Abstract
Low-level vancomycin-resistant Staphylococcus aureus (vancomycin-intermediate S. aureus [VISA] and heterogenous VISA [hVISA]) is increasingly reported and leads to glycopeptide treatment failure. Various phenotypic features have been reported for these isolates, but the genetic changes leading to hVISA and VISA have yet to be clearly determined. We assessed phenotypic, antibiotic resistance, and genomic changes by using genomic DNA microarray comparison and sequencing of selected loci in five pairs of clinical hVISA/VISA strains and the initial methicillin-resistant Staphylococcus aureus (MRSA) isolates obtained prior to vancomycin therapy. The isolates were from adult patients in Australia and New Zealand who had persistent MRSA bacteremia (>7 days) while receiving vancomycin therapy. In all cases, the initial isolates were found to be fully vancomycin-susceptible Staphylococcus aureus (VSSA). The hVISA/VISA phenotype was associated with increased cell wall thickness, reduced autolytic activity in four of five hVISA/VISA strains, and a striking reduction in biofilm formation compared to the parent strains in all pairs. All five pairs appeared to be isogenic, and genomic DNA microarray comparison suggested that major genetic changes are not required for the development of the resistant phenotype in these strains. No sequence differences were found in the agr locus or the tcaRA genes for any pair, but a marked reduction in RNAIII expression was found in four pairs. In summary, hVISA/VISA arises from fully VSSA during persistent infection that fails to respond to glycopeptide therapy and is associated with significant phenotypic changes, including a marked reduction in biofilm-forming ability. These clinically derived pairs of isolates will be a useful resource to elucidate the genetic mechanism of resistance in hVISA/VISA strains.
Low-level vancomycin resistance in Staphylococcus aureus (vancomycin-intermediate S. aureus [VISA] and heterogenous VISA [hVISA]) has become a significant clinical problem in many parts of the world (19, 44, 48). VISA and hVISA strains are associated with serious infections that can lead to glycopeptide treatment failure (5, 14, 22, 29). VISA strains with a vancomycin MIC of 8 μg/ml by broth dilution have been the focus of much of the concern. However, strains of VISA with an MIC of 4 μg/ml (according to new CLSI definitions [6]) and hVISA (vancomycin MIC, ≤2 μg/ml; but with a resistant subpopulation able to grow at higher vancomycin concentrations) now appear very frequently (48). These strains that develop during continued glycopeptide exposure are thought to be precursors of VISA with a vancomycin MIC of 8 μg/ml (20, 29, 33, 34, 42, 43). A complete understanding of the mechanisms and factors leading to resistance remains elusive. Resistance may develop by different pathways (48), but a consistent observation appears to be a thickened cell wall with reduced peptidoglycan cross-linking leading to cell wall “clogging” with vancomycin (8, 10, 48).
A marked reduction in autolytic activity and reduced cell wall turnover have been found in hVISA/VISA strains from the United States (3, 33, 41). Similarly, reduced whole-cell autolytic activity was recently confirmed in the Japanese VISA strain Mu50 (45) after initial reports suggesting increased cell wall autolytic activity (15). Studies assessing the genetic changes in VISA isolates have demonstrated a number of metabolic pathways and regulatory genes that may contribute to resistance (9, 25, 28, 38, 48). In particular, the agr two-component regulatory system has been linked to low-level vancomycin resistance in S. aureus, with reports suggesting that agr type II strains and loss of agr function are associated with VISA (37, 38, 39). Loss of agr function could have significant effects on exoprotein and adhesion expression and could potentially promote biofilm formation (31, 47). It has previously been noted that many reported VISA infections have involved biomedical devices (37), and biofilm formation on these devices could be an important initial step in the pathway to vancomycin resistance. The tcaRAB operon has also been linked to glycopeptide resistance in S. aureus. In particular, inactivation of tcaA is associated with increased teicoplanin resistance (4, 27). Two clinical VISA strains with truncated tcaA genes have been described previously (27); however, further analysis revealed that the tcaA sequences of the resistant strains were identical to those of vancomycin-susceptible strains N315 and COL (50). Nonsense mutations and frameshift mutations of agr have also previously been described for some hVISA and VISA strains (37).
We have previously noted that the hVISA/VISA phenotype was detected in the laboratory only after many days of failed vancomycin therapy in most patients (5, 22). We were able to retrieve the initial blood culture or clinical isolates obtained prior to the initiation of vancomycin therapy in a number of patients from these studies. These isolates were all from patients in Australia and New Zealand, a region geographically distinct from the other parts of the world where work to describe and understand the resistant phenotype has been done. In this study, we characterized five clinical pairs of vancomycin-susceptible S. aureus (VSSA) and hVISA/VISA (vancomycin MICs, 2 to 4 μg/ml) isolates obtained before and after failed vancomycin therapy to better understand the changes associated with this more common level of vancomycin resistance.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Staphylococcus aureus strains are listed in Table 1. The five pairs of clinical strains were isolates from patients with persistent methicillin-resistant S. aureus bacteremia (persistently blood culture positive after >7 days of vancomycin therapy). The initial isolates from each strain pair were recovered prior to the commencement of vancomycin therapy in all patients. Vancomycin was the only glycopeptide used to treat these patients, with serum levels monitored in all patients. The serum trough levels were generally targeted at 15 μg/ml. Strains were stored in glycerol broth at −80°C and subcultured twice onto Columbia blood agar (Oxoid) for 48 h before being used for any experiment. Unless otherwise indicated, all isolates were grown in brain heart infusion broth (BHIB; Oxoid).
TABLE 1.
Study isolates, susceptibilities, clinical features, and typing results
| Isolate | Origina | Infection type (days of bacteremia on vancomycin) | Source | Phenotypeb | Vancomycin exposure (days)c | Avg vancomycin trough level (μg/ml) (range) | MIC (μg/ml) ford:
|
spa type | Source/reference and comments | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| VCM | TEIC | OXA | |||||||||
| Pair 1 | |||||||||||
| JKD6000 | MEL | Endocarditis (13) | BCf | VSSA | 13 | 15.0 (4.0-26.0) | 2.0 | 0.5 | >256 | Type 3 | This study |
| JKD6001 | BC | VISA | 4.0 | 8.0 | >256 | Type 3 | 22 | ||||
| Pair 2 | |||||||||||
| JKD6009 | NZ | Endocarditis (8) | Wound | VSSA | 42 | 11.6 (10.8-12.3) | 1.0 | 0.5 | >256 | Type 3 | This study |
| JKD6008 | BC | VISA | 4.0 | 2.0 | >256 | Type 3 | 22 | ||||
| Pair 3 | |||||||||||
| JKD6021 | MEL | Liver abscess (15) | BC | VSSA | 15 | 5.4 (5.0-5.8) | 1.0 | 0.25 | >256 | Type 3 | This study |
| JKD6023 | BC | VISA | 4.0 | 8.0 | >256 | Type 3 | 22 | ||||
| Pair 4 | |||||||||||
| JKD6052 | BRIS | Endocarditis (32) | BC | VSSA | 32 | NAg (>10.0) | 1.0 | 0.5 | >256 | Type 3 | This study |
| JKD6051 | BC | hVISA | 2.0 | 4.0 | >256 | Type 3 | This study; previous VCM MIC, 4 μg/ml | ||||
| Pair 5 | |||||||||||
| JKD6004 | BRIS | PPMe infection (8) | BC | VSSA | 8 | 16.1 (5.3-30.5) | 1.0 | 0.5 | >256 | Type 574 | This study |
| JKD6005 | BC | hVISA | 2.0 | 4.0 | >256 | Type 574 | 22; previous VCM MIC, 4 μg/ml | ||||
| ATCC 25923 | VSSA | 1.0 | 0.5 | 2 | |||||||
| Mu3 (ATCC 700698) | hVISA | 2.0 | 4.0 | 15 | |||||||
MEL, Melbourne, Australia; BRIS, Brisbane, Australia; NZ, New Zealand.
Defined by population analysis profile (see Materials and Methods).
Number of days of vancomycin exposure between the first and last isolates in each pair.
VCM, vancomycin; TEIC, teicoplanin; OXA, oxacillin.
PPM, permanent pacemaker.
BC, blood culture.
NA, incomplete data available; however, all vancomycin serum levels were above 10 μg/ml.
Antibiotic susceptibility testing.
Vancomycin and teicoplanin MICs were determined by broth microdilution in Mueller-Hinton broth and read at 24 h according to CLSI (formerly NCCLS) criteria (6). Using the new criteria, S. aureus strains with a vancomycin MIC of 4 to 8 μg/ml were defined as VISA. The oxacillin MIC was determined by Etest (AB Biodisk) according to the manufacturer's instructions.
Population analysis.
A vancomycin population analysis profile (PAP) was performed by serial dilution of an overnight BHIB culture and inoculation of BHI agar containing 0 to 8 μg/ml vancomycin. Colonies were counted after incubation for 48 h in air at 35°C and plotted on a graph of the number of CFU/ml versus vancomycin concentration. The hVISA strain Mu3 (ATCC 700698) and the vancomycin-susceptible S. aureus strain ATCC 25923 were tested in parallel as positive and negative controls, respectively. An isolate was defined as hVISA if the vancomycin MIC was ≤2 μg/ml and the PAP area under the curve (AUC) of the test strain to Mu3 was ≥0.9 (49).
Molecular typing.
For pulsed-field gel electrophoresis, agarose plugs of genomic DNA were prepared, digested with SmaI, and then subjected to electrophoresis as described previously (30). Images were assessed visually to determine the number of bands of difference between the VSSA and hVISA/VISA strains from each pair. Isolates were considered clonal if there were ≤3 bands of difference. Amplification and sequencing of the protein A gene repeat region (spa typing) were performed using primers 1095F and 1517R as previously described (16). The spa sequences were analyzed using eGenomics software to define the spa type (24). The agr type of each strain was determined after sequencing the whole agr locus (see below) and defined by performing a BLAST search using the hypervariable region of the sequence (3′ end of agrB and agrD and 5′ end of agrC) (51) and matching the region to known agr type strains.
DNA techniques.
Genomic DNA was extracted using the GenElute bacterial genomic DNA kit (Sigma) according to the manufacturer's instructions. PCR amplification of DNA was carried out using Taq DNA polymerase (Roche Molecular Biochemicals). DNA sequencing was performed using BigDye Terminator version 3.1 cycle sequencing kits (Applied Biosystems), and the reaction mixtures were analyzed with the 3730 DNA analyzer (Applied Biosystems). A PCR for enterococcal vanA and vanB genes was performed using previously described primers (1). Enterococcus faecalis vanB strain ATCC 51299 and an Enterococcus faecium vanA clinical isolate were used as positive controls, and vancomycin-susceptible E. faecalis strain ATCC 29212 was used as a negative control. Primers for amplifying and sequencing the whole agr locus were agr1 and agr2 as previously described (35). Primers for the tcaRA genes were designed using the methicillin-resistant S. aureus (MRSA) COL strain genome sequence (GenBank accession number CP000046). They were tcaRA1 (5′-CAATCCCTTCAAAGTAATTCACA-3′) and tcaRA2 (5′ TGCGATACAATGATTGCTGAG-3′).
Electron microscopy.
Staphylococcus aureus strains were subcultured onto Columbia blood agar plates and grown at 37°C for 24 h. A sample of each strain was resuspended in phosphate-buffered saline, pelleted by centrifugation, and resuspended in fixative as previously described (12). Briefly, prefixation was in 75 mM l-lysine (L-5626; Sigma) in 0.075% ruthenium red, 2% paraformaldehyde, and 2.5% glutaraldehyde in 0.1 M cacodylate buffer for 20 min. This was followed by fixation in the above-described solution, excluding the lysine, for 2 h. From this point, cells were then handled as pellets. After three 10-min washes in cacodylate buffer, the cells were postfixed in 1% OsO4 for 2 h, followed again by three 10-min cacodylate buffer washes. Dehydration was through a graded acetone series, followed by infiltration and embedding in Epon-araldite epoxy resin. Thin sections were cut and stained with uranyl acetate and lead citrate before being viewed with a Philips CM10 electron microscope at 60 kV. For cell wall thickness analysis, 100 measurements of equatorially cut cells were recorded for each strain and expressed as mean and 95% confidence interval, with the operator blinded with respect to the resistance status of the strain. The statistical significance of the data was evaluated by Student's t test using GraphPad Prism 4.0 (GraphPad Software Inc., San Diego, CA).
Autolytic assay.
Triton X-100-induced autolysis was measured as previously described, with some modifications (40). Briefly, after overnight growth in BHIB, 500 μl was added to 50 ml BHIB and grown to an optical density at 600 nm (OD600) of ∼0.5. Twenty milliliters of culture was rapidly chilled, then washed in ice-cold distilled water, and resuspended in 50 mM glycine-0.01% Triton X-100 (pH 8.0) to an OD600 of ∼1.0. Samples were incubated at 37°C with gentle agitation, and the OD600 was measured hourly using a 552 UV-VIS spectrophotometer (PerkinElmer).
Genomic DNA microarrays.
Genomic DNA arrays were performed to compare the VSSA and hVISA/VISA strains from each clinical pair. Two micrograms of AluI-digested genomic DNA from each isolate was used in a random primer/reaction buffer mix (125 mM Tris, pH 7.0, 750 ng/μl random primers, 25 mM mercaptoethanol, 12.5 mM MgCl2), and the sample was boiled for 5 min and then placed on ice. Five microliters of deoxynucleoside triphosphate mix (1.2 mM each of dATP, dGTP, and dCTP and 0.6 mM dTTP), 3 μl of Cy3- or Cy5-labeled dUTP (1 mM) (Amersham Bioscience), and 1 μl of high-concentration Klenow fragment (50 units) (Biolabs) were added. The mixture was incubated at 37°C for 2 h, and the reaction was stopped by the addition of 5 μl 0.5 M EDTA (pH 8.0). The cDNA probe was purified on a Microcon-30 filter (Millipore). The Cy3 and Cy5 probes were combined and then dried to completion in a Speed Vac.
Version 2 Staphylococcus aureus microarray slides (The Institute for Genomic Research, Maryland) were used. These amplicon-based arrays contain 2,592 open reading frames from S. aureus strain COL and 617 unique open reading frames from strains Mu50, MW2, and N315, all in triplicate.
Thirty microliters of hybridization mixture containing Cy3- and Cy5-labeled probes, 15 μl formamide (Sigma), 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.1% sodium dodecyl sulfate, and 22.5 μg herring sperm DNA (Promega) was denatured at 95°C, applied to the microarray slide, and incubated overnight at 42°C.
After being washed, the microarrays were scanned with a GMS418 array scanner (Genetic MicroSystems, California) and signal intensity was analyzed with ImaGene version 5.1 (Biodiscovery, Louisiana). The data were normalized and analyzed using BioArray Software Environment (available at http://base.thep.lu.se) (36). The data from spots where there was a signal intensity of less than 250 in both channels were filtered out. The data were then subjected to a global median ratio and Lowess transformation. The ratio (n-fold) of the background-corrected mean signal intensities for each gene identification remaining in the data set was calculated. The Wilcoxon signed-rank t test was used to assess the probability that the background-corrected mean signal intensities differed between the hVISA/VISA and VSSA genomic preparations. A cutoff of ≥3-fold change and a P value of <0.05 were considered significant and used to assess genomic changes between the pairs.
Preparation of total RNA.
Exponential-phase culture (20 ml) was added to 10 ml RNAlater RNA stabilization reagent (QIAGEN) and allowed to stand for 10 min. RNA was isolated using the RNeasy midi kit (QIAGEN) with on-column DNase I digestion (QIAGEN). The quality and quantity of total RNA were confirmed by agarose gel electrophoresis and spectrophotometry.
Quantitative real-time PCR for RNAIII expression.
After a second DNase I treatment, cDNA was synthesized using SuperScript II RNase H reverse transcriptase (Invitrogen) at 42°C with 5 μg of RNA. mRNA levels for RNAIII and 16S rRNA were determined using previously described primers and cycling conditions (35). Each assay mixture contained 10 μl of SYBR green PCR master mix (Applied Biosystems), 1.6 μl of each primer (final concentration, 0.2 μM), and 2.4 μl of template. Fluorescence emission was detected with an ABI 7700 sequence detection system (Applied Biosystems), and RNAIII expression was normalized on the basis of 16S rRNA levels as previously described (35). Each assay was performed in triplicate and repeated twice.
Biofilm assay.
The ability of strains to adhere to a polystyrene microtiter plate was assessed as previously described, with some modifications (18). After overnight growth in 10 ml BHIB with aeration, the culture was diluted 1:200 in BHIB (with no additives) and then 200 μl was used to inoculate a well of a 96-well polystyrene microtiter plate (Sarstedt), with eight replicates per experiment. The starting inoculum was calculated for all isolates and was the same (∼107 CFU/ml). The tray was incubated at 37°C with shaking for 24 h. Unbound cells were removed by inversion of the microtiter plate and tapping on absorbent paper. The adherent cells were stained with 200 μl of 0.1% crystal violet, and excess stain was removed by multiple washes with phosphate-buffered saline. After being dried, the crystal violet was solubilized by adding 200 μl of ethanol-acetone (80:20, wt/wt) and the absorbance at 570 nm was measured with an enzyme-linked immunosorbent assay reader. Each assay was performed five times.
Microarray data accession number.
The microarray data have been deposited in GEO with the series accession number GSE5047.
RESULTS
Isolate details.
The five pairs of clinical strains (Table 1) were isolated from patients with methicillin-resistant S. aureus infections who had persistent bacteremia. The duration of bacteremia was between 8 and 32 days while receiving vancomycin. All of the initial isolates were obtained prior to the commencement of vancomycin therapy, and the isolates were obtained from two states of Australia and also from New Zealand. All isolates were blood culture isolates except JKD6009, which was a swab isolate from an infected wound which was the source of subsequent bacteremia. The wound isolate was used because all other available isolates for this patient were obtained after the commencement of vancomycin therapy.
Antibiotic susceptibility results.
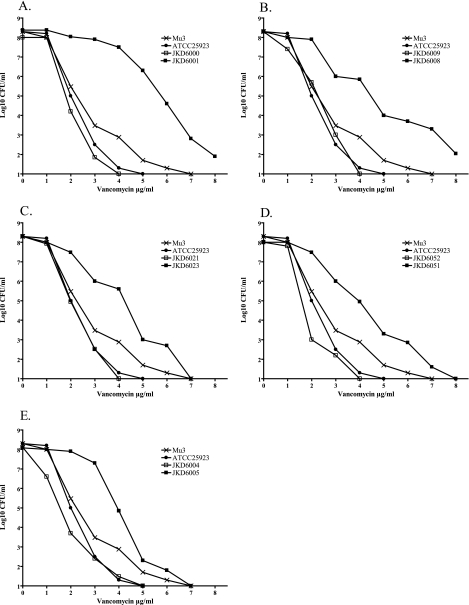
PAP confirmed that the initial isolates from all five pairs were VSSA (vancomycin PAP AUC ratio, <0.9 compared to that of Mu3), while the later isolates were either hVISA (n = 2; vancomycin PAP AUC ratio, >0.9 compared to that of Mu3) or true VISA (n = 3; vancomycin microbroth MIC, 4.0 μg/ml) (Fig. 1). The vancomycin and teicoplanin MICs increased in all five pairs between the initial and later isolates. Of note, the two hVISA isolates with a vancomycin MIC of 2 μg/ml had previously been documented to have an MIC of 4 μg/ml.
FIG. 1.
Vancomycin population analysis profiles of initial patient isolates prior to vancomycin therapy (JKD6000, JKD6009, JKD6021, JKD6052, and JKD6004) and later blood culture isolates after persistent bacteremia despite vancomycin therapy (JKD6001, JKD6008, JKD6023, JKD6051, and JKD6005). Also included are the hVISA reference strain Mu3 (ATCC 700698) and VSSA strain ATCC 25923.
Molecular typing.
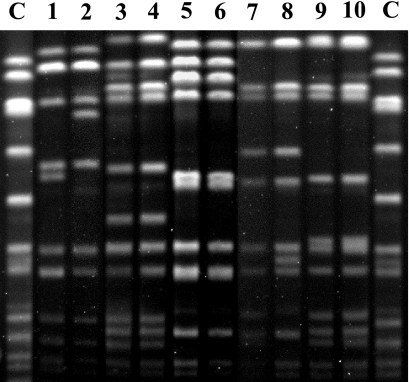
Pulsed-field gel electrophoresis revealed identical banding patterns for three of the isolate pairs (Fig. 2), while a two-band difference was noted for the other pairs, indicating that the hVISA/VISA strains were clonally related to the initial VSSA isolates in all patients. The spa typing confirmed these results and identified identical spa types for the VSSA and hVISA isolates from all pairs. Of note, four of the pairs had the same spa type (type 3) while the last pair (JKD6004 and JKD6005) had a previously unrecognized spa type (type 574). Comparison of the sequence of the hypervariable region of the agr locus for all 10 clinical isolates revealed a 99% match to that of the same region of the MRSA COL genome, which contains a type I agr locus (51). All 10 isolates were therefore agr type I strains.
FIG. 2.
Pulsed-field gel electrophoresis of SmaI-restricted genomic DNA for five VSSA and hVISA/VISA pairs. Banding patterns are identical for strains JKD6004 and JKD6005 (lanes 3 and 4), JKD6008 and JKD6009 (lanes 5 and 6), and JKD6051 and JKD6052 (lanes 9 and 10). A two-band difference was noted for strain pairs JKD6000 and JKD6001 (lanes 1 and 2) and JKD6021 and JKD6023 (lanes 7 and 8). C, control strain (NCTC8325).
Electron microscopy.
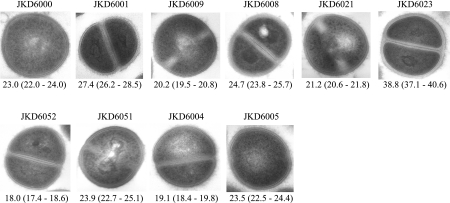
Electron microscopy was performed to compare the cell wall thicknesses of each pair of strains. All hVISA/VISA isolates had a thicker cell wall than the parent strains (Fig. 3). Student's t test demonstrated that the increase in cell wall thickness was statistically significant in all cases (P < 0.001).
FIG. 3.
Transmission electron microscopy of parent and hVISA/VISA strains. Magnification, ×30,000. The values given under each image are the mean and 95% confidence interval of the cell wall thickness of the cells in nanometers.
Autolytic assay.
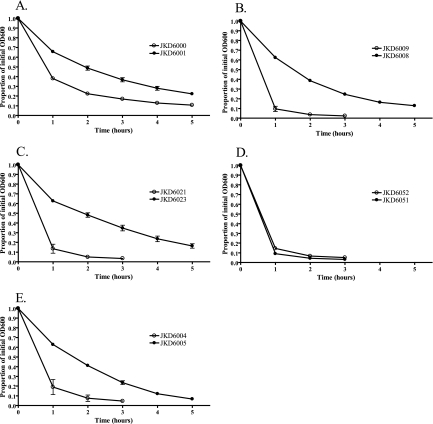
Previous studies have demonstrated various results for autolytic activity in VISA strains, although reduced autolysis is more common. We therefore tested our strains for autolytic activity compared to that of the parent strain. Results of the autolytic analysis revealed a reduction in autolytic activity for all hVISA/VISA isolates compared to that of the parent VSSA strains, except for one pair (JKD6051 and JKD6052) (Fig. 4). No difference was observed between the autolytic rates for this last pair.
FIG. 4.
Autolytic assay results for five VSSA and hVISA/VISA pairs. The results are expressed as the averages of two independent experiments.
Analysis of genetic determinants of resistance.
To exclude acquisition of vanA or vanB as the mechanism of resistance, a PCR for these resistance genes was performed. PCR for vanA and vanB genes was negative in all hVISA/VISA isolates. Because mutations in the agr locus have previously been described for VISA strains, we compared the sequence of the whole agr locus (∼3,500 bp) for each hVISA/VISA isolate to that of the parent strain. No difference was found between the VSSA and hVISA/VISA isolates for each pair, with only one or two base pair differences found compared to the MRSA COL agr region, none of which were predicted to affect function. In addition, no sequence change was detected in the tcaR and tcaA genes in any of the isolate pairs.
DNA genomic microarray comparison.
No genomic differences were detected using genomic DNA microarray comparison between the initial VSSA and hVISA/VISA isolates for each pair, suggesting that the pairs were isogenic and that major genomic changes, such as deletion of one or more genes, had not occurred during the development of the resistant phenotype.
Reduced expression of the agr transcript RNAIII in hVISA/VISA strains.
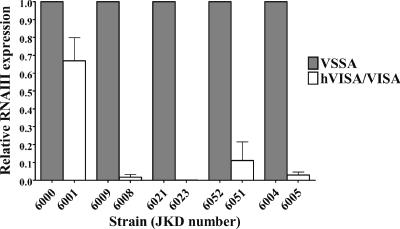
Although no mutations were detected in the agr loci of the hVISA/VISA strains, we assessed agr expression and found a marked reduction in RNAIII expression in four hVISA/VISA strains compared to that of the parent, vancomycin-susceptible strains (Fig. 5).
FIG. 5.
Relative RNAIII expression of hVISA/VISA strains compared to that of the parent strains, determined by quantitative real-time PCR and normalized to 16S rRNA expression.
Biofilm assay.
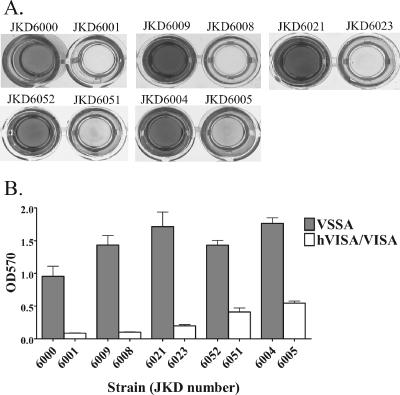
Because hVISA/VISA commonly develops in patients with prosthetic devices, we tested the hypothesis that clinical hVISA/VISA isolates would have an increased propensity for biofilm formation. Analysis of the ability of strains to adhere to a 96-well polystyrene microtiter plate revealed a marked difference between the VSSA and hVISA/VISA strains for all pairs (Fig. 6); however, the adherence ability was significantly reduced in all hVISA/VISA strains compared to that of the parent VSSA strains.
FIG. 6.
(A) Analysis of biofilm formation by five VSSA and hVISA/VISA pairs, using a 24-h polystyrene microtiter plate adherence assay. (B) Quantification of biofilm formation determined by crystal violet staining and reading with an enzyme-linked immunosorbent assay plate reader (OD570). Results are expressed as the averages of five independent experiments.
DISCUSSION
Infections caused by strains of hVISA/VISA are increasingly being reported worldwide and are often associated with glycopeptide treatment failure, but a better understanding of the clinical correlates of hVISA/VISA infection are limited by problems with definitions and laboratory detection (21, 26, 48). Understanding how hVISA and, subsequently, VISA develops may lead to more-rapid diagnostic tests and methods to prevent further emergence of resistance. We assessed phenotypic and genotypic changes in clinically derived pairs of VSSA and hVISA/VISA strains isolated from a region of the world geographically removed from regions where previous studies have been done.
We have confirmed that isolates of S. aureus with reduced glycopeptide susceptibility develop during failed therapy and arise from fully susceptible VSSA isolates. Molecular typing and genomic DNA microarray analysis suggested that the isolate pairs were isogenic. This has important implications for laboratory testing of clinical isolates for hVISA, as a negative hVISA test early in a clinical infection does not exclude the later development of hVISA/VISA, and repeat testing of clinical isolates should be performed for patients who are failing glycopeptide therapy. It is interesting that none of the patients developed VISA with a higher vancomycin MIC (8 μg/ml) despite up to 32 days of persistent bacteremia with ongoing vancomycin therapy. It may be that some clones of S. aureus have a limit to the level of resistance that can be generated during a clinical infection.
Although our isolates come from Australia and New Zealand, a number of the findings were consistent with previous reports. In particular, a significant increase in cell wall thickness was demonstrated in all hVISA/VISA strains, supporting the hypothesis that cell wall thickening and vancomycin “clogging” are the final phenotype responsible for resistance even in geographically diverse regions (8). Also, a reduction in autolytic activity was observed in four of the five hVISA/VISA strains, a feature repeatedly reported from the United States (3, 33) and recently confirmed in the Japanese VISA strain Mu50 (45). One of the hVISA strains did not demonstrate a reduction in autolytic activity compared to the VSSA parent strain, and it appears that reduced autolytic activity is not an absolute requirement for strains of S. aureus that are hVISA. This may reflect the possible heterogeneous nature of the changes leading to low-level glycopeptide resistance in S. aureus, or alternatively, reduced autolytic activity may be one result of the multiple selective pressures on the organism and could therefore be independent of the glycopeptide resistance. The agr typing results again indicate that a number of agr type strains can be associated with hVISA. Previous reports suggested an overrepresentation of agr II in hVISA and VISA strains (37), but our data and those of others have demonstrated that agr type I strains appear to be commonly associated with clinical hVISA generation (23, 46).
The enterococcal vancomycin resistance genes vanA and vanB were not detected in the hVISA/VISA strains in this study. It was important to exclude vanB as a possible cause of low-level vancomycin resistance in S. aureus in our region, where vanB vancomycin-resistant enterococci are much more common than vanA (1, 32). Given the incomplete expression of the vanA resistance determinant in S. aureus (44), the level of resistance mediated by vanB in S. aureus, if it were to occur, could be low. We chose to sequence the whole agr locus because of reports linking changes in expression or documented mutations or insertions with hVISA or VISA (37) but were unable to demonstrate any changes in our strains. However, using quantitative real-time PCR, a significant decrease in RNAIII expression was demonstrated in the hVISA/VISA strain in four pairs, with a minor relative reduction in the fifth pair, reinforcing the potential importance of the agr locus in the expression of low-level vancomycin resistance. As noted by Renzoni et al. (35), the marked reduction in agr transcripts without mutations in the agr locus itself suggests that upstream regulators that repress agr expression have an effect in these strains.
Some studies have linked the tcaRAB locus with teicoplanin resistance in S. aureus (4). Despite an increase in teicoplanin resistance, no changes in tcaA or tcaR gene sequences were detected in any of our strains. Also, using genomic microarray analysis, we did not demonstrate any significant changes between isolate pairs. The genetic changes leading to resistance in our strains are clearly associated with changes that do not lead to differences on the genomic array (such as single-base mutations) or, less likely, acquisition or deletion of genes which are not present on the arrays. These data support the notion that multiple genetic pathways may lead to a common phenotypic end point, given that our strains demonstrate similar phenotypes but not some of the reported genetic changes associated with reduced glycopeptide susceptibility in S. aureus. Elucidation of the molecular mechanisms of resistance in our strains will require further study.
The biofilm assay was performed in this study because many patients with hVISA or VISA infections have infection associated with prosthetic materials (22, 37), and biofilm formation plays an important role in the pathogenesis of these infections (11). In addition, the loss of agr function, as was demonstrated in our strains, has also been associated in vitro with increased propensity for biofilm formation (47). We were surprised to find a marked and consistent reduction in the biofilm-forming ability of clinical hVISA/VISA strains compared to that of their parent VSSA strain. To our knowledge, biofilm assays have not previously been performed with clinical hVISA or VISA strains. Sakoulas et al. tested the biofilm-forming ability of a laboratory-induced hVISA strain and demonstrated increased biofilm formation compared to that of the parent strain by using a method similar to that described here (37). Our results suggest that the findings of increased biofilm formation by laboratory-derived strains of hVISA or VISA may not be relevant to clinical strains. There are two important steps in biofilm formation: (i) early attachment of the bacterial cells to a surface, mediated by surface proteins, including microbial surface components that recognize adhesive matrix molecules (7, 13), and (ii) accumulation of bacterial cell clusters in layers, which involves intercellular adhesion (7, 17). It seems plausible that the significant cell wall changes occurring in strains of hVISA/VISA could limit biofilm formation by interfering with initial attachment or later intercellular adhesion. Further work is required to understand the mechanisms of altered biofilm formation in our strains.
In summary, hVISA and VISA emerge during failed glycopeptide therapy in patients with serious MRSA infections. Many of the phenotypic changes are consistent with previous reports; however, reduced autolytic activity is not essential for the expression of low-level vancomycin resistance in S. aureus. The potential importance of reduced agr expression in hVISA/VISA is also supported by our results. In contrast to previous reports, we demonstrated a significant reduction in the biofilm-forming ability of hVISA/VISA strains. These pairs of clinically derived, isogenic strains of VSSA and hVISA/VISA will be a valuable resource for understanding the mechanisms of low-level glycopeptide resistance in S. aureus.
Acknowledgments
Benjamin P. Howden was supported by a Postgraduate Medical and Dental Scholarship from the National Health and Medical Research Council, Australia. This work was supported by the Australian Bacterial Pathogenesis Program from the National Health and Medical Research Council, Australia.
Staphylococcus aureus microarray slides were kindly supplied by The Institute for Genomic Research (TIGR), Rockville, MD. We thank Vicki Bennett-Wood, University of Melbourne, for performing the electron microscopy.
No author has any conflict of interest related to the manuscript.
REFERENCES
- 1.Bell, J. M., J. C. Paton, and J. Turnidge. 1998. Emergence of vancomycin-resistant enterococci in Australia: phenotypic and genotypic characteristics of isolates. J. Clin. Microbiol. 36:2187-2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boyle, V. J., M. E. Fancher, and R. W. Ross, Jr. 1973. Rapid, modified Kirby-Bauer susceptibility test with single, high-concentration antimicrobial disks. Antimicrob. Agents Chemother. 3:418-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyle-Vavra, S., M. Challapalli, and R. S. Daum. 2003. Resistance to autolysis in vancomycin-selected Staphylococcus aureus isolates precedes vancomycin-intermediate resistance. Antimicrob. Agents Chemother. 47:2036-2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brandenberger, M., M. Tschierske, P. Giachino, A. Wada, and B. Berger-Bachi. 2000. Inactivation of a novel three-cistronic operon tcaR-tcaA-tcaB increases teicoplanin resistance in Staphylococcus aureus. Biochim. Biophys. Acta 1523:135-139. [DOI] [PubMed] [Google Scholar]
- 5.Charles, P. G., P. B. Ward, P. D. Johnson, B. P. Howden, and M. L. Grayson. 2004. Clinical features associated with bacteremia due to heterogeneous vancomycin-intermediate Staphylococcus aureus. Clin. Infect. Dis. 38:448-451. [DOI] [PubMed] [Google Scholar]
- 6.Clinical and Laboratory Standards Institute. 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 7th ed. Approved standard. CLSI document M7-A7. Clinical and Laboratory Standards Institute, Wayne, Pa.
- 7.Cucarella, C., C. Solano, J. Valle, B. Amorena, I. Lasa, and J. R. Penades. 2001. Bap, a Staphylococcus aureus surface protein involved in biofilm formation. J. Bacteriol. 183:2888-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cui, L., A. Iwamoto, J. Q. Lian, H. M. Neoh, T. Maruyama, Y. Horikawa, and K. Hiramatsu. 2006. Novel mechanism of antibiotic resistance originating in vancomycin-intermediate Staphylococcus aureus. Antimicrob. Agents Chemother. 50:428-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cui, L., J. Q. Lian, H. M. Neoh, E. Reyes, and K. Hiramatsu. 2005. DNA microarray-based identification of genes associated with glycopeptide resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 49:3404-3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cui, L., X. Ma, K. Sato, K. Okuma, F. C. Tenover, E. M. Mamizuka, C. G. Gemmell, M. N. Kim, M. C. Ploy, N. El-Solh, V. Ferraz, and K. Hiramatsu. 2003. Cell wall thickening is a common feature of vancomycin resistance in Staphylococcus aureus. J. Clin. Microbiol. 41:5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donlan, R. M., and J. W. Costerton. 2002. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15:167-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fassel, T. A., P. E. Mozdziak, J. R. Sanger, and C. E. Edmiston. 1997. Paraformaldehyde effect on ruthenium red and lysine preservation and staining of the staphylococcal glycocalyx. Microsc. Res. Tech. 36:422-427. [DOI] [PubMed] [Google Scholar]
- 13.Foster, T. J., and M. Hook. 1998. Surface protein adhesins of Staphylococcus aureus. Trends Microbiol. 6:484-488. [DOI] [PubMed] [Google Scholar]
- 14.Fridkin, S. K., J. Hageman, L. K. McDougal, J. Mohammed, W. R. Jarvis, T. M. Perl, F. C. Tenover, and the Vancomycin-Intermediate Staphylococcus aureus Epidemiology Study Group. 2003. Epidemiological and microbiological characterization of infections caused by Staphylococcus aureus with reduced susceptibility to vancomycin, United States, 1997-2001. Clin. Infect. Dis. 36:429-439. [DOI] [PubMed] [Google Scholar]
- 15.Hanaki, H., K. Kuwahara-Arai, S. Boyle-Vavra, R. S. Daum, H. Labischinski, and K. Hiramatsu. 1998. Activated cell-wall synthesis is associated with vancomycin resistance in methicillin-resistant Staphylococcus aureus clinical strains Mu3 and Mu50. J. Antimicrob. Chemother. 42:199-209. [DOI] [PubMed] [Google Scholar]
- 16.Harmsen, D., H. Claus, W. Witte, J. Rothgänger, H. Claus, D. Turnwald, and U. Vogel. 2003. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J. Clin. Microbiol. 41:5442-5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heilmann, C., C. Gerke, F. Perdreau-Remington, and F. Gotz. 1996. Characterization of Tn917 insertion mutants of Staphylococcus epidermidis affected in biofilm formation. Infect. Immun. 64:277-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heilmann, C., O. Schweitzer, C. Gerke, N. Vanittanakom, D. Mack, and F. Gotz. 1996. Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol. Microbiol. 20:1083-1091. [DOI] [PubMed] [Google Scholar]
- 19.Hiramatsu, K. 2001. Vancomycin-resistant Staphylococcus aureus: a new model of antibiotic resistance. Lancet Infect. Dis. 1:147-155. [DOI] [PubMed] [Google Scholar]
- 20.Hiramatsu, K., K. Okuma, X. X. Ma, M. Yamamoto, S. Hori, and M. Kapi. 2002. New trends in Staphylococcus aureus infections: glycopeptide resistance in hospital and methicillin resistance in the community. Curr. Opin. Infect. Dis. 15:407-413. [DOI] [PubMed] [Google Scholar]
- 21.Howden, B. P., P. B. Ward, P. D. Johnson, P. G. Charles, and M. L. Grayson. 2005. Low-level vancomycin resistance in Staphylococcus aureus—an Australian perspective. Eur. J. Clin. Microbiol. Infect. Dis. 24:100-108. [DOI] [PubMed] [Google Scholar]
- 22.Howden, B. P., P. B. Ward, P. G. Charles, T. M. Korman, A. Fuller, P. du Cros, E. A. Grabsch, S. A. Roberts, J. Robson, K. Read, N. Bak, J. Hurley, P. D. Johnson, A. J. Morris, B. C. Mayall, and M. L. Grayson. 2004. Treatment outcomes for serious infections caused by methicillin-resistant Staphylococcus aureus with reduced vancomycin susceptibility. Clin. Infect. Dis. 38:521-528. [DOI] [PubMed] [Google Scholar]
- 23.Howe, R. A., A. Monk, M. Wootton, T. R. Walsh, and M. C. Enright. 2004. Vancomycin susceptibility within methicillin-resistant Staphylococcus aureus lineages. Emerg. Infect. Dis. 10:855-857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koreen, L., S. V. Ramaswamy, E. A. Graviss, S. Naidich, J. M. Musser, and B. N. Kreiswirth. 2004. spa typing method for discriminating among Staphylococcus aureus isolates: implications for use of a single marker to detect genetic micro- and macrovariation. J. Clin. Microbiol. 42:792-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuroda, M., K. Kuwahara-Arai, and K. Hiramatsu. 2000. Identification of the up- and down-regulated genes in vancomycin-resistant Staphylococcus aureus strains Mu3 and Mu50 by cDNA differential hybridisation method. Biochem. Biophys. Res. Commun. 269:485-490. [DOI] [PubMed] [Google Scholar]
- 26.Liu, C., and H. F. Chambers. 2003. Staphylococcus aureus with heterogeneous resistance to vancomycin: epidemiology, clinical significance, and critical assessment of diagnostic methods. Antimicrob. Agents Chemother. 47:3040-3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maki, H., N. McCallum, M. Bischoff, A. Wada, and B. Berger-Bachi. 2004. tcaA inactivation increases glycopeptide resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 48:1953-1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mongodin, E., J. Finan, M. W. Climo, A. Rosato, S. Gill, and G. L. Archer. 2003. Microarray transcription analysis of clinical Staphylococcus aureus isolates resistant to vancomycin. J. Bacteriol. 185:4638-4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moore, M. R., F. Perdreau-Remington, and H. F. Chambers. 2003. Vancomycin treatment failure associated with heterogeneous vancomycin-intermediate Staphylococcus aureus in a patient with endocarditis and in the rabbit model of endocarditis. Antimicrob. Agents Chemother. 47:1262-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murchan, S., M. E. Kaufmann, A. Deplano, R. de Ryck, M. Struelens, C. E. Zinn, V. Fussing, S. Salmenlinna, J. Vuopio-Varkila, N. El Solh, C. Cuny, W. Witte, P. T. Tassios, N. Legakis, W. van Leeuwen, A. van Belkum, A. Vindel, I. Laconcha, J. Garaizar, S. Haeggman, B. Olsson-Liljequist, U. Ransjo, G. Coombes, and B. Cookson. 2003. Harmonization of pulsed-field gel electrophoresis protocols for epidemiological typing of strains of methicillin-resistant Staphylococcus aureus: a single approach developed by consensus in 10 European laboratories and its application for tracing the spread of related strains. J. Clin. Microbiol. 41:1574-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Novick, R. P. 2003. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 48:1429-1449. [DOI] [PubMed] [Google Scholar]
- 32.Padiglione, A. A., R. Wolfe, E. A. Grabsch, D. Olden, S. Pearson, C. Franklin, D. Spelman, B. Mayall, P. D. Johnson, and M. L. Grayson. 2003. Risk factors for new detection of vancomycin-resistant enterococci in acute-care hospitals that employ strict infection control procedures. Antimicrob. Agents Chemother. 47:2492-2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pfeltz, R. F., V. K. Singh, J. L. Schmidt, M. A. Batten, C. S. Baranyk, M. J. Nadakavukaren, R. K. Jayaswal, and B. J. Wilkinson. 2000. Characterization of passage-selected vancomycin-resistant Staphylococcus aureus strains of diverse parental backgrounds. Antimicrob. Agents Chemother. 44:294-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ploy, M. C., C. Grelaud, C. Martin, L. de Lumley, and F. Denis. 1998. First clinical isolate of vancomycin-intermediate Staphylococcus aureus in a French hospital. Lancet 351:1212. [DOI] [PubMed] [Google Scholar]
- 35.Renzoni, A., P. Francois, D. Li, W. L. Kelley, D. P. Lew, P. Vaudaux, and J. Schrenzel. 2004. Modulation of fibronectin adhesins and other virulence factors in a teicoplanin-resistant derivative of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 48:2958-2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saal, L. H., C. Troein, J. Vallon-Christersson, S. Gruvberger, A. Borg, and C. Peterson. 2002. BioArray Software Environment (BASE): a platform for comprehensive management and analysis of microarray data. Genome Biol. 3:1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sakoulas, G., G. M. Eliopoulos, R. C. Moellering, Jr., C. Wennersten, L. Venkataraman, R. P. Novick, and H. S. Gold. 2002. Accessory gene regulator (agr) locus in geographically diverse Staphylococcus aureus isolates with reduced susceptibility to vancomycin. Antimicrob. Agents Chemother. 46:1492-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sakoulas, G., G. M. Eliopoulos, R. C. Moellering, Jr., R. P. Novick, L. Venkataraman, C. Wennersten, P. C. DeGirolami, M. J. Schwaber, and H. S. Gold. 2003. Staphylococcus aureus accessory gene regulator (agr) group II: is there a relationship to the development of intermediate-level glycopeptide resistance? J. Infect. Dis. 187:929-938. [DOI] [PubMed] [Google Scholar]
- 39.Sakoulas, G., G. M. Eliopoulos, V. G. Fowler, Jr., R. C. Moellering, Jr., R. P. Novick, N. Lucindo, M. R. Yeaman, and A. S. Bayer. 2005. Reduced susceptibility of Staphylococcus aureus to vancomycin and platelet microbicidal protein correlates with defective autolysis and loss of accessory gene regulator (agr) function. Antimicrob. Agents Chemother. 49:2687-2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sieradzki, K., and A. Tomasz. 1997. Inhibition of cell wall turnover and autolysis by vancomycin in a highly vancomycin-resistant mutant of Staphylococcus aureus. J. Bacteriol. 179:2557-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sieradzki, K., and A. Tomasz. 2003. Alterations of cell wall structure and metabolism accompany reduced susceptibility to vancomycin in an isogenic series of clinical isolates of Staphylococcus aureus. J. Bacteriol. 185:7103-7110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sieradzki, K., R. B. Roberts, S. W. Haber, and A. Tomasz. 1999. The development of vancomycin resistance in a patient with methicillin-resistant Staphylococcus aureus infection. N. Engl. J. Med. 340:517-523. [DOI] [PubMed] [Google Scholar]
- 43.Sieradzki, K., T. Leski, J. Dick, L. Borio, and A. Tomasz. 2003. Evolution of a vancomycin-intermediate Staphylococcus aureus strain in vivo: multiple changes in the antibiotic resistance phenotypes of a single lineage of methicillin-resistant S. aureus under the impact of antibiotics administered for chemotherapy. J. Clin. Microbiol. 41:1687-1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tenover, F. C., and L. C. McDonald. 2005. Vancomycin-resistant staphylococci and enterococci: epidemiology and control. Curr. Opin. Infect. Dis. 18:300-305. [DOI] [PubMed] [Google Scholar]
- 45.Utaida, S., R. F. Pfeltz, R. K. Jayaswal, and B. J. Wilkinson. 2006. Autolytic properties of glycopeptide-intermediate Staphylococcus aureus Mu50. Antimicrob. Agents Chemother. 50:1541-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Verdier, I., M. E. Reverdy, J. Etienne, G. Lina, M. Bes, and F. Vandenesch. 2004. Staphylococcus aureus isolates with reduced susceptibility to glycopeptides belong to accessory gene regulator group I or II. Antimicrob. Agents Chemother. 48:1024-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vuong, C., H. L. Saenz, F. Gotz, and M. Otto. 2000. Impact of the agr quorum-sensing system on adherence to polystyrene in Staphylococcus aureus. J. Infect. Dis. 182:1688-1693. [DOI] [PubMed] [Google Scholar]
- 48.Walsh, T. R., and R. A. Howe. 2002. The prevalence and mechanisms of vancomycin resistance in Staphylococcus aureus. Annu. Rev. Microbiol. 56:657-675. [DOI] [PubMed] [Google Scholar]
- 49.Wootton, M., R. A. Howe, R. Hillman, T. R. Walsh, P. M. Bennett, and A. P. MacGowan. 2001. A modified population analysis profile (PAP) method to detect hetero-resistance to vancomycin in Staphylococcus aureus in a UK hospital. J. Antimicrob. Chemother. 47:399-403. [DOI] [PubMed] [Google Scholar]
- 50.Wootton, M., A. P. MacGowan, and T. R. Walsh. 2005. Expression of tcaA and mprF and glycopeptide resistance in clinical glycopeptide-intermediate Staphylococcus aureus (GISA) and heteroGISA strains. Biochim. Biophys. Acta 1726:326-327. [DOI] [PubMed] [Google Scholar]
- 51.Wright, J. S., III, K. E. Traber, R. Corrigan, S. A. Benson, J. M. Musser, and R. P. Novick. 2005. The agr radiation: an early event in the evolution of staphylococci. J. Bacteriol. 187:5585-5594. [DOI] [PMC free article] [PubMed] [Google Scholar]