Abstract
Currently the shikimate pathway is reported as a metabolic feature of prokaryotes, ascomycete fungi, apicomplexans, and plants. The plant shikimate pathway enzymes have similarities to prokaryote homologues and are largely active in chloroplasts, suggesting ancestry from the plastid progenitor genome. Toxoplasma gondii, which also possesses an alga-derived plastid organelle, encodes a shikimate pathway with similarities to ascomycete genes, including a five-enzyme pentafunctional arom. These data suggests that the shikimate pathway and the pentafunctional arom either had an ancient origin in the eukaryotes or was conveyed by eukaryote-to-eukaryote horizontal gene transfer (HGT). We expand sampling and analyses of the shikimate pathway genes to include the oomycetes, ciliates, diatoms, basidiomycetes, zygomycetes, and the green and red algae. Sequencing of cDNA from Tetrahymena thermophila confirmed the presence of a pentafused arom, as in fungi and T. gondii. Phylogenies and taxon distribution suggest that the arom gene fusion event may be an ancient eukaryotic innovation. Conversely, the Plantae lineage (represented here by both Viridaeplantae and the red algae) acquired different prokaryotic genes for all seven steps of the shikimate pathway. Two of the phylogenies suggest a derivation of the Plantae genes from the cyanobacterial plastid progenitor genome, but if the full Plantae pathway was originally of cyanobacterial origin, then the five other shikimate pathway genes were obtained from a minimum of two other eubacterial genomes. Thus, the phylogenies demonstrate both separate HGTs and shared derived HGTs within the Plantae clade either by primary HGT transfer or secondarily via the plastid progenitor genome. The shared derived characters support the holophyly of the Plantae lineage and a single ancestral primary plastid endosymbiosis. Our analyses also pinpoints a minimum of 50 gene/domain loss events, demonstrating that loss and replacement events have been an important process in eukaryote genome evolution.
Chorismate is the common precursor of aromatic compounds including the folates, ubiquinone, and aromatic amino acids. It is synthesized via the shikimate pathway, which catalyzes the step-wise conversion of erythrose-4-phosphate and phosphoenolpyruvate to chorismate. This pathway is present in numerous prokaryotic lineages and in some eukaryotes, including plants and fungi. More recently, evidence of this pathway has been identified in some alveolates (6, 10, 39), including the medically important and plastid bearing Apicomplexa (26, 28), where a full shikimate pathway has been identified in Toxoplasma gondii (6) and an incomplete pathway has been detected in the genomes of several plasmodia species (6, 23, 27). The pathway is a potential therapeutic target, inhibited by the herbicide glyphosate (which targets the shikimate pathway enzyme 5-enolpyruvylshikimate-3-phospate synthase) and fluoroshikimate analogues (that act as subversive substrates), and has been demonstrated to curtail the growth of Plasmodium falciparum in vitro (26, 39). Understanding the evolution of this metabolic pathway, which in some organisms appears both conserved and a key metabolic pathway while in other organisms the opposite is evident, is critical. Furthermore, the absence of the pathway in the Metazoa (38) and the possibility that the shikimate pathway may constitute a therapeutic target makes a better understanding of the distribution and evolution of the shikimate pathway in eukaryotes important.
The pathway consists of seven steps, with several isoenzymes and multiple naming conventions. For simplicity, we adopted the Escherichia coli (Aro-) naming system where appropriate but have used other names where equivalent orthologous genes are absent from the E. coli genome. Step one is performed by 3-deoxy-d-arabino-heptulosonate 7-phosphate (DAHP) synthase, of which there are two nonhomologous types or isoenzymes named DAHP synthase class I (DAHP I) and class II (DAHP II). Step 2 is catalyzed by 3-dehydroquinate synthase, or AroB. Step 3 is also catalyzed by two isoenzymes, 3-dehydroquinate dehydratase types I and II (named here AroD and DHQase II, respectively). Steps 4 to 7 are catalyzed by shikimate 5-dehydrogenase, shikimate kinase, 5-enolpyruvylshikimate-3-phosphate synthase, and chorismate synthase, or AroE, AroL/K, AroA, and AroC, respectively.
The molecular organization and structure of the shikimate pathway enzymes varies considerably between taxonomic groups (8), with a differential distribution of shikimate fusion genes throughout both prokaryotes and eukaryotes. In most prokaryotes examined to date, the seven shikimate pathway enzymes are encoded as separate polypeptides. The plant enzymes are also encoded in separate polypeptides, with the exception of AroD and AroE, which are fused and encode a bifunctional polypeptide (31). Plant shikimate pathway enzymes are nucleus encoded, but many possess N-terminal plastid targeting peptides. Thus, they are assumed to function within the plastid, although this has only been experimentally demonstrated for a selection of shikimate pathway enzymes (32, 42). In contrast, all ascomycetes sampled and T. gondii possess monofunctional DAHP II and AroC genes and a pentafunctional supergene, the arom pathway (6, 9) which includes conserved domains homologous to the genes (in fusion order) AroB, AroA, AroL/K, AroD, and AroE (details of the pathway and naming convention used are also summarized in Fig. 8B).
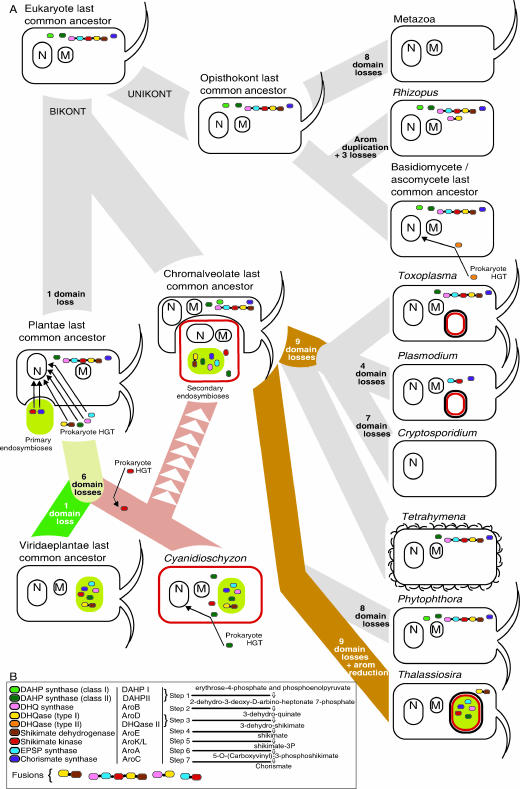
FIG. 8.
(A) Schematic of evolution of the eukaryotes and their shikimate pathway genes using the bikont/unikont eukaryote root (37, 48, 49). Shikimate pathway genes are colored and are in the gene fusion order as indicated in panel B. Mitochondria and nuclear organelles are marked M and N, respectively. Apicoplastid organelles are illustrated as a red capsule, plastids are colored green, and plastids of secondary endosymbiotic origin are colored green with a red border. Secondary endosymbiotic events, hypothetical loss events, and horizontal gene transfers are labeled and are given an arrow to infer the direction and target of the gene transfer. Note that the shikimate pathway genes are localized in the cell only to reflect the likely associated origin, i.e., not to demonstrate genome localization or functional localization. For example, the arom pathway probably arose at a similar time as the eukaryotic cytoplasm, so it is drawn in the cytoplasm; the Plantae shikimate pathway is likely to have arisen at a similar time to the plastid endosymbiosis, so it is drawn associated with the plastid, although in several cases origin is not directly from the plastid progenitor genome. (B) Key for the color coding of the shikimate pathway genes and illustration of the alternative naming for each gene/enzyme. A brief outline of the metabolic pathway is given.
The identification of an arom gene structure in T. gondii and the phylogenetic analyses of individual components of the arom pathway have demonstrated that the T. gondii arom domains group with the ascomycete arom domains, suggesting two potential explanations for the evolutionary origin of the arom gene (6): (i) the arom gene could have formed in one eukaryote lineage and then transferred by horizontal gene transfer (HGT) between the ascomycetes and T. gondii or (ii) the arom pathway could be vertically inherited from the last common eukaryotic ancestor and then lost and/or replaced in other eukaryotic lineages, including the Plantae. Thus, the shikimate pathway potentially represents a rich source of data for understanding eukaryotic evolution including plastid acquisition in the Plantae.
Herein we report the evolutionary relationships of the shikimate pathway genes to address their origin and evolution in eukaryotes. These analyses include sampling from recently complete prokaryotic and eukaryotic genomes, notably expanding the sampling to include seven additional divergent eukaryotic taxa: the oomycete Phytophthora ramorum, the green alga Chlamydomonas reinhardtii, the red alga Cyanidoschyzon merolae, the basidiomycete fungi Ustilago maydis and Cryptococcus neoformans, the zygomycete Rhizopus oryzae, the diatom Thalassiosira pseudonana, and the ciliate Tetrahymena thermophila. We confirm the presence of a fused arom supergene in T. thermophila by cloning and sequencing from cDNA. The majority of the phylogenetic analysis reveals both a unified fungal-chromalveolate clade and a pattern of multiple prokaryote-to-eukaryote gene transfers in both the red and green Plantae lineages by both HGT and endosymbiotic gene transfer (EGT).
MATERIALS AND METHODS
Shikimate pathway gene sampling.
Ascomycete and plant shikimate pathway protein sequences of all nine possible proteins (where available) were used as seeds for BLASTp and tBLASTn searches of all of the eukaryote genome projects available at NCBI genome BLAST server (http://www.ncbi.nlm.nih.gov/sutils/genom_tree.cgi?organism=euk); additional BLAST searches were performed on the Phytophthora, Chlamydomonas, and Cyanidioschyzon genome projects. PSI-BLAST (3 iterations) and BLASTp analyses were conducted against the GenBank nonredundant database to identify additional eukaryote and putative prokaryote homologues. A diverse taxonomic selection (and where possible, consistent between alignments) of putative homologues were aligned using CLUSTAL-X (52) and then manually refined using the alignment program SE-AL (http://evolve.zoo.ox.ac.uk/software.html?id=seal). Sequence data were obtained from The Institute for Genomic Research (TIGR) website at http://www.tigr.org, the Department of Energy Joint Genome Institute at http://www.jgi.doe.gov, and the C. merolae genome website at http://merolae.biol.s.u-tokyo.ac.jp/. The Paramecium tetraurelia and Perkinsus marinus databases at Genoscope (http://www.cns.fr/) and TIGR, respectively, were also searched, but only incomplete sequence reads could be recovered and so were judged unreliable for phylogenetic reconstruction at this stage.
Sequencing of shikimate pathway genes from T. thermophila.
To determine if the five putative T. thermophila components of the arom pathway were transcribed as a true pentafusion gene rather than a gene cluster, we amplified the appropriate regions from cDNA. T. thermophila (a kind gift from Aaron Turkewitz, University of Chicago) were grown at 30°C with gentle rotation in SPP media (1% Proteose peptone, 0.2% dextrose, 0.1% yeast extract, 0.009% ferric EDTA). RNA was extracted from T. thermophila cells using Trizol reagent (Invitrogen) and used to generate cDNA using Superscript III (Invitrogen) according to the manufacturer's instructions. A series of overlapping clones were amplified by PCR, and the oligonucleotide primers are listed in Table S1 in the supplemental material. Amplicons were cloned into the pDRIVE vector by using the QIAGEN PCR cloning kit (QIAGEN) according to manufacturer's instructions. Clones were sequenced by MWG Biotech, Milton Keynes, United Kingdom, and assembled using Sequencher (Gene Codes, Ann Arbor, MI). This revealed a single open reading frame of 4,734 bp encoding a polypeptide of 1,578 amino acids.
Alignment and phylogenetic analyses of shikimate pathway genes.
Nine alignments were made for all shikimate pathway enzyme domains using CLUSTAL-X (52) and then manually corrected using SE-AL (http://evolve.zoo.ox.ac.uk/software.html?id=seal). These alignments include homologues from a wide diversity of prokaryote taxa with consistent taxon sampling between alignments where possible. Nonhomologous insertions and sequence characters that could not be aligned with confidence were removed from the alignments. The AroD enzyme domain was, in our opinion, too divergent to permit confident alignment and phylogenetic analyses across a broad sampling of taxa. We also excluded the hypothetical Plasmodium 5-enolpyruvylshikimate-3-phosphate synthase-shikimate kinase gene (aroA-K/L) (27) because we could not align this sequence for phylogenetic analyses with confidence. Since so many genes were included in the proceeding analyses, a table with GenBank accession numbers is not provided. However, all masked alignments, which contain GenBank accession numbers, are available from the corresponding author upon request.
Prior to tree reconstruction, each alignment was analyzed using MODELGENERATOR (http://bioinf.nuim.ie/software/modelgenerator) to find the best model for each alignment (see Table S2 in the supplemental material). MRBAYES 3.1.2 (41) was run with two separate MCMCMC analyses for 1,000,000 generations at a sampling frequency of 100 generations. Each MCMCMC run had four MCMC chains (three heated and one cold; heat parameter, 0.2). Comparisons of likelihood score and model parameter values and topologies within and between the two independent runs confirmed that the tree log-likelihood scores and parameters had reached a plateau and converged by 110,000 generations at the latest. Consequently, a maximum of 1,100 samples were excluded as a burnin.
Maximum likelihood (ML) distance bootstrap values (from 1,000 replicates) were obtained using Tree-Puzzle 5.1 (43) for parameter estimation (substitution model, eight multivariant plus invariant sites or only eight multivariant dependent on MODELGENERATOR analyses; see Table S2 in the supplemental material) and in coordination with Puzzleboot (21) to obtain distance matrices. Programs from the PHYLIP package (13) were used to create pseudoreplicate data sets (SEQBOOT), calculate distance trees (NEIGHBOR), and assemble a bootstrap consensus tree (CONSENSE). In addition, 100 fast ML (PHYML) (17) bootstrap replicates were run for each alignment with the model as before. In the case of the AroDE fusion gene, the analyses demonstrated contrasting results with moderate support. Consequently, we completed 1,000 parsimony bootstrap analyses using PROTPARS (13) with 3× jumbling to offer an additional investigation of topology support.
The phylogeny results of the AroDE fusion gene analyses and the phylogeny of AroE on its own produced ambiguous results, with the branching order between the plants, red algae, and Rhodopirellula baltica. Consequently, we took the MRBAYES tree and flipped the red algae-R. baltica branching order and, enforcing Plantae monophyly, recalculated the branch lengths using ML in P4 (14), using the model specified using the MODELGENERATOR results (see Table S2 in the supplemental material). This process was repeated for both the large taxa AroE data sets (see Fig. 4A) and the AroDE fusion gene data set (see Fig. 4B). The alternative topologies were evaluated using Shimodaira-Hasegawa (SH) and approximately unbiased (AU) (44) using the program CONSEL (45) invoked through P4 (14).
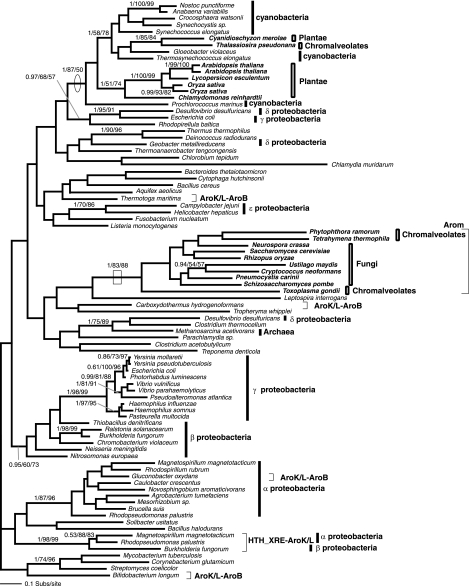
FIG. 4.
Phylogeny of AroE. (A) Phylogeny of AroE was calculated from an amino acid alignment of 97 sequences and 163 characters. The node demonstrating monophyly of the arom-possessing taxa and suggesting the reduction of arom in T. pseudonana is boxed. Note also the PheA domain (chorismate mutase EC 5.4.99.5) is fused to the AroE in Clostridium acetobutylicum. The node supporting the close relationship of the R. baltica and land plants is circled. Note that the presence of C. merolae as the immediate out-group to this clade prompted the analysis in panel B. (B) Phylogeny of the AroDE fusion gene calculated from an amino acid alignment of 16 sequences and 419 characters. In this analysis, 1,000 protein parsimony bootstrap analyses were also conducted to provide more data in view of the uncertainties of where C. merolae branches. The fourth support value is the result of the 1,000 protein parsimony bootstraps. Branching support within the land plants and the Chlamydiales is not shown, to reduce figure complexity. Alternative C. merolae branching position and bootstrap support is shown using the gray hatched line.
In addition, the AroE phylogeny showed weak support for the separation of the Plantae clade from all the other eukaryotes. To test the possibility that the Plantae clade may be a sister to the other eukaryotes, monophyly of the eukaryotes with R. baltica was constrained. This represents a radical topological alteration over the MrBayes result, and consequently, alternative topologies were optimized under the constraint using parsimony methods implicated in PAUP (51). The branch lengths of these synthetic phylogenies were optimized using ML in P4 (14), with the model specified using the MODELGENERATOR results (see Table S2 in the supplemental material). The alternative topologies were evaluated using SH (45) and AU (44) using the program CONSEL (45) invoked through P4 (14).
Ribosomal operon structural comparisons.
Comparisons of ribosomal operon structure had previously indicated a close evolutionary relationship between Chlamydia, cyanobacteria, and the plastid progenitor, consistent with past analyses of the AroDE fusion gene (5). Given that our analysis does not support the previous findings for this particular data set (5), we extended the ribosomal operon structure analysis previously used to support the Chlamydia-plastid sisterhood to include other prokaryotes, such as R. baltica. This was done to test if a different prokaryote may also be closely related to the eubacterial plastid progenitor under the same criteria used by Brinkman et al. (5). To compare ribosomal operon structures, initial tables were made using the genome annotation reported to GenBank. If a gene was missing from the operon and not present in the remaining genome annotation, we used BLASTp to determine if the protein was correctly predicted but has not been given a standard annotation. If we could not identify the protein with a BLASTp search, we then used a tBLASTn search against the genome nucleotide sequence to see if the protein was incorrectly predicted or classified as a pseudogene. If we could not find the gene with either of these methods, we reported the gene as missing.
Nucleotide sequence accession number.
The sequences of arom and DAHP II have been deposited in GenBank under the accession numbers AY601281 and AY787836, respectively.
RESULTS
Comparative genomic and molecular biological sampling of shikimate genes.
No shikimate homologues were identified from the present Entamoeba histolytica, Dictyostelium discoideum, Trypanosoma cruzi, Trypanosoma brucei, Leishmania major, Trichomonas vaginalis, or Giardia intestinalis genome projects (last sampled December 2005). Putative arom homologues, as well as DAHP II and AroC genes, were identified in T. thermophila, P. ramorum, and the basidiomycetous and zygomycetous fungi. In addition, R. oryzae possessed an extra fusion gene as a reduced form of the arom gene (see Fig. 2). Analyses of T. pseudonana, C. reinhardtii, and C. merolae revealed homologues for all seven steps of the shikimate pathway, but no arom structure was present and all five functionally equivalent genes had a domain fusion arrangement similar to that seen in land plants. The C. reinhardtii sequence data recovered for DAHP II, the AroDE fusion gene, and the AroC gene were partial and so were excluded from the respective phylogenies.
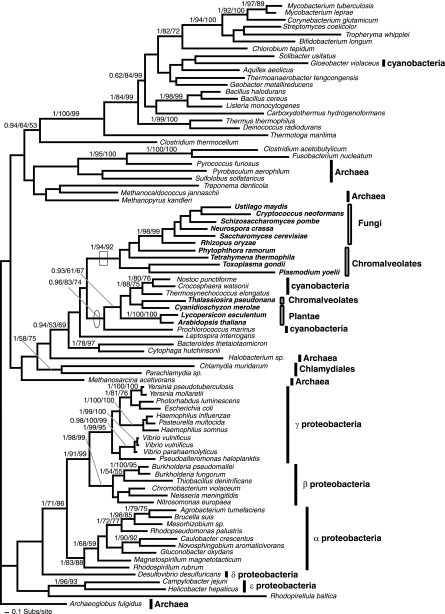
FIG. 2.
Phylogeny of AroB. This phylogeny was calculated from an amino acid alignment of 87 sequences and 209 characters. The node showing separation of the Plantae away from the arom-possessing eukaryotes, indicative of an HGT from the β/γ-proteobacteria is circled. The node supporting monophyly of arom-possessing taxa, with the exception of T. thermophila and T. gondii, and demonstrating arom reduction in R. oryzae is boxed. The figure is illustrated using the same conventions as Fig. 1, with the addition that fusion genes are noted using square brackets and the shikimate gene fusion order is illustrated using the Aro convention. This convention for fusion genes will be used in all subsequent relevant figures.
The T. thermophila shikimate AroB, AroA, AroK, AroD, and AroE genes were amplified by PCR from cDNA as eight overlapping fragments and sequenced in their entirety (see “Nucleotide sequence accession number” above). This confirmed that these shikimate genes are transcribed as a single arom supergene and not simply as separate genes tandemly arrayed in the T. thermophila genome.
Genes of the alternative shikimate pathway enzymes DAHP I and DHQase II were detected in a number of different eukaryote lineages. DAHP I genes were found in the honeybee Apis melifera, all three fungus groups surveyed, and P. ramorum. The honeybee DAHP I sequence was very similar to the β-proteobacteria sequences (Fig. 1a), but no other shikimate pathway genes could be detected from metazoan genome projects; consequently, we suspect that this gene maybe a genome project contamination artifact. DHQase II was detected in the ascomycetes and the basidiomycetes only.
FIG. 1.
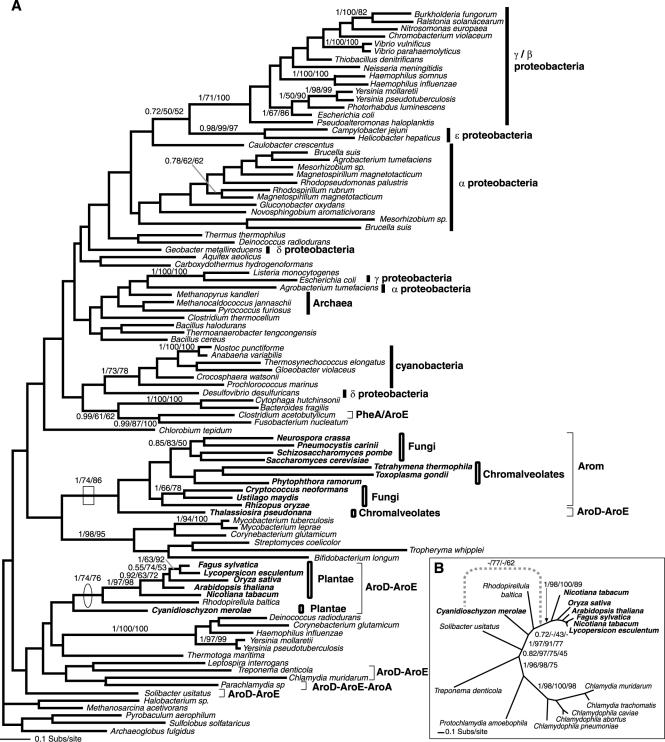
Phylogeny of DAHP I and DAHP II genes. (A) Phylogeny of DAHP I gene, calculated from an amino acid alignment of 45 sequences and 327 characters. The node demonstrating oomycete and fungal monophyly is shown with a box. (B) Phylogeny of DAHP II, calculated from an amino acid alignment of 35 sequences and 372 characters. The node demonstrating the monophyly of arom possessing taxa and the putative HGT with S. usitatus is shown with a box; the node showing separation of the land plants/diatom from the other eukaryotes is circled. This node also represents the best support for a putative HGT between Plantae and a proteobacterium. The topologies shown are the results of the Bayesian topology search arbitrarily rooted on a eubacterial branch. Topology support values for this and all subsequent phylogeny figures are illustrated when both bootstrap results were above 49% and are shown in the order Bayesian posterior probability/% 1,000 ML distance bootstraps/% 100 fast ML-PHYML bootstraps. A selection of eubacteria and the eukaryotes are labeled according to higher taxonomic classification (prokaryote taxa using line bars and eukaryotes using box bars). In addition, the eukaryotes are named in bold. These labeling conventions are also used in all subsequent figures.
To examine the evolutionary relationships of the shikimate pathway enzymes, phylogenetic analysis was performed. The results are presented in Fig. 1 to 7 following the order of the pathway, with the exception of AroD which was deemed too divergent for phylogenetic analysis.
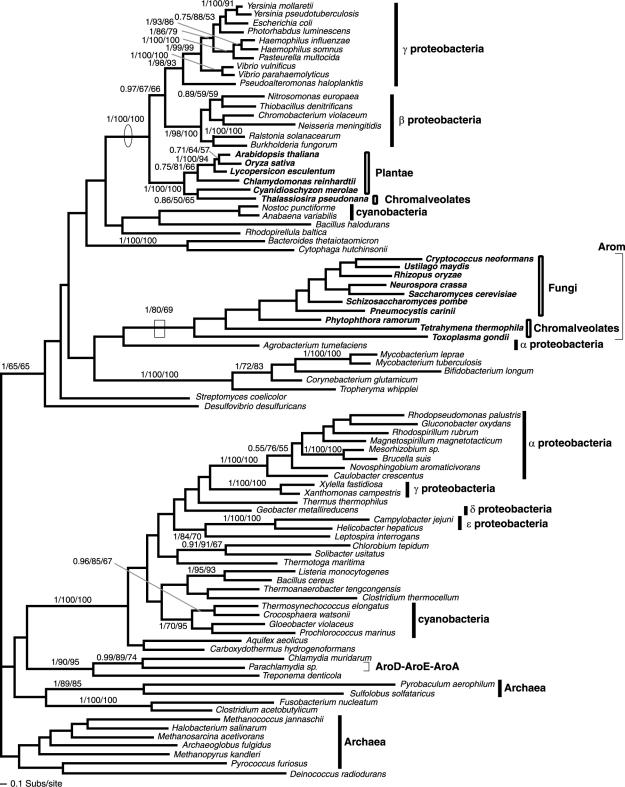
FIG. 7.
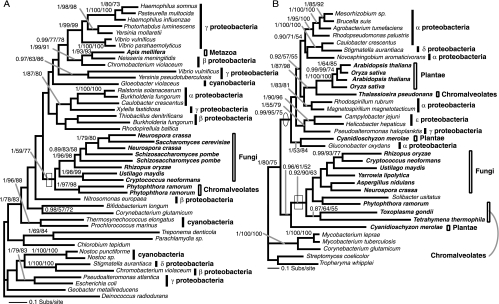
Phylogeny of AroC. The AroC phylogeny was calculated from an amino acid alignment of 84 sequences and 267 characters. The node showing monophyly of arom-possessing taxa, despite this gene not being part of arom, is illustrated with a box. The node showing separation of the Plantae away from the remaining eukaryotes and grouping with the cyanobacteria is circled.
Monophyly of arom and the ancestral eukaryotic shikimate pathway.
The four phylogenies for the component domains of the arom pathway demonstrated that, in three cases, the arom genes and the likely related reduced fusion forms were monophyletic with robust support of 1/74/86, 1/83/88, and 1/80/69 (here, and in all subsequent examples, topology support values are listed in the following order: posterior probability values, percent bootstrap values for ML-corrected distance analyses, and percent bootstrap values for PHYML analyses) (Fig. 4A, 5, and 6). In the fourth case, some chromalveolate AroB sections of the arom pathway grouped separately from the other arom sequences but with bootstrap values below 50% (Fig. 2), suggesting that this section of the phylogeny was unresolved. All other phylogenies and the homologous pentadomain gene structure suggest that the arom gene evolved as a monophyletic character (6).
FIG. 5.
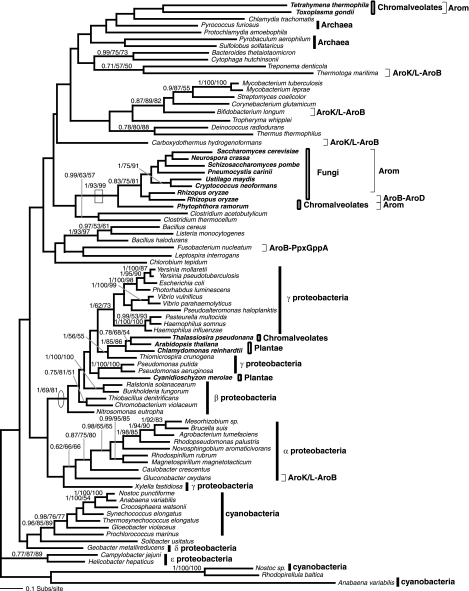
Phylogeny of AroK/L. The AroK/L phylogeny was calculated from an amino acid alignment of 88 sequences and 123 characters. The node supporting monophyly of arom-possessing taxa is boxed. The node showing separation of the Plantae away from the remaining eukaryotes and grouping with the cyanobacteria is circled. Note also that the HTH_XRE domain (Helix-turn-helix motif pfam01381) is fused to AroK in some proteobacteria.
FIG. 6.
Phylogeny of AroA. The AroA phylogeny was calculated from an amino acid alignment of 90 sequences and 259 characters. The node supporting monophyly of arom-possessing taxa is boxed. The node showing separation of the Plantae away from the arom-possessing eukaryotes and indicating an HGT from the β/γ-proteobacteria to the Plantae is circled.
arom and Plantae shikimate genes have different evolutionary histories.
While phylogeny of the arom genes and DAHP I and II showed the eukaryotes possessing the pentadomain arom to be monophyletic, the red algae and the Viridaeplantae grouped separately. In five of the six data sets which contained gene sampling from both the Plantae and other eukaryotes (Fig. 1b, 2, 5, 6, and 7), the Plantae grouped with a cluster of eubacteria, separate from the other eukaryotes, with 0.99/95/75, 1/69/81, 1/87/50, 1/100/100, and 0.96/83/74 support. Although the trees have many weakly supported nodes (a common feature of single-gene trees dealing with ancient evolutionary relationships), in five cases, at least one bootstrap method demonstrated separate origin of Plantae genes with 81% or more bootstrap support, robustly consistent with different evolutionary histories of the shikimate pathway in eukaryotes. Only in the AroE phylogeny (Fig. 4a) did the tree bootstrap support values for the separation of the Plantae genes from the other eukaryotes remain below 50% and not reach a value above 80% bootstrap support, as in the other data sets. However, AU and SH tests rejected the monophyly of the Plantae with the other eukaryote AroE genes with statistical significance at the 0.02 level (data not shown). This, together with the data detailed below, suggests that the Plantae AroDE fusion gene was derived from a different prokaryote ancestral genome than the paralogous arom.
Eukaryote-prokaryote HGT.
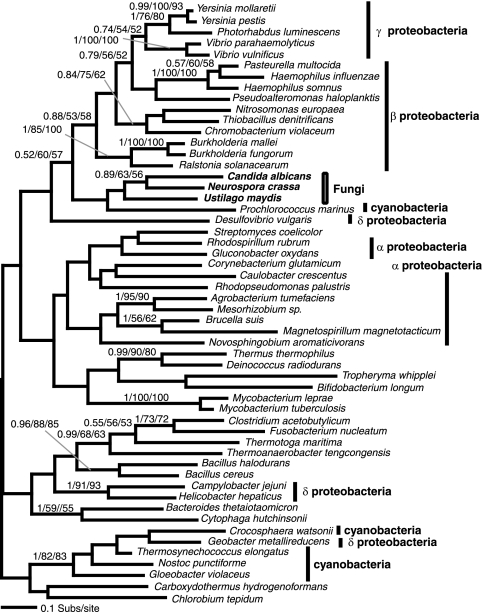
The complicated shikimate evolutionary history is due in part to HGT between eukaryotic and prokaryotic groups. The DAHP II phylogeny (Fig. 1B) revealed two evolutionary complications: (i) the acidobacterium Solibacter usitatus grouped within the fungus-chromalveolate clade with at least one robust node (0.92/90/63), suggesting horizontal gene transfer from a eukaryote into the S. usitatus lineage; (ii) C. merolae possesses two DAHP II homologues, one that groups with the fungus-Chromalveolate clade with 0.96/61/52 support, while the second groups within the proteobacteria clade with 0.99/95/75 and 1/53/84 support (Fig. 1B). These data are consistent with the red alga lineage possessing two paralogues of DAHP II, one derived from a HGT event arising from a proteobacterial genome into the C. merolae lineage, while the second appears to be an anciently derived character of many eukaryote groups. Additionally, the DHQase II gene is found only in the ascomycetes, basidiomycetes, and a diverse selection of eubacteria. Its apparent absence in all other eukaryotic lineages suggests an HGT from a eubacterial lineage into the fungi prior to the division of the basidiomycete and ascomycete lineages. The phylogeny of DHQase II produced no resolution of the fungal group with any given prokaryote group, so it is not possible to pinpoint the source of the putative HGT (Fig. 3). However, this analysis demonstrates that the evolutionary history of the shikimate pathway in the fungi as well as the Plantae appears to have been influenced by prokaryote-to-eukaryote gene transfer events.
FIG. 3.
Phylogeny of DHQase II, which encodes step 3 of the shikimate pathway. Type I 3-dehydroquinate dehydratase could not be reliably aligned and analyzed so is not included here. Phylogeny of DHQase II gene was calculated from an amino acid alignment of 54 sequences and 139 characters. The figure is illustrated using the same conventions as Fig. 1.
Plantae shikimate pathway genes are derived from multiple prokaryotic origins.
Since the plant genes have a different ancestry than the rest of the eukaryote shikimate pathway genes, it is germane to specifically examine their evolutionary origin. Gosset et al. (16) suggested that the plant shikimate pathway genes were acquired from a bacterial source, which, based on other analyses (for examples, see reference 25), might be presumed to be the cyanobacterial progenitor of the chloroplast. However, our phylogenetic results are consistent with the prokaryotic origin of the Plantae shikimate genes deriving from at least three different prokaryotic sources: (i) the cyanobacterial plastid progenitor, (ii) a proteobacterium related to the γ/β group, and (iii) a proteobacterium that groups within or near the α-proteobacteria. In these five examples, the genes from the diatom T. pseudonana group with the Plantae sequences, suggestive of a secondary EGT from the engulfed Plantae cell which became the diatom's plastid endosymbiont (Fig. 1B, 2, 5, 6, and 7).
First, the Plantae AroK and AroC genes grouped with a clade of cyanobacteria with 1/87/50 and 0.96/83/74 support, respectively (Fig. 5 and 7), consistent with a gene origin from the progenitor of the plastid. Second, the AroB and AroA phylogenies (Fig. 2 and 6) demonstrate that the Plantae taxa group with the β/γ-proteobacteria with 1/69/81 and 1/100/100 support, respectively. In both of these cases, the α-proteobacteria sampled branch elsewhere on the phylogeny, suggesting that the Plantae AroB and AroA genes were acquired from separate gene transfers from a proteobacterial lineage and not from the progenitor genome of the mitochondria. The AroA phylogeny (Fig. 6) specifically demonstrates strong support for Plantae monophyly in the form of a shared derived HGT.
The DAHP II (Fig. 1B) phylogeny again shows the Viridaeplantae grouping separately from the other eukaryotes but within a clade of proteobacteria supported by 0.99/95/75 bootstrap support. The red alga C. merolae branches elsewhere in the proteobacterial clade with 1/53/84 support (Fig. 1B). These phylogenetic relationships suggest that the Plantae DAHP II genes were acquired from two separate horizontal gene transfers from proteobacteria. The absence of any other eukaryote orthologues suggests that these transfers were recent HGT events rather than anciently acquired as an endosymbiotic gene transfer from the genome of the mitochondrial progenitor.
AroDE in plants is an HGT and is not indicative of a planctomycete-plastid progenitor sisterhood.
The final possible example of HGT in the Plantae shikimate pathway comes from the phylogeny of AroE domain which showed the Plantae forming a weakly supported clade with R. baltica, with the land plants grouping with R. baltica with 1/74/76 support (Fig. 4A). All members of this clade possess an AroDE fusion gene consistent with the monophyly of the Plantae and R. baltica and an HGT between these unrelated taxa. Relationships between the Plantae and the R. baltica branches were weakly resolved within this AroDE clade (Fig. 4A). Interestingly, the AroDE fusion characteristic and phylogeny, along with many other data sets, was initially used to demonstrate a close sisterhood of the Chlamydiales and the cyanobacterial progenitor of the chloroplast (5). Our analyses did not show a sisterhood relationship for the Plantae and the Chlamydiales for the AroDE fusion gene (Fig. 4A); consequently, we investigated its evolutionary history more closely. To pinpoint the branching relationship between the Plantae and R. baltica, we created an alignment of all AroDE fusion genes and included prokaryote AroDE fusion genes that branched elsewhere in the tree but were not separated from the Plantae-R. baltica clade with bootstrap support above 50%. While analyses of these fusion genes continued to resolve a clade of R. baltica plus Plantae sequences with strong support (1/97/91/77), it did not resolve the internal relationships of this clade (Fig. 4B). The Bayesian search and PHYML searches demonstrated R. baltica grouping within the Plantae (0.72 and 43% topology support). However, two other bootstrap analyses (ML distance and parsimony) found the Plantae to be monophyletic with 77% and 62% support, respectively (Fig. 4B). This inconsistency is important, as it makes polarizing the direction of the proposed eukaryote-prokaryote HGT difficult and suggests four possible evolutionary scenarios: (i) a planctomycete-to-Plantae HGT, (ii) a plastid-progenitor-to-Plantae EGT followed by an HGT to the planctomycetes, (iii) the planctomycetes may be closely related to the primary plastid progenitor and represent a surrogate branch for the cyanobacteria, as suggested for Chlamydia (5), or (iv) an HGT from the eubacterial plastid progenitor genome to the planctomycetes consistent with frequent HGT events between prokaryotes (4).
Analyses of the diversity of the shikimate pathway genes in cyanobacteria showed patterns of gene loss and putative HGT events in the cyanobacteria (as illustrated by the resolved separation of cyanobacterial representatives in Fig. 1A, 6, and 7) consistent with instability in the vertical evolution of this pathway in the eubacteria most closely related to the plastid progenitor (25). Consequently, it is possible that other prokaryote genomes represent appropriate surrogates for the plastid progenitor genome, as Brinkman et al. (5) hypothesized for the Chlamydia. To test the third hypothesis, that “the planctomycetes may be closely related to the primary plastid progenitor and represent a surrogate branch for the cyanobacteria,” we analyzed the planctomycete R. baltica genome (15) for the genetic characteristics previously used to infer sisterhood of the Chlamydia, cyanobacteria, and the plant plastid progenitor. While the Chlamydiales, chloroplasts, and mitochondria possess an unspliced intron in the 23S rRNA gene (11), R. baltica does not. Analysis of the ribosomal operon structures had showed three further characters unique to Chlamydiales, cyanobacteria, and chloroplasts: (i) loss of S10 protein from the ribosomal superoperon, (ii) S10 found as part of a suboperon with S7/S12 ribosomal proteins, and (iii) loss of S14 protein (5). None of these three synapomorphies are present in the R. baltica genome (15). Although Chlamydiales, cyanobacteria, and R. baltica all possess L30 and S4 deletion/rearrangement, these characteristics are not unique to these three eubacterial groups.
Consequently, our data, along with published phylogenetic analyses (for examples, see references 15 and 22), suggests that the Chlamydiales and cyanobacteria do not form a closely related group with the planctomycetes. The close relatedness of the Plantae and R. baltica AroDE gene is therefore likely to be a product of planctomycete-Plantae or planctomycete-plastid progenitor HGT rather than vertical ancestry through the plastid endosymbiosis coupled with loss in sampled cyanobacteria. It is currently not possible to polarize the direction of transfer with complete confidence. Given that comparative topology tests strongly support the separate branching of the Plantae clade from the other eukaryotes and the placement of the Plantae clade among the eubacteria, we favor a HGT from R. baltica, or a close relative into the ancestral Plantae lineage.
DISCUSSION
The shikimate pathway is widespread but lost in some eukaryotes.
Our identification of shikimate homologues in three of the six eukaryotic supergroups (47), including plants, fungi, and chromalveolates, suggests that it may be a more general feature of eukaryotes than previously reported. Nonetheless, the pathway is apparently absent from some taxa, notably metazoans, making it an attractive antimicrobial target in pathogens that possess the pathway (38). We note that the human pathogens C. neoformans and Pneumocystis carinii may be vulnerable to inhibition by glyphosate and/or other inhibitors of the shikimate pathway. While the absence of shikimate pathway homologues in metazoan genomes is biologically valid, the absence of identifiable homologues in some of the other genomes examined may be due to an artifact of partial genomic coverage or possession of highly divergent homologues. Sensitive homology-searching algorithms should be used to search for shikimate genes once these projects are nearer to completion. Nonetheless, our PSI-BLAST searches of the complete Dictyostelium or trypanosomatid genomes revealed no plausible candidate shikimate pathway genes. In addition, we could not detect any more candidate genes in the Plasmodium genome sequences searched, confirming that currently only AroC and a highly divergent AroA-K/L gene fusion (6, 23, 26, 39) have been identified. This means that Plasmodium only have three identifiable putative shikimate pathway genes/enzymes but does mean that Plasmodium is potentially capable of converting shikimate to chorismate, the very section of the shikimate pathway which glyphosate inhibits.
The shikimate pathway is ancient in eukaryotes.
The description of the T. gondii arom structure, only previously demonstrated in ascomycetes, has been interpreted as evidence that arom was present in the last common eukaryotic ancestor (6). However, an alternative explanation is possible involving the acquisition of arom in either the fungal or chromalveolate lineage followed by intraeukaryotic gene transfer. In all four phylogenies of the arom domains, the relationship between the fungi and chromalveolates, within the arom clade, was weakly supported. However, in the three phylogenies with a monophyletic arom clade (Fig. 4A, 5, and 6), the chromalveolates or the fungi were rendered paraphyletic. This, in our opinion, does not constitute support for chromalveolate-fungus HGT because all of these branching relationships were supported by less than 50% bootstrap values. It is, of course, possible that the arom gene was conveyed by a single transfer predating either the diversification of the chromalveolates (7, 12, 18, 56) or the fungi sampled. Particularly, the chromalveolate branch is considered an ancient eukaryotic event that occurred shortly after the divergence of extant eukaryotic supergroups (55), making arom an ancient innovation. However, we prefer the alternative hypothesis that the arom, AroC, and DAHP II genes represent an ancestral form of the eukaryote pathway, present in the last common eukaryote ancestor and inherited vertically in both the fungi and the chromalveolates (47). We suggest that this particular form of the shikimate pathway is ancestral to all eukaryotes, since the last common ancestor of chromalveolates and fungi is thought to represent a close approximation of the last common eukaryotic ancestor (3, 24, 40, 48). We prefer the vertical origin hypothesis because it is more parsimonious than the horizontal hypothesis requiring one less transfer event. However, ultimately, increased genome sampling, specifically among the Excavata, Cercozoa/Rhizaria, and the Amoebozoa supergroups (47), will enable a thorough test of these two alternative hypotheses.
The presence of DAHP I homologue in fungi and the chromalveolates and their monophyly on the trees reported here (Fig. 1A) also suggests that the DAHP I in addition to DAHP II, arom, and AroC could have been present in the last common ancestor of the eukaryotes (Fig. 8A). This hypothesis infers that all of the Plantae shikimate pathway genes that do not group with the fungus-chromalveolate genes represent evolutionary replacements. However, the replacement hypothesis relies on the theory that the Plantae branch groups above the fungus-chromalveolate bifurcation in the eukaryote tree (37, 48, 49). The only possible alternative is if the Plantae branched away from the eukaryotic trunk prior to the last common fungus-chromalveolate ancestor, rendering the bikonts paraphyletic (37, 48, 49). Even so, two of the seven shikimate pathway genes are likely to represent gene replacements or later additions, as they have been derived from the plastid progenitor genome (Fig. 5 and 7), which has to have occurred after the Plantae branched from the ancestral trunk of the eukaryote tree (37, 40, 48) (Fig. 8A).
It follows that the shikimate pathway, along with arom, has been lost in many eukaryotic lineages examined to date and that the seven gene domains of the Plantae shikimate pathway have been serially acquired by EGT and HGT, with the notable exception of DAHP II in the red algae, which possesses two paralogues, one of which is placed with weak bootstrap support as a putatively ancestral eukaryotic paralogue (0.96/61/52 topology support) (Fig. 1B). Since the excavate (46) Euglena is known to possess both a cytosolic and plastid-associated shikimate pathway (36), investigation into the shikimate gene arrangement in this taxon would be valuable in determining the evolution of the pathway.
Selection for gene fusions.
Comparative analyses demonstrated six different protein domain fusion arrangements for the five functional domains contained within arom. This includes the arom arrangement (i.e., AroB-AroA-AroK/L-AroD-AroE) and five other fusion arrangements: (i) the AroK/L-B fusion found in a number of prokaryotes (Fig. 2 and 5), (ii) the AroDE fusion found in the Plantae, some eubacteria, and the diatom (Fig. 4A and B), (iii) the AroDEA fusion found in Parachlamydia (Fig. 4A and B and 6), (iv) the AroBD fusion found in Rhizopus (Fig. 2), and (v) the AroA-K/L putative fusion gene found in Plasmodium (6, 27), not analyzed here. The data reported here present some evidence for the separate fusion events of the shikimate pathway genes. The independent and convergent occurrence of these fusion genes can be confirmed in two ways: (i) they represent different fusion patterns with distinct domain compositions and orders or (ii) one example of each fusion order is polyphyletic in at least one tree supported by moderate to strong bootstrap support. Examples of the first type are shown in the cases of the AroK/L-B fusion (Fig. 2), the AroDEA fusion (Fig. 4A), and fusion of non-shikimate pathway genes to AroE and AroK/L (Fig. 4A and 5). In several cases, there is also moderate to strong bootstrap support for the separate branching of fusion genes. This includes 1/90/95 support for the separation of the Parachlamydia trifusion and arom (Fig. 6) and 1/87/96 support for the grouping of the Gluconobacter AroK/L-B fusion gene with other nonfused prokaryotic versions separate from other AroK/L-B fusion genes (Fig. 5).
There is also evidence of arom reduction in two lineages. The diatom possesses an AroDE fusion gene, exactly the same order as other photosynthetic eukaryotes. However, in this case, the diatom AroDE fusion is orthologous to arom (1/74/86) (Fig. 4A). Therefore, the diatom is likely to have acquired a bifunctional arrangement as a reduction of the ancestral arom, while the Plantae acquired the same bifunctional arrangement separately (polyphyly of eukaryotes confirmed by AU and SH tests of constrained alternative tree topologies) (Fig. 4A). In addition, Rhizopus has an AroBD fusion gene orthologous to the arom clade (1/93/99) and grouping specifically with the full Rhizopus arom, suggesting an arom duplication followed by an additional incidence of arom reduction (Fig. 2) in the zygomycetes.
Interestingly, it is only the five genes found in the arom that are found in different gene fusion arrangements. This suggests that the functional/genetic linking of these five shikimate pathway enzymes may be selectively favorable (54) and has occurred on numerous occasions by patterns of convergent evolution. A number of prokaryotic organisms have clustering of shikimate pathway genes, which in some cases function as operons (33, 34). We hypothesize that clustering of genes in operons would make the independent occurrence of gene fusions more likely as the genes are placed in close genomic proximity over evolutionary time. In other data sets, gene fusions have been used as characters for pinpointing ancient evolutionary relationships (48, 49). The analysis of the shikimate pathway suggests that patterns of gene fusion may be affected by convergent evolution and fission, and so it is prudent to apply phylogenetic analysis to ensure monophyly of the fusion forms when using gene fusions as evolutionary informative characters.
Serial gene transfer between prokaryotes and the Plantae beyond cyanobacterial endosymbiosis.
Our data suggest that the shikimate pathway genes in the Plantae and some other photosynthetic eukaryotes were acquired from a combination of EGT and HGT events. The presence of a non-arom pathway in photosynthetic eukaryotes, which appears to be plastid located in the land plants (32) would suggest some advantage to a non-arom and putatively plastid-targeted pathway in photosynthetic lineages. This may include increased availability of precursors and cofactors, including erythrose-4-phosphate, phosphoenolpyruvate, NADH, and ATP, which are produced in chloroplasts from photosynthesis. As a likely consequence of this, the shikimate pathway has been reported to be light regulated in some plants (19, 53). Furthermore, Euglena gracilis has been reported to have a cytosolic pathway used when grown in darkness but a plastidic pathway used when grown in light (36). Interestingly, some photosynthetic chromalveolates (Thallasiosira) appear to have acquired five of the seven component genes of this pathway by EGT, presumably concordantly with the acquisition of the photosynthetic plastid organelle by secondary endosymbiosis, while the nonphotosynthetic chromalveolates have retained their arom cytosolic pathway. For example, T. gondii, which is nonphotosynthetic, has retained the arom and putatively cytosolic pathways. This assumes (i) that the chromalveolates are a monophyletic clade (7, 12, 18, 35, 56) and (ii) that both the arom and non-arom pathways were retained during the diversification of the chromalveolates. The second assumption is supported by the observation that the diatom Thallasiosira possesses a chimera of the arom and non-arom pathways, possessing five Plantae non-arom-like genes and an AroDE fusion gene orthologous to a two-domain section of the arom pathway found in the fungi and the alveolates (Fig. 4A).
The number of primary endosymbiotic acquisitions of plastids has been a topic of debate (30, 50, 29). Although nuclear gene phylogenies and conservation of plastid machinery (29, 30) support monophyly of the Plantae group, discrete evolutionary events such as HGT events (1, 20) and gene replacements (for examples, see references 18 and 35) also provide crucial evidence in two ways: shared derived HGT between lineages from nonplastid sources strongly supports the holophyly of the Plantae clade, while shared plastid transfers confirm the monophyly of the primary plastid. Our analysis of shikimate pathway genes has revealed gene transfer events of both types. Three Plantae gene phylogenies (AroB, AroDE, and AroA) support shared derived prokaryote-to-Plantae HGT events (Fig. 2, 4A and B, and 6), while the AroC gene phylogeny supports a shared derived cyanobacterium-to-Plantae EGT (Fig. 7).
There are at least two scenarios that could explain Plantae shikimate gene relationships: (i) a combination of endosymbiotic and HGT gene acquisition and (ii) endosymbiotic gene acquisition followed by replacement in cyanobacteria. In the first scenario, the primary plastid endosymbiosis equipped the ancestral Plantae genome with cyanobacterial shikimate genes. Then genes from at least two other eubacteria have replaced cyanobacterium-derived genes prior to the divergence of the Plantae into the red algae and the Viridaeplantae. The alternative hypothesis is that the HGT events occurred in the common ancestor of extant and sampled cyanobacteria, essentially leaving the Plantae genes to resemble the ancestral cyanobacterial genes while the cyanobacterial genes thus far sequenced resemble other eubacterial genes.
Irrespective of the above alternative explanations, shared prokaryote-to-Plantae HGTs which also demonstrate Plantae monophyly (AroA represents the best example) (Fig. 6), strongly implies holophyly of the red algae and the Viridaeplantae. Although, as a word of caution, we found possible cases of separate origins of shikimate pathway genes in the red algae and the Viridaeplantae (DAHP II [Fig. 1B], AroB [Fig. 2], and AroK/L [Fig. 5]). However, only in the AroK/L phylogeny was the bootstrap support for Plantae polyphyly even moderate (1/58/78) (Fig. 5). These are unlikely to count as robust evidence against Plantae monophyly and, in the case of AroK/L, may simply be a further case of HGT and/or hidden parology and therefore represent additional evolutionary noise.
It is possible that intraeukaryotic HGT may have occurred between the red alga and Viridaeplantae lineages, invalidating support for monophyly of the Plantae lineages. However, Archibald et al. suggest that such intereukaryotic HGT between photosynthetic eukaryotes is rare (2). In addition, one of the shikimate pathway genes demonstrates shared derived origin from the plastid progenitor genome (Fig. 7). Each shared transfer into the common ancestor of the red and green photosynthetic eukaryotes implies monophyly of the Plantae nuclear genome and, in the case of plastid transfers, represents a unique evolutionary event from a single shared plastid ancestor.
The present study demonstrates that the shikimate pathway is an ancient eukaryotic pathway that has been subject to diverse evolutionary pressures. This has resulted in its entire loss from a number of lineages, including the Metazoa (38), that derive their aromatic compounds from diet. Furthermore, the entire pathway has been transferred to the Plantae from a range of different eubacterial sources. In fungi and the red algae, additional discrete HGT of shikimate pathway genes have occurred.
We present a candidate ancestral pathway (Fig. 8A) given the bikont/unikont eukaryote root (37, 48, 49) and demonstrate that the shikimate pathway has been retained in many eukaryote lineages, now known to comprise the zygomycete, basidiomycete, and ascomycete fungi, apicomplexa, ciliates, and the oomycetes. Interestingly, the photosynthetic chromalveolate Thalassiosira and the red alga Cyanidioschyzon possess a chimeric pathway in part composed of the Plantae non-arom pathway and in other parts composed of component of the arom type pathway. Our model of the shikimate pathway evolution in the eukaryotes (Fig. 8A) demonstrates, at a conservative estimate, over 50 independent loss events in the evolution of the eukaryotic shikimate pathway. With its implications as both a practical antimicrobial therapeutic target and as critical biochemical pathway in diverse eukaryotic organisms, the shikimate pathway represents a rich data set for studying the evolution and regulation of biochemical pathways, endosymbiosis, and processes of gene loss, gene fusion, replacement, and gene transfer.
Supplementary Material
Acknowledgments
This work was supported by NIH grants R01 AI43228 (R.M. and C.W.R.) and AI27530 (R.M.) and the Research to Prevent Blindness Foundation. T.A.R. is supported by a BBSRC studentship. J.L.B. was supported by a University of Massachusetts Healey Endowment Grant. J.B.D. was supported by a joint CIHR/Welcome Trust Traveling research fellowship. J.B.D. also thanks St. Johns College, Oxford University for hosting him as a Visiting Scholar during part of the preparation of the manuscript.
Preliminary sequence data were obtained from The Institute for Genomic Research website at http://www.tigr.org and Department of Energy Joint Genome Institute http://www.jgi.doe.gov. We thank TIGR and DOE JGI for making their data publicly available. We also thank Aaron Turkewitz for the Tetrahymena cultures.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Andersson, J. O., S. W. Sarchfield, and A. J. Roger. 2005. Gene transfers from nanoarchaeota to an ancestor of diplomonads and parabasalids. Mol. Biol. Evol. 22:85-90. [DOI] [PubMed] [Google Scholar]
- 2.Archibald, J. M., M. B. Rogers, M. Toop, K. Ishida, and P. J. Keeling. 2003. Lateral gene transfer and the evolution of plastid-targeted proteins in the secondary plastid-containing alga Bigelowiella natans. Proc. Natl. Acad. Sci. USA 24:7678-7683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baldauf, S. L., A. J. Roger, I. Wenk-Siefert, and W. F. Doolittle. 2000. A kingdom-level phylogeny of eukaryotes based on combined protein data. Science 290:972-977. [DOI] [PubMed] [Google Scholar]
- 4.Boucher, Y., C. J. Douady, R. T. Papke, D. A. Walsh, M. E. Boudreau, C. L. Nesbo, R. J. Case, and W. F. Doolittle. 2003. Lateral gene transfer and the origins of prokaryotic groups. Annu. Rev. Genet. 37:283-328. [DOI] [PubMed] [Google Scholar]
- 5.Brinkman, F. S., J. L. Blanchard, A. Cherkasov, Y. Av-Gay, R. C. Brunham, R. C. Fernandez, B. B. Finlay, S. P. Otto, B. F. Ouellette, P. J. Keeling, A. M. Rose, R. E. Hancock, S. J. Jones, and H. Greberg. 2002. Evidence that plant-like genes in Chlamydia species reflect an ancestral relationship between Chlamydiaceae, cyanobacteria, and the chloroplast. Genome Res. 12:1159-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campbell, S. A., T. A. Richards, E. J. Mui, B. U. Samuel, J. R. Coggins, R. McLeod, and C. W. Roberts. 2004. A complete shikimate pathway in Toxoplasma gondii: an ancient eukaryotic innovation. Int. J. Parasitol. 34:5-13. [DOI] [PubMed] [Google Scholar]
- 7.Cavalier-Smith, T. 1999. Principles of protein and lipid targeting in secondary symbiogenesis: Euglenoid, Dinoflagellate, and sporozoan plastid origins and the eukaryote family tree. J. Eukaryot. Microbiol. 46:347-366. [DOI] [PubMed] [Google Scholar]
- 8.Coggins, J. R., K. Duncan, I. A. Anton, M. R. Boocock, S. Chaudhuri, J. M. Lambert, A. Lewendon, G. Millar, D. M. Mousdale, and D. D. Smith. 1987. The anatomy of a multifunctional enzyme. Biochem. Soc. Trans. 15:754-759. [DOI] [PubMed] [Google Scholar]
- 9.Duncan, K., R. M. Edwards, and J. R. Coggins. 1987. The pentafunctional arom enzyme of Saccharomyces cerevisiae is a mosaic of monofunctional domains. Biochem. J. 246:375-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elandalloussi, L. M., P. M. Rodrigues, R. Afonso, R. B. Leite, P. A. Nunes, and M. L. Cancela. 2005. Shikimate and folate pathways in the protozoan parasite, Perkinsus olseni. Mol. Biochem. Parasitol. 142:106-109. [DOI] [PubMed] [Google Scholar]
- 11.Everett, K. D., S. Kahane, R. M. Bush, and M. G. Friedman. 1999. An unspliced group I intron in 23S rRNA links chlamydiales, chloroplasts, and mitochondria. J. Bacteriol. 181:4734-4740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fast, N. M., J. C. Kissinger, D. S. Roos, and P. J. Keeling. 2001. Nuclear-encoded, plastid-targeted genes suggest a single common origin for apicomplexan and dinoflagellate plastids. Mol. Biol. Evol. 18:418-426. [DOI] [PubMed] [Google Scholar]
- 13.Felsenstein, J. 1995. PHYLIP (Phylogeny Inference Package), 3.57 ed. Department of Genetics, University of Washington, Seattle.
- 14.Foster, P. G. 2006. p4. [Online.] http://www.nhm.ac.uk/zoology/external/p4.html.
- 15.Glockner, F. O., M. Kube, M. Bauer, H. Teeling, T. Lombardot, W. Ludwig, D. Gade, A. Beck, K. Borzym, K. Heitmann, R. Rabus, H. Schlesner, R. Amann, and R. Reinhardt. 2003. Complete genome sequence of the marine planctomycete Pirellula sp. strain 1. Proc. Natl. Acad. Sci. USA 100:8298-8303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gosset, G., C. A. Bonner, and R. A. Jensen. 2001. Microbial origin of plant-type 2-keto-3-deoxy-d-arabino-heptulosonate 7-phosphate synthases, exemplified by the chorismate- and tryptophan-regulated enzyme from Xanthomonas campestris. J. Bacteriol. 183:4061-4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guindon, S., F. Lethiec, P. Duroux, and O. Gascuel. 2005. PHYML Online-a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res. 33:W557-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harper, J. T., and P. J. Keeling. 2003. Nucleus-encoded, plastid-targeted glyceraldehyde-3-phosphate dehydrogenase (GAPDH) indicates a single origin of chromalveolate plastids. Mol. Biol. Evol. 20:1730-1735. [DOI] [PubMed] [Google Scholar]
- 19.Henstrand, J. M., K. F. McCue, K. Brink, A. K. Handa, K. M. Herrmann, and E. E. Conn. 1992. Light and fungal elicitor induce 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase mRNA in suspension cultured cells of parsley (Petroselinum crispum L.). Plant Physiol. 98:761-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henze, K., D. S. Horner, S. Suguri, D. V. Moore, L. B. Sanchez, M. Muller, and T. M. Embley. 2001. Unique phylogenetic relationships of glucokinase and glucosephosphate isomerase of the amitochondriate eukaryotes Giardia intestinalis, Spironucleus barkhanus and Trichomonas vaginalis. Gene 281:123-131. [DOI] [PubMed] [Google Scholar]
- 21.Holder, M., and A. J. Roger. 2006. PUZZLEBOOT, version 1.03. [Online.] http://rogerlab.biochemistryandmolecularbiology.dal.ca/Software/Software.htm#puzzleboot.
- 22.Jenkins, C., and J. A. Fuerst. 2001. Phylogenetic analysis of evolutionary relationships of the planctomycete division of the domain bacteria based on amino acid sequences of elongation factor Tu. J. Mol. Evol. 52:405-418. [DOI] [PubMed] [Google Scholar]
- 23.Keeling, P. J., J. D. Palmer, R. G. Donald, D. S. Roos, R. F. Waller, and G. I. McFadden. 1999. Shikimate pathway in apicomplexan parasites. Nature 397:219-220. [DOI] [PubMed] [Google Scholar]
- 24.Lang, B. F., C. O'Kelly, T. Nerad, M. W. Gray, and G. Burger. 2002. The closest unicellular relatives of animals. Curr. Biol. 12:1773-1778. [DOI] [PubMed] [Google Scholar]
- 25.Martin, W., T. Rujan, E. Richly, A. Hansen, S. Cornelsen, T. Lins, D. Leister, B. Stoebe, M. Hasegawa, and D. Penny. 2002. Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc. Natl. Acad. Sci. USA 99:12246-12251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McConkey, G. A. 1999. Targeting the shikimate pathway in the malaria parasite Plasmodium falciparum. Antimicrob. Agents Chemother. 43:175-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McConkey, G. A., J. W. Pinney, D. R. Westhead, K. Plueckhahn, T. B. Fitzpatrick, P. Macheroux, and B. Kappes. 2004. Annotating the Plasmodium genome and the enigma of the shikimate pathway. Trends Parasitol. 20:60-65. [DOI] [PubMed] [Google Scholar]
- 28.McFadden, G. I., M. E. Reith, J. Munholland, and N. Lang-Unnasch. 1996. Plastid in human parasites. Nature 381:482. [DOI] [PubMed] [Google Scholar]
- 29.McFadden, G. I., and G. G. van Dooren. 2004. Evolution: red algal genome affirms a common origin of all plastids. Curr. Biol. 14:R514-R516. [DOI] [PubMed] [Google Scholar]
- 30.Moreira, D., H. Le Guyader, and H. Philippe. 2000. The origin of red algae and the evolution of chloroplasts. Nature 405:69-72. [DOI] [PubMed] [Google Scholar]
- 31.Mousdale, D. M., M. S. Campbell, and J. R. Coggins. 1987. Purification and characterisation of a bi-functional dehydroquinase-shikimate: NADP oxidoreductase from pea seedlings. Phytochemistry 26:2665-2670. [Google Scholar]
- 32.Mousdale, D. M., and J. R. Coggins. 1985. Subcellular localization of the common shikimate-pathway enzymes in Pisum sativum L. Planta 163:241-249. [DOI] [PubMed] [Google Scholar]
- 33.Panina, E. M., A. G. Vitreschak, A. A. Mironov, and M. S. Gelfand. 2003. Regulation of biosynthesis and transport of aromatic amino acids in low-GC Gram-positive bacteria. FEMS Microbiol. Lett. 222:211-220. [DOI] [PubMed] [Google Scholar]
- 34.Parish, T., and N. G. Stoker. 2002. The common aromatic amino acid biosynthesis pathway is essential in Mycobacterium tuberculosis. Microbiology 148:3069-3077. [DOI] [PubMed] [Google Scholar]
- 35.Patron, N. J., M. B. Rogers, and P. J. Keeling. 2004. Gene replacement of fructose-1,6-bisphosphate aldolase supports the hypothesis of a single photosynthetic ancestor of the chromalveolates. Eukaryot. Cell 3:1169-1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reinbothe, C., B. Ortel, B. Parthier, and S. Reinbothe. 1994. Cytosolic and plastid forms of 5-enolpyruvylshikimate-3-phosphate synthase in Euglena gracilis are differentially expressed during light-induced chloroplast development. Mol. Gen. Genet. 245:616-622. [DOI] [PubMed] [Google Scholar]
- 37.Richards, T. A., and T. Cavalier-Smith. 2005. Myosin domain evolution and the primary divergence of eukaryotes. Nature 436:1113-1118. [DOI] [PubMed] [Google Scholar]
- 38.Roberts, C. W., F. Roberts, R. E. Lyons, M. J. Kirisits, E. J. Mui, J. Finnerty, J. J. Johnson, D. J. Ferguson, J. R. Coggins, T. Krell, G. H. Coombs, W. K. Milhous, D. E. Kyle, S. Tzipori, J. Barnwell, J. B. Dame, J. Carlton, and R. McLeod. 2002. The shikimate pathway and its branches in apicomplexan parasites. J. Infect. Dis. 185(Suppl. 1):S25-S36. [DOI] [PubMed] [Google Scholar]
- 39.Roberts, F., C. W. Roberts, J. J. Johnson, D. E. Kyle, T. Krell, J. R. Coggins, G. H. Coombs, W. K. Milhous, S. Tzipori, D. J. Ferguson, D. Chakrabarti, and R. McLeod. 1998. Evidence for the shikimate pathway in apicomplexan parasites. Nature 393:801-805. [DOI] [PubMed] [Google Scholar]
- 40.Rodriguez-Ezpeleta, N., H. Brinkmann, S. C. Burey, B. Roure, G. Burger, W. Loffelhardt, H. J. Bohnert, H. Philippe, and B. F. Lang. 2005. Monophyly of primary photosynthetic eukaryotes: green plants, red algae, and glaucophytes. Curr. Biol. 15:1325-1330. [DOI] [PubMed] [Google Scholar]
- 41.Ronquist, F., and J. P. Huelsenbeck. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572-1574. [DOI] [PubMed] [Google Scholar]
- 42.Schmid, J., A. Schaller, N. Leibinger, W. Boll, and N. Amrhein. 1992. The in vitro synthesized tomato shikimate kinase precursor is enzymatically active and is imported and processed to the mature enzyme by chloroplasts. Plant J. 2:375-383. [PubMed] [Google Scholar]
- 43.Schmidt, H. A., K. Strimmer, M. Vingron, and A. von Haeseler. 2002. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 18:502-504. [DOI] [PubMed] [Google Scholar]
- 44.Shimodaira, H. 2002. An approximately unbiased test of phylogenetic tree selection. Syst. Biol. 51:492-508. [DOI] [PubMed] [Google Scholar]
- 45.Shimodaira, H., and M. Hasegawa. 2001. CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics 17:1246-1247. [DOI] [PubMed] [Google Scholar]
- 46.Simpson, A. G. 2003. Cytoskeletal organization, phylogenetic affinities and systematics in the contentious taxon Excavata (Eukaryota). Int. J. Syst. Evol. Microbiol. 53:1759-1777. [DOI] [PubMed] [Google Scholar]
- 47.Simpson, A. G., and A. J. Roger. 2004. The real ‘kingdoms’ of eukaryotes. Curr. Biol. 14:R693-696. [DOI] [PubMed] [Google Scholar]
- 48.Stechmann, A., and T. Cavalier-Smith. 2003. The root of the eukaryote tree pinpointed. Curr. Biol. 13:R665-666. [DOI] [PubMed] [Google Scholar]
- 49.Stechmann, A., and T. Cavalier-Smith. 2002. Rooting the eukaryote tree by using a derived gene fusion. Science 297:89-91. [DOI] [PubMed] [Google Scholar]
- 50.Stiller, J. W., and B. D. Hall. 1997. The origin of red algae: implications for plastid evolution. Proc. Natl. Acad. Sci. USA 94:4520-4525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Swofford, D. L., P. J. Waddell, J. P. Huelsenbeck, P. G. Foster, P. O. Lewis, and J. S. Rogers. 2001. Bias in phylogenetic estimation and its relevance to the choice between parsimony and likelihood methods. Syst. Biol. 50:525-539. [PubMed] [Google Scholar]
- 52.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weaver, L. M., and K. M. Herrmann. 1997. Dynamics of the shikimate pathway in plants. Trends Plant Sci. 2:346-351. [Google Scholar]
- 54.Welch, G. R., and F. H. Gaertner. 1975. Influence of an aggregated multienzyme system on transient time: kinetic evidence for compartmentation by an aromatic-amino-acid synthesizing complex of Neurospora crassa. Proc. Natl. Acad. Sci. USA 72:4218-4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yoon, H. S., J. D. Hackett, C. Ciniglia, G. Pinto, and D. Bhattacharya. 2004. A molecular timeline for the origin of photosynthetic eukaryotes. Mol. Biol. Evol. 21:809-818. [DOI] [PubMed] [Google Scholar]
- 56.Yoon, H. S., J. D. Hackett, G. Pinto, and D. Bhattacharya. 2002. The single, ancient origin of chromist plastids. Proc. Natl. Acad. Sci. USA 99:15507-15512. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.