Abstract
Johne's disease, caused by infection with Mycobacterium avium subsp. paratuberculosis, causes significant economic losses to the livestock farming industry. Improved investigative and diagnostic tools—necessary to understand disease processes and to identify subclinical infection—are much sought after. Here, we describe the production of single-chain antibodies with defined specificity for M. avium subsp. paratuberculosis surface proteins. Single-chain antibodies (scFv) were generated from sheep with Johne's disease by cloning heavy-chain and lambda light-chain variable regions and expressing these in fusion with gene III of filamentous phages. Two scFv clones (designated SurfS1.2 and SurfS2.2) were shown to be immunoreactive against M. avium subsp. paratuberculosis surface targets by flow cytometry, and immunoblotting identified specificity for a 34-kDa proteinase-susceptible determinant. Both antibodies were cross-reactive against Mycobacterium avium subsp. avium but nonreactive against Mycobacterium bovis or Mycobacterium phlei cells and were shown to be capable of enriching M. avium subsp. paratuberculosis cells by a factor of approximately 106-fold when employed in magnetic bead separation of mixed Mycobacterium sp. cultures. Further, magnetic bead separation using SurfS1.2 and SurfS2.2 was capable of isolating as few as 103 M. avium subsp. paratuberculosis cells from ovine fecal samples, indicating the diagnostic potential of these reagents. Finally, inclusion of SurfS1.2 or SurfS2.2 in in vitro broth culture with M. avium subsp. paratuberculosis indicated that surface binding activity did not impede bacterial growth, although colony clumping was prevented. These results are discussed in terms of the potential use of single-chain phage display monoclonal antibodies as novel diagnostic reagents.
The Mycobacterium avium complex (MAC) includes major pathogens for humans, birds, and ruminants. The most important members of this complex are Mycobacterium avium subsp. avium, as it causes disseminated infections in patients with AIDS that do not receive treatment or where modern anti-human immunodeficiency virus therapy fails (7), and Mycobacterium avium subsp. paratuberculosis, the causative agent of Johne's disease (JD). Johne's disease is a chronic enteritis of ruminants that produces major economic losses in livestock and dairy industries worldwide. Infection due to M. avium subsp. paratuberculosis manifests as a chronic inflammatory gastroenteritis, with progressive infiltration of inflammatory leukocytes into lesion sites in the intestinal mucosa and submucosa, which eventually produce extensive granuloma in the affected sites and draining lymphatic tissues (20). The underlying pathology leads to epithelial thickening in the lower intestine, causing malabsorption of nutrients, leading to wasting and eventual death in affected animals. Ruminants affected by JD develop strong immunological reactivity against M. avium subsp. paratuberculosis antigens, comprising activation of peripheral blood CD4+ lymphocytes capable of secreting gamma interferon, along with the production of immunoglobulin G (IgG) class antibodies specific for mycobacterial surface glycolipids, particularly during the later stages of disease. However, it can take several months to years for clinical symptoms of JD to present, highlighting the chronic nature of the disease. It is during the chronic, subclinical, and clinical stages of JD that affected animals lose condition, causing the major economic losses to industry through reduced production.
Detection of subclinical JD is reliant on the screening of feces via PCR for bacterial shedding (27) or on the identification of serological reactivity (24). However, improved tools for rapid and more cost-effective diagnosis of subclinical JD are much needed. In this regard, the development of monoclonal antibodies to M. avium subsp. paratuberculosis was identified as an unmet need at the 7th and 8th International Colloquia on Paratuberculosis (Spain, 2002, and Denmark, 2005, respectively). Furthermore, basic disease processes (such as the role of anti-M. avium subsp. paratuberculosis antibody in contributing to protection or disease pathogenesis in vivo) remain incompletely described, and further progress in this area could be expedited by the development of refined investigative tools such as monoclonal antibodies with defined target specificity.
Sheep infected with M. avium subsp. paratuberculosis develop serological reactivity to the pathogen, predominated by IgG antibodies (25). Utilizing this serological response, it may be possible to design and develop pathogen-specific monoclonal antibody probes that can fulfill the role of improved diagnostic and investigative reagents. As an alternative to hybridoma technology, reverse transcription-PCR amplification of the variable region of both the light and heavy chains of large animal host immunoglobulins has enabled the recombination of functional antibody fragments in Escherichia coli expression systems (16). The translational fusion of these to genes of filamentous phages has further made the selection of single-chain antibody fragments (scFv) by phage display possible. Antibody phage display technology has begun to replace standard hybridoma technology (29) and has allowed the generation of monoclonal antibody fragments from species other than rodents, including humans, rabbits, chickens, camels, and sheep (13).
The development of antibody tools that recognize microbial surface components is of particular interest in investigative research and diagnostic assay development for several reasons. It facilitates the identification of surface-exposed epitopes (23), enables the investigation of potential antibody-mediated bactericidal effects (4), provides a potential tool for cell separation (14), and enables differentiation of closely related species (22). Surface proteins mediate important pathogen-host interactions and are interesting targets for antimicrobial chemotherapy and vaccination. In this study, we isolated scFv from M. avium subsp. paratuberculosis-infected sheep scFv against members of the MAC by antibody phage display. We have characterized the antigenic specificity of these antibodies by enzyme-linked immunosorbent assay (ELISA), immunoblotting, flow cytometry, immunofluorescence microscopy, and immunomagnetic separation. Furthermore, the ability of these antibodies to modulate mycobacterial growth characteristics in vitro was also investigated.
MATERIALS AND METHODS
Bacterial cultures and media.
The following M. avium subsp. paratuberculosis isolates were used in this study: K10 (17), the vaccine strain 316F (AquaVax Ltd.), and two field isolates (JD3 and strain W) from clinical cases of Johne's disease in sheep from New Zealand (3). Bacteria were grown on Middlebrook plates supplemented with mycobactin J (Allied Monitor, Fayette, MO) (28). In addition, the IS901-positive M. avium subsp. avium strain T264 was obtained from AgResearch (Wallaceville Animal Research Centre, Upper Hutt, New Zealand) and grown under the same culture conditions without the addition of mycobactin J. Where necessary, M. avium subsp. avium was grown in liquid culture as described below. Escherichia coli XL1-Blue (Stratagene, La Jolla, CA) was used as a host for cloning, and E. coli JM83 (ATCC) was used as a host for protein expression.
Construction of ovine scFv library from diseased sheep.
Sheep were infected, diagnosed, and culled as described previously (3). In brief, 1-year-old desexed male sheep were managed under conventional New Zealand farming conditions in open paddocks. Sheep were infected via the oral route with two doses of 109 CFU of a low-passage M. avium subsp. paratuberculosis strain W. The animals' condition and serological response was monitored longitudinally as disease developed, and animals losing 18% or more of their maximum weight were culled. The spleen, blood, and bone marrow of 3 sheep that showed this weight loss (and also exhibited a high IgG1 antibody titer against M. avium subsp. paratuberculosis antigens by screening ELISA) (9) were removed at necropsy and immediately frozen in liquid nitrogen. Total RNA was extracted from these samples using TRIzol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. cDNA was synthesized using oligo(dT) and Superscript II reverse transcriptase (Invitrogen, Carlsbad, CA). The variable regions were amplified by PCR using previously published primers (18). Vl and Vh fragments were fused by splicing by overlapping extension PCR (SOE-PCR) as described previously (15), resulting in a PCR product of approximately 800 bp. PCR products were digested with SfiI overnight and cloned into the vector pAK100.
Panning of scFv library.
To prepare antigenic targets for screening, mycobacterial cell suspensions were harvested from culture, cleared of clumps by low-speed centrifugation, then resuspended in phosphate-buffered saline (PBS), and washed three times prior to use. To produce a cell lysate, M. avium subsp. paratuberculosis strain 316F cells were disrupted by mechanical treatment as previously described (12). The M. avium subsp. paratuberculosis cell lysate and intact cell suspensions of M. avium subsp. paratuberculosis 316F, M. avium subsp. avium, and Mycobacterium phlei in PBS-4% skim milk powder were used to screen homologous and cross-reactive antibodies. Phagemid particles were rescued from the scFv library and used to pan against mycobacterial antigen targets that had been immobilized on Immunotubes (MaxiSorb, Nunc, Denmark) as described previously (15). The expression library was panned with two experimental designs. One part of the library was panned twice against an M. avium subsp. paratuberculosis lysate and once against M. avium subsp. avium whole cells. Another part of the library was panned four times against M. avium subsp. paratuberculosis 316F whole cells. Both sublibraries were preabsorbed against M. phlei cells (1011 to 1012 scFv displaying phage were incubated with 200 μl of M. phlei cells for 1 h). Bound phages were released by the addition of 100 mM glycine (pH 2.2) followed by neutralization with 2 M Tris (pH 7.0). Rescued single clones were tested by phage ELISA using 96-well ELISA plates (MaxiSorb, Nunc, Denmark) that had been coated with M. avium subsp. paratuberculosis strain 316F cell lysate. Bound phages were detected with monoclonal antibody anti-M13 conjugated with horseradish peroxidase (Amersham Biosciences, Castle Hill, NSW, Australia) and visualized with TMB substrate (Sigma, Australia). BstN1 (New England Biolabs, Beverly, MA) fingerprinting was undertaken as described previously (10).
Soluble expression and immobilized metal affinity chromatography of scFv fragments.
For periplasmic expression of scFv antibody fragments, 250 ml of 2× yeast-tryptone medium, supplemented with 25 μg/ml chloramphenicol, was inoculated overnight with a culture of E. coli JM83 harboring the expression plasmid pAK500 (containing the scFv insert). Expression was induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) when the culture optical density at 600 nm (OD600) reached 0.5 to 0.6. After 18 h of incubation at 25°C with vigorous aeration, the cells were collected by centrifugation and resuspended to 5% (vol/vol) of the initial culture volume in TES bugbuster (Novagen, Madison, WI), followed by incubation with 1 mg/ml lysozyme (Sigma) and 5 units/ml DNase I (New England Biolabs) for 20 min. Purification of scFv protein by immobilized metal affinity chromatography was done with Ni-nitrilotriacetic acid resin slurry (QIAGEN, Hildne, Germany) packed into a PD-10 column (Pharmacia Ltd., Bucks, United Kingdom) according to the manufacturer's recommendation. Protein levels were determined via BCA assay (Bio-Rad Laboratories, Auckland, New Zealand).
SDS-polyacrylamide gel electrophoresis (PAGE) and immunoblot analysis.
The molecular masses and purity of the scFv were monitored by electrophoresis on a 10% (wt/vol) sodium dodecyl sulfate (SDS)-polyacrylamide gel under reducing conditions. The gels were stained with Coomassie blue staining solution (0.25% [wt/vol] Coomassie blue R250 in 45:45:10 methanol-water-glacial acetic acid) and destained under the same solvent system.
Mycobacterial lysate was generated as described above. Approximately 20 μg of the lysate either was treated with proteinase K for 30 min at 50°C or remained untreated. The lysate was separated on an SDS-polyacrylamide gel and transferred to a nitrocellulose membrane by electrophoresis using a wet blotter (Bio-Rad). The membrane was blocked using QIAGEN blocking solution overnight at 4°C, washed five times with PBS supplemented with 0.05% Tween 20 and incubated with purified antibody fragments (5 μg/ml) for 1 h at room temperature. After 4 washes with PBS-0.05% Tween 20, antibody fragments were detected with a 1:1,000 dilution of an horseradish peroxidase-conjugated anti-His antibody (Penta His; QIAGEN), and the signal was developed with SuperSignal Pico substrate (Pierce Chemical Co., Rockford, IL).
Flow cytometric analysis.
Single-cell suspensions were generated by resuspending approximately 100 mg of mycobacterial culture (grown on agar plates) in 1 ml PBS and conducting 3 to 6 centrifugation steps at 100 × g (5 min at 4°C). The pellets were discarded, and the supernatant was adjusted to an OD600 of 1.0 after the last centrifugation step. Two hundred microliters of this cell suspension was centrifuged at 1,000 × g for 20 min at 4°C, and the bacterial pellet was further analyzed. For inactivation experiments, cells were heat inactivated for 1 h at 80°C, fixed with 4% paraformaldehyde for 15 min at room temperature, or left untreated. All of the following steps were performed on ice, and washes were done in PBS, pH 7.4, supplemented with 10% heat-inactivated fetal calf serum. Cells were blocked for 1 h, washed 3 times, and stained with 10-μg/ml purified antibody fragments. Penta-His Alexa Fluor 488 (QIAGEN) was used as the secondary antibody (1:10) for 1 h. Thirty thousand cells were acquired and analyzed using a FACScan flow cytometer (Becton Dickinson, San Jose, CA) without gating.
Stress response.
Studies were conducted to assess the influence of intrinsic factors (stage of growth phase) and extrinsic stressors (heat, pH, and oxidation) on patterns of surface labeling by scFv against mycobacterial cells. To assess the influence of growth phase, M. avium subsp. avium cells were harvested at an early phase of culture (4 days postseeding) or at a late phase (3 weeks), then washed, prepared, and labeled with scFv antibodies for flow cytometry as above. To assess the influence of extrinsic stressors, mid-log-phase M. avium subsp. avium cells were harvested and washed as above and then resuspended in Middlebrook broth. Broth was supplemented with 0.2 or 2 mM H2O2 (Sigma) to assess the oxidative effect, or the pH was adjusted to 4.0 or 10 to assess the pH effect. After 2 h of incubation at 37°C, cells were washed with fresh Middlebrook broth and incubated for an additional 2 h at 37°C in Middlebrook broth without additives. Prior to labeling and analysis, cells were centrifuged at 1,000 × g for 15 min. To assess the heat-shock effect, cells were heat stressed at 52°C for 20 min, then cooled to room temperature, incubated for 2 h in fresh Middlebrook broth, centrifuged, labeled, and analyzed by flow cytometry. In all cases, patterns of surface labeling by scFv antibodies were compared against nonstressed mid-log-phase growth cells.
Effect of recombinant antibodies on mycobacterial cell growth characteristics in vitro.
M. avium subsp. avium was grown in Middlebrook medium supplemented with glycerol and without Tween 80. A 2-week-old culture was serial diluted from 1/100 to 1/400 with fresh medium in a 96-well culture plate (Becton Dickinson), and scFv were added at a final concentration of 5 μg/ml. Cells were grown for 48 h and then observed with an inverted microscope to describe culture growth characteristics. Subsequently, cell cultures were resuspended by vigorous pipetting to create uniform single-cell suspensions, and the OD600 was measured via spectrophotometry.
Bacterial cell separation and isolation from fecal samples.
To investigate the relative binding of antibodies to M. avium subsp. paratuberculosis in a sample predominated by a different mycobacterial species, M. avium subsp. paratuberculosis and M. phlei cells were labeled either with N-hydroxysulfosuccinimide-fluorescein (Pierce) or tetramethylrhodamine B isothiocyanate (TRITC) (Sigma) as described previously (11). Quantification of cell numbers was determined by visually counting labeled cells in a Neubauer hematocytometer chamber under fluorescence. M. avium subsp. paratuberculosis cells were mixed with M. phlei at a calculated ratio of 1:1,000 (density, 2 × 106 cells/ml). Magnetic beads (Dynabeads Talon; Dynal, Oslo, Norway) were washed 5 times with PBS before purified antibody fragments were bound by incubating approximately 0.6 mg of beads in 0.5 ml antibody fragment solution (0.5 mg/ml) at 4°C overnight. Magnetic beads were separated from unbound proteins by 5 washes with PBS. Antibody-bound beads (0.6 mg) were then incubated for 1 h with 2 × 106 total mycobacterial cells in 1 ml PBS. Beads and bound bacteria were washed 5 times with PBS, resuspended in 50 μl PBS, and counted under fluorescence.
In a separate experiment to determine the ability of antibodies to recognize M. avium subsp. paratuberculosis cells in a natural sample, 1 g of feces from either red deer (Cervus elaphus) or sheep was first diluted in 3 ml PBS and heat-inactivated for 1 h at 80°C. Fibrous material was removed by centrifugation at 100 × g for 3 min. The supernatant was diluted 1:2 in PBS, and this separation step was repeated once. The heat-treated fecal samples were then spiked with a serial dilution of heat-inactivated M. avium subsp. paratuberculosis cells, ranging from 102 to 106 cells/sample. scFv were coupled to beads as described above. Approximately 0.6 mg of coupled beads was incubated with spiked fecal sample for 1 h at room temperature, with constant gentle agitation of the tubes. Beads and bound bacteria were then removed from the samples via magnetic field separation and washed 10 times with PBS. Finally, bacteria were eluted from the beads with glycine, pH 2.2, and neutralized with Tris, pH 7.0. Three microliters of these eluted samples was mixed with 47 μl PCR mix bearing IS900-specific primers to identify M. avium subsp. paratuberculosis cells. IS900 PCR was done as previously described (14).
Statistical analysis.
Results were compared by using two-way analysis of variance, and significant differences were tested by the Bonferroni-Dunn post hoc test. All statistical analysis was performed using InStat software (GraphPad, Inc., San Diego, CA).
Nucleotide sequence accession numbers.
The GenBank accession numbers for the SurfS1.2 and SurfS2.2 sequences are DQ858289 and DQ858290, respectively.
RESULTS
Isolation of M. avium subsp. paratuberculosis-specific ovine antibody fragments.
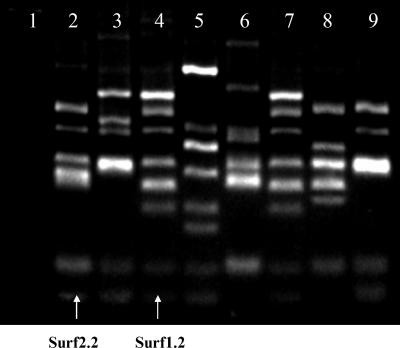
Sheep were infected with M. avium subsp. paratuberculosis to provide a source of biased immunoglobulin genes for library construction. An scFv library containing approximately 5 × 106 independent clones was constructed from immunoglobulin genes amplified by reverse transcription-PCR from the spleens, blood, and bone marrow of three sheep with high antibody titers against M. avium subsp. paratuberculosis and significant weight loss. The resulting phage display library was panned against MAC antigens. The first subset of the library was panned twice against an M. avium subsp. paratuberculosis cell lysate and once against living M. avium subsp. avium cells with 6 preabsorption steps against M. phlei. Seventy-one clones were analyzed by phage ELISA; 38 of these clones gave ELISA signals approximately three times higher than background (i.e., signal generated against nonrecombinant E. coli lysate). Of these, 8 different clones were identified by BstNI fingerprints (Fig. 1). The second subset of this library was panned 4 times against M. avium subsp. paratuberculosis whole cells with 6 preabsorption steps against M. phlei. Twelve of 62 clones analyzed by this procedure gave positive signals by ELISA. Three of these clones gave a distinct BstNI fingerprint (data not shown). Sequence analysis identified that two clones were common to both library subsets. These two clones were designated Surf1.2 and Surf2.2 and are indicated in Fig. 1.
FIG. 1.
BstNI fingerprinting of scFv genes from the first subset of an ovine M. avium subsp. paratuberculosis-specific phage-displayed antibody. Clones were selected for reactivity against M. avium subsp. paratuberculosis cells via three rounds of panning against M. avium subsp. paratuberculosis cell lysate and confirmed positive by ELISA. The selected antibody genes were amplified by PCR, digested for 3 h with BstNI, and analyzed by electrophoresis after resolution through a 4% (wt/vol) agarose gel. Lane 1, negative control; lane 2, clone SurfS2.2; lane 4, SurfS1.2; lanes 3 and 5 to 9, other scFv clones.
Expression and characterization of ovine scFv against mycobacterial antigens.
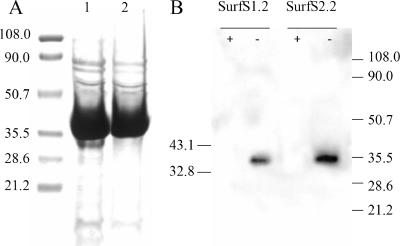
All scFv genes from seven unique positive clones were subcloned from the phage display vector pAK100 to the periplasmic expression vector pAK500, resulting in a 35.5-kDa scFv protein. One of these clones was not expressed in E. coli at a detectable level, despite successful cloning and absence of mutations (data not shown). Yields of the other purified antibody fragments were in the range of 1.5 to 2.5 mg/liter culture broth, with acceptable purity similar to that shown for SurfS1.2 and SurfS2.2 (Fig. 2A). SDS-PAGE-Western blotting identified target specificity for a 34-kDa determinant by two cloned antibody sources (Fig. 2B); the signal was abolished by pretreatment of mycobacterial cell lysate with proteinase K.
FIG. 2.
Purification of two selected scFv clones and antigen recognition in M. avium subsp. paratuberculosis cell lysate. (A) Purification of scFv His-tagged fusion proteins SurfS1.2 (lane 1) and SurfS2.2 (lane 2). scFv were expressed using the vector pAK500 in JM83 E. coli host cells for 18 h at 25°C. The cells were lysed, the supernatant collected, and the fusion protein purified by immobilized metal affinity chromatography. Approximately 1% of purified scFv from a 200-ml culture was resolved by 10% SDS-PAGE and stained with Coomassie-brilliant blue. (B) Immunoblotting of M. avium subsp. paratuberculosis lysate and detection with SurfS1.2 or SurfS2.2. Approximately 20 μg of mycobacterial proteins were either digested with proteinase K (+) or remained untreated (−) and resolved on a 10% SDS-PAGE gel, and the products were transferred to nitrocellulose and probed with peroxidase-conjugated anti-His antibodies. The figure depicts strong binding of both antibodies to a proteinase-susceptible 34-kDa determinant.
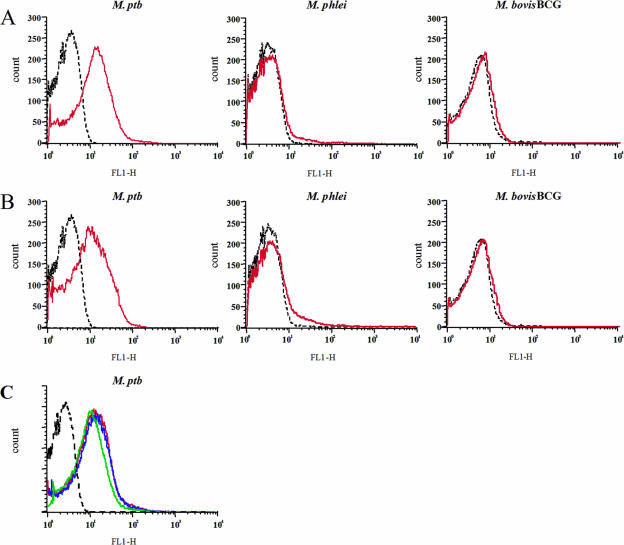
Purified antibody fragments were used to label living M. avium subsp. avium cells, and surface staining was analyzed by flow cytometry. Only 2 of the 7 expressed clones gave a detectable signal in fluorescence-activated cell sorter analysis, these being designated SurfS1.2 and SurfS2.2. Both antibody fragments also stained M. avium subsp. paratuberculosis cells but did not cross-react with M. bovis BCG cells or with M. phlei cells (Fig. 3A and B). Simultaneous labeling of M. avium subsp. paratuberculosis cells with both SurfS1.2 and SurfS2.2 indicated that the surface-binding signal was similar to that obtained using either antibody in isolation (Fig. 3C). Heat inactivation of the bacterium (80°C/1 h) led to a minor increase in the fluorescence signal after staining with either SurfS1.2 or SurfS2.2, while fixation with 4% paraformaldehyde did not alter the signal (data not shown), indicating that the antibody fragments were robust to fixation processes used in flow cytometry.
FIG. 3.
Flow cytometry to detect Mycobacterium sp. surface antigen recognition by selected scFv clones SurfS1.2 and SurfS2.2. Data represent fluorescence intensity histograms generated by the surface binding of scFv clones SurfS1.2 and SurfS2.2 to different Mycobacterium sp. targets. Each histogram depicts 30,000 acquired mycobacterial targets. Red lines represent signals generated using SurfS1.2 (A) or SurfS2.2 (B); black lines represent signals from coculture of bacteria with a non-target-specific scFv. Note the specific signal generated against M. avium subsp. paratuberculosis targets only. (C) M. avium subsp. paratuberculosis (M. ptb) cells were stained with either SurfS1.2 (red line) or SurfS2.2 (green line) or with both antibodies (blue line). Controls were stained with an irrelevant scFv (dashed black line). No increase in the fluorescence intensity was observed by the addition of both scFv simultaneously over the signal observed using either antibody alone.
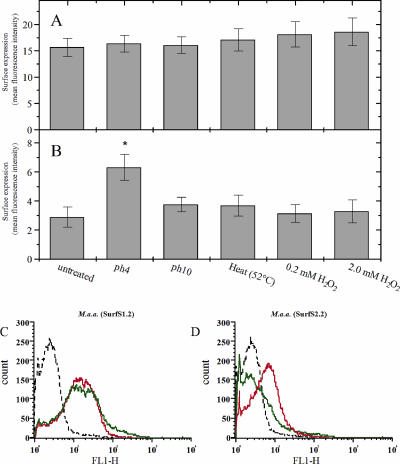
Further flow cytometric analysis using SurfS1.2 and SurfS2.2 indicated that they both displayed surface binding activity against M. avium subsp. avium cells (Fig. 4). The patterns of antibody surface binding were largely unaffected by prestressing bacteria due to high pH, temperature shock, or oxidative stress (Fig. 4), with the only exception being preexposure of M. avium subsp. avium cells to pH 4 conditions, which increased surface binding of SurfS2.2 (Fig. 4B). The surface labeling intensity due to SurfS1.2 binding appeared similar in early- and late-growth-phase samples of M. avium subsp. avium cells (Fig. 4C); in contrast, the signal provided by SurfS2.2 appeared diminished in the early-phase samples (Fig. 4D).
FIG. 4.
Influence of extrinsic stressors or the intrinsic growth phase of M. avium subsp. avium (M. a.a.) on the subsequent patterns of antibody binding by scFv clones SurfS1.2 and SurfS2.2. (A and B) Live M. avium subsp. avium cells were subjected to pH stress (pH 4 and pH 10, 2 h), heat stress (52°C, 15 min) or oxidative stress (0.2 mM and 2.0 mM H2O2, 2 h) prior to incubation with scFv clones SurfS1.2 and SurfS2.2. Patterns of surface antigen expression were examined by flow cytometry, and data were expressed as units of fluorescence intensity due to the binding of SurfS1.2 (A) or SurfS2.2 (B). The asterisk indicates a significant increase in signal generation by pH 4-treated M. avium subsp. avium cells exposed to SurfS2.2 in comparison to nontreated control cells (P < 0.05). (C, D) M. avium subsp. avium cells were harvested at different phases of the growth cycle and labeled with scFv clone SurfS1.2 (C) or SurfS2.2 (D). Green lines represent antibodies incubated with fresh cultures (incubated for 4 days), red lines represent antibodies incubated with late-phase cultures (3-week incubation), and black lines represent signal obtained using a non-target-specific antibody. Note the down-regulated surface binding by SurfS2.2 in late-phase cultures (D).
Influence of surface-bound scFv on the growth of M. avium subsp. avium.
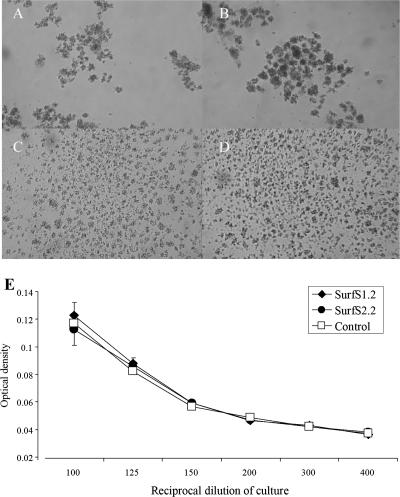
To determine whether surface-bound SurfS1.2 or SurfS2.2 could influence mycobacterial growth, M. avium subsp. avium was grown in Middlebrook broth without dispersal reagents such as Tween 80 in the presence or absence of SurfS1.2 or SurfS2.2. In the absence of dispersal reagents, mycobacterial cells readily formed clumps (Fig. 5A); clumping was clearly reduced in cultures by the inclusion of SurfS1.2 (Fig. 5C) or SurfS2.2 (Fig. 5D). Inclusion of an irrelevant scFv control did not alter the clumping characteristics of M. avium subsp. avium (Fig. 5B). Following coculture of target cells with antibodies, samples were resuspended and OD measurements taken; over a serial dilution range, there was no obvious difference in the end-point growths of cells that had been incubated with and without Surf antibodies (Fig. 5E).
FIG. 5.
Effect on bacterial growth of the inclusion of SurfS1.2 and SurfS2.2 with M. avium subsp. avium cells. One-week-old cultures of M. avium subsp. avium were harvested from flasks and further cultured for 48 h in the presence of PBS (control) (A), an irrelevant scFv (B), or 5 μg/ml of SurfS1.2 (C) or SurfS2.2 (D). Cell colony morphology was examined visually under a phase-contrast microscope (A to D). Note the prevention of bacterial cell clumping by the inclusion of antibodies. All bacterial cell suspensions were then resuspended by vigorous pipetting, and the OD600 was assessed (E) (data refer to mean OD ± standard errors of the means for 1/100 to 1/400 dilutions of the cultures).
Immunomagnetic separation of M. avium subsp. paratuberculosis from M. phlei.
Magnetic beads were coated with either SurfS1.2, SurfS2.2, or an irrelevant scFv control and used for cell separation. To visually distinguish M. avium subsp. paratuberculosis from M. phlei, cells were fluorescently labeled with either fluorescein isothiocyanate or TRITC as described above. An influence of the fluorescent dye was excluded by repeating the experiment and labeling mycobacterial species with the opposite dye. Magnetic bead separation with both SurfS1.2 and SurfS2.2 resulted in an enrichment of M. avium subsp. paratuberculosis by a factor of approximately 106 from a cell suspension with a ratio of 1:1,000 M. avium subsp. paratuberculosis to M. phlei cells (Fig. 6). Magnetic beads coated with an unrelated antibody fragment or with an antibody fragment that was negative in fluorescence-activated cell sorter analysis (SurfS1.23) led to loss of bacterial cells and no enrichment of M. avium subsp. paratuberculosis (data not shown).
FIG. 6.
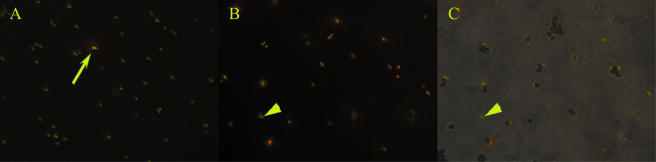
Use of scFv antibodies to isolate M. avium subsp. paratuberculosis cells from mixed Mycobacterium sp. samples. Magnetic beads were coated with SurfS1.2 antibodies and used to separate mycobacterial cells from a mixture comprising M. phlei and M. avium subsp. paratuberculosis cells at a ratio of 100:1. To differentiate, M. phlei cells were labeled with fluorescein isothiocyanate (green) and M. avium subsp. paratuberculosis cells with TRITC (red), and cells were examined at ×400 magnification using a UV microscope. (A) The arrow indicates a single M. avium subsp. paratuberculosis cell among several M. phlei cells in the preseparation mixture. (B) After immunomagnetic separation, M. avium subsp. paratuberculosis cells were highly enriched, with only a few M. phlei cells present (arrowheads). (C) Direct binding of M. avium subsp. paratuberculosis cells to SurfS1.2-bearing magnetic beads (the arrowhead identifies a single unbound M. phlei cell).
Immunomagnetic separation of M. avium subsp. paratuberculosis from fecal samples and detection by PCR.
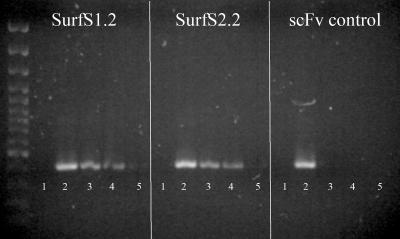
The utility of the isolated scFv antibody fragments for detection of M. avium subsp. paratuberculosis in fecal samples was evaluated by first using the antibodies to capture bacterial cells and then determining whether DNA from these cells would remain amplifiable by IS900 PCR. PBS-diluted fecal samples were spiked with heat-killed M. avium subsp. paratuberculosis cells ranging from 102 to 106 cells per g of feces, which were subsequently recovered by scFv antibody magnetic separation and subject to PCR amplification. A minor reduction in the PCR product yield was observed after the addition of magnetic beads (data not shown). Detection of the lower limit of M. avium subsp. paratuberculosis cells (102/g) was possible, but the result was shown to be inconsistent; in contrast, detection of samples with 103 M. avium subsp. paratuberculosis cells/g feces or higher was positive and consistent (Fig. 7). Beads coated with irrelevant antibody fragments did not result in positive IS900 PCR (Fig. 7).
FIG. 7.
Recovery and identification of IS900-positive cells from M. avium subsp. paratuberculosis-spiked fecal samples, using scFv antibodies as the primary purification method. Magnetic beads were coated with SurfS1.2 or SurfS2.2 monoclonal antibodies and employed as the primary purification method to isolate target bacterial cells from ovine fecal samples that had been spiked with various numbers of M. avium subsp. paratuberculosis cells. A monoclonal antibody with no specificity for M. avium subsp. paratuberculosis was used as a control. Following purification, captured cells were subjected to PCR amplification for the IS900 conserved region. The figure represents an ethidium bromide-stained gel bearing the PCR-amplified products. Lane 1, unspiked fecal sample; lane 2, 104 M. avium subsp. paratuberculosis cells alone (positive control); lanes 3 to 5, fecal sample spiked with 104 to 102 M. avium subsp. paratuberculosis cells.
DISCUSSION
The present study has used phage display technology to develop single-chain antibodies with specificity against surface determinants of M. avium subsp. paratuberculosis. This is the first report of the successful cloning of scFv with high affinity from a Mycobacterium-infected ruminant and raises the possibility that phage display technology could be utilized to further characterize antibody reactivity in other ruminant species (including cattle and deer) that are economically important and naturally susceptible hosts for virulent mycobacteria, including M. avium subsp. paratuberculosis and M. bovis. Our previous studies have identified serological reactivity of deer infected with M. avium subsp. paratuberculosis against a panel of recombinant M. avium subsp. paratuberculosis antigens (9), although a similar screening of scFv against recombinant antigens was not employed in the present study. Nevertheless, it is known that in M. avium subsp. paratuberculosis-infected cattle, serological responses are predominantly directed against an immunodominant 34-kDa protein of the A36 complex (26), while in the present study, Western blotting identified scFv specificity against a proteinase-susceptible 34-kDa determinant. In contrast, Cummings et al. (5) reported that among a panel of recombinant single-chain antibodies developed from mice immunized with killed M. tuberculosis cells, none showed reactivity against the immunodominant 65-kDa antigen of that bacterium. This differential development of antibody reactivity against immunodominant B-cell epitopes may well reflect the original derivation of immunoglobulin hypervariable genes, viz., cDNA derived from mice primed by injection with killed mycobacteria (5) versus that derived from sheep infected with live mycobacteria (present study).
Flow cytometry in the present study identified that scFv binding activity was detected strongly at the mycobacterial cell surface and that this activity was common against both M. avium subsp. paratuberculosis and M. avium subsp. avium cells, but not against M. bovis BCG or M. phlei. The common reactivity of SurfS1.2 and SurfS2.2 against M. avium subsp. paratuberculosis and M. avium subsp. avium surface antigens is not surprising, given the close phylogenetic relatedness of these two species. In a practical sense, this common reactivity allowed further study of the interaction of scFv with live mycobacteria using M. avium subsp. avium cells as the target, thus overcoming the inherent difficulties in culturing M. avium subsp. paratuberculosis in vitro (1). To that end, SurfS1.2 or SurfS2.2 was included in cultures of live, actively metabolizing M. avium subsp. avium cells. The noted clumping of mycobacterial cells (which is evident unless a dispersal agent such as Tween 80 is included in the growth medium) was prevented by the coinclusion of either antibody, suggesting that surface binding by SurfS1.2 and SurfS2.2 impedes cell aggregation. Despite this, there was no obvious effect on bacterial culture optical density, indicating that these antibodies did not impair mycobacterial growth rates in vitro, as has been shown for scFv against Helicobacter pylori (4). The fact that SurfS1.2 and SurfS2.2 do not inhibit the performance of live mycobacterial cells makes them suitable as tools for target cell separation and subsequent culturing, as a toxic effect is not desirable prior to culturing when using antibodies in a diagnostic context. Interestingly, antigen recognition in live M. avium subsp. avium was unaffected by intrinsic bacterial growth characteristics or by extrinsic stressors of the bacteria when using SurfS1.2 as a probe, suggesting that the epitope recognized by this antibody is expressed constitutively; in contrast, preexposure of live M. avium subsp. avium cells to low pH resulted in an up-regulation of the surface binding activity of SurfS2.2, while in contrast, cells harvested during the early phase of their log growth showed a reduced labeling intensity with SurfS2.2, which may indicate that the epitope determinant recognized by SurfS2.2 is under regulatory control by the bacterium.
A previous study has reported the potential of polyclonal rabbit anti-M. avium subsp. paratuberculosis antibodies to be used as a separation tool to detect M. avium subsp. paratuberculosis cells in milk or fecal samples (14). Here, we have reported that SurfS1.2 and SurfS2.2 single-chain antibodies can bind to and selectively isolate target M. avium subsp. paratuberculosis cells, both from culture samples bearing mixed mycobacterial species and from feces artificially spiked with M. avium subsp. paratuberculosis cells. The use of polyclonal antibodies for target cell recognition suffers from the potential for cross-reactivity against environmental species, including environmental mycobacteria like M. phlei (8), and methods based on polyclonal antibodies may be very difficult to standardize. Therefore, in this initial study, we have shown that ovine scFv can be used to specifically separate M. avium subsp. paratuberculosis from M. phlei and to isolate M. avium subsp. paratuberculosis from fecal samples. As the scFv did not cross-react with M. phlei or M. bovis BCG, the immunomagnetic separation method may be unaffected by the presence of environmental mycobacteria; however, since both scFv cross-reacted with M. avium subsp. avium, an ancillary verification test for M. avium subsp. paratuberculosis (such as specific IS900 PCR) is essential.
In this study, the detection limit of target mycobacterial cells was approximately 103 bacteria/4 ml fecal sample using SurfS1.2 or SurfS2.2 scFv, which is higher than that described for immunomagnetic bead separation with polyclonal antibodies (14). While that may be a result of an initial centrifugation step employed here to separate bacteria from fibrous material present in sheep feces (and the consequent loss of some bacteria), the important fact remains that isolated M. avium subsp. paratuberculosis cells could be further identified using IS900 PCR (without prior DNA amplification), indicating the compatibility of these two screening methodologies in tandem. In a practical sense, the opportunity to undertake diagnostic PCR without DNA purification, as has been routine for diagnosis of M. tuberculosis (2), would reduce the cost and time of M. avium subsp. paratuberculosis detection in field practice.
In summary, we have demonstrated here the utility of two scFv, SurfS1.2 and SurfS2.2, in several different applications, like immunoblotting, flow cytometry, and immunomagnetic separation of the M. avium subsp. paratuberculosis pathogen from mixed cultures and fecal samples. A recent review (21) has highlighted the utility of employing phage display technology in the study of microbial infectious diseases, including defining protein-ligand interactions, epitope mapping, and screening for receptor agonists or antagonists. Further, recent studies have reported the use of recombinant phage technology to establish scFv libraries with defined specificity for pathogens, such as coronavirus (19) and Clostridium difficile toxin (6), that may have applications in vaccine development or diagnostics for human use. The work reported here has further demonstrated that it may be possible to develop scFv as rapid and cost-effective tools for research in (and diagnosis of) ruminant Johne's disease, applicable to livestock husbandry.
Acknowledgments
We thank the following individuals for their contributions to this work: Phil Farquhar and Colin Mackintosh (AgResearch Invermay) for animal husbandry and necropsies and the staff at the Disease Research Laboratory (University of Otago) and Gary Yates (AgResearch Wallaceville) for culture of M. avium subsp. paratuberculosis. We gratefully acknowledge the assistance of Frank Cross in the preparation of the manuscript.
This research was supported by grants from the Foundation of Research, Science, and Technology (FoRST) and the Disease Research Laboratory. We also acknowledge funding from USDA-ARS and a USDA-NRI-CAP grant to J.P.B.
REFERENCES
- 1.Bannantine, J. P., Q. Zhang, L. L. Li, and V. Kapur. 2003. Genomic homogeneity between Mycobacterium avium subsp. avium and Mycobacterium avium subsp. paratuberculosis belies their divergent growth rates. BMC Microbiol. 3:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barnes, P. F., and M. D. Cave. 2003. Molecular epidemiology of tuberculosis. N. Engl. J. Med. 349:1149-1156. [DOI] [PubMed] [Google Scholar]
- 3.Begg, D. J., R. O'Brien, C. G. Mackintosh, and J. F. Griffin. 2005. Experimental infection model for Johne's disease in sheep. Infect. Immun. 73:5603-5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cao, J., Y. Sun, T. Berglindh, B. Mellgard, Z. Li, B. Mardh, and S. Mardh. 2000. Helicobacter pylori-antigen-binding fragments expressed on the filamentous M13 phage prevent bacterial growth. Biochim. Biophys. Acta 1474:107-113. [DOI] [PubMed] [Google Scholar]
- 5.Cummings, P. J., N. E. Hooper, and S. S. Rowland. 1998. Generation of a recombinant bacteriophage antibody library to Mycobacterium tuberculosis. Hybridoma 17:151-156. [DOI] [PubMed] [Google Scholar]
- 6.Deng, X. K., L. A. Nesbit, and K. J. Morrow, Jr. 2003. Recombinant single-chain variable fragment antibodies directed against Clostridium difficile toxin B produced by use of an optimized phage display system. Clin. Diagn. Lab. Immunol. 10:587-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Falkinham, J. O., III. 1996. Epidemiology of infection by nontuberculous mycobacteria. Clin. Microbiol. Rev. 9:177-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grant, I. R., H. J. Ball, and M. T. Rowe. 1998. Isolation of Mycobacterium paratuberculosis from milk by immunomagnetic separation. Appl. Environ. Microbiol. 64:3153-3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Griffin, J. F., E. Spittle, C. R. Rodgers, S. Liggett, M. Cooper, D. Bakker, and J. P. Bannantine. 2005. Immunoglobulin G1 enzyme-linked immunosorbent assay for diagnosis of Johne's Disease in red deer (Cervus elaphus). Clin. Diagn. Lab. Immunol. 12:1401-1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Griffiths, A. D., S. C. Williams, O. Hartley, I. M. Tomlinson, P. Waterhouse, W. L. Crosby, R. E. Kontermann, P. T. Jones, N. M. Low, T. J. Allison, et al. 1994. Isolation of high affinity human antibodies directly from large synthetic repertoires. EMBO J. 13:3245-3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hayashi, T., S. P. Rao, and A. Catanzaro. 1997. Binding of the 68-kilodalton protein of Mycobacterium avium to αvβ3 on human monocyte-derived macrophages enhances complement receptor type 3 expression. Infect. Immun. 65:1211-1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Homuth, M., P. Valentin-Weigand, M. Rohde, and G. F. Gerlach. 1998. Identification and characterization of a novel extracellular ferric reductase from Mycobacterium paratuberculosis. Infect. Immun. 66:710-716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoogenboom, H. R., A. P. de Bruine, S. E. Hufton, R. M. Hoet, J. W. Arends, and R. C. Roovers. 1998. Antibody phage display technology and its applications. Immunotechnology 4:1-20. [DOI] [PubMed] [Google Scholar]
- 14.Khare, S., T. A. Ficht, R. L. Santos, J. Romano, A. R. Ficht, S. Zhang, I. R. Grant, M. Libal, D. Hunter, and L. G. Adams. 2004. Rapid and sensitive detection of Mycobacterium avium subsp. paratuberculosis in bovine milk and feces by a combination of immunomagnetic bead separation-conventional PCR and real-time PCR. J. Clin. Microbiol. 42:1075-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krebber, A., S. Bornhauser, J. Burmester, A. Honegger, J. Willuda, H. R. Bosshard, and A. Pluckthun. 1997. Reliable cloning of functional antibody variable domains from hybridomas and spleen cell repertoires employing a reengineered phage display system. J. Immunol. Methods 201:35-55. [DOI] [PubMed] [Google Scholar]
- 16.Laffly, E., and R. Sodoyer. 2005. Monoclonal and recombinant antibodies, 30 years after. Hum. Antibodies 14:33-55. [PubMed] [Google Scholar]
- 17.Li, L., J. P. Bannantine, Q. Zhang, A. Amonsin, B. J. May, D. Alt, N. Banerji, S. Kanjilal, and V. Kapur. 2005. The complete genome sequence of Mycobacterium avium subspecies paratuberculosis. Proc. Natl. Acad. Sci. USA 102:12344-12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li, Y., J. Kilpatrick, and G. C. Whitelam. 2000. Sheep monoclonal antibody fragments generated using a phage display system. J. Immunol. Methods 236:133-146. [DOI] [PubMed] [Google Scholar]
- 19.Liu, Z. X., G. H. Yi, Y. P. Qi, Y. L. Liu, J. P. Yan, J. Qian, E. Q. Du, and W. F. Ling. 2005. Identification of single-chain antibody fragments specific against SARS-associated coronavirus from phage-displayed antibody library. Biochem. Biophys. Res. Commun. 329:437-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manning, E. J., and M. T. Collins. 2001. Mycobacterium avium subsp. paratuberculosis: pathogen, pathogenesis and diagnosis. Rev. Sci. Tech. 20:133-150. [DOI] [PubMed] [Google Scholar]
- 21.Mullen, L. M., S. P. Nair, J. M. Ward, A. N. Rycroft, and B. Henderson. 2006. Phage display in the study of infectious diseases. Trends Microbiol. 14:141-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paoli, G. C., C. Y. Chen, and J. D. Brewster. 2004. Single-chain Fv antibody with specificity for Listeria monocytogenes. J. Immunol. Methods 289:147-155. [DOI] [PubMed] [Google Scholar]
- 23.Sabarth, N., R. Hurvitz, M. Schmidt, U. Zimny-Arndt, P. R. Jungblut, T. F. Meyer, and D. Bumann. 2005. Identification of Helicobacter pylori surface proteins by selective proteinase K digestion and antibody phage display. J. Microbiol. Methods 62:345-349. [DOI] [PubMed] [Google Scholar]
- 24.Sergeant, E. S., D. J. Marshall, G. J. Eamens, C. Kearns, and R. J. Whittington. 2003. Evaluation of an absorbed ELISA and an agar-gel immuno-diffusion test for ovine paratuberculosis in sheep in Australia. Prev. Vet. Med. 61:235-248. [DOI] [PubMed] [Google Scholar]
- 25.Stewart, D. J., J. A. Vaughan, P. L. Stiles, P. J. Noske, M. L. Tizard, S. J. Prowse, W. P. Michalski, K. L. Butler, and S. L. Jones. 2004. A long-term study in Merino sheep experimentally infected with Mycobacterium avium subsp. paratuberculosis: clinical disease, faecal culture and immunological studies. Vet. Microbiol. 104:165-178. [DOI] [PubMed] [Google Scholar]
- 26.Vannuffel, P., P. Gilot, B. Limbourg, B. Naerhuyzen, C. Dieterich, M. Coene, L. Machtelinckx, and C. Cocito. 1994. Development of species-specific enzyme-linked immunosorbent assay for diagnosis of Johne's disease in cattle. J. Clin. Microbiol. 32:1211-1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vary, P. H., P. R. Andersen, E. Green, J. Hermon-Taylor, and J. J. McFadden. 1990. Use of highly specific DNA probes and the polymerase chain reaction to detect Mycobacterium paratuberculosis in Johne's disease. J. Clin. Microbiol. 28:933-937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whittington, R. J., I. Marsh, S. McAllister, M. J. Turner, D. J. Marshall, and C. A. Fraser. 1999. Evaluation of modified BACTEC 12B radiometric medium and solid media for culture of Mycobacterium avium subsp. paratuberculosis from sheep. J. Clin. Microbiol. 37:1077-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Winter, G., A. D. Griffiths, R. E. Hawkins, and H. R. Hoogenboom. 1994. Making antibodies by phage display technology. Annu. Rev. Immunol. 12:433-455. [DOI] [PubMed] [Google Scholar]