Abstract
G protein-gated inwardly rectifying K+ (Kir) channels are found in neurones, atrial myocytes, and endocrine cells and are involved in generating late inhibitory postsynaptic potentials, slowing the heart rate and inhibiting hormone release. They are activated by G protein-coupled receptors (GPCRs) via the inhibitory family of G protein, Gi/o, in a membrane-delimited fashion by the direct binding of Gβγ dimers to the channel complex. In this study we are concerned with the kinetics of deactivation of the cloned neuronal G protein-gated K+ channel, Kir3.1 + 3.2A, after stimulation of a number of GPCRs. Termination of the channel activity on agonist removal is thought to solely depend on the intrinsic hydrolysis rate of the G protein α subunit. In this study we present data that illustrate a more complex behavior. We hypothesize that there are two processes that account for channel deactivation: agonist unbinding from the GPCR and GTP hydrolysis by the G protein α subunit. With some combinations of agonist/GPCR, the rate of agonist unbinding is slow and rate-limiting, and deactivation kinetics are not modulated by regulators of G protein-signaling proteins. In another group, channel deactivation is generally faster and limited by the hydrolysis rate of the G protein α subunit. G protein isoform and interaction with G protein-signaling proteins play a significant role with this group of GPCRs.
Keywords: G protein α subunit‖inward rectifier‖cell signaling‖drug development‖G protein-signaling protein
Members of the family of inwardly rectifying K+ (Kir) channels gated by G proteins were first identified in atrial myocytes, where they are activated through stimulation of M2 muscarinic receptors by acetylcholine (1). Physiologically, activation of this current is partly responsible for slowing of the heart rate in response to vagal-nerve stimulation (2, 3). It is now known that channel activation is membrane-delimited (4), mimicked by nonhydrolyzable GTP analogues (5), and sensitive to pertussis toxin (PTx), implicating the inhibitory family of G proteins (Gi/o) (6). Channel activation occurs because of direct binding of Gβγ dimers, released from Gi/oα-containing heterotrimers, to domains on the channel (7–9). G protein-gated Kir channels are also expressed in many central neurones, where they can be activated by a large variety of neurotransmitters acting at Gi/o-coupled receptors (10) including γ-aminobutyric acid (GABA) at the GABA type B (GABAB) receptor complex and adenosine at A1 receptors, and they mediate postsynaptic inhibitory events (9, 11, 12). The molecular counterparts of these currents have now been identified by cloning techniques (13–16): the channel is a heterotetramer of members of the Kir3.0 family of K+ channels. Coexpression of Kir3.1 with Kir3.2 or Kir3.4 in heterologous expression systems results in currents that show many of the basic characteristics of the native channels in neurones and atria, respectively.
The kinetic behavior of these channels after agonist application and withdrawal has been a subject of intense investigation. To date, these issues have largely been addressed by using the cloned atrial channel, Kir3.1 + 3.4, and the muscarinic M2 receptor expressed in Xenopus laevis oocytes (17, 18). In particular, a number of studies have sought to explain why these channels when expressed in X. laevis oocytes deactivate more slowly than the native atrial current following stimulation of M2 receptors. The identification of the family of G protein-signaling (RGS) proteins can account, in part, for this discrepancy (17, 18). RGS proteins interact with Gi/o and Gq/11α subunits to increase the intrinsic GTPase rate of the Gα subunit (19–23). Overexpression of RGS4, for example, accelerates the channel-deactivation kinetics and changes other kinetic parameters such that the measured time constants are more consistent with those occurring after stimulation of native channels in atrial cells (17, 18, 24–26). Because this family of channels can be activated by a wide variety of Gi/o-coupled G protein-coupled receptors (GPCRs) in heterologous and native conditions, it is important to establish how general such processes are, and it is these issues that we address in the current study.
Materials and Methods
Molecular Biology, Cell Culture, and Transfection.
We generated and used a series of PTx-resistant Gi/o mutant α subunits and cyan fluorescent protein (CFP)-tagged PTx-resistant Gi/oα subunits as described (27, 28). In addition, we also made RGS-insensitive Giα1 and GoαA where the PTx-resistant Giα1C351G and GoαAC351G were mutated further to G183S and G184S, respectively (29). Mutations were introduced by using the QuikChange kit (Stratagene) and confirmed by using automated DNA sequencing (Cytomyx, Cambridge, U.K.). GABA-B1b and GABA-B2 were expressed in the dual-promoter vector pBudCE4.1 (Invitrogen) following the use of standard molecular cloning techniques to excise the clones from the previous vector, pcDNA3.1/neo/(+). The excised GABA-B1b (PmeI/XhoI) and GABA-B2 (KpnI/XhoI) then were introduced into the two polylinkers in pBudCE4.1 (GABA-B1b, ScaI/SalI sites; GABA-B2, KpnI/XhoI sites).
Cell-culture methods and the generation of stable cell lines were as described (30, 31). In addition to the stable lines wae have described previously [Kir3.1+3.2A channel plus either the A1 adenosine receptor (HKIR3.1/3.2/A1) or the D2S dopaminergic receptor (HKIR3.1/3.2/D2)], we used a further three-dual-receptor + channel stable lines that were denoted as α2A adrenergic receptor (HKIR3.1/3.2/α2), GABA-B1b/2 receptor (HKIR3.1/3.2/GGB), and M4-muscarinic receptor (HKIR3.1/3.2/M4). Monoclonal cell lines were established by picking single colonies of cells after transfection and growth under selective pressure. For all of the dual-receptor and channel-expressing lines we used a dual-selection strategy with 727 μg/ml G418 and 364 μg/ml Zeocin (Invitrogen). Transiently transfected cells suitable for patch-clamping were identified by epifluorescence from cotransfection of 100 ng of the enhanced variant of the GFP (pEGFP-N1; CLONTECH). Data were obtained from at least two independent transfections.
Electrophysiology.
Whole-cell membrane currents were recorded by using an Axopatch 200B amplifier (Axon Instruments, Foster City, CA). Patch pipettes were pulled from filamented borosilicate glass (Clark Electromedical Instruments, Pangbourne, U.K.) and had a resistance of 1.5–2.5 MΩ when filled with pipette solution (see below). Before filling, tips of patch pipettes were coated with a Parafilm/mineral oil suspension. Data were acquired and analyzed by using a Digidata 1200B interface (Axon Instruments) and PCLAMP 6.0 software (Axon Instruments). Cell capacitance was ≈15 pF, and series resistance (<10 MΩ) was at least 75% compensated by using the amplifier circuitry. Recordings of membrane current were commenced after an equilibration period of ≈5 min. Immediately after patch rupture a current–voltage relationship was performed to establish that currents were inwardly rectifying. Thereafter cells were voltage-clamped at −60 mV, and agonist-induced currents were measured at this potential. For current–voltage relationships, records were filtered at 1 kHz and digitized at 5 kHz. For continual data acquisition where cells were voltage-clamped at −60 mV, records were digitized at 100 Hz. Drugs were applied by using a “sewer-pipe” system (Rapid Solution Changer RSC-160; Biologic, Grenoble, France) whereby an array of perfusion capillaries was placed in the bath ≈40 μm from the recorded cell. This system allowed rapid solution switching between capillary tubes and localized application of drugs due to the laminar flow over the studied cell from the pipes as described (32). After agonist application, current activated with an initial delay (“lag”) followed by a rapid rise to peak amplitude (“time to peak”). In some figures we illustrate this as lag + time to peak. During continued agonist application currents exhibited desensitization. However, in this study we particularly focused on the deactivation kinetics, the decay of current back to baseline after removal of agonist. This declining phase was fitted to a single exponential decay function, A⋅exp(−t/τ) + C (where A is the current amplitude at the start of the fit, t is time, τ is the deactivation time constant, and C is the steady-state asymptote, using a Simplex iterative procedure where the sum of squared errors was minimized). F tests indicated that the deactivation phase was best fitted by a single exponential time constant. In some recordings of HKIR3.1/3.2/A1 cells, we observed a transient increase in current after removal of agonist. In these cells channel-deactivation rates were measured during the declining phase of the current after the peak of current reactivation.
For each cell we assessed whether there were any flow artifacts resulting from the pressure of drug application. We did this by applying bath solution from one of the sewer pipes and recording any flow-induced currents. If any such current was observed, then the position of the perfusion head was moved to minimize it. Furthermore, to control for variations in positioning of the sewer-pipe system relative to the cell, we calibrated this system by using the kinetics of channel block by barium. The cell was positioned in the center of the field by using cross hairs in the microscope eyepieces. Barium (1 mM) was applied to the cell in the presence of agonist when the agonist-induced current had reached a plateau phase. Block of the current occurred with an initial delay before reaching equilibrium. It was assumed that this lag reflected the intrinsic delivery time to the cell. A barium calibration was performed before the start of experiments to ascertain correct positioning of the sewer pipe and was repeated on several cells during each recording session. In general the results were highly reproducible (the lag time for barium block was 237.3 ± 11.68 ms; n = 73).
Membrane currents were measured at −60 mV, and all data are presented as mean ± SEM where n indicates the number of cells recorded from. Time measurements were reciprocated before statistical analysis because the reciprocal of time is normally distributed. Data are shown untransformed. We determined statistical significance using either Student's t test or one-way repeated-measures ANOVA tests with Bonferroni correction as appropriate (∗, P ≤ 0.05; ∗∗, P ≤ 0.01; ∗∗∗, P ≤ 0.001).
Materials and Drugs.
The solutions used were pipette solution [107 mM KCl/1.2 mM MgCl2/1 mM CaCl2/10 mM EGTA/5 mM Hepes/2 mM MgATP/0.3 mM Na2GTP (KOH to pH 7.2)/≈140 mM total K+) and bath solution (140 mM KCl/2.6 mM CaCl2/1.2 mM MgCl2/5 mM Hepes, pH 7.4). Cell-culture materials were from GIBCO/BRL and Invitrogen. All chemicals were from Sigma or Calbiochem. Drugs were made up as concentrated stock solutions and kept at −20°C.
Results
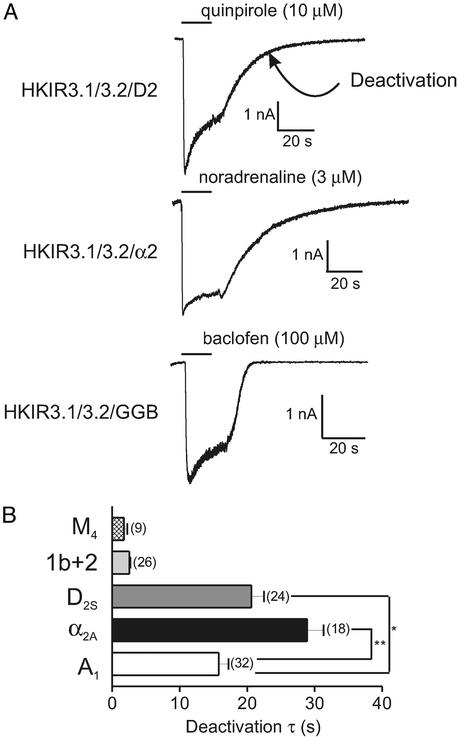
Using the whole-cell configuration of the patch-clamp technique we studied receptor-mediated G protein-gated Kir channel currents in a HEK293 stable cell line robustly expressing the Kir3.1+3.2A channel complex (31). To apply agonists we used a rapid and localized drug-perfusion system that enabled us to apply and remove agonist in under 0.25 s. We used five different dual-receptor and channel stable lines to investigate receptor-mediated Kir3.1+3.2A currents (A1, HKIR3.1/3.2/A1; α2A, HKIR3.1/3.2/α2; D2S, HKIR3.1/3.2/D2; M4, HKIR3.1/3.2/M4; and GABA-B1b/2, HKIR3.1/3.2/GGB). Radioligand binding performed on the HKIR3.1/3.2/A1, HKIR3.1/3.2/α2, and HKIR3.1/3.2/D2 cell lines revealed similar levels of receptor expression (data not shown). Agonists, which were applied for 20 s, were used at concentrations likely to lead to full receptor occupancy. Fig. 1A shows representative current traces obtained from HKIR3.1/3.2/D2, HKIR3.1/3.2/α2, and HKIR3.1/3.2/GGB cells. In this study we measured channel-deactivation rates (τ) as currents return to baseline after the removal of agonist (indicated in Fig. 1A Top). The mean deactivation data for all five cell lines is shown in Fig. 1B; strikingly there is a wide variation in deactivation rates ranging from ≈2 s in the HKIR3.1/3.2/M4 cell line to ≈30 s in the HKIR3.1/3.2/α2A cells. Other clonal isolates of α2A in which binding was not characterized have faster deactivation rates (data not shown). We used the agonists 5′-N-ethylcarboxamidoadenosine (NECA) for A1, carbachol for M4, baclofen for GABA-B1b/2, quinpirole for D2S, and noradrenaline for α2A. We also consistently observed in the HKIR3.1/3.2/A1 receptor line a transient reactivation in current after removal of agonist, which could often reach a similar magnitude to the initial potentiation. This never occurred in the HKIR3.1/3.2/M4, HKIR3.1/3.2/GGB, and HKIR3.1/3.2/D2 cells but occasionally occurred in the HKIR3.1/3.2/α2 line (≈30% of cells).
Figure 1.
Receptor-mediated kinetics. This figure illustrates the basic characteristics of the receptor-mediated responses and a summary of the mean data obtained. (A) Representative current traces of cells from three different receptor + channel cell lines (D2S receptor, HKIR3.1/3.2/D2; α2A receptor, HKIR3.1/3.2/α2; GABA-B1b/2 receptor, HKIR3.1/3.2/GGB) voltage-clamped at −60 mV and exposed to agonist for 20 s (as indicated by the horizontal bar). We calculated a deactivation time constant (τ) as a measurement of channel-deactivation kinetics (as indicated) after the removal of agonist (see Materials and Methods). (B) Summary of deactivation data obtained from five cell lines expressing the Kir3.1 + 3.2A channel complex and the receptor indicated. Radioligand binding revealed similar levels of receptor expression in the HKIR3.1/3.2/A1, HKIR3.1/3.2/α2, and HKIR3.1/3.2/D2 cell lines (not shown); thus, we compared deactivation time constants in these three cell lines and found that deactivation was significantly faster in the HKIR3.1/3.2/A1 cell line than in the HKIR3.1/3.2/α2 and HKIR3.1/3.2/D2 cell lines (P < 0.01 and P < 0.05, respectively).
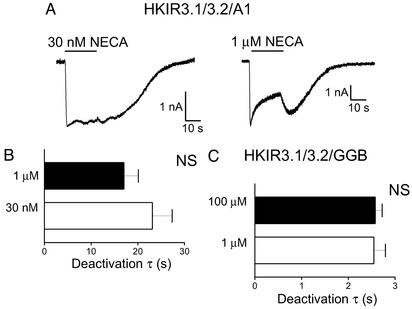
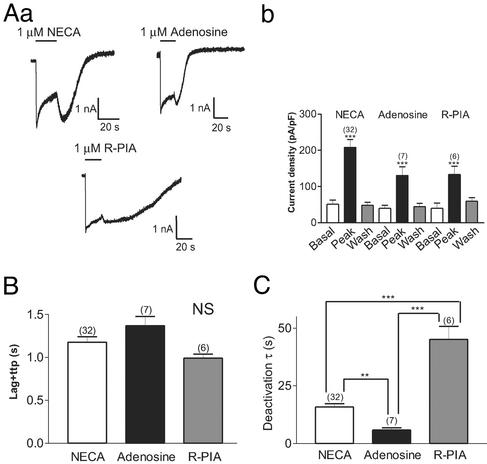
We first examined the effects of receptor occupancy on channel deactivation by varying the concentration of agonist used. Two agonist concentrations were chosen: a low concentration (approximately the EC50 value) and a high, saturating concentration. These experiments were performed by using the HKIR3.1/3.2/GGB and HKIR3.2/3.2/A1 cell lines, and representative traces from the HKIR3.2/3.2/A1 cell lines with 30 nM and 1 μM NECA are shown in Fig. 2A. As would be expected for a bimolecular reaction, agonist concentration did not influence channel deactivation after stimulation of either of these receptors (Fig. 2 B and C). We next examined whether different agonists would affect channel deactivation, and for these experiments we used three different A1 adenosine receptor agonists applied to HKIR3.1/3.2/A1 cells. We found that although NECA, adenosine, and N6-(R-phenylisopropyl)adenosine (R-PIA) (1 μM) all produced agonist-induced currents of similar magnitudes (Fig. 3Ab), the kinetics of the channel response were clearly different (Fig. 3Aa). Although channel activation kinetics (lag + time to peak) were not affected by the nature of the agonist (Fig. 3B), a striking difference in deactivation kinetics among the three agonists was observed such that R-PIA caused the slowest deactivation, whereas adenosine induced the fastest deactivation (Fig. 3C).
Figure 2.
Effects of receptor occupancy on channel deactivation. Using the HKIR3.1/3.2/A1 cell line we studied the effects of agonist concentration on channel-deactivation rates. (A) Representative examples of these experiments with 30 nM and 1 μM NECA. The bar charts that summarize the data obtained from these experiments show that channel deactivation was unaffected by agonist concentration with both the A1 receptor (B) and the GABA-B1b/2 receptor (C). NS, not significant.
Figure 3.
Effects of agonist type on channel kinetics and desensitization. (Aa) Representative traces recorded from HKIR3.1/3.2/A1 cells voltage-clamped at −60 mV in response to 20-s applications of NECA, adenosine, and R-PIA (all at 1 μM). Mean data are summarized in the bar chart shown (Ab), where each agonist caused a significant and reversible potentiation of currents (P < 0.001). (B) Channel activation kinetics were not significantly different between the agonists. NS, not significant. (C) Channel-deactivation time constants were markedly affected by the type of agonist used. Deactivation was accelerated with adenosine but slowed with R-PIA when compared with NECA.
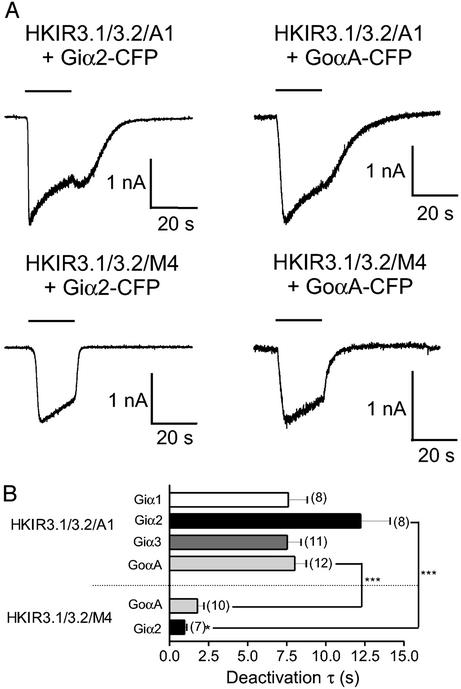
The use of engineered PTx-resistant Gi/oα subunits has made it possible to look exclusively at coupling between a number of Gi/o-coupled receptors and the channel via different Gi/oα isoforms. We found previously that receptors exhibit different G protein-coupling profiles to activate the channel (27). Recently we extended this approach to include a series of constructs of PTx-resistant Gi/oα subunits that are fused to the CFP. Such chimeric G proteins are membrane-targeted and functional (they can interact with Gβγ dimers and inhibit adenylate cyclase activity); furthermore we established conditions under which these constructs are expressed at equivalent levels (28). We compared deactivation rates via Giα1–3-CFP and Goα-CFP at equivalent concentrations with the A1 receptor in the HKIR3.1/3.2/A1 line and via Giα2-CFP and Goα-CFP at equivalent concentrations with the M4 receptor in the HKIR3.1/3.2/M4 line. Example traces are shown in Fig. 4A. There are two major points to note with these data. First, there is a large difference in the observed deactivation rates between M4-mediated and A1-mediated currents via both Giα2-CFP and GoαA-CFP. Second, after M4-receptor stimulation we noticed a difference in channel deactivation between Giα2-CFP and GoαA-CFP: channels deactivated significantly faster through Giα2-CFP than GoαA-CFP (Fig. 4B).
Figure 4.
Effects of the Gα isoform on channel deactivation. (A) Representative examples of current profiles obtained from HKIR3.1/3.2/A1 and HKIR3.1/3.2/M4 cell lines transiently transfected with Giα2-CFP and GoαA-CFP and voltage-clamped at −60 mV. The maximal concentration of agonist (1 μM NECA and 10 μM carbachol, respectively) was applied for 20 s (as indicated by the horizontal bar). (B) Summary of data obtained. Channel deactivation via both Giα2 and GoαA was significantly faster after stimulation of the M4 than the A1 receptor.
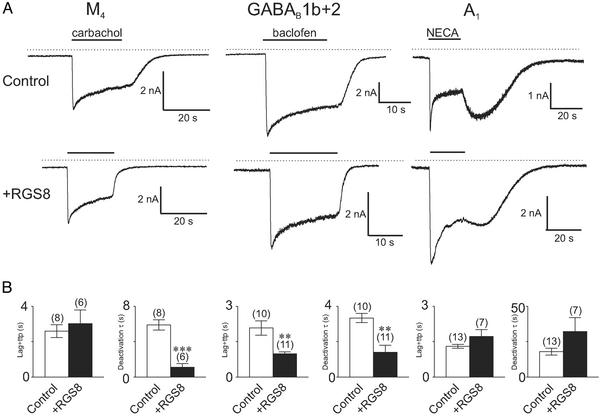
We further investigated this phenomenon by looking at the role of RGS proteins in modulating the channel response to receptor stimulation. It has become apparent recently that the kinetics of Kir3.0 channel response can be influenced significantly by this family of proteins (17, 18). Overexpression of RGS proteins leads to an increase in both the deactivation and, perhaps paradoxically, activation rates of Kir3.0 currents in heterologous expression systems (17, 18). We examined whether one of the RGS proteins, RGS8, could differentially modulate the dynamics of current activation. We first examined this by overexpressing RGS8 in dual-receptor and channel stable lines and found marked differences in channel kinetics after RGS8 expression between different receptors. Although RGS8 had little effect on either activation or deactivation kinetics of A1-mediated currents (Fig. 5 A and B Right), we found that overexpression of RGS8 solely increased deactivation rates after M4-receptor stimulation (Fig. 5 A and B Left) but increased both activation and deactivation rates of GABAB-mediated currents (Fig. 5 A and B Center).
Figure 5.
Differential effects of RGS8 on channel kinetics in response to receptor stimulation. (A) We recorded currents in HKIR3.1/3.2/M4 (Left), HKIR3.1/3.2/GGB (Center), and HKIR3.1/3.2/A1 (Right) cells voltage-clamped at −60 mV in response to 20-s agonist applications. (Upper) Control responses. (Lower) Responses in the presence of RGS8 (1 μg of cDNA) transiently transfected. (B) Summary of kinetic data in the absence (Control) and presence (+RGS8) of RGS8. In the HKIR3.1/3.2/M4 cell line, RGS8 affects only the deactivation kinetics but not the activation kinetics, whereas in the HKIR3.1/3.2/GGB cell line it significantly accelerates both activation and deactivation parameters; in the HKIR3.1/3.2/A1 cell line RGS8 has no effects at all. NS, not significant; ttp, time to peak.
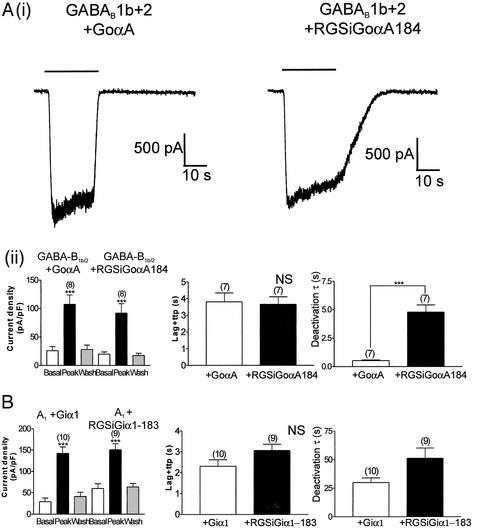
Finally, we investigated the role of RGS proteins by expressing a PTx-resistant G protein with an additional point mutation, G184S (designated RGSiGoαA) rendering it resistant to the actions of RGS proteins (29, 33). When transiently transfected into the HKIR3.1/3.2/GGB line we found that this Gα subunit was able to support GABA-B1b/2-mediated channel activation with similar activation properties to GoαAC351G but with very much slowed channel-deactivation kinetics (Fig. 6 Ai and Aii). In contrast, the A1 receptor cell line showed no statistically different changes in kinetic parameters (Fig. 6B) when constrained to signal via Giα1C351G compared with the engineered RGS-insensitive, RGSiGiα1 (a PTx-resistant G protein with point mutation G183S).
Figure 6.
Effects of RGS-insensitive GoαA on channel kinetics in response to GABA-B1b/2 receptor stimulation. (Ai) Examples of current traces recorded from HKIR3.1/3.2/GGB cells transiently transfected with either GoαA (Left) or RGSiGoαA (Right). Baclofen (100 μM) was applied for 20 s as indicated. (Aii) Mean data obtained from these experiments. Current densities are shown (Left). Current density was measured at −60 mV before (basal), during (peak), and after (wash) application of 100 μM baclofen for 20 s in the HKIR3.1/3.2/GGB cell line transiently expressing either the PTx-resistant GoαA or the RGS-insensitive, PTx-resistant GoαA (RGSiGoαA). Channel activation (Center) and deactivation (Right) kinetics are shown. We saw no significant difference in channel activation between GoαA and RGSiGoαA (P > 0.05), whereas deactivation via the RGS-insensitive G protein was dramatically slower than GoαA (P < 0.001). (B) Summary of data from similar experiments with the HKIR3.1/3.2/A1 cell line and transiently expressing either Giα1C351G or RGSiGiα1. (Left) Current densities were comparable in both groups, but no significant differences were seen between the PTx-resistant Giα1 and RGSiGiα1 in terms of channel activation and deactivation. ttp, time to peak.
Discussion
After the initial cloning of the components constituting the atrial G protein-gated Kir channel (13–15), a major discrepancy remained. The deactivation rate after M2-muscarinic receptor activation was much slower with heterologous expression of Kir3.1 and Kir3.4 in X. laevis oocytes in comparison with that of the native atrial channel under analogous conditions. Subsequently it became apparent that this was due to the actions of a previously uncharacterized class of protein (the family of RGS proteins) (17, 34). The preeminent hypothesis in the field is that channel-deactivation kinetics are determined by the Gα-GTP hydrolysis rates, and this process dictates how fast Gβγ is sequestered by Gα-GDP. In general, the Gα-GTP hydrolysis rate is determined by the particular G protein, and this rate is modulated by RGS proteins and effectors with intrinsic GTPase-activating activity such as phospholipase Cβ (21, 22). Furthermore, it is now clear that Kir3.0 subunits are widely distributed in neuronal and neuroendocrine tissues and in principle can be regulated by a large variety of Gi/o-coupled receptors (9–11, 16, 35–40). The heterotetramer of Kir3.1 and Kir3.2A used in this study is likely to be equivalent to the channel found in many neuronal populations. Indeed the nature and numerical details of channel-activation and -deactivation kinetics we observe in our heterologous expression system are comparable to those observed with the native channel in hippocampal neurones. For example, baclofen-mediated responses in hippocampal neurones deactivate in ≈1 s after relatively prolonged agonist exposure, which is similar to our observations especially with RGS8 overexpression (12, 41). With this much broader choice of heptahelical receptors, we question whether channel-deactivation kinetics are determined solely by the GTP hydrolysis rate of the G protein α subunit.
In this study we investigated the factors that influence the deactivation of Kir3.1+3.2A currents after agonist removal through a number of receptors and report data that are incompatible with the above hypothesis of channel-deactivation kinetics being solely determined by Gα-GTP hydrolysis. It is apparent that deactivation rates vary widely between the different GPCRs studied and are independent of receptor occupancy (i.e., agonist concentration). At a simplistic level it is possible to propose that either the channel, G protein, or receptor may be the major limiting factor in the deactivation phase of the kinetic response. Our initial observations of the widely varying deactivation rates between different receptors make it unlikely that Gβγ binding and unbinding are the rate-limiting steps for channel activation and deactivation. However, it is paradoxical that binding assays with purified Gβγ and channel domains can be performed with high nM/low μM affinities, and channel activation by Gβγ in inside-out patches occurs with a low nM EC50 (9, 42–44). Generally, nanomolar affinities are consistent with slow unbinding rates, which is discrepant with the observed deactivation rates. Speculatively, it is possible that association and dissociation of G protein α subunits occurs with Gβγ bound to channel, and residues important on Gβγ for activation are masked by the α subunit (45). We have data that support the division of agonist/receptor combinations into two classes. For one class (such as the GABA-B1b/2 receptor with baclofen or the M4 receptor with carbachol) the data are compatible with the intrinsic hydrolysis rate of the G protein being rate-limiting. In this case deactivation kinetics are enhanced by RGS8 overexpression and slowed by constraining signaling to occur via an RGS-insensitive G protein. Signaling constrained via different Gi/o isoforms is fast, but there are significant differences between Giα2 and Goα subunits. In another group, for example the A1 receptor with NECA as an agonist, deactivation kinetics are slower. Increasing or decreasing the intrinsic hydrolysis rate of the G protein by overexpression of RGS or signaling via an RGS-resistant G protein has little effect on overall kinetics. In addition, constraining channel activation via a series of different Gi/o isoforms results in similarly slow kinetics with no significant difference between the various isoforms. Finally, deactivation kinetics vary with the chemical nature of the agonist at the A1 receptor. The rank order of potency (R-PIA > NECA > adenosine) is similar to that determined for the high-affinity state with displacement radioligand binding (46). These data are consistent with the idea that agonist unbinding is rate-limiting.
It is noticeable that the ligands we use in these studies are largely synthetic agonists developed for enhanced affinity to their cognate receptor. However, other investigators have noticed significantly different deactivation rates between the α2A and α2C receptors with physiological agonists and postulated that agonist unbinding may be limiting with the latter (47). In this regard our deactivation kinetics for noradrenaline at the α2A receptor, in the clonal isolate used here, are slower than those of others (33, 47). Data (unpublished observations) indicate that it is possible to modulate the deactivation rate with RGS overexpression, and thus slow kinetics per se do not imply that agonist unbinding is rate-limiting. The endogenous peptide ligands for the opioid receptors are another potentially important group. However, the appreciation of such issues may have important consequences. In synaptic transmission for example, some agonist/GPCR combinations may have more prolonged post-synaptic inhibitory effects. It also means that some receptor pathways can follow a rapidly changing stimulus, whereas others will time-integrate the signal to a new steady-state level. The fact that the channel essentially acts as a biosensor for membrane Gβγ concentrations makes it likely that these observations can be extrapolated to other effectors activated by these receptor pathways.
It is clear that HEK293 cells contain endogenous RGS proteins; thus it is difficult to make categorical statements about the intrinsic hydrolysis rates of G protein α isoforms in living cells. Our data do show significant differences for Giα2 and GoαA with the M4 receptor; however, this could reflect an intrinsic preference for certain G protein subunits by endogenous RGS proteins. Second, our data show that endogenous RGS proteins predominantly affect deactivation kinetics, whereas overexpression of RGS8 additionally accelerates the activation rate for GABA-B1b/2. In addition, overexpression of RGS8 seems to selectively accelerate activation kinetics for GABA-B1b/2 but not the M4 receptor. Generally the acceleration of the activation kinetics has been explained by physical or kinetic scaffolding, but this issue is still controversial (21, 48). Our data show potential layers of selectivity and a role for level of RGS expression.
In summary, we propose that there are two processes that account for channel deactivation: agonist unbinding from the GPCR and GTP hydrolysis by the G protein α subunit. With some combinations of agonist/GPCR, the rate of agonist unbinding is slow and rate-limiting, and in another group deactivation is generally faster and is determined by the hydrolysis rate of the G protein α subunit. The G protein isoform and interaction with RGS proteins play a significant role with this group of GPCRs.
Acknowledgments
We are grateful to Dr. F. Marshall for providing the GABA-B receptor clones. This work was supported by the Wellcome Trust, Royal Society, and the British Heart Foundation. J.L.L. is a Royal Society Dorothy Hodgkin Fellow.
Abbreviations
- Kir
inwardly rectifying K+
- PTx
pertussis toxin
- GABA
γ-aminobutyric acid
- GABAB
GABA type B
- RGS
G protein-signaling
- GPCR
G protein-coupled receptor
- CFP
cyan fluorescent protein
- NECA
5′-N-ethylcarboxamidoadenosine
- R-PIA
N6-(R-phenylisopropyl)adenosine
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Noma A, Trautwein W. Pflügers Arch. 1978;377:193–200. doi: 10.1007/BF00584272. [DOI] [PubMed] [Google Scholar]
- 2.Wickman K, Nemec J, Gendler S J, Clapham D E. Neuron. 1998;20:103–114. doi: 10.1016/s0896-6273(00)80438-9. [DOI] [PubMed] [Google Scholar]
- 3.Drici M D, Diochot S, Terrenoire C, Romey G, Lazdunski M. Br J Pharmacol. 2000;131:569–577. doi: 10.1038/sj.bjp.0703611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Soejima M, Noma A. Pflügers Arch. 1984;400:424–431. doi: 10.1007/BF00587544. [DOI] [PubMed] [Google Scholar]
- 5.Breitwieser G E, Szabo G. Nature. 1985;317:538–540. doi: 10.1038/317538a0. [DOI] [PubMed] [Google Scholar]
- 6.Pfaffinger P J, Martin J M, Hunter D D, Nathanson N M, Hille B. Nature. 1985;317:536–538. doi: 10.1038/317536a0. [DOI] [PubMed] [Google Scholar]
- 7.Logothetis D E, Kurachi Y, Galper J, Neer E J, Clapham D E. Nature. 1987;325:321–326. doi: 10.1038/325321a0. [DOI] [PubMed] [Google Scholar]
- 8.Reuveny E, Slesinger P A, Inglese J, Morales J M, Iniguez Lluhi J A, Lefkowitz R J, Bourne H R, Jan Y N, Jan L Y. Nature. 1994;370:143–146. doi: 10.1038/370143a0. [DOI] [PubMed] [Google Scholar]
- 9.Yamada M, Inanobe A, Kurachi Y. Pharmacol Rev. 1998;50:723–757. [PubMed] [Google Scholar]
- 10.North R A. Br J Pharmacol. 1989;98:13–28. doi: 10.1111/j.1476-5381.1989.tb16855.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luscher C, Jan L Y, Stoffel M, Malenka R C, Nicoll R A. Neuron. 1997;19:687–695. doi: 10.1016/s0896-6273(00)80381-5. [DOI] [PubMed] [Google Scholar]
- 12.Sodickson D L, Bean B P. J Neurosci. 1998;18:8153–8162. doi: 10.1523/JNEUROSCI.18-20-08153.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kubo Y, Reuveny E, Slesinger P A, Jan Y N, Jan L Y. Nature. 1993;364:802–806. doi: 10.1038/364802a0. [DOI] [PubMed] [Google Scholar]
- 14.Dascal N, Schreibmayer W, Lim N F, Wang W, Chavkin C, DiMagno L, Labarca C, Kieffer B L, Gaveriaux-Ruff C, Trollinger D, et al. Proc Natl Acad Sci USA. 1993;90:10235–10239. doi: 10.1073/pnas.90.21.10235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krapivinsky G, Gordon E A, Wickman K, Velimirovic B, Krapivinsky L, Clapham D E. Nature. 1995;374:135–141. doi: 10.1038/374135a0. [DOI] [PubMed] [Google Scholar]
- 16.Lesage F, Duprat F, Fink M, Guillemare E, Coppola T, Lazdunski M, Hugnot J P. FEBS Lett. 1994;353:37–42. doi: 10.1016/0014-5793(94)01007-2. [DOI] [PubMed] [Google Scholar]
- 17.Saitoh O, Kubo Y, Miyatani Y, Asano T, Nakata H. Nature. 1997;390:525–529. doi: 10.1038/37385. [DOI] [PubMed] [Google Scholar]
- 18.Doupnik C A, Davidson N, Lester H A, Kofuji P. Proc Natl Acad Sci USA. 1997;94:10461–10466. doi: 10.1073/pnas.94.19.10461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berman D M, Gilman A G. J Biol Chem. 1998;273:1269–1272. doi: 10.1074/jbc.273.3.1269. [DOI] [PubMed] [Google Scholar]
- 20.Koelle M R. Curr Opin Cell Biol. 1997;9:143–147. doi: 10.1016/s0955-0674(97)80055-5. [DOI] [PubMed] [Google Scholar]
- 21.Ross E M, Wilkie T M. Annu Rev Biochem. 2000;69:795–827. doi: 10.1146/annurev.biochem.69.1.795. [DOI] [PubMed] [Google Scholar]
- 22.De Vries L, Zheng B, Fischer T, Elenko E, Farquhar M G. Annu Rev Pharmacol Toxicol. 2000;40:235–271. doi: 10.1146/annurev.pharmtox.40.1.235. [DOI] [PubMed] [Google Scholar]
- 23.Dohlman H G, Thorner J. J Biol Chem. 1997;272:3871–3874. doi: 10.1074/jbc.272.7.3871. [DOI] [PubMed] [Google Scholar]
- 24.Inanobe A, Fujita S, Makino Y, Matsushita K, Ishii M, Chachin M, Kurachi Y. J Physiol (London) 2001;535:133–143. doi: 10.1111/j.1469-7793.2001.t01-1-00133.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fujita S, Inanobe A, Chachin M, Aizawa Y, Kurachi Y. J Physiol (London) 2000;526:341–347. doi: 10.1111/j.1469-7793.2000.00341.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chuang H H, Yu M, Jan Y N, Jan L Y. Proc Natl Acad Sci USA. 1998;95:11727–11732. doi: 10.1073/pnas.95.20.11727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leaney J L, Tinker A. Proc Natl Acad Sci USA. 2000;97:5651–5656. doi: 10.1073/pnas.080572297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leaney J L, Benians A, Graves F M, Tinker A. J Biol Chem. 2002;277:28803–28809. doi: 10.1074/jbc.M204683200. [DOI] [PubMed] [Google Scholar]
- 29.Jeong S W, Ikeda S R. J Neurosci. 2000;20:4489–4496. doi: 10.1523/JNEUROSCI.20-12-04489.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Giblin J P, Leaney J L, Tinker A. J Biol Chem. 1999;274:22652–22659. doi: 10.1074/jbc.274.32.22652. [DOI] [PubMed] [Google Scholar]
- 31.Leaney J L, Milligan G, Tinker A. J Biol Chem. 2000;275:921–929. doi: 10.1074/jbc.275.2.921. [DOI] [PubMed] [Google Scholar]
- 32.Leaney J L, Dekker L V, Tinker A. J Physiol (London) 2001;534:367–379. doi: 10.1111/j.1469-7793.2001.00367.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jeong S W, Ikeda S R. J Physiol (London) 2001;535:335–347. doi: 10.1111/j.1469-7793.2001.00335.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Delmas P, Abogadie F C, Dayrell M, Haley J E, Milligan G, Caulfield M P, Brown D A, Buckley N J. Eur J Neurosci. 1998;10:1654–1666. doi: 10.1046/j.1460-9568.1998.00170.x. [DOI] [PubMed] [Google Scholar]
- 35.Lesage F, Guillemare E, Fink M, Duprat F, Heurteaux C, Fosset M, Romey G, Barhanin J, Lazdunski M. J Biol Chem. 1995;270:28660–28667. doi: 10.1074/jbc.270.48.28660. [DOI] [PubMed] [Google Scholar]
- 36.Inanobe A, Yoshimoto Y, Horio Y, Morishige K-I, Hibino H, Matsumoto S, Tokunaga Y, Maeda T, Hata Y, Takai Y, et al. J Neurosci. 1999;19:1006–1017. doi: 10.1523/JNEUROSCI.19-03-01006.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liao Y J, Jan Y N, Jan L Y. J Neurosci. 1996;16:7137–7150. doi: 10.1523/JNEUROSCI.16-22-07137.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ikeda K, Kobayashi T, Kumanishi T, Yano R, Sora I, Niki H. Neurosci Res. 2002;44:121–131. doi: 10.1016/s0168-0102(02)00094-9. [DOI] [PubMed] [Google Scholar]
- 39.Takano K, Yasufuku-Takano J, Kozasa T, Nakajima S, Nakajima Y. J Physiol (London) 1997;502:559–567. doi: 10.1111/j.1469-7793.1997.559bj.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Velimirovic B M, Koyano K, Nakajima S, Nakajima Y. Proc Natl Acad Sci USA. 1995;92:1590–1594. doi: 10.1073/pnas.92.5.1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sodickson D L, Bean B P. J Neurosci. 1996;16:6374–6385. doi: 10.1523/JNEUROSCI.16-20-06374.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang C L, Slesinger P A, Casey P J, Jan Y N, Jan L Y. Neuron. 1995;15:1133–1143. doi: 10.1016/0896-6273(95)90101-9. [DOI] [PubMed] [Google Scholar]
- 43.Huang C L, Jan Y N, Jan L Y. FEBS Lett. 1997;405:291–298. doi: 10.1016/s0014-5793(97)00197-x. [DOI] [PubMed] [Google Scholar]
- 44.Wickman K D, Iniguez-Lluhi J A, Davenport P A, Taussig R, Krapivinsky G B, Linder M E, Gilman A G, Clapham D E. Nature. 1994;368:255–257. doi: 10.1038/368255a0. [DOI] [PubMed] [Google Scholar]
- 45.Ford C E, Skiba N P, Bae H, Daaka Y, Reuveny E, Shekter L R, Rosal R, Weng G, Yang C-S, Iyengar R, et al. Science. 1998;280:1271–1274. doi: 10.1126/science.280.5367.1271. [DOI] [PubMed] [Google Scholar]
- 46.Fredholm B B, IJzerman A P, Jacobson K A, Klotz K-N, Linden J. Pharmacol Rev. 2001;53:527–552. [PMC free article] [PubMed] [Google Scholar]
- 47.Bunemann M, Bucheler M M, Philipp M, Lohse M J, Hein L. J Biol Chem. 2001;276:47512–47517. doi: 10.1074/jbc.M108652200. [DOI] [PubMed] [Google Scholar]
- 48.Zhong H, Wade S M, Woolf P J, Linderman J J, Traynor J R, Neubig R R. J Biol Chem. 2003;278:7278–7284. doi: 10.1074/jbc.M208819200. [DOI] [PubMed] [Google Scholar]