Abstract
G-protein-coupled receptors (GPCRs) control important aspects of asexual and sexual development in eukaryotic organisms. We have identified a predicted GPCR in the filamentous fungus Neurospora crassa with similarity to cyclic AMP-receptor like GPCRs from Dictyostelium discoideum and GCR1 from Arabidopsis thaliana. Expression of gpr-1 is highest in female reproductive structures, and deletion of gpr-1 leads to defects during sexual development. Unfertilized female structures (protoperithecia) from Δgpr-1 strains are weakly pigmented, small, and submerged in the agar. The perithecia produced after fertilization have deformed beaks that lack ostioles, the openings through which ascospores are discharged. Localization studies using a GPR-1-green fluorescent protein fusion protein showed that GPR-1 is targeted to female reproductive structures. Genetic epistasis experiments with the three Gα genes were inconclusive due to the early block in mating exhibited by Δgna-1 strains. Phenotypic analysis of mutants from a high-throughput N. crassa knockout project allowed identification of BEK-1, a homeodomain transcription factor that is a potential target of GPR-1. The perithecial defects of Δbek-1 strains are similar to those of the Δgpr-1 strain, and epistasis analysis indicates that bek-1 could function downstream of gpr-1 during postfertilization events. The effect must be posttranscriptional, as bek-1 transcript levels are not affected in Δgpr-1 strains. The lack of ostioles in Δgpr-1 and Δbek-1 mutants has an undesirable effect on the ability to spread progeny (ascospores) by the normal ejection mechanism and would severely compromise the fitness of these strains in nature.
In the multicellular fungus Neurospora crassa, sexual development is a complex process essential for survival during difficult environmental circumstances, including desiccation, heat, and nutrient deprivation. It has been shown in previous studies that formation of female reproductive structures, protoperithecia, is triggered by nitrogen starvation in N. crassa (reviewed in reference 62). Protoperithecia contain specialized chemotropic hyphae, termed trichogynes. Trichogynes grow toward male cells (conidia or other vegetative cells) of opposite mating type and fuse with them. The molecular mechanism of trichogyne chemotropism toward male cells is based on an interaction between pheromones secreted from male cells and cognate pheromone receptors localized in the plasma membrane of trichogynes (44, 45; H. Kim and K. Borkovich, unpublished observations). Fusion is followed by recruitment of a male nucleus to the base of the protoperithecium. The nuclei from the male and female parent recognize one another and undergo mitosis to form ascogenous hyphae. Subsequent fusion of male and female nuclei is followed by two meiotic divisions and one postmeiotic mitosis. Each resulting ascus contains eight homokaryotic, haploid ascospores. About 200 to 400 asci are encapsulated in each mature fruiting body (perithecium). Perithecia are large flask-shaped, melanized structures with a neck (beak) and ostiole (pore) at the tip through which ascospores are ejected (35). Perithecial beaks exhibit positive phototropism towards blue light (34), thus ensuring that ascospores are ejected upwards and can be spread effectively in the environment.
G-protein-coupled receptors (GPCRs) are seven-transmembrane helix proteins that are the sensory components of heterotrimeric G protein signaling pathways. GPCRs are crucial for the swift response to extracellular stimuli, including light, odors, hormones, neurotransmitters, and other signals (48, 53). In fungi, heterotrimeric G proteins mediate responses to environmental agents (including nutrients and pheromones) to regulate growth, colony formation, cell-cell recognition and fusion, chemotropism, mating, and cell differentiation. The early stage of mating (pheromone attraction and fusion) in N. crassa is regulated by pheromone receptors localized in the plasma membrane of trichogynes that sense pheromones secreted from male cells of opposite mating type (44, 45; Kim and Borkovich, unpublished). N. crassa has two pheromone receptors, PRE-1, expressed in mat A cells and PRE-2, produced by mat a strains (44, 45). PRE-1 and PRE-2 share homology with the Saccharomyces cerevisiae pheromone receptors Ste2p and Ste3p (10, 31). Pheromone receptor GPCRs are well conserved and have been reported in other fungi, including Schizosaccharomyces pombe (66), Aspergillus nidulans (65), Ustilago maydis (11), Cryptococcus neoformans (18, 21), Schizophyllum commune (72), and Coprinus cinereus (54, 56). Besides the pheromone receptors, few other groups of GPCRs have been studied in detail in fungi. Carbon source and amino acid sensing receptors have been identified and characterized in S. cerevisiae (75, 78, 79), Candida albicans (49, 50, 52), S. pombe (71), C. neoformans (74), and N. crassa (48a). In the homothalic fungus A. nidulans, a GPCR (GprD) was found to function as a negative regulator of fruiting body development (32).
Ten putative GPCRs, three G protein α subunits (GNA-1, GNA-2, and GNA-3), one Gβ subunit (GNB-1), and one Gγ subunit (GNG-1) have been identified in N. crassa (12, 28, 43, 47, 68, 77). Loss of gna-1 leads to trichogynes that grow randomly and are blind to males, causing female (not male) sterility in both mating types (39, 44). These observations indicate that GNA-1 is involved in the pheromone response pathway and is presumably coupled to the pheromone receptors. In contrast to Δpre-1 mutants which form normal protoperithecia, Δgna-1 strains produce fewer protoperithecia, which have abundant fringe hyphae (39). This shows that GNA-1 is also required for normal protoperithecial development. The observation that protoperithecia are normal in Δpre-1 mutants suggests that regulation of protoperithecial formation by GNA-1 occurs through coupling to a nonpheromone receptor GPCR. Deletion of gna-2 or gna-3 does not result in female sterility (4, 43, 44), and the female infertility of Δgnb-1 and Δgng-1 strains very likely results from reduced protein levels of all three Gα subunits (47, 77).
In this study, we report isolation and characterization of a putative GPCR, GPR-1, in N. crassa. GPR-1 is a member of a GPCR family consisting of three closely related receptors, GPR-1, GPR-2, and GPR-3 (12, 28). Deletion of gpr-1 leads to pleiotropic phenotypic defects during sexual development. Δgpr-1 mutants form small, pale protoperithecia, and perithecia are frequently ruptured, lack ostioles (pores), and have defective beaks. Epistatic analysis between gpr-1 and the three Gα genes indicates that GNA-1 may function downstream of GPR-1. Using a GPR-1-green fluorescent protein (GFP) fusion construct, we demonstrate that GPR-1 is localized in protoperithecia. In addition, we show that a homeodomain transcription factor, BEK-1, may function downstream of GPR-1 during perithecial development.
MATERIALS AND METHODS
N. crassa culture conditions.
N. crassa strains (Table 1) were maintained in Vogel's minimal (VM) medium (70). For vegetative growth, strains were cultured in either liquid VM medium with shaking (submerged cultures) or on solid VM medium, while synthetic crossing medium (SCM) (73) was used to induce formation of female reproductive structures on plates. Sorbose-containing medium (FIGS [fructose-inositol-glucose-sorbose]) was used to facilitate colony formation on plates (24).Transformants were plated using regeneration agar (17). For phosphinothricin-containing medium, the ammonium nitrate in VM or FIGS medium was replaced with potassium nitrate and 0.5% proline, and phosphinothricin was added to a final concentration of 400 μg/ml (57).
TABLE 1.
N. crassa strains used in this study
| Strain | Relevant genotype | Commentsa | Sourcea |
|---|---|---|---|
| 74A-OR23-1A | Wild type, mat A | FGSC 987 | FGSC |
| 74a-OR8-1a | Wild type, mat a | FGSC 988 | FGSC |
| his-3A | his-3 mat A | FGSC 6103 | FGSC |
| his-3a | his-3 mat a | This study | |
| 3B10 | Δgna-1::hph mat a | gna-1 homokaryon | 40 |
| A33-4 | Δgna-2::pyrG+pyr-4 mat a | gna-2 homokaryon | 4 |
| 31c2 | Δgna-3::hphmat A | gna-3 homokaryon | 43 |
| 20-1 | Δgpr-1::bar+mat a | gpr-1 homokaryon | This study |
| 20-2 | Δgpr-1::bar+matA | gpr-1 homokaryon | This study |
| 20-3 | Δgpr-1::bar+mat a | gpr-1 homokaryon | This study |
| 28-3 | Δgpr-1::bar+mat a | gpr-1 homokaryon | This study |
| 28-4 | Δgpr-1::bar+mat a | gpr-1 homokaryon | This study |
| 28-6 | Δgpr-1::bar+mat A | gpr-1 homokaryon | This study |
| 20-1-1 | Δgpr-1::bar+his-3 mat A | 20-1 × his-3 A progeny | This study |
| 28-6-1 | Δgpr-1::bar+his-3 mat a | 28-6 × his-3 a progeny | This study |
| R-28-6 | Δgpr-1::bar+gpr-1+::his-3+mat A | Complemented strain | This study |
| GP1GN1 | Δgpr-1::bar+ Δgna-1::hph+mat a | 20-1 × 3B10 progeny | This study |
| GP1GN2 | Δgpr-1::bar+ Δgna-2::pyrG+pyr-4 mat A | 28-6 × A33-4 progeny | This study |
| GP1GN3 | Δgpr-1::bar+ Δgna-3::hph+mat a | 20-1 × 31c2 progeny | This study |
| G1AC1 | Δgpr-1::bar+gna-1(Q204L)::his-3+mat A | gna-1(Q204L) in Δgpr-1 background | This study |
| 21-3-7 | gna-1::mtr+gna-1(Q204L)::his-3+pdx-1 mat a | gna-1 (Q204L) allele | 76 |
| NG22 | H1-GFP::his-3+mat a | H1-GFP fusion | This study |
| N2276 | H1-GFP::his-3+mat A | H1-GFP fusion | 27 |
| H1GFP | Δgpr-1::bar+ H1-GFP::his-3+mat A | H1-GFP fusion | This study |
| 6103GFP | gpr-1-GFP::his-3+mat A | GPR-1-GFP fusion | This study |
| 28-6-GFP | Δgpr-1::bar+gpr-1-GFP::his-3+mat A | GPR-1-GFP fusion | This study |
| H4-2-2 | Δbek-1::hph+mat A | bek-1 homokaryon | 22 |
| (16)A | Δpre-1::hph+mat A | pre-1 homokaryon | 44 |
| (16)a | Δpre-1::hph+mat a | pre-1 homokaryon | 44 |
| GR1PRE1A | Δgpr-1::bar+ Δpre-1::hph+mat A | (16)a × 28-1 progeny | This study |
| GR1PRE1a | Δgpr-1::bar+ Δpre-1::hph+mat a | (16)a × 28-1 progeny | This study |
FGSC, Fungal Genetics Stock Center, Kansas City, MO.
The tissue samples for RNA and protein extractions were obtained from submerged cultures, vegetative tissues grown on solid VM medium, and fertilized (perithecia) and nonfertilized (protoperithecia) female reproductive structures formed on SCM. For submerged cultures, 500 ml of liquid VM medium was inoculated with conidia from 7-day-old flask cultures at a final concentration of 1 × 106 cells/ml. Cultures were incubated in the dark at 30°C with shaking at 200 rpm for 8 or 16 h, as indicated in the figure legends. The vegetative tissues were grown in the dark at 30°C for 3 days on solid VM medium overlaid with cellophane (Bio-Rad, Hercules, CA). Protoperithecial tissues were grown on cellophane-overlaid SCM plates in constant light at 25°C for 6 days. For perithecial tissues, 6-day-old cultures grown on cellophane-overlaid SCM plates were fertilized either with the wild-type strain (heterozygous cross) or the corresponding mutant (homozygous cross) of opposite mating type and incubated for an additional 3 days under the same conditions as those used for unfertilized SCM plates.
gpr-1 isolation and gene structure confirmation.
The N. crassa gpr-1 gene was identified during homology searches (BLAST [1]) of the Munich Information Center for Protein Sequences (MIPS) Neurospora database (http://mips.gsf.de/projects/fungi/neurospora), using the Dictyostelium discoideum cAR1 protein sequence, and corresponds to hypothetical gene 90c4_060. The predicted GPR-1 protein sequence is designated NCU00786.2 at the Broad Institute Neurospora genome database (http://www.broad.mit.edu/annotation/fungi/neurospora_crassa_7/index.html). A gpr-1 genomic clone was isolated from the pMOcosX cosmid library (55) using PCR amplification with specific primers (GPR-1-FW and GPR-1-RV) as the screening procedure. One positive clone (G11B20) was obtained and shown to contain the gpr-1 gene by sequence analysis (Core Sequencing Facility, Department of Microbiology and Molecular Genetics, University of Texas-Houston Medical School). Plasmids were maintained in Escherichia coli strain DH5α (33).
Construction of N. crassa strains.
The gpr-1 gene was mutated by gene replacement using the selectable marker bar. The gpr-1 gene replacement vector, pSVK21, was constructed as follows. Cosmid G11B20 was digested with XbaI, and a 9-kb fragment containing the gpr-1 open reading frame (ORF) was cloned into pBSSKII+ (Stratagene, La Jolla, CA), generating pSVK19. pSVK19 was subsequently digested with NcoI and SacI, leaving the 3-kb 3′ flank and removing the gpr-1 ORF and 5′ region. pSVK20 contains the 1.1-kb bar gene (58) under the control of the A. nidulans trpC promoter. pSVK20 was constructed by digestion of pBARGEM-7 (58) using XbaI and SpeI to release the bar gene cassette. The ends of the bar cassette fragment were filled in using DNA polymerase I (Klenow; Promega, Madison, WI) and the blunt-end fragment was subsequently cloned into pGEM5zf digested with EcoRV to yield pSVK20. The bar gene cassette was excised from pSVK20 using NcoI and SacI and a 1.5-kb 5′ region of gpr-1 from pSVK19 using SacI. The bar gene cassette and 1.5-kb 5′ flank of gpr-1 were ligated into pSVK19 digested with NcoI and SacI, yielding pSVK21. The correct orientation of the gpr-1 5′ region was verified by sequencing.
Conidia from 10-day-old cultures of N. crassa wild-type strain 74a (Table 1) were electroporated with 1 μg of pSVK21 linearized with EcoRI, as described previously (39, 69). Transformants were selected on FIGS medium supplemented with phosphinothricin. To identify homologous and ectopic integrations, genomic DNA was isolated from transformants, digested with KpnI, and subjected to Southern analysis (39). Genomic DNA was extracted using a Puregene kit as described by the manufacturer (Gentra Systems, Minneapolis, MN) (47). The 1.5-kb 5′ SacI-XbaI fragment from pSVK19 was labeled using the Prime-A-Gene method (Promega, Madison, WI) and used as a probe. The wild-type gpr-1 allele produced a 6-kb hybridizing fragment during Southern analysis, while a 2.7-kb fragment was detected only in strains with homologous insertion of the barR cassette at the gpr-1 locus (data not shown). Heterokaryotic strains with homologous recombination events were crossed to the wild-type strain 74A (Table 1). The progeny were selected on FIGS medium with phosphinothricin, and homokaryotic status of strains was confirmed by Southern analysis as described above.
To complement the Δgpr-1 mutation in trans, the gpr-1 genomic clone was inserted into vector pRAUW122 and targeted to the his-3 locus (3). The rescue plasmid pSVK22 was generated by cloning of the 9-kb XbaI fragment from pSVK19 into pRAUW122 digested with XbaI. In order to isolate a Δgpr-1 his-3 recipient strain, a Δgpr-1 mat A strain (28-6) was crossed to a his-3 mat a strain (his-3a), and progeny were plated on FIGS medium supplemented with histidine and phosphinothricin. The progeny were tested for his-3 prototrophy using VM medium without histidine. The Δgpr-1 his-3 strain (28-6-1) was transformed by electroporation with 1 μg of pSVK22, and transformants were plated on histidine-free FIGS medium. Genomic DNA from heterokaryons was digested with HindIII, and strains containing the wild-type gpr-1 allele integrated at the his-3 locus were identified by Southern analysis using the HindIII 8.8-kb his-3 fragment excised from pRAUW122 as a probe (data not shown). Homokaryotic Δgpr-1::bar+ gpr-1+::his-3+ rescued strains were isolated using the microconidiation procedure (25).
Vector pMF280 contains the N. crassa histone H1 protein as a carboxy-terminal GFP fusion, under the control of the ccg-1 promoter (27). Strain N2276 contains this vector targeted to the his-3 locus in an otherwise wild-type background (27). pMF280 was also electroporated into the Δgpr-1 his-3 strain 28-6-1 (Table 1). Transformants were subjected to Southern analysis using the 8.8-kb HindIII fragment from pRAUW122 as a probe (see above) (data not shown), and strains with the vector integrated at the his-3 locus were purified using the microconidiation procedure (25).
The Δgpr-1 his-3 strain 28-6-1 and his-3 strains were used as recipients to produce strains expressing a GPR-1-GFP fusion protein. The pSVK52 gpr-1-GFP vector was constructed as follows. A 2,388-bp PCR product was amplified from pSVK19 using Turbo Pfu DNA polymerase (Stratagene) and Gpr1gfp-Xba-FW and Gpr1gfp-Bgl-RV oligomers designed with XbaI (5′ end) and BglII (3′ end) restriction sites. The primary blunt-end PCR product was cloned into pGEM5zf digested with EcoRV (Promega) yielding pSVK51. The 2,338-bp gpr-1 fragment was subsequently released from pSVK51 with XbaI and BglII and cloned into his-3 targeting vector pMF272 (27) (described above) digested with XbaI and BamHI. Ten-day-old conidia from Δgpr-1 his-3 strain 28-6-1 were transformed by electroporation with 1 μg of pSVK52, and transformants were plated on FIGS medium. Strains with homologous insertion of the his-3::gpr-1-GFP fragment were identified by Southern analysis of genomic DNA using the 8.8-kb HindIII fragment from pRAUW122 as a probe (data not shown). Homokaryons were purified using the microconidiation technique (25).
Western analysis.
For Western analysis, plasma membrane fractions from various tissues (see above) were isolated as described previously (68). The protein concentration was determined using the Bradford protein assay (Bio-Rad). Samples containing 25 μg of total protein were denatured and solubilized in sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis sample buffer (62.5 mM Tris-HCl, pH 6.8, 10% glycerol, 2% SDS, 1% β-mercaptoethanol, 0.005% bromophenol blue) by boiling for 5 min. To detect GNA-1, GNA-2, GNA-3, and GNB-1, protein samples were resolved using 10% SDS-polyacrylamide gel electrophoresis and transferred to a nitrocellulose membrane (39, 68). The primary polyclonal rabbit antibodies against GNA-1, GNA-2, GNA-3, and GNB-1 were used at dilutions of 1:1,000, 1:3,000, 1:1,000, and 1:3,000, respectively (4, 39, 43, 77). A horseradish peroxidase conjugate (Bio-Rad) was used as the secondary antibody at a dilution of 1:10,000. The blocking solution contained 5% nonfat dry milk, 25 mM Tris-Cl (pH 7.6), 140 mM NaCl, 3 mM KCl, and 0.2% Tween-20. The membranes were incubated with the primary antibody in the blocking solution for 3 h at room temperature and for 1 h with the secondary antibody. The membranes were washed four times for 5 min each time between incubations with the primary and secondary antibodies and before the application of chemiluminescence detection reagents. Detection was performed using a Biochemi system (UVP, Upland, CA) with chemiluminescence detection reagents used according to the manufacturer's protocol (Pierce, Rockford, IL).
RT-PCR analysis.
In order to investigate the expression profile of gpr-1, total RNA was extracted from various tissues (5), and 1 μg was used in quantitative reverse transcriptase PCR (RT-PCR) (43) using the Access RT-PCR System (Promega). RT-PCR products were electrophoresed on 1.5% agarose gels, blotted onto nylon membranes, and subjected to Southern analysis as described above. The 2.4-kb gpr-1 fragment excised from pSVK51 (XbaI-BglII) was used as the probe. The tissues used were conidia (harvested from 7-day-old flask cultures), 8- and 16-h submerged cultures, and cultures from 3-day-old VM medium plates and 6-day-old SCM plates prior to (protoperithecia) and 3 days after fertilization with opposite mating type conidia (perithecia).
Four sets of primers (Table 2) were designed to confirm the gpr-1 gene structure predicted by the Broad Institute automated gene caller using RT-PCR (Access RT-PCR; Promega). Total RNA was extracted from 6-day-old SCM plate cultures as previously described (5), and 1 μg was used in RT-PCRs (43). Genomic intron-containing DNA controls were amplified by PCR using LA Taq (Takara) with cosmid G11B20 as a template. The 2.4-kb gpr-1 fragment excised from pSVK51 (XbaI-BglII) was used as a probe.
TABLE 2.
Oligonucleotides used in this study
| Name | Sequence |
|---|---|
| GPR1-INT-1-FW | 5′-CGAGTTCGGACAGTACCCAACA-3′ |
| GPR1-INT-1-RV | 5′-AACCAGAGGTAGTCGCGAAAAG-3′ |
| GPR1-INT-2-FW | 5′-CTTTTCGCGACTACCTCTGGTT-3′ |
| GPR1-INT-2-RV | 5′-GCTGATGAAACACTTGATAGCC-3′ |
| GPR1-INT-3-FW | 5′-GGCTATCAAGTGTTTCATCAGC-3′ |
| GPR1-INT-3-RV | 5′-AAGAGGAAGACGAGGAGTAGTT-3′ |
| GPR1-INT-4-FW | 5′-CCAAAGATACCAGTTACCCGCT-3′ |
| GPR1-INT-4-RV | 5′-CATAACGAGCCATCCCTAATGA-3′ |
| Gpr1gfp-Xba-FW | 5′-GGTCTAGAACATFFACGACTTCATC-3′ |
| Gpr1gfp-Bgl-RV | 5′-GCAGATTAACGAGCCATCCCTACC-3′ |
| GPR-1-FW | 5′-ATCTCTCCAATCACCCAGGCTGCT-3′ |
| GPR-1-RV | 5′-TAACGAGCCATCCCTAATGACTCG-3′ |
Phenotypic analysis and microscopy.
To analyze phenotypes in submerged cultures, liquid VM medium was inoculated with conidia at a final concentration of 1 × 106 cells/ml and incubated with shaking at 200 rpm for 16 h at 30°C. Apical extension rates were determined by measuring colony diameters after inoculation of 1 μl of a conidial suspension in the center of VM medium plates. The plates were incubated at 30°C in the dark, and the colony diameter was measured at 2-h intervals. Cultures were then viewed and photographed using a BX41 fluorescent microscope and C-4040 digital camera (Olympus, Lake Success, NY). Unfertilized and fertilized female reproductive tissues were cultured on SCM plates in constant light and were observed using a SZX9 stereomicroscope with an ACH 1× objective lens outfitted with the C-4040 digital camera (Olympus). For microscopic observation of GFP fluorescence in strains expressing GFP fusion proteins, a BX41 fluorescent microscope with a WIB long-pass fluorescence cube (excitation 460 to 490 nm; emission 515 to 550 nm) and UM Plan Fluorite objective lenses (Olympus) was used.
For trichogyne pheromone attraction assays (7, 8, 44), cultures were grown for 6 days on 2% water agar. Chemoattraction between trichogynes and microconidia from wild-type strains was observed using a BX41 fluorescent microscope with UM Plan Fluorite objective lenses as described (44).
RESULTS
gpr-1 identification, gene structure analysis, and expression profile.
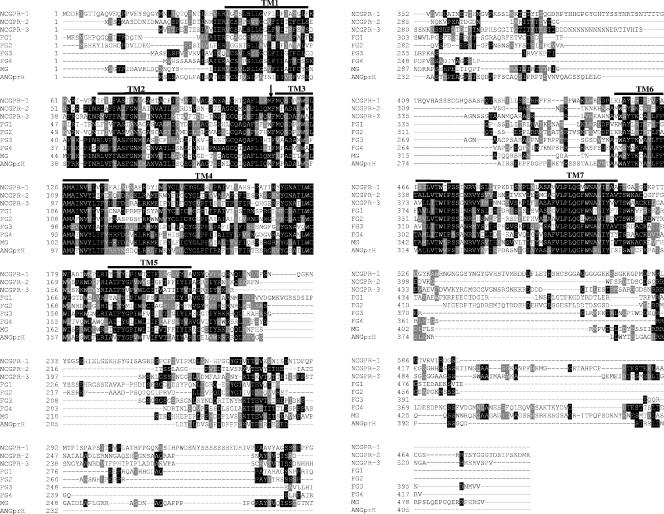
The gpr-1 gene was identified during BLAST (1) searches of the N. crassa genome at the MIPS Neurospora database (http://mips.gsf.de/projects/fungi/neurospora) using the cyclic AMP (cAMP) binding receptor cAR1 from D. discoideum. The MIPS designation is 90c4_060, while that of the Broad Institute Neurospora genome database (http://www.broad.mit.edu/annotation/fungi/neurospora_crassa_7/index.html)is NCU00786.2 (28). The predicted GPR-1 protein contains 596 amino acids and has a secondary structure characteristic of GPCRs, with seven transmembrane helices and a long third cytoplasmic loop and carboxy terminus (Fig. 1). GPR-1 is distantly related to the four cAMP receptors (cAR1 to cAR4) (46) and three cAMP receptor-like proteins (CrlA to CrlC) (61) from D. discoideum, as well as GCR1 from Arabidopsis thaliana (60).
FIG. 1.
Alignment of GPR-1 with other predicted fungal cAMP receptor-like GPCRs. ClustalW (http://www.ch.embnet.org/software/ClustalW.html) was used to align cAMP receptor-like GPCR protein sequences from N. crassa (NCGPR-1, NCU00786.2; NCGPR-2, NCU04626.2; and NCGPR-3, NCU09427.2) with predicted proteins from M. grisea (MG; MG06738.4), F. graminearum (FG1, FG09693.1; FG2, FG05239.1; FG3, FG07716.1; and FG4, FG03023.1) and A. nidulans (ANGprH, AN8262.2). Boxshade was subsequently used to indicate identical (black shading) and similar (gray shading) amino acid residues (http://www.ch.embnet.org/software/BOX_form.html). The bars above the sequences indicate predicted seven-helix transmembrane regions, while the arrow indicates the position of the first intron.
Additional BLAST searches against the N. crassa and other fungal genomes were used to identify other ORFs with homology to GPR-1. In this way, two additional hypothetical proteins GPR-2 (NCU04626.2) and GPR-3 (NCU09427.2) were identified in N. crassa. The position of the second intron in GPR-2 was predicted incorrectly by the automated gene caller (data not shown). Seven amino acids (188 to 195) were removed, and the resulting protein sequence was used in the alignment. The relatively high identity shared between GPR-1 and GPR-2 (25%) and between GPR-1 and GPR-3 (29%) indicates that these three proteins very likely form a gene family (Fig. 1).
GPCRs homologous to GPR-1 are absent from the genomes of the ascomycete yeasts S. cerevisiae and S. pombe (28). Among ascomycete filamentous fungi whose genome sequences are available, only Fusarium graminearum has a GPCR gene family similar to GPR-1, -2, and -3 and contains four members (24 to 36% identical to GPR-1) (Fig. 1). In contrast, Magnaporthe grisea and A. nidulans have only a single GPCR similar to GPR-1 (28% and 25% identical, respectively). The ClustalW alignment (20) (Fig. 1) shows that these proteins are similar in all seven transmembrane helices as well as in the first and second extracellular loops that are important for ligand binding. The proteins are less homologous in the third intracellular loop and the carboxy-terminal tail that are required for the physical interaction with Gα subunits. A GPCR with 27% identity to GPR-1 has been described in the basidiomycete yeast C. neoformans (Gpr4) (74) and is implicated in sensing the amino acids proline and methionine.
The predicted gene structure of gpr-1 contains five exons and four introns (Fig. 2B). The presence of the introns was confirmed by RT-PCRs using four sets of oligonucleotides (Table 2) designed within the exons that flank each intron. Total RNA was used as the template in RT-PCR, whereas the genomic cosmid clone G11B20 was used for amplification of genomic fragment controls using the same sets of primers. The RT-PCR and genomic DNA PCR products were subjected to Southern analysis. The analysis confirmed the presence and sizes of the four introns as predicted by the automated gene caller (Fig. 2B) (data not shown). The A. nidulans gprH gene (Fig. 1) also contains four predicted introns (http://www.broad.mit.edu/annotation/fungi/aspergillus). In contrast, other GPR-1-related GPCRs have two predicted introns in their ORFs at very similar or identical positions as GPR-1. In particular, the position of the first intron is highly conserved in all GPR-1-related GPCRs and is located at the junction of the first extracellular loop and the third transmembrane helix (Fig. 1).
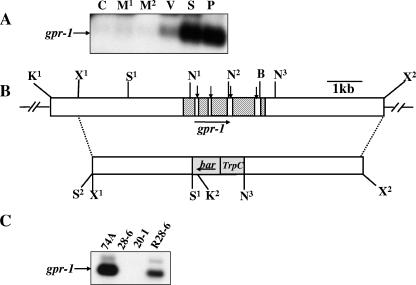
FIG. 2.
Structure of the N. crassa gpr-1 genomic region and construction of a Δgpr-1 strain. (A) Expression of gpr-1 during the N. crassa life cycle. Total RNA was isolated from cultures of wild-type strain 74A and subjected to quantitative RT-PCR analysis as described in Materials and Methods. The tissues used were the following: C, conidia; M1, 8-h submerged (mycelial) cultures; M2, 16-h submerged (mycelial) cultures; V, asexual vegetative cultures grown for 3 days at 30°C in the dark on solid VM medium; S, cultures grown for 6 days on SCM plates at 25°C under light (protoperithecial cultures); and P, SCM plate cultures 3 days after fertilization with wild-type strain 74a conidia (perithecial cultures). (B) gpr-1 genomic clone and gene replacement vector. The hatched region (top bar) indicates the gpr-1 exons, while the vertical arrows indicate introns (white areas between exons). The gray area (bottom bar) corresponds to the phosphinothricin-resistance gene, bar, under control of the A. nidulans trpC promoter. The dashed lines illustrate the homologous recombination event leading to replacement of the gpr-1 ORF with the bar cassette between the SalI (S1) and the third NcoI sites. The horizontal arrows show the direction of transcription of gpr-1 and bar. Restriction sites are as follows: N, NcoI; S, SalI; K, KpnI; B, BamHI; and X, XbaI. The second SalI site (S2) is an artifact of cloning. (C) Verification of the gpr-1 deletion mutant and complemented strains. Total RNA was isolated from 6-day-old protoperithecial tissues of wild type (74A), homokaryotic Δgpr-1 mutants (28-6 and 20-1), and a Δgpr-1 + gpr-1+ complemented strain (R28-6). Levels of gpr-1 mRNA were quantitated using RT-PCR as described in Materials and Methods. The faint, larger molecular mass species in the 74A and R28-6 samples is due to amplification of residual genomic DNA in the RNA preparations.
gpr-1 mRNA levels are too low to detect by Northern analysis (data not shown). Therefore, quantitative RT-PCR was performed to determine the expression of gpr-1 during various developmental stages. Detectable levels of gpr-1 mRNA were present in three tissues: vegetative plates and protoperithecial and perithecial tissues (Fig. 2A). Highest expression was observed in the two sexually differentiated tissues. The relative amounts of gpr-1 in protoperithecial and perithecial cultures were comparable, suggesting that gpr-1 may function during pre- and postfertilization events in N. crassa.
Δgpr-1 strains exhibit phenotypic defects during sexual development.
A Δgpr-1 mutant was isolated after electroporation of a wild-type strain with a construct in which the gpr-1 coding region was replaced with a phosphinothricin resistance gene cassette (Fig. 2B) (58). Heterokaryotic primary transformants were obtained by selection on medium containing phosphinothricin (see Materials and Methods for details). Strains with the proper homologous recombination event were crossed to a wild-type strain of opposite mating type in order to produce homokaryotic Δgpr-1 mutant ascospore progeny (data not shown). The purity of homokaryons was verified by Southern analysis (data not shown), and RT-PCR analysis confirmed that homokaryotic Δgpr-1 strains lack gpr-1 mRNA (Fig. 2C). The Δgpr-1 mutation was complemented in trans with the original 9-kb gpr-1 genomic fragment targeted to the his-3 locus (see Materials and Methods). The restoration of gpr-1 mRNA expression in Δgpr-1 + gpr-1+ complemented strains was confirmed by RT-PCR (Fig. 2C).
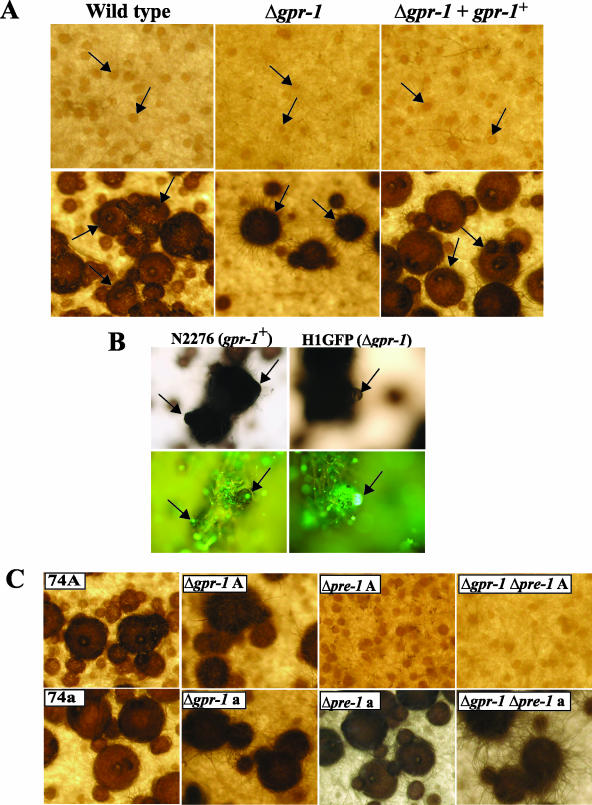
Δgpr-1 strains were analyzed for phenotypes during the life cycle. Δgpr-1 mutants do not possess any obvious defects during asexual growth or development (data not shown). Traits tested included hyphal extension rate under normal and hyperosmotic conditions and various aspects of macroconidiation. Consistent with the results from expression profile analysis showing that gpr-1 mRNA levels are highest in unfertilized and fertilized female tissues, deletion of gpr-1 leads to several defects during sexual development. Protoperithecia are weakly pigmented, and a significant number are small and submerged in the agar during growth on solid medium (Fig. 3A).Perithecia from Δgpr-1 mutants possess a significantly greater number of fringe hyphae on the surface in comparison to the wild-type strain, giving them a “hairy” appearance. The abundant formation of fringe hyphae has been reported previously in Δgna-1 strains (39) (see below). The underlying cause of this phenomenon, which has been observed in other mutants with defects in sexual reproduction, is not known. The Δgpr-1 perithecia have deformed beaks and lack ostioles (pores) at the tips that are essential for appropriate ejection of ascospores. Moreover, they are frequently ruptured, leading to the release of the perithecial contents (Fig. 3A and B; see also Fig. 6B). In contrast to wild type, significantly fewer perithecia reach maturity in Δgpr-1 mutants (Fig. 3A). Those Δgpr-1 perithecia that are fully developed produce a similar number of ascospores as wild-type strains. The viability of ascospores was identical to that of wild type during both homozygous and heterozygous crosses (data not shown). Δgpr-1 + gpr-1+ rescued strains are phenotypically identical to wild type (Fig. 3A and B), confirming that the defects of Δgpr-1 mutants result from loss of the gpr-1 gene product.
FIG. 3.
Phenotypic characterization during the sexual cycle. (A) Unfertilized (protoperithecia) and fertilized (perithecia) structures. Strains were cultured on solid SCM at 25°C for 6 days in light (top panels; protoperithecia). At this time, half of the plate was fertilized with wild-type conidia of opposite mating type (74a or 74A) and photographed 3 days after fertilization (bottom panels; perithecia). For the analysis, wild-type (74A), Δgpr-1 (28-6 and 20-1), and Δgpr-1 + gpr-1+ (R28-6) strains were used. Arrows indicate protoperithecia (top panels) or perithecia (bottom panels; enlarged dark bodies). Photographs were taken at ×25 magnification. (B) Microscopic images of perithecial beaks. Strains expressing a histone H1-GFP fusion protein, N2276 (gpr-1+ strain) and H1GFP (Δgpr-1 strain), were cultured on SCM plates for 6 days and then fertilized with opposite mating type conidia. Seven days later, perithecia were subjected to phase-contrast microscopy (upper panels) and detection of GFP fluorescence (bottom panels) (see Materials and Methods). Images are shown at ×200 magnification. The arrows indicate perithecial beaks (left panels) or ruptured perithecia (right panels). (C) Epistasis analysis between gpr-1 and pre-1. Strains were cultured to produce perithecia as indicated in panel A. Wild-type mat A (74A) and mat a (74a), Δgpr-1 mat A (28-6; Δgpr-1A), Δgpr-1 mat a (20-1; Δgpr-1 a), Δpre-1 mat A [(16)A; Δpre-1 A], Δpre-1 mat a [(16)a; Δpre-1 a], Δgpr-1 Δpre-1 mat A (GR1PRE1A; Δgpr-1 Δpre-1 A), and Δgpr-1 Δpre-1 mat a (GR1PRE1a; Δgpr-1 Δpre-1 a) strains were used for analysis. Photographs were taken at ×25 magnification.
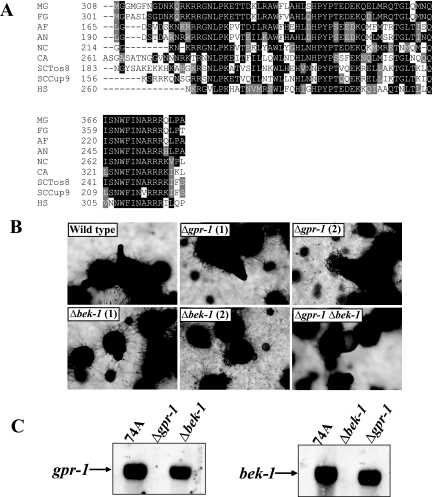
FIG. 6.
Epistasis analysis between gpr-1 and bek-1. (A) Alignment of BEK-1 with related homeodomain transcription factors. ClustalW (http://www.ch.embnet.org/software/ClustalW.html) was used to align N. crassa BEK-1 (NC; NCU00097.2) with putative homeodomain transcription factors from the fungi F. graminearum (FG; FG05475.1), A. fumigatus (AF; Afu4g10110), A. nidulans (AN; AN2020.2), and C. albicans (CA; CaJ7.0235) and previously characterized homeodomain proteins identified as M. grisea Pth12p (MG; DQ060925.1), S. cerevisiae Tos8p (SCTos8; YGL096W) and Cup9p (SCCup9; YPL177C), and human PKNOX1 (HS; accession number AAC51243). Boxshade was used to indicate identical (black shading) and similar (gray shading) amino acid residues (http://www.ch.embnet.org/software/BOX_form.html). (B) Perithecial development. Perithecia from wild-type (74A), Δgpr-1 (28-6), Δbek-1 (BK1), and Δgpr-1 Δbek-1 (GP1BK1) strains were analyzed microscopically 6 days after fertilization. Images were captured at ×52 magnification. Two images were taken for Δgpr-1 and Δbek-1 strains, showing perithecial defects (1) and ruptured perithecia (2). (C) Expression of gpr-1 and bek-1. Total RNA was isolated from perithecial tissues of wild-type (74A), Δgpr-1 (28-6), and Δbek-1 (BK1) strains 3 days after fertilization and subjected to quantitative RT-PCR as described in Materials and Methods.
In order to better observe ascospore emergence from perithecia, we analyzed strains containing a histone H1-GFP fusion protein under the control of ccg-1 promoter (see Materials and Methods). Previous studies have shown that this GFP fusion allows clear visualization of ascospores, due to the strong fluorescence of the H1-GFP protein in their nuclei (27). Microscopic analysis of strains expressing the H1-GFP fusion protein confirmed the morphological differences observed in wild-type gpr-1+ (N2276) and Δgpr-1 mutant (H1GFP) strains with regard to missing ostioles and ruptures in the perithecial wall (Fig. 3B). Fluorescence from ascospores within intact perithecia was observed through the ostiole in wild type (Fig. 3B). In contrast, perithecia from Δgpr-1 mutants are ruptured, with ascospores oozing from the fissures in the structure (Fig. 3B).
GPR-1 is not the only GPCR that is highly expressed during sexual development in N. crassa. A previous study has shown that the pheromone receptor gene pre-1 is present at high levels in both protoperithecial and perithecial tissues (44). Deletion of pre-1 in a mat A background leads to defective chemoattraction towards mat a males, subsequently resulting in a complete block in cell fusion and perithecial development (44). The trichogyne attraction assay was used to investigate the possibility that Δgpr-1 strains might also be affected during early stages of mating. Chemotropism of trichogynes toward male cells of opposite mating type, as well as the ability of male cells (conidia) to attract trichogynes of opposite mating type, were not disrupted in Δgpr-1 strains (data not shown). This result indicates that although PRE-1 and GPR-1 are highly expressed during sexual development, they very likely regulate different events. Furthermore, contrary to pre-1, gpr-1 expression is not mating-type specific (data not shown). We further probed the relationship between pre-1 and gpr-1 through analysis of Δgpr-1 Δpre-1 double mutants (Fig. 3C). Δgpr-1 Δpre-1 strains possessed the defects of both single mutants, confirming independent roles for these receptors during sexual development (Fig. 3C). The Δgpr-1 Δpre-1 double mutants formed fewer protoperithecia than wild-type or Δgpr-1 strains, and protoperithecia from the double mutants were “hairy.” The mat A Δgpr-1 Δpre-1 mutants did not form perithecia after fertilization with mat a males, reflecting the absence of PRE-1. Conversely, the mat a Δgpr-1 Δpre-1 strains formed perithecia without ostioles after fertilization with mat A males, indicating the loss of GPR-1. Thus, PRE-1 is essential for cell-cell recognition and fusion, while GPR-1 is required for normal development of unfertilized (protoperithecial) and fertilized (perithecial) female structures.
Epistasis analysis between gpr-1 and the three N. crassa Gα subunits.
Previous work has demonstrated that the Gα subunit GNA-1 (39, 44) and Gβγ dimer GNB-1/GNG-1 (44, 47, 77) are essential for female (but not male) fertility in N. crassa. The functions of these proteins are mating type independent, since they are required downstream of both pheromone receptors (PRE-1 and PRE-2) in mat A and mat a strains, respectively (44; Kim and Borkovich, unpublished observations). Trichogynes of Δgna-1, Δgnb-1, and Δgng-1 mutants exhibit severe defects in chemoattraction and fusion with male cells, similar to those of Δpre-1 mat A strains (44, 47). It has been hypothesized that GNA-1 is the Gα subunit that communicates the pheromone signal from PRE-1 to downstream effectors (44). During homozygous crosses, Δgna-3 strains form a large number of perithecia with short beaks that are embedded in the agar medium (43; data not shown). Such crosses also produce a large proportion of nonviable ascospores. The Δgna-2 mutant has no obvious defects during either asexual or sexual development, but Δgna-1 Δgna-2 and Δgna-2 Δgna-3 double mutants have more severe defects than strains lacking only gna-1 or gna-3 (4, 42).
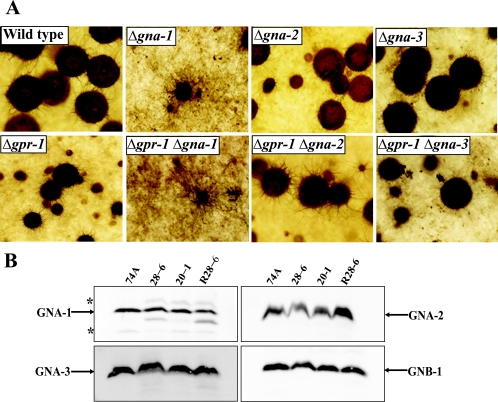
The results demonstrate that gpr-1 is required for normal perithecial development, but the phenotype of Δgpr-1 strains differs from that observed in the single Gα mutants. Therefore, we explored functional relationships between GPR-1 and the three Gα proteins through examination of protoperithecial and perithecial development in Δgpr-1 Gα single and double mutants. Δgna-1 strains form “hairy” protoperithecia that enlarge slightly after fertilization but fail to develop mature perithecia. This phenotype is similar to that observed for fringe hyphae production in perithecia of Δgpr-1 mutants (Fig. 4A). The Δgpr-1 Δgna-1 double mutant exhibits phenotypic defects identical to those of the Δgna-1 mutant during protoperithecial and perithecial development, leaving open the possibility that GNA-1 is coupled to GPR-1 (Fig. 4A). However, since the other two Gα subunits are also implicated in sexual development, we could not exclude the possibility that either GNA-2 or GNA-3 might be coupled to GPR-1 at a certain time during sexual development. The Δgpr-1 Δgna-2 strain has defects identical to those observed in Δgpr-1 single mutants (Fig. 4A), but the epistatic relationship is unclear due to the absence of phenotypes in Δgna-2 single mutants. Similar to Δgpr-1 strains, Δgpr-1 Δgna-3 mutants form small perithecia with no ostioles. However, perithecia from Δgpr-1 Δgna-3 mutants also have short beaks, like Δgna-3 strains (Fig. 4A). The phenotype of Δgpr-1 Δgna-3 mutants is a combination of both gene mutations, and therefore the epistatic analysis does not support coupling between GNA-3 and GPR-1 during perithecial development.
FIG. 4.
Relationship between GPR-1 and G protein subunits. (A) Epistasis analysis between gpr-1 and the three Gα genes. Strains were cultured to produce perithecia as indicated in the legend of Fig. 3A. Wild-type (74A), Δgpr-1 (28-6), Δgna-1 (3B10), Δgna-2 (A33-4), Δgna-3 (31c2), Δgpr-1 Δgna-1 (GP1GN1), Δgpr-1 Δgna-2 (GP1GN2), and Δgpr-1 Δgna-3 (GP1GN3) strains were used for analysis. Photographs were taken at ×25 magnification. (B) Analysis of Gα and Gβ protein levels. The plasma membrane fraction was isolated from SCM plate cultures, and samples containing 25 μg of protein were subjected to Western analysis using specific antisera (see Materials and Methods). Wild-type (74A), Δgpr-1 (28-6 and 20-1), and Δgpr-1 + gpr-1+ (R28-6) strains were used for analysis. The asterisks indicate nonspecific bands.
Since the epistasis analysis between gpr-1 and the Gα genes ruled out GNA-2 and GNA-3 as downstream effectors of GPR-1 but left open the possibility that GNA-1 is coupled to GPR-1, we examined whether constitutive activation of gna-1 could bypass the defects of the Δgpr-1 mutation. The predicted GTPase-deficient, dominant-activated gna-1(Q204L) allele construct (76) was targeted to the his-3 locus of a Δgpr-1 his-3 strain, and transformants were selected and purified as described in Materials and Methods. Δgpr-1 strains expressing the dominant-activated gna-1 allele showed an even greater reduction in perithecial formation than observed in Δgpr-1 strains, but ostiole production was not recovered (data not shown). This outcome indicates that activation of GNA-1 alone cannot rescue the perithecial defects of strains lacking GPR-1.
To elucidate whether deletion of gpr-1 affects G protein levels, we performed Western analysis using specific antibodies raised against individual G protein subunits (4, 39, 43, 77). The results show that the three Gα proteins, GNA-1, GNA-2, and GNA-3, and the Gβ protein, GNB-1, are present at the same level in Δgpr-1 and wild-type strains (Fig. 4B). Thus, the presence of gpr-1 has no effect on the concentration of G protein subunits associated with the plasma membrane, supporting the proposal that the aberrations observed during sexual development in Δgpr-1 mutants result from loss of GPR-1.
GPR-1 is localized in female reproductive structures.
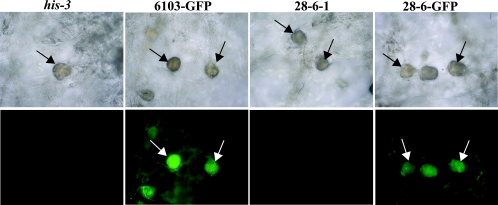
The phenotypic defects identified during sexual development in Δgpr-1 strains are consistent with the high expression of gpr-1 in female reproductive structures. To determine whether GPR-1 localization correlates with the presence of gpr-1 mRNA, we constructed strains that express a carboxy-terminal fusion of GFP to GPR-1. The fusion vector was transformed into his-3 and Δgpr-1 his-3 strains, and transformants with homologous recombination events at the his-3 locus were isolated (see Materials and Methods). The GPR-1-GFP fusion protein did not have any obvious effect on sexual development in the wild-type background and was able to partially complement the defects of the Δgpr-1 mutant (data not shown). Since perithecia are heavily melanized, we focused on microscopic observation of unfertilized female reproductive tissues. Fluorescence was observed in protoperithecia of strains containing the GPR-1-GFP fusion, consistent with localization of GPR-1 in this structure (Fig. 5).
FIG. 5.
Localization of a GPR-1-GFP fusion protein. Phase-contrast (upper panels) and GPR-1-GFP localization (bottom panels) images from 6-day-old unfertilized SCM tissues were obtained as described in the legend of Fig. 3B. Strains carrying gpr-1+ (his-3) and Δgpr-1 (28-6-1) untransformed controls or those expressing a GPR-1-GFP fusion protein in the gpr-1+ (6103-GFP) or Δgpr-1 (28-6-GFP) strain background were used for analysis. Images are shown at ×200 magnification. Arrows indicate protoperithecia.
BEK-1 is a homeodomain transcription factor that functions downstream of GPR-1.
BEK-1 is a homeodomain transcription factor protein (NCU00097.2) previously annotated in the N. crassa genome sequence (12). Typically, homeobox proteins contain a ∼60-amino-acid long homeodomain sequence (30). BEK-1 is similar to members of the TALE (three-amino-acid loop extension) superfamily of atypical homeodomain transcription factors characterized by an extension of three amino acids between two α helices within the homeodomain (16). This extension almost always consists of proline-tyrosine-proline (PYP) and is followed by a serine or threonine and several acidic residues (16). These amino acids are crucial for the interaction with other homeobox proteins, such as PBX and HOX (29). Members of the TALE family include MEIS (myeloid ecotropic viral insertion sites), TGIF, PBX, and IRO (Iroquois) in animals, KNOX and BEL in plants, and M-ATYP and CUP in fungi (16). Alignment analysis of the homeodomain regions of BEK-1 with members of the TALE family revealed that BEK-1 shares high identity (53 to 77%) with the homeodomains of these proteins and confirmed that BEK-1 contains the PYP motif in the same position as other proteins in the TALE superfamily (Fig. 6A).
A Δbek-1 N. crassa mutant was created during a high-throughput reverse genetics project (22). In contrast to results with Δgpr-1 mutants, there were no visible protoperithecial defects in Δbek-1 strains. However, the Δbek-1 mutant shares several defects in common with Δgpr-1 strains during perithecial development. The bek-1 mutant forms small, “hairy” perithecia that are frequently ruptured, and these perithecia also lack ostioles (Fig. 6B). However loss of bek-1 results in more severe deformation of perithecial beaks than observed in Δgpr-1 strains. Perithecia from Δbek-1 strains have very short beaks (Fig. 6B), while those from Δgpr-1 mutants form volcano-shaped beaks during later stages of perithecial development. The more severe defects observed in the Δbek-1 mutant could result from a convergence of multiple signal transduction pathways, including GPR-1, that regulate BEK-1 function. To corroborate a possible functional relationship between GPR-1 and BEK-1, Δgpr-1 Δbek-1 double mutants were isolated. Δgpr-1 Δbek-1 strains exhibit defects very similar to those observed in the Δbek-1 mutant, consistent with BEK-1 functioning downstream of GPR-1 during perithecial development. The mechanism by which GPR-1 regulates BEK-1 is not currently known. However, deletion of gpr-1 does not greatly affect expression of bek-1, and loss of bek-1 does not lead to a significant reduction in gpr-1 mRNA levels, indicating that any regulation of BEK-1 function by GPR-1 is posttranscriptional (Fig. 6C).
DISCUSSION
N. crassa GPR-1 is the first cAMP receptor-like (CRL) GPCR characterized in ascomycete fungi. GPR-1 is differentially expressed, and the highest expression was detected during development of female reproductive structures, before and after fertilization. Deletion of gpr-1 leads to several phenotypic defects during sexual development. Although Δgpr-1 strains are not female sterile, their ability to disperse progeny is severely compromised. This is caused by the absence of ostioles at the tip of the perithecium through which mature ascospores are discharged.
Fungal GPCRs have been divided into four major groups: (i) the pheromone-sensing receptors Ste2 and Ste3 of S. cerevisiae, (ii) the glucose sensing Gpr1 receptor of S. cerevisiae, (iii) the putative nutrient sensing receptor Stm1 of S. pombe, and (iv) proteins related to the CRL receptors of D. discoideum (12, 28, 32). A fifth group of seven-helix proteins, the microbial opsins, have not been implicated in G protein signaling in fungi (6; data not shown). The CRL receptors constitute a functionally divergent group of GPCRs that are similar to cAMP receptor GPCRs from D. discoideum. In N. crassa, this receptor group consists of three proteins (GPR-1, GPR-2, and GPR-3). A. nidulans and M. grisea have only one CRL-related protein, while F. graminearum contains four CRL receptors (Fig. 1). The related protein from A. nidulans, GprH, has been described previously, but no function has yet been assigned (32).
CRL GPCRs have been characterized phenotypically in slime mold D. discoideum (61) and the model plant A. thaliana (19, 23, 41, 60). Although CRL GPCRs are absent from both budding and fission yeasts, proteins with weak similarity to this group of receptors have been identified in the basidiomycete C. neoformans (74). Despite their sequence homology, CRL receptors appear to have very diverse functions in different species. For example, although N. crassa GPR-1, GPR-2, and GPR-3 possess significant amino acid identity, our analysis indicates that GPR-1 does not share any obvious functions with GPR-2 or GPR-3 (S. Krystofova and K. A. Borkovich, unpublished observations). In D. discoideum, the CrlA receptor functions as a negative regulator of cell growth and appears to be required for prestalk cell differentiation (61). GCR1 is the only characterized GPCR from A. thaliana and physically interacts with the lone A. thaliana Gα protein, GPA1 (59). Previous studies investigating loss-of-function or overexpression gcr1 A. thaliana lines, as well as in vitro experiments with BY2 tobacco cell cultures, showed that GCR1 is required for seed germination and that GCR1 regulates DNA synthesis through activation of phosphatidylinositol-specific phospholipase C (2, 19, 23). Deletion of the CRL-related gene gpr4 in C. neoformans leads to modest defects in cell fusion during mating and reduced capsule formation (74).
The shared sequence homology of CRL proteins in ascomycete fungi is distributed in both extracellular and transmembrane regions, suggesting that these GPCRs share structural or functional similarity, e.g., by responding to similar stimuli (ligands) or coupling to comparable downstream components. In addition, we examined the correlation between intron positions and secondary protein structure in this group of GPCRs. In our previous studies, we reported that the position of introns in heterotrimeric G protein genes (Gα and Gγ subunits) was highly conserved in mammals and N. crassa (47, 68). Since mammalian GPCRs are for the most part intronless (14) and do not contain receptors related to GPR-1, we compared gene structure only within the cAMP receptor-like family present in ascomycete fungi (Fig. 1). These receptors share a very similar gene structure in which introns are found only in nontransmembrane regions. It is worthwhile to point out that the position of the first intron in all genes presented in our study is highly conserved and located at the junction of the first extracellular loop and the third transmembrane region (Fig. 1). The evolutionary significance of the presence of an intron in a highly conserved region of these proteins has not yet been established. The large number of GPCRs identified in N. crassa and other fungal and nonfungal organisms provides a unique opportunity to examine the evolution of a specific intron on a larger scale.
It has been demonstrated that Δgna-1 strains form fewer protoperithecia than wild type and that these protoperithecia are “hairy,” with an abundance of fringe hyphae on their surfaces. Importantly, no ascospore progeny are produced after application of opposite mating type conidia (39). The failure of Δgna-1 protoperithecia to develop into mature perithecia with ascospores could be explained by involvement of GNA-1 in a signaling pathway required for perithecial development. In this aspect, Δgna-1 strains display a defect similar to Δpre-1 mutants during development of perithecia (44). PRE-1 is one of two pheromone receptors required for chemotropism of female trichogynes toward male cells in N. crassa. In comparison to Δpre-1 mutants, the complexity of phenotypic defects during protoperithecial development and the block in perithecia formation observed in Δgna-1 strains indicate that multiple receptors must be coupled to GNA-1 during sexual development. The epistasis analysis between gpr-1 and Gα genes rules out GNA-2 and GNA-3 acting downstream of GPR-1, leaving GNA-1 as a potential candidate for coupling to GPR-1. The genetic analysis using single and double mutants in the Δgna-1 background is complicated by the fact that GNA-1 is coupled to the pheromone receptors, and therefore deletion of gna-1 causes a simultaneous block in the pheromone response pathway (39, 44). Defects in protoperithecial development in Δgpr-1 Δgna-1 strains were identical to those observed in Δgna-1 mutants, consistent with GNA-1 acting downstream of GPR-1 for this trait. It is also possible that GPR-1 exerts its action in N. crassa independently of heterotrimeric G proteins, particularly during perithecial development. Accumulating evidence points to G-protein-independent activities for GPCRs in several systems (13, 15). Along these lines, it is noteworthy that a heterotrimeric G-protein-independent function has been proposed for A. thaliana GCR1 with respect to hormonal regulation of seed germination (19). Determination of possible G-protein-independent functions for N. crassa GPR-1 will require additional study.
Six putative homeodomain transcription factors have been annotated in N. crassa (12). Homeodomain proteins form a large superfamily of transcription factors fundamental to cellular proliferation, differentiation, and cell death in various species. Interestingly, deletion of the homeodomain transcription factor gene bek-1 led to several defects in common with Δgpr-1 mutants, including the formation of ruptured perithecia lacking ostioles. Three TALE superclass homeobox genes have been identified in the genome of S. cerevisiae and have been grouped into the M-ATYPE and the CUP classes (16). One of the yeast homeobox proteins, Tos8p, targets promoter regions of genes that are involved in bud growth during G1/S events (36). A TALE member, Pth12p, has been previously identified in the filamentous fungus M. grisea and has been shown to be involved in appressorium maturation (P. Hauck et al. [http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?db = protein&val = 67005921]).
Some members of the TALE homeodomain protein family have been implicated as downstream targets of signal transduction pathways. It has been reported that protein kinase A strongly activates expression through PBX-HOX binding sites in HEK293 cells (human embryonic kidney epithelial cell line) (64), and protein kinase A transactivation domains were mapped to the MEIS1A and MEIS1B C termini (38). The MEIS protein forms stable heterodimers with the PBX homeodomain protein and binds to DNA in cooperation with a HOX partner (51). In a recent study, the ERK1/2 mitogen-activated protein kinase was shown to phosphorylate and negatively regulate the Arix/Phox2a homedomain protein (37). The epistasis analysis between gpr-1 and bek-1 indicates that BEK-1 possibly acts downstream of GPR-1 in N. crassa. However, the relationship between these two proteins remains unclear. We propose that the effect must be posttranscriptional, as the bek-1 transcript level was not greatly affected in Δgpr-1 strains or vice versa. Further experimental work will be required to establish the molecular mechanism by which BEK-1 is regulated by the GPR-1 signaling pathway, including identification of potential partners, upstream regulators, and genes targeted by BEK-1.
Sexual development in N. crassa is a complex process for which molecular mechanisms are not well understood. Unfertilized and fertilized female reproductive structures consist of numerous different cell types (9). The mature perithecium is filled with asci (unbranched nonseptate hyphae) that contain ascospores and paraphyses (branched, multinucleate nonascogenous hyphae). The internal contents of the perithecium are enveloped by the outer perithecial wall, consisting of thick-walled pseudoparenchymatous cells (63). The cell types of the perithecial neck differ from the cells of the perithecial body. The ostiolar canal is lined with short branched cells (periphyses) which differentiate into pseudoparenchymatous neck wall cells (63). Upon maturation, a single ascus extends to the ostiole to discharge the ascospores (35). The actual mechanism of ascospore ejection has not been determined either in N. crassa or other ascomycetes. A recent study in Giberella zeae showed that three major osmolytes, mannitol, K+, and Cl−, were present in the epiplasmic fluid that is discharged along with ascospores but that potassium and chloride ions are likely the main osmolytes driving ascospore ejection (67).
Δgpr-1 strains display unique morphological defects (deformed beaks with no ostioles) that have not been previously reported in N. crassa. However, the isolation of two mutants that lack ostioles, m and n, has been reported for the ascomycete filamentous fungus Sordaria macrospora (26). The loss of ostioles dramatically compromises the ability of these fungi to disseminate progeny. In addition, Δgpr-1 mutants form beaks that are defective in phototropism (unpublished data). We speculate that the absence of ostioles stems from defects in development of periphyses and wall cells of the perithecial neck (63). Moreover, we cannot exclude the possibility that the volcano-shaped beaks and ruptured perithecia observed in Δgpr-1 mutants could result from an excess of turgor pressure inside perithecia of this strain. Therefore, future studies will require a multipronged approach to identify the molecular mechanism(s) underlying GPR-1 action. This includes identification and characterization of the cell types affected by loss of gpr-1 during protoperithecial and perithecial development by using microscopic approaches, determination of the genes regulated by GPR-1 and BEK-1 during sexual development, and analysis of possible changes in concentration of osmolytic compounds in developing perithecia.
Acknowledgments
We thank Nick Read and members of the Borkovich laboratory for many helpful discussions. We thank Michael Freitag and Eric Selker for plasmids pMF272 and pMF280. We acknowledge Carol Jones, Liande Li, Sara Martinez, and Gyungsoon Park for comments on the manuscript.
This work was supported by grant GM48626 from the National Institutes of Health to K.A.B.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Apone, F., N. Alyeshmerni, K. Wiens, D. Chalmers, M. J. Chrispeels, and G. Colucci. 2003. The G-protein-coupled receptor GCR1 regulates DNA synthesis through activation of phosphatidylinositol-specific phospholipase C. Plant Physiol. 133:571-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aramayo, R. 1996. Gene replacement at the his-3 locus of Neurospora crassa. Fungal Genet. Newsl. 43:9-13. [Google Scholar]
- 4.Baasiri, R. A., X. Lu, P. S. Rowley, G. E. Turner, and K. A. Borkovich. 1997. Overlapping functions for two G protein alpha subunits in Neurospora crassa. Genetics 147:137-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bell-Pedersen, D., J. C. Dunlap, and J. J. Loros. 1996. Distinct cis-acting elements mediate clock, light, and developmental regulation of the Neurospora crassa eas (ccg-2) gene. Mol. Cell. Biol. 16:513-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bieszke, J. A., E. L. Braun, L. E. Bean, S. Kang, D. O. Natvig, and K. A. Borkovich. 1999. The nop-1 gene of Neurospora crassa encodes a seven transmembrane helix retinal-binding protein homologous to archaeal rhodopsins. Proc. Natl. Acad. Sci. USA 96:8034-8039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bistis, G. N. 1981. Chemotropic interactions between trichogynes and conidia of opposite mating-type in Neurospora crassa. Mycologia 73:959-975. [Google Scholar]
- 8.Bistis, G. N. 1983. Evidence for diffusible, mating-type-specific trichogyne attractants in Neurospora crassa. Exp. Mycol. 7:292-295. [Google Scholar]
- 9.Bistis, G. N., D. D. Perkins, and N. D. Read. 2003. Different cell types in Neurospora crassa. Fungal Genet. Newsl. 50:17-19. [Google Scholar]
- 10.Blumer, K. J., J. E. Reneke, and J. Thorner. 1988. The STE2 gene product is the ligand-binding component of the alpha-factor receptor of Saccharomyces cerevisiae. J. Biol. Chem. 263:10836-10842. [PubMed] [Google Scholar]
- 11.Bolker, M., M. Urban, and R. Kahmann. 1992. The a mating type locus of U. maydis specifies cell signaling components. Cell 68:441-450. [DOI] [PubMed] [Google Scholar]
- 12.Borkovich, K. A., L. A. Alex, O. Yarden, M. Freitag, G. E. Turner, N. D. Read, S. Seiler, D. Bell-Pedersen, J. Paietta, N. Plesofsky, M. Plamann, M. Goodrich-Tanrikulu, U. Schulte, G. Mannhaupt, F. E. Nargang, A. Radford, C. Selitrennikoff, J. E. Galagan, J. C. Dunlap, J. J. Loros, D. Catcheside, H. Inoue, R. Aramayo, M. Polymenis, E. U. Selker, M. S. Sachs, G. A. Marzluf, I. Paulsen, R. Davis, D. J. Ebbole, A. Zelter, E. R. Kalkman, R. O'Rourke, F. Bowring, J. Yeadon, C. Ishii, K. Suzuki, W. Sakai, and R. Pratt. 2004. Lessons from the genome sequence of Neurospora crassa: tracing the path from genomic blueprint to multicellular organism. Microbiol. Mol. Biol. Rev. 68:1-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brady, A. E., and L. E. Limbird. 2002. G protein-coupled receptor interacting proteins: emerging roles in localization and signal transduction. Cell Signal. 14:297-309. [DOI] [PubMed] [Google Scholar]
- 14.Bryson-Richardson, R. J., D. W. Logan, P. D. Currie, and I. J. Jackson. 2004. Large-scale analysis of gene structure in rhodopsin-like GPCRs: evidence for widespread loss of an ancient intron. Gene 338:15-23. [DOI] [PubMed] [Google Scholar]
- 15.Brzostowski, J. A., and A. R. Kimmel. 2001. Signaling at zero G: G-protein-independent functions for 7-TM receptors. Trends Biochem. Sci. 26:291-297. [DOI] [PubMed] [Google Scholar]
- 16.Burglin, T. R. 1997. Analysis of TALE superclass homeobox genes (MEIS, PBC, KNOX, Iroquois, TGIF) reveals a novel domain conserved between plants and animals. Nucleic Acids Res. 25:4173-4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Case, M., M. Schweizer, S. Kushner, and N. Giles. 1979. Efficient transformation of Neurospora crassa by utilizing hybrid plasmid DNA. Proc. Natl. Acad. Sci. USA 76:5259-5263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang, Y. C., G. F. Miller, and K. J. Kwon-Chung. 2003. Importance of a developmentally regulated pheromone receptor of Cryptococcus neoformans for virulence. Infect. Immun. 71:4953-4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen, J. G., S. Pandey, J. Huang, J. M. Alonso, J. R. Ecker, S. M. Assmann, and A. M. Jones. 2004. GCR1 can act independently of heterotrimeric G-protein in response to brassinosteroids and gibberellins in Arabidopsis seed germination. Plant Physiol. 135:907-915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chenna, R., H. Sugawara, T. Koike, R. Lopez, T. J. Gibson, D. G. Higgins, and J. D. Thompson. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31:3497-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chung, S., M. Karos, Y. C. Chang, J. Lukszo, B. L. Wickes, and K. J. Kwon-Chung. 2002. Molecular analysis of CPRα, a MATα-specific pheromone receptor gene of Cryptococcus neoformans. Eukaryot. Cell 1:432-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Colot, H., G. Park, G. Turner, C. Ringelberg, C. Crew, L. Litvinkova, R. Weiss, K. Borkovich, and J. Dunlap. 2006. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc. Natl. Acad. Sci. USA 103:10352-10357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Colucci, G., F. Apone, N. Alyeshmerni, D. Chalmers, and M. J. Chrispeels. 2002. GCR1, the putative Arabidopsis G-protein-coupled receptor gene is cell cycle-regulated, and its overexpression abolishes seed dormancy and shortens time to flowering. Proc. Natl. Acad. Sci. USA 99:4736-4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davis, R. H., and F. J. deSerres. 1970. Genetic and microbiological research techniques for Neurospora crassa. Methods Enzymol. 71A:79-143. [Google Scholar]
- 25.Ebbole, D. J., and M. S. Sachs. 1990. A rapid and simple method for isolation of Neurospora crassa homokaryons using microconidia. Fungal Genet. Newsl. 37:17-18. [Google Scholar]
- 26.Esser, K., and J. Straub. 1958. Genetic studies on Sordaria macrospora Auersw., compensation and induction in gene-dependent developmental defects. Z. Vererbungsl. 89:729-746. (In German.) [PubMed] [Google Scholar]
- 27.Folco, H. D., M. Freitag, A. Ramon, E. D. Temporini, M. E. Alvarez, I. Garcia, C. Scazzocchio, E. U. Selker, and A. L. Rosa. 2003. Histone H1 Is required for proper regulation of pyruvate decarboxylase gene expression in Neurospora crassa. Eukaryot. Cell 2:341-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Galagan, J. E., S. E. Calvo, K. A. Borkovich, E. U. Selker, N. D. Read, D. Jaffe, W. FitzHugh, L. J. Ma, S. Smirnov, S. Purcell, B. Rehman, T. Elkins, R. Engels, S. Wang, C. B. Nielsen, J. Butler, M. Endrizzi, D. Qui, P. Ianakiev, D. Bell-Pedersen, M. A. Nelson, M. Werner-Washburne, C. P. Selitrennikoff,J. A. Kinsey, E. L. Braun, A. Zelter, U. Schulte, G. O. Kothe, G. Jedd, W. Mewes, C. Staben, E. Marcotte, D. Greenberg, A. Roy, K. Foley, J. Naylor, N. Stange-Thomann, R. Barrett, S. Gnerre, M. Kamal, M. Kamvysselis, E. Mauceli, C. Bielke, S. Rudd, D. Frishman, S. Krystofova, C. Rasmussen, R. L. Metzenberg, D. D. Perkins, S. Kroken, C. Cogoni, G. Macino, D. Catcheside, W. Li, R. J. Pratt, S. A. Osmani, C. P. DeSouza, L. Glass, M. J. Orbach, J. A. Berglund, R. Voelker, O. Yarden, M. Plamann, S. Seiler, J. Dunlap, A. Radford, R. Aramayo, D. O. Natvig, L. A. Alex, G. Mannhaupt, D. J. Ebbole, M. Freitag, I. Paulsen, M. S. Sachs, E. S. Lander, C. Nusbaum, and B. Birren. 2003. The genome sequence of the filamentous fungus Neurospora crassa. Nature 422:859-868. [DOI] [PubMed] [Google Scholar]
- 29.Geerts, D., N. Schilderink, G. Jorritsma, and R. Versteed. 2003. The role of the MEIS homeobox genes in neuroblastoma. Cancer Lett. 197:87-92. [DOI] [PubMed] [Google Scholar]
- 30.Gehring, W. J., M. Affolter, and T. Burglin. 1994. Homeodomain proteins. Annu. Rev. Biochem. 63:487-526. [DOI] [PubMed] [Google Scholar]
- 31.Hagen, D. C., G. McCaffrey, and G. F. Sprague, Jr. 1986. Evidence the yeast STE3 gene encodes a receptor for the peptide pheromone a-factor: gene sequence and implications for the structure of the presumed receptor. Proc. Natl. Acad. Sci. USA 83:1418-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Han, K. H., J. A. Seo, and J. H. Yu. 2004. A putative G-protein-coupled receptor negatively controls sexual development in Aspergillus nidulans. Mol. Microbiol. 51:1333-1345. [DOI] [PubMed] [Google Scholar]
- 33.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 34.Harding, R. W., and S. Melles. 1983. Genetic analysis of phototropism of Neurospora crassa perithecial beaks using white collar and albino mutants. Plant Physiol. 72:996-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harris, J. L., H. B. Howe, Jr., and I. L. Roth. 1975. Scanning electron microscopy of surface and internal features of developing perithecia of Neurospora crassa. J. Bacteriol. 122:1239-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Horak, C. E., N. M. Luscombe, J. Qian, P. Bertone, S. Piccirrillo, M. Gerstein, and M. Snyder. 2002. Complex transcriptional circuitry at the G1/S transition in Saccharomyces cerevisiae. Genes Dev. 16:3017-3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hsieh, M. M., G. Lupas, J. Rychlik, S. Dziennis, B. A. Habecker, and E. J. Lewis. 2005. ERK1/2 is a negative regulator of homeodomain protein Arix/Phox2a. J. Neurochem. 94:1719-1727. [DOI] [PubMed] [Google Scholar]
- 38.Huang, H., M. Rastegar, C. Bodner, S. L. Goh, I. Rambaldi, and M. Featherstone. 2005. MEIS C termini harbor transcriptional activation domains that respond to cell signaling. J. Biol. Chem. 280:10119-10127. [DOI] [PubMed] [Google Scholar]
- 39.Ivey, F. D., P. N. Hodge, G. E. Turner, and K. A. Borkovich. 1996. The G alpha i homologue gna-1 controls multiple differentiation pathways in Neurospora crassa. Mol. Biol. Cell 7:1283-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ivey, F. D., Q. Yang, and K. A. Borkovich. 1999. Positive regulation of adenylyl cyclase activity by a Gαi homolog in Neurospora crassa. Fungal Genet. Biol. 26:48-61. [DOI] [PubMed] [Google Scholar]
- 41.Josefsson, L. G., and L. Rask. 1997. Cloning of a putative G-protein-coupled receptor from Arabidopsis thaliana. Eur. J. Biochem. 249:415-420. [DOI] [PubMed] [Google Scholar]
- 42.Kays, A. M., and K. A. Borkovich. 2004. Severe impairment of growth and differentiation in a Neurospora crassa mutant lacking all heterotrimeric G alpha proteins. Genetics 166:1229-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kays, A. M., P. S. Rowley, R. A. Baasiri, and K. A. Borkovich. 2000. Regulation of conidiation and adenylyl cyclase levels by the Gα protein GNA-3 in Neurospora crassa. Mol. Cell. Biol. 20:7693-7705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim, H., and K. A. Borkovich. 2004. A pheromone receptor gene, pre-1, is essential for mating type-specific directional growth and fusion of trichogynes and female fertility in Neurospora crassa. Mol. Microbiol. 52:1781-1798. [DOI] [PubMed] [Google Scholar]
- 45.Kim, H., and K. A. Borkovich. 2006. Pheromones are essential for male fertility and sufficient to direct chemotropic polarized growth of trichogynes during mating in Neurospora crassa. Eukaryot. Cell 5:544-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Klein, P. S., T. J. Sun, C. L. Saxe III, A. R. Kimmel, R. L. Johnson, and P. N. Devreotes. 1988. A chemoattractant receptor controls development in Dictyostelium discoideum. Science 241:1467-1472. [DOI] [PubMed] [Google Scholar]
- 47.Krystofova, S., and K. A. Borkovich. 2005. The heterotrimeric G-protein subunits GNG-1 and GNB-1 form a Gβγ dimer required for normal female fertility, asexual development, and Gα protein levels in Neurospora crassa. Eukaryot. Cell 4:365-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lengeler, K. B., R. C. Davidson, C. D'Souza, T. Harashima, W. C. Shen, P. Wang, X. Pan, M. Waugh, and J. Heitman. 2000. Signal transduction cascades regulating fungal development and virulence. Microbiol. Mol. Biol. Rev. 64:746-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48a.Li, L., and K. A. Borkovich. 2006. GPR-4 is a predicted G-protein-coupled receptor required for carbon source-dependent asexual growth and development in Neurospora crassa. 5:1287-1300. [DOI] [PMC free article] [PubMed]
- 49.Maidan, M. M., L. De Rop, J. Serneels, S. Exler, S. Rupp, H. Tournu, J. M. Thevelein, and P. Van Dijck. 2005. The G-protein-coupled receptor Gpr1 and the Gα protein Gpa2 act through the cAMP-protein kinase A pathway to induce morphogenesis in Candida albicans. Mol. Biol. Cell 16:1971-1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maidan, M. M., J. M. Thevelein, and P. Van Dijck. 2005. Carbon source induced yeast-to-hypha transition in Candida albicans is dependent on the presence of amino acids and on the G-protein-coupled receptor Gpr1. Biochem. Soc. Trans. 33:291-293. [DOI] [PubMed] [Google Scholar]
- 51.Mann, R. S., and M. Affolter. 1998. Hox proteins meet more partners. Curr. Opin. Genet. Dev. 8:423-429. [DOI] [PubMed] [Google Scholar]
- 52.Miwa, T., Y. Takagi, M. Shinozaki, C. W. Yun, W. A. Schell, J. R. Perfect, H. Kumagai, and H. Tamaki. 2004. Gpr1, a putative G-protein-coupled receptor, regulates morphogenesis and hypha formation in the pathogenic fungus Candida albicans. Eukaryot. Cell 3:919-931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Neves, S. R., P. T. Ram, and R. Iyengar. 2002. G protein pathways. Science 296:1636-1639. [DOI] [PubMed] [Google Scholar]
- 54.Olesnicky, N. S., A. J. Brown, S. J. Dowell, and L. A. Casselton. 1999. A constitutively active G-protein-coupled receptor causes mating self-compatibility in the mushroom Coprinus. EMBO J. 18:2756-2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Orbach, M. J. 1994. A cosmid with a HyR marker for fungal library construction and screening. Gene 150:159-162. [DOI] [PubMed] [Google Scholar]
- 56.O'Shea, S. F., P. T. Chaure, J. R. Halsall, N. S. Olesnicky, A. Leibbrandt, I. F. Connerton, and L. A. Casselton. 1998. A large pheromone and receptor gene complex determines multiple B mating type specificities in Coprinus cinereus. Genetics 148:1081-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pall, M. L. 1993. The use of ignite (basta; glufosinate; phosphinothricin) to select transformants of bar-containing plasmids in Neurospora crassa. Fungal Genet. Newsl. 40:58. [Google Scholar]
- 58.Pall, M. L., and J. P. Brunelli. 1993. A series of six compact fungal transformation vectors containing polylinkers with multiple unique restriction sites. Fungal Genet. Newsl 40:59-62. [Google Scholar]
- 59.Pandey, S., and S. M. Assmann. 2004. The Arabidopsis putative G-protein-coupled receptor GCR1 interacts with the G protein alpha subunit GPA1 and regulates abscisic acid signaling. Plant Cell 16:1616-1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Plakidou-Dymock, S., D. Dymock, and R. Hooley. 1998. A higher plant seven-transmembrane receptor that influences sensitivity to cytokinins. Curr. Biol. 8:315-324. [DOI] [PubMed] [Google Scholar]
- 61.Raisley, B., M. Zhang, D. Hereld, and J. A. Hadwiger. 2004. A cAMP receptor-like G protein-coupled receptor with roles in growth regulation and development. Dev. Biol. 265:433-445. [DOI] [PubMed] [Google Scholar]
- 62.Raju, N. B. 1992. Genetic control of the sexual cycle in Neurospora. Mycol. Res. 96:241-262. [Google Scholar]
- 63.Read, N., and A. Beckett. 1985. The anatomy of the mature perithecium in Sordaria humana and its significance for fungal multicellular development. Can. J. Bot. 63:281-296. [Google Scholar]
- 64.Saleh, M., I. Rambaldi, X. J. Yang, and M. S. Featherstone. 2000. Cell signaling switches HOX-PBX complexes from repressors to activators of transcription mediated by histone deacetylases and histone acetyltransferases. Mol. Cell. Biol. 20:8623-8633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Seo, J. A., K. H. Han, and J. H. Yu. 2004. The gprA and gprB genes encode putative G protein-coupled receptors required for self-fertilization in Aspergillus nidulans. Mol. Microbiol. 53:1611-1623. [DOI] [PubMed] [Google Scholar]
- 66.Tanaka, K., J. Davey, Y. Imai, and M. Yamamoto. 1993. Schizosaccharomyces pombe map3+ encodes the putative M-factor receptor. Mol. Cell. Biol. 13:80-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Trail, F., I. Gaffoor, and S. Vogel. 2005. Ejection mechanics and trajectory of the ascospores of Gibberella zeae (anamorph Fusarium graminearum). Fungal Genet. Biol. 42:528-533. [DOI] [PubMed] [Google Scholar]
- 68.Turner, G. E., and K. A. Borkovich. 1993. Identification of a G protein alpha subunit from Neurospora crassa that is a member of the Gi family. J. Biol. Chem. 268:14805-14811. [PubMed] [Google Scholar]
- 69.Vann, D. 1995. Electroporation-based transformation of freshly harvested conidia of Neurospora crassa. Fungal Genet. Newsl. 42A:53. [Google Scholar]
- 70.Vogel, H. 1964. Distribution of lysine pathways among fungi: evolutionary implications. Am. Nat. 98:435-446. [Google Scholar]
- 71.Welton, R. M., and C. S. Hoffman. 2000. Glucose monitoring in fission yeast via the gpa2 Gα, the git5 Gβ and the git3 putative glucose receptor. Genetics 156:513-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wendland, J., L. J. Vaillancourt, J. Hegner, K. B. Lengeler, K. J. Laddison, C. A. Specht, C. A. Raper, and E. Kothe. 1995. The mating-type locus B alpha 1 of Schizophyllum commune contains a pheromone receptor gene and putative pheromone genes. EMBO J. 14:5271-5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Westergaard, M., and H. Mitchell. 1947. Neurospora V. A synthetic medium favoring sexual reproduction. Am. J. Bot. 34:573-577. [Google Scholar]
- 74.Xue, C., Y. S. Bahn, G. M. Cox, and J. Heitman. 2006. G protein-coupled receptor Gpr4 senses amino acids and activates the cAMP-PKA pathway in Cryptococcus neoformans. Mol. Biol. Cell 17:667-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xue, Y., M. Battle, and J. P. Hirsch. 1998. GPR1 encodes a putative G protein-coupled receptor that associates with the Gpa2p Galpha subunit and functions in a Ras-independent pathway. EMBO J. 17:1996-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang, Q., and K. A. Borkovich. 1999. Mutational activation of a Gαi causes uncontrolled proliferation of aerial hyphae and increased sensitivity to heat and oxidative stress in Neurospora crassa. Genetics 151:107-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yang, Q., S. I. Poole, and K. A. Borkovich. 2002. A G-protein β subunit required for sexual and vegetative development and maintenance of normal Gα protein levels in Neurospora crassa. Eukaryot. Cell 1:378-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yun, C. W., H. Tamaki, R. Nakayama, K. Yamamoto, and H. Kumagai. 1997. G-protein coupled receptor from yeast Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 240:287-292. [DOI] [PubMed] [Google Scholar]
- 79.Yun, C. W., H. Tamaki, R. Nakayama, K. Yamamoto, and H. Kumagai. 1998. Gpr1p, a putative G-protein-coupled receptor, regulates glucose-dependent cellular cAMP level in yeast Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 252:29-33. [DOI] [PubMed] [Google Scholar]