
- 29.Oren, Z., and Y. Shai. 2000. Cyclization of a cytolytic amphipathic alpha-helical peptide and its diastereomer: effect on structure, interaction with model membranes, and biological function. Biochemistry 39:6103-6114. [DOI] [PubMed] [Google Scholar]
- 30.Otvos, L., Jr., I. O, M. E. Rogers, P. J. Consolvo, B. A. Condie, S. Lovas, P. Bulet, and M. Blaszczyk-Thurin. 2000. Interaction between heat shock proteins and antimicrobial peptides. Biochemistry 39:14150-14159. [DOI] [PubMed] [Google Scholar]
- 31.Papo, N., and Y. Shai. 2003. Can we predict biological activity of antimicrobial peptides from their interactions with model phospholipid membranes? Peptides 24:1693-1703. [DOI] [PubMed] [Google Scholar]
- 32.Patrzykat, A., C. L. Friedrich, L. Zhang, V. Mendoza, and R. E. Hancock. 2002. Sublethal concentrations of pleurocidin-derived antimicrobial peptides inhibit macromolecular synthesis in Escherichia coli. Antimicrob. Agents Chemother. 46:605-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peschel, A., M. Otto, R. W. Jack, H. Kalbacher, G. Jung, and F. Gotz. 1999. Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J. Biol. Chem. 274:8405-8410. [DOI] [PubMed] [Google Scholar]
- 34.Pollock, W. K., T. J. Rink, and R. F. Irvine. 1986. Liberation of [3H]arachidonic acid and changes in cytosolic free calcium in fura-2-loaded human platelets stimulated by ionomycin and collagen. Biochem. J. 235:869-877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rinaldi, A. C., M. L. Mangoni, A. Rufo, C. Luzi, D. Barra, H. Zhao, P. K. Kinnunen, A. Bozzi, A. Di Giulio, and M. Simmaco. 2002. Temporin L: antimicrobial, haemolytic and cytotoxic activities, and effects on membrane permeabilization in lipid vesicles. Biochem. J. 368:91-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sal-Man, N., Z. Oren, and Y. Shai. 2002. Preassembly of membrane-active peptides is an important factor in their selectivity toward target cells. Biochemistry 41:11921-11930. [DOI] [PubMed] [Google Scholar]
- 37.Savage, P. B., C. Li, U. Taotafa, B. Ding, and Q. Guan. 2002. Antibacterial properties of cationic steroid antibiotics. FEMS Microbiol. Lett. 217:1-7. [DOI] [PubMed] [Google Scholar]
- 38.Scott, M. G., M. R. Gold, and R. E. Hancock. 1999. Interaction of cationic peptides with lipoteichoic acid and gram-positive bacteria. Infect. Immun. 67:6445-6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sleep, J., D. Wilson, R. Simmons, and W. Gratzer. 1999. Elasticity of the red cell membrane and its relation to hemolytic disorders: an optical tweezers study. Biophys. J. 77:3085-3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tao, R., R. J. Jurevic, K. K. Coulton, M. T. Tsutsui, M. C. Roberts, J. R. Kimball, N. Wells, J. Berndt, and B. A. Dale. 2005. Salivary antimicrobial peptide expression and dental caries experience in children. Antimicrob. Agents Chemother. 49:3883-3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tjabringa, G. S., J. Aarbiou, D. K. Ninaber, J. W. Drijfhout, O. E. Sorensen, N. Borregaard, K. F. Rabe, and P. S. Hiemstra. 2003. The antimicrobial peptide LL-37 activates innate immunity at the airway epithelial surface by transactivation of the epidermal growth factor receptor. J. Immunol. 171:6690-6696. [DOI] [PubMed] [Google Scholar]
- 42.Utsugi, T., A. J. Schroit, J. Connor, C. D. Bucana, and I. J. Fidler. 1991. Elevated expression of phosphatidylserine in the outer membrane leaflet of human tumor cells and recognition by activated human blood monocytes. Cancer Res 51:3062-3066. [PubMed] [Google Scholar]
- 43.Xian, W., R. Vegners, P. A. Janmey, and W. H. Braunlin. 1995. Spectroscopic studies of a phosphoinositide-binding peptide from gelsolin: behavior in solutions of mixed solvent and anionic micelles. Biophys. J. 69:2695-2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zanetti, M., R. Gennaro, M. Scocchi, and B. Skerlavaj. 2000. Structure and biology of cathelicidins. Adv. Exp. Med. Biol. 479:203-218. [DOI] [PubMed] [Google Scholar]
- 45.Zasloff, M. 2002. Antimicrobial peptides of multicellular organisms. Nature 415:389-395. [DOI] [PubMed] [Google Scholar]
- 46.Zhang, L., and T. J. Falla. 2004. Cationic antimicrobial peptides—an update. Exp. Opin. Investig. Drugs 13:97-106. [DOI] [PubMed] [Google Scholar]
- 47.Zhao, H., and P. K. Kinnunen. 2003. Modulation of the activity of secretory phospholipase a(2) by antimicrobial peptides. Antimicrob. Agents Chemother. 47:965-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zwaal, R. F., P. Comfurius, and E. M. Bevers. 2005. Surface exposure of phosphatidylserine in pathological cells. Cell Mol. Life Sci. 62:971-988. [DOI] [PMC free article] [PubMed] [Google Scholar]