Abstract
In the present study, we sought to elucidate the contribution of the Cryptococcus neoformans catalase gene family to antioxidant defense. We employed bioinformatics techniques to identify four members of the C. neoformans catalase gene family and created mutants lacking single or multiple catalase genes. Based on a phylogenetic analysis, CAT1 and CAT3 encode putative spore-specific catalases, CAT2 encodes a putative peroxisomal catalase, and CAT4 encodes a putative cytosolic catalase. Only Cat1 exhibited detectable biochemical activity in vitro, and Cat1 activity was constitutive in the yeast form of this organism. Although they were predicted to be important in spores, neither CAT1 nor CAT3 was essential for mating or spore viability. Consistent with previous studies of Saccharomyces cerevisiae, the single (cat1, cat2, cat3, and cat4) and quadruple (cat1 cat2 cat3 cat4) catalase mutant strains exhibited no oxidative-stress phenotypes under conditions in which either exogenous or endogenous levels of reactive oxygen species were elevated. In addition, there were no significant differences in the mean times to mortality between groups of mice infected with C. neoformans catalase mutant strains (the cat1 and cat1 cat2 cat3 cat4 mutants) and those infected with wild-type strain H99. We conclude from the results of this study that C. neoformans possesses a robust antioxidant system, composed of functionally overlapping and compensatory components that provide protection against endogenous and exogenous oxidative stresses.
Fungi, like many other organisms, rely on antioxidant defense mechanisms for protection against oxidative damage. These antioxidant defense mechanisms have evolved as a result of several factors, including adaptation to growth in aerobic environments, utilization of oxidative phosphorylation for energy production, and protection against exogenous oxidants encountered in the environment. A prerequisite for the success of human pathogenic fungi is their ability to defend against reactive oxygen species (ROS) elicited by host effector cells during the course of an infection. Catalase contributes to the pathogenesis of several human and plant pathogens, including Campylobacter jejuni, Mycobacterium tuberculosis, and Agrobacterium tumefaciens (9,32, 57), and there has been much interest in ascertaining whether catalase provides a similar protective function to pathogenic fungi.
Cryptococcus neoformans is an opportunistic fungal pathogen and a well-established model organism utilized for the study of mechanisms that contribute to fungal pathogenesis (38). The initial host defense against infection by C. neoformans is mediated by alveolar macrophages, which contribute to the mobilization of a cellular immune response (16). Alveolar macrophages also appear to provide a unique niche for C. neoformans cells, which can survive within these immune cells (14, 15, 29, 30, 42, 50). This observation implies that C. neoformans can survive within the harsh environment of the phagolysosome, suggesting the presence of an antioxidant defense system that is capable of providing protection against host-derived ROS. Consistent with this hypothesis, several studies have demonstrated a correlation between virulence and the ability of C. neoformans strains to resist oxidative stress in vitro (2, 6, 56). For example, ROS elicited by human polymorphonuclear neutrophils have been shown to kill C. neoformans (10). Furthermore, polymorphonuclear neutrophils and mononuclear cells from patients with chronic granulomatous disease, in which NADPH oxidase activity is defective, exhibited minimal fungicidal activity against C. neoformans (34). Cumulatively, the results of these studies suggest that the survival of C. neoformans in the host environment is dependent in part on its ability to defend against damage by host-derived ROS.
Several recent studies have greatly expanded our understanding of the contribution of the enzymatic constituents of the C. neoformans antioxidant defense system to protect against oxidative damage. We have demonstrated in vitro that the C. neoformans cytosolic copper-zinc superoxide dismutase (Sod1) (6), mitochondrial manganese superoxide dismutase (Sod2) (19), cytochrome c peroxidase (Ccp1) (20), and alternative oxidase (Aox1) (2) contribute to resistance against oxidative stress. Aox1 and Sod1 also contribute to the pathogenesis of C. neoformans, and aox1 and sod1 null strains have exhibited diminished virulence in a murine cryptococcosis inhalation model (2, 6). Interestingly, we found that Sod2 is essential for high-temperature growth (37°C), demonstrating an important link between the regulation of endogenously produced ROS and adaptation to host environmental conditions (19). Narasipura et al. demonstrated that Sod1 and Sod2 exhibited similar antioxidant functions in C. neoformans var. gattii (40, 41), which can cause disease in immunocompetent individuals (17). Missall et al. demonstrated that TSA1, one of three C. neoformans thiol peroxidase genes, and the glutathione peroxidase genes GPX1 and GPX2 contribute to protection against oxidative stress in vitro (36, 37). In addition, TSA1 has contributed to virulence in a murine model of cryptococcosis (36, 37). The results of these studies suggest that the C. neoformans antioxidant system is composed of several functionally overlapping and compensatory components that provide protection against endogenous and exogenous oxidative stresses.
In the present study, we sought to elucidate the contribution of catalase to the C. neoformans antioxidant defense system. We employed bioinformatics techniques to identify four members of the C. neoformans catalase gene family, the largest antioxidant gene family thus far identified for C. neoformans. We then utilized a molecular genetics approach to construct a series of mutants lacking single or multiple catalase genes. We hypothesized that the catalases might contribute to resistance against oxidative stress via one of two models: the activities of individual catalases might contribute to resistance against oxidative stress independently, or the catalase gene family members might function cooperatively. In addition, the virulence potential of the strains lacking catalase was assessed in a murine model of cryptococcosis.
MATERIALS AND METHODS
Strains and media.
Cryptococcus neoformans strains H99 (serotype A, mating type α) and H99R were recovered from 15% glycerol stocks stored at −80°C prior to use in this study. H99R is a spontaneous ura5 auxotroph isolated by plating strain H99 on 5-fluoroorotic agar as described previously (20, 28). Transformants were selected on synthetic complete medium without uracil and maintained on yeast extract-peptone-dextrose (YPD; 1% yeast extract, 2% peptone, and 2% dextrose) agar. Dopamine agar and Christensen's broth were made as described previously (28). Prior to their use in the mouse studies, the yeast strains were grown for 18 to 20 h at 30°C with shaking in YPD broth and then harvested, washed three times with sterile phosphate-buffered saline, and counted with a hemacytometer to determine the cell number. The inoculum sizes for mouse experiments were confirmed by plating dilutions of cells on YPD agar plates. The growth rate for each strain was quantified by determining the numbers of CFU at specified time points.
Identification of C. neoformans catalase genes.
The amino acid sequences of the two Saccharomyces cerevisiae catalases (Cta1, NCBI protein database [GenBank] accession no. NP_010542; and Ctt1, GenBank accession no. NP_011602) were used to query the Cryptococcus neoformans H99 genome sequencing project database (Duke Center for Genome Technology; http://cgt.genetics.duke.edu/). TBLASTN analysis revealed the presence of four unique C. neoformans catalase genes in the genome, which we named CAT1, CAT2, CAT3, and CAT4. The intron-exon boundaries of each of the catalase genes were established by comparing cDNA sequences (Oklahoma University Cryptococcus neoformans cDNA Sequencing Project [http://www.genome.ou.edu/cneo.html] and The Institute for Genomic Research [http://www.tigr.org/tdb/e2k1/cna1/]) and genomic sequences and by identifying the 5′- and 3′-splice sites of GTNNGY and YAG, respectively, in the genomic sequence.
Phylogenetic and sequence analyses.
Catalase homologs in fungi, animals, plants, bacteria, and Archaea were identified by searching the NCBI (http://www.ncbi.nih.gov/) nonredundant protein database with BLASTP (3). Additionally, FASTP (45) searches of the authors' annotations of Podospora anserina, Coprinus cinereus, Phanerochaete chrysosporium (33), Ustilago maydis, Aspergillus fumigatus, and Ajellomyces capsulatus genomes were performed. Gene annotations for Fusarium graminearum, Magneporthe grisea, Aspergillus nidulans, and Neurospora crassa (18) were obtained from the Fungal Genome Initiative at the Broad Institute (http://www.broad.mit.edu/annotation/fungi/fgi). Sequence data for A. fumigatus were obtained from The Institute for Genomic Research website (http://www.tigr.org). Genome sequence data for C. cinereus and U. maydis were obtained from the Fungal Genome Initiative at the Broad Institute (http://www.broad.mit.edu/annotation/fungi/fgi/). Genome sequence data for P. anserina were obtained from the Podospora anserina Genome Project (http://podospora.igmors.u-psud.fr/).
The protein sequences corresponding to these homologous genes were aligned using MUSCLE (12). The resulting multiple sequence alignment was used as input into the ProtML program part of the MOLPHY software package (http://bioweb.pasteur.fr/seqanal/interfaces/MolPhy.html) (1). Perl scripts using the Bioperl package (49) were used to convert the data into suitable formats for the phylogenetic analysis programs and to generate summary reports about the data.
The catalase homologs were also analyzed with the HMMER package (11) and the Pfam database (4) to identify protein domains encoded by the sequences. Pair-wise alignments of the catalases to produce both global and local alignments were performed with the “needle” and “water” applications, respectively, which are parts of the EMBOSS toolkit (46).
Construction of catalase mutant strains.
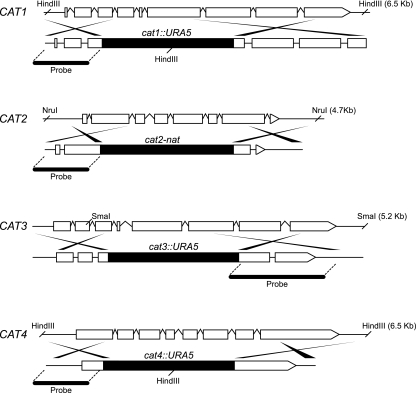
Deletion constructs for each of the C. neoformans catalases were created (Fig. 2) by overlap PCR as described previously (8). Each linear overlap PCR construct contained a genomic sequence that flanked the 5′ and 3′ regions of the respective catalase open reading frame and URA5 from the serotype D strain B3501 (53) or the nourseothricin (nat) selectable marker (22). The primer sequences used to generate each of the overlap constructs are listed in Table 1. The overlap PCR constructs were used to transform either ura5 auxotrophic strain H99R or wild-type strain H99 using biolistic DNA delivery, as described previously (51). Transformants were selected on synthetic medium lacking uracil or YPD medium containing nourseothricin (100 μg/ml), and mutants were identified by colony PCR.
FIG. 2.
Constructionof catalase null mutant strains. Overlap PCR was performed to create the cat1::URA5, cat2::NAT, cat3::URA5, and cat4::URA5 deletion constructs. Allele-specific integration of constructs at the native catalase loci resulted in the deletion of 39%, 50%, 54%, and 74% of the Cat1, Cat2, Cat3, and Cat4 catalase domains, respectively.
TABLE 1.
Primers used in this study
| Primer name | Use(s)a | Sequence (5′-3′) |
|---|---|---|
| sgCAT1 OL-1 | A, C | TGAGGGGACAATGTTAAAGAGGAT |
| sgCAT1 OL-2 | A, C | GGTCGAGCAACTTCGCTCTGCCGGTCAGAGGAGTATGC |
| sgCAT1 OL-3 | A | CCACCTCCTGGAGGCAAGCAATCGAAGTCGGGGCATAC |
| sgCAT1 OL-4 | A | GACCACCGGCGAAGACAATA |
| sgCAT1 OL-5 | A | GCATACTCCTCTGACCGGCAGAGCGAAGTTGCTCGACC |
| sgCAT1 OL-6 | A | GTATGCCCCGACTTCGATTGCTTGCCTCCAGGAGGTGG |
| sgCAT1 RT-1 | B | TCAATGGCAAGCACCAAG |
| sgCAT1 RT-2 | B | AAGGAATCATCGGGACCA |
| sgCAT2 OL-1 | A, C | GACCCACAAATCTACCCGTTC |
| sgCAT2 OL-2 | A, C | GCTCACCTCCCGCAGCCTTTCAGCACCCATACAACCA |
| sgCAT2 OL-3 | A | CTCGTTTCTACATCTCTTCCGCCCTACCAGCGTGA |
| sgCAT2 OL-4 | A | CCCTTACGACCAGGCAAGTAT |
| sgCAT2 OL-5 | A | TGGTTGTATGGGTGCTGAAAGGCTGCGGGAGGTGAGC |
| sgCAT2 OL-6 | A | TCACGCTGGTAGGGCGGAAGAGATGTAGAAACGAG |
| sgCAT2 RT-1 | B | CGCCTCGGTGTCAACTAC |
| sgCAT2 RT-2 | B | CGACCCATTCATCGTGTTT |
| sgCAT3 OL-1 | A | CACAAGCCCCTCGACGGACATCAA |
| sgCAT3 OL-2 | A | GGTCGAGCAACTTCGCTCCAAACCCACGGACATCACGCACTG |
| sgCAT3 OL-3 | A, C | CCACCTCCTGGAGGCAAGGACTTTACCGATGATCCTCTCCTA |
| sgCAT3 OL-4 | A, C | ACGCCATTCTCTTGCACCACACTA |
| sgCAT3 OL-5 | A | CAGTGCGTGATGTCCGTGGGTTTGGAGCGAAGTTGCTCGACC |
| sgCAT3 OL-6 | A | TAGGAGAGGATCATCGGTAAAGTCCTTGCCTCCAGGAGGTGG |
| sgCAT3 RT-1 | B | TCATTCCTCACAAGCCACTC |
| sgCAT3 RT-2 | B | CTTCAGGCAGGCTCAGATG |
| sgCAT4 OL-1 | A, C | CCGCCTACTGATGTTGGGTTTTATT |
| sgCAT4 OL-2 | A, C | GGTCGAGCAACTTCGCTCTGTACTCCAAGCCCGACTGTTCTCT |
| sgCAT4 OL-3 | A | CCACCTCCTGGAGGCAAGTCGTCCGCCAGAGTGTTGCTATTAT |
| sgCAT4 OL-4 | A | GGGGACATGCCTTCGGGTGGTTGC |
| sgCAT4 OL-5 | A | AGAGAACAGTCGGGCTTGGAGTACAGAGCGAAGTTGCTCGACC |
| sgCAT4 OL-6 | A | ATAATAGCAACACTCTGGCGGACGACTTGCCTCCAGGAGGTGG |
| sgCAT4 RT-1 | B | GAAGACCCGCTTATCTCTCC |
| sgCAT4 RT-2 | B | CTCGTCGCTAAACACTTTCTG |
Primers were used for creating deletion constructs by overlap PCR (A), catalase transcript detection (B), and probes for Southern blot analysis (C).
Deletion of the native catalase alleles was confirmed by Southern blot analysis. Genomic DNA was isolated from the cat1, cat2, cat3, and cat4 mutant strains and wild-type strain H99 as described previously (47). Restriction digestion, gel electrophoresis, DNA transfer, prehybridization, hybridization, and autoradiography were performed as described previously (47). The primers listed in Table 1 were used to generate probes that hybridized near the 5′ or 3′ region of the appropriate catalase open reading frame. A random-primed DNA labeling kit (Boehringer Mannheim) and [32P]dCTP (Amersham) were used to label the probe.
Total RNA was isolated from C. neoformans strain H99 using TRIzol reagent (Life Technologies). cDNA was generated from total RNA using a SuperScript first-strand synthesis system for reverse transcription-PCR (Invitrogen). PCRs were performed with gene-specific primers to detect CAT1, CAT2, CAT3, and CAT4 transcripts.
Genetic crosses.
To generate MATa catalase mutant strains (Table 2), each MATα catalase mutant strain was crossed with JF99a (MATa ura5), generating strains SG54 (MATa cat1::URA5 ura5), strain SG55 (MATa cat2::NAT ura5), strain SG56 (MATa cat3::URA5 ura5), and strain SG57 (MATa cat4::URA5 ura5). Strain SG58 (MATα cat1::URA5 cat2::NAT ura5) was generated by crossing strain SG50 (MATα cat1::URA5 ura5) with strain SG55 (MATa cat2::NAT ura5). Strain SG59 (MATa cat3::URA5 cat4::URA5 ura5) was generated by crossing strain SG52 (MATα cat3::URA5 ura5) with strain SG57 (MATa cat4::URA5 ura5). To generate a quadruple catalase mutant strain, strain SG58 (MATα cat1::URA5 cat2::NAT ura5) was crossed with strain 59 (MATa cat3::URA5 cat4::URA5 ura5). Over 20 viable spores were analyzed, and no quadruple mutants were generated. However, a MATα cat1::URA5 cat2::NAT cat3::URA5 ura5 mutant strain (SG60) was identified. This mutant strain was used to generate a MATα cat1::URA5 cat2::NAT cat3::URA5 cat4::NEO ura5 quadruple mutant strain (SG61) by deletion of the CAT4 gene by biolistic transformation. For each cross, strains were cocultured on V8 agar medium for 14 to 28 days until basidiospores were produced. The basidiospores were dissected by micromanipulation onto YPD agar medium and allowed to germinate at 25°C. The resulting colonies were replicated onto SD-Ura (synthetic dextrose), YPD-NAT (natamycin at 0. mg/ml), and YPD-NEO (neomycin at 0.2 mg/ml), media tomonitor the segregation of the ura5 and the catalase mutant alleles. The genotype of each strain was confirmed by PCR or Southern blot analysis or both.
TABLE 2.
Strains
| Strain | Genotype | Source/reference |
|---|---|---|
| H99 | MATα | 45a |
| Kn99a | MATa | 43b |
| JF99 | MATaura5 | 43a |
| SG50 | MATαcat1::URA5ura5 | This study |
| SG51 | MATαcat2::NAT | This study |
| SG52 | MATαcat3::URA5ura5 | This study |
| SG53 | MATαcat4::URA5ura5 | This study |
| SG54 | MATacat1::URA5ura5 | This study |
| SG55 | MATacat2::NAT | This study |
| SG56 | MATacat3::URA5ura5 | This study |
| SG57 | MATacat4::URA5ura5 | This study |
| SG58 | MATαcat1::URA5cat2::NAT ura5 | This study |
| SG59 | MATacat3::URA5 cat4::URA5 ura5 | This study |
| SG60 | MATαcat1::URA5 cat2::NAT cat3::URA5 ura5 | This study |
| SG61 | MATαcat1::URA5 cat2::NAT cat3::URA5 ura5 cat3::URA5 cat4::NEO ura5 | This study |
Oxidative-stress phenotype.
Sensitivity to exogenous and endogenous oxidative stress was assessed by a disc diffusion assay and by growth on yeast nitrogen base medium supplemented with various fatty acids as the sole carbon source, respectively. For disc diffusion assays, cat1, cat2, cat3, and cat4 mutant strains and wild-type strain H99 were grown on YPD medium at 30°C overnight with shaking, washed with sterile phosphate-buffered saline, and diluted in fresh YPD medium to an optical density at 600 nm of 0.2 (Bio-Rad Smart Spec 3100). Each strain was then diluted 1:10 in molten YPD agar medium, and media were poured into plates and allowed to solidify. Sterile paper discs (6-mm diameter) saturated with 10 μl of either 8% or 16% H2O2 were added to the center of each plate. The plates were incubated at 30°C or 37°C for 48 h and then photographed, and the diameter of each zone of inhibition was determined. Results were confirmed by determining the number of CFU by using a liquid culture method. Experiments were repeated a minimum of three times.
Catalase activity assay.
cat1, cat2, cat3, and cat4 mutant strains and wild-type strain H99 were grown in YPD medium at 30°C, collected by centrifugation, and lysed by glass bead disruption. A protease inhibitor (P8340; Sigma) was used to prevent degradation of the catalase polypeptides. Cell lysates (20 μg protein/lane) were separated by native gel electrophoresis (10% Tris-HCl Criterion ready-cast gels; Bio-Rad), and proteins with catalase activity were visualized by ferricyanide staining as described previously (54). Briefly, the gels were soaked in 0.01% hydrogen peroxide for 10 min with gentle shaking. They were then stained with a solution of potassium ferricyanide (1.0%, wt/vol) and ferric chloride (1.0%, wt/vol) until bands were visible, usually within 5 or 10 min. The gels were destained in distilled water overnight and photographed.
In vivo testing.
Female A/Jcr mice (NCI/Charles River Laboratories; 20 to 24 g each) were used to compare the virulence of the cat1 and cat1 cat2 cat3 cat4 mutant strains to that of wild-type strain H99. Groups consisting of 10 A/Jcr mice each were infected via intranasal inhalation with 5 × 105 CFU of either catalase mutant strains (cat1 or cat1 cat2 cat3 cat4) or wild-type strain H99 (in a volume of 50 μl). Mice that appeared lethargic or exhibited rapid weight loss were euthanized. Mice were monitored twice daily. The Duke University Animal Use Committee approved the animal protocol used for these experiments. The Mann-Whitney U test was used to evaluate survival data for statistical significance.
Nucleotide sequence accession numbers.
The C. neoformans var. grubii CAT1, CAT2, CAT3, and CAT4 sequences have been submitted to GenBank and assigned accession numbers DQ468109, DQ468110, DQ468111, and DQ468112, respectively.
RESULTS
Identification of catalase homologs.
The newly available genomic DNA sequence for the C. neoformans serotype A strain H99 (Duke University C. neoformans H99 genome database; http://cneo.genetics.duke.edu/) allowed us to utilize bioinformatics techniques to rapidly identify C. neoformans catalase homologs. TBLASTN analyses of the C. neoformans serotype A strain H99 genome sequence database (Duke University C. neoformans H99 genome database; http://cneo.genetics.duke.edu/) were performed using two Saccharomyces cerevisiae catalase protein sequences (Ctt1p and Cta1p) as the queries. Four C. neoformans catalase genes were identified, each with a highly conserved catalase domain, and named CAT1, CAT2, CAT3, and CAT4. One of these genes, CAT3, is located on chromosome 1, while the rest are located on chromosome 4, within 700 kb of each other. We determined the coding sequences for each of the catalases by comparing cDNA (Oklahoma University Cryptococcus neoformans cDNA Sequencing Project; http://www.genome.ou.edu/cneo.html) and genomic DNA (Duke University C. neoformans H99 genome database; http://cneo.genetics.duke.edu/) sequences.
Sequence analysis of the C. neoformans catalases.
After obtaining the predicted protein sequences of the four C. neoformans catalases, we analyzed the sequences for conserved protein domains using the hmmpfam (http://pfam.wustl.edu/) tool. We searched the proteins against the Pfam database of conserved protein domains for significantly similar sequence matches. The analysis revealed the expected conserved catalase domains, and in addition, the Cat1 and Cat3 catalases contained a DJ-1/PfpI family domain, to which is ascribed several putative functions, including transcriptional regulation. Table 3 shows the average percentages of identity and similarity of pair-wise alignments of the four C. neoformans serotype A H99 catalases.
TABLE 3.
Pair-wise calculation of amino acid similarity and identitya
| Catalase | % Amino acid similarity (% identity)
|
|||
|---|---|---|---|---|
| Cat1 | Cat2 | Cat3 | Cat4 | |
| Cat1 | 50.7 (34.0) | 84.8 (75.8) | 47.1 (32.6) | |
| Cat2 | 44.7 (27.8) | 59.8 (42.5) | ||
| Cat3 | 42.1 (27.3) | |||
| Cat4 | ||||
Pair-wise calculation of percentages of amino acid similarity and identity of the four C. neoformans catalase based on a local alignment using the “water” application of EMBOSS.
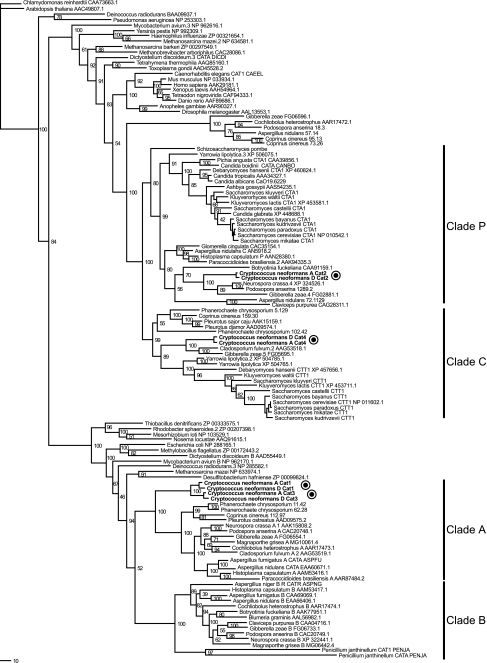
We performed a phylogenetic analysis of multiple fungal catalases, revealing four clades, consistent with previous findings (24, 27): clades P (peroxisomal catalases), C (cytoplasmic catalases), A (spore-specific catalases), and B (secreted catalases) (Fig. 1). The animal, plant, protist, archaeal, and bacterial homologs of these catalases cluster into distinct groups relative to the fungal catalases. This analysis revealed a single catalase form in animals and at least two different bacterial forms. The phylogenetic analysis revealed multiple copies of catalase genes for most fungal species, with several deeply branching clades. However, a few hemiascomycete yeasts such as Ashbya gossypii and Candida albicans have retained only a single catalase gene. Interestingly, translated database searches and searches of the predicted protein sets of the basidiomycete Ustilago maydis revealed that this fungus did not possess any identifiable catalases. Clade A contains only euascomycete and basidiomycete genes, including C. neoformans CAT1 and CAT3, and the proteins encoded by them are distinguished by having a strong similarity to the catalase-related and DJ-1/PfpI (Pfam; PF0165) domains. catA has been shown to be conidium specific to Aspergillus fumigatus and Aspergillus nidulans (44). Clade B is made up exclusively of euascomycete genes, many of which have been shown to encode secreted catalases. Clade C includes the S. cerevisiae cytosolic catalase CTT1 gene and several basidiomycete catalase genes, including C. neoformans CAT4. Cladosporium fulvuman and Gibberella zeae (F. graminearum) are the only euascomycetes found within clade C. The MIPS F. graminearum database (http://mips.gsf.de/genre/proj/fusarium; also see http://www.broad.mit.edu/annotation/fungi/fusarium/) lists the G. zeae FG06595 gene as a probable cytosolic catalase gene. Some proteins in this clade also have a weak similarity (hmmsearch; 10−5 < E < 0.1) to the catalase-related domain (Pfam; PF06628). Clade P is composed of peroxisomal catalase genes and includes the S. cerevisiae CTA1 and C. neoformans CAT2 genes.
FIG. 1.
Phylogenetic analysis of the C. neoformans catalases. The phylogenetic tree of fungal catalases and selected animal, protist, bacterial, and archaeal catalases is rooted with two plant catalases. Homologs were identified with BLASTP searches of C. neoformans Cat1, Cat2, Cat3, and Cat4 proteins against the nonredundant protein database from NCBI. A multiple sequence alignment was performed automatically with MUSCLE, and the tree was constructed via NJDIST and PROTML (available in MOLPHY). Numbers on branches indicate the bootstrap values produced by PROTML running with the -R option and starting with an input neighbor-joining tree calculated from NJDIST. Some bootstrap values were removed at the tips of the tree for clarity in visualizing the tree. There are four distinct clades of fungal catalases: clade P, the peroxisomal catalases; clade C, the cytoplasmic catalases; clade A, spore-specific catalases; and clade B, primarily secreted catalases. C. neoformans possesses catalases in three of the four clades.
Construction of catalase mutant strains.
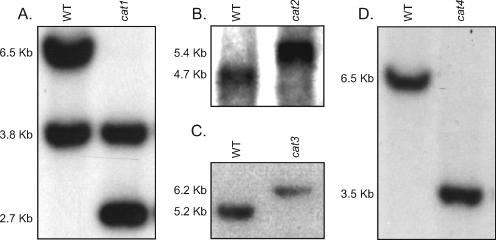
Deletion mutant strains were created to assess the individual contribution of each of the C. neoformans catalases to antioxidant defense. Mutant strains were constructed via the allele-specific homologous integration of deletion constructs at the native locus of each of the four catalase genes. A single allele-specific integration of the cat1::URA5, cat2::NAT, cat3::URA5, and cat4::URA5 deletion constructs at the CAT1, CAT2, CAT3, and CAT4 native loci disrupted each of these genes, deleting approximately 39%, 50%, 54%, and 74% of the Cat1, Cat2, Cat3, and Cat4 catalase domains, respectively (Fig. 2). Southern blot analysis of genomic DNA from each strain confirmed that, in each case, a single allele-specific homologous integration event had occurred (Fig. 3). With one exception (Fig. 3A), only two bands appeared on the individual Southern blots. The 3.8-kb bands present in the lanes corresponding to the wild type and the cat1 mutant are due to cross-hybridization of the probe with the CAT3 gene. One of the two restriction sites required to generate the observed bands was deliberately chosen outside of the deletion construct. Mating reactions were performed to create catalase double, triple, and quadruple mutants, as described in Materials and Methods and shown in Table 2. The genotypes of all of the catalase quadruple mutants were confirmed by PCR.
FIG. 3.
Southern blot analysis confirmed deletion of the native catalase alleles. (A to D) Genomic DNA was isolated from cat1, cat2, cat3, and cat4 mutant strains and wild-type strain H99 and used to perform Southern blot analysis, as described previously. Southern blot analysis confirmed that a single allele-specific integration event occurred at each catalase locus. As shown in Fig. 2, restriction enzymes were chosen so that one of the two restriction sites used to digest genomic DNA was outside of the deletion construct. WT, wild type.
Catalase activity.
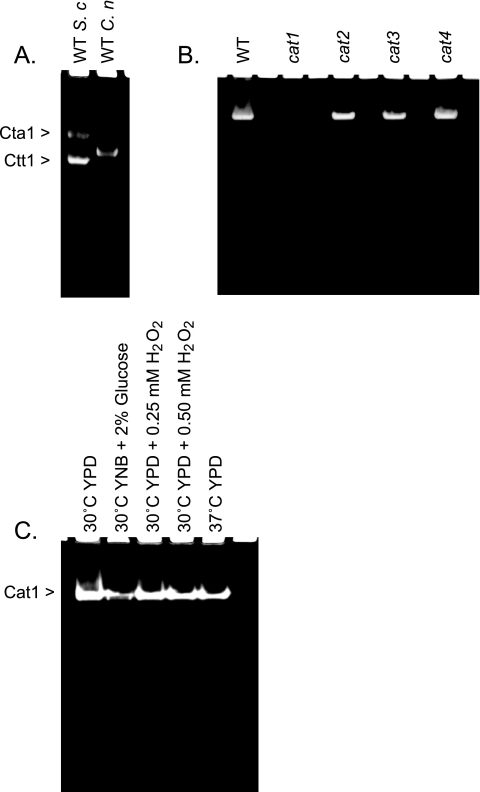
Cell lysates were prepared from C. neoformans cat1, cat2, cat3, and cat4 mutant strains, from wild-type strain H99, and from Saccharomyces cerevisiae to assess the total catalase activity. Lysates (20 μg total protein) were separated by electrophoresis on 7% polyacrylamide gels under nondenaturing and nonreducing conditions. Catalase activity was visualized by ferricyanide-negative staining as described previously by Wayne and Diaz (54). As anticipated, two bands representing Cta1 and Ctt1 were observed in the lane containing lysate from S. cerevisiae (Fig. 4A). Given that C. neoformans possesses four catalases, we anticipated that four bands would be present in the lane containing lysates from wild-type strain H99. However, we observed only one band (Fig. 4A). This single band was present in lanes containing lysate from the cat2, cat3, and cat4 mutants (Fig. 4B); however, no activity was detected in the lane containing cell lysate from the cat1 mutant, suggesting that Cat1 was the only functionally active catalase detected under these conditions. Consistent with the results of our phylogenetic analysis, we were unable to detect secreted catalase activity from the conditioned supernatants of cultures for any of the mutant strains or for wild-type strain H99 (data not shown), demonstrating that C. neoformans does not possess a secreted catalase.
FIG. 4.
Cat1 is the sole catalase with activity in vitro. (A) Cell lysates from S. cerevisiae (S. c) and C. neoformans wild-type (WT) strain H99 (C. n) grown at 30°C in YPD medium were separated on a 10% acrylamide gel under nondenaturing conditions. Catalase activity was visualized by potassium ferricyanide-negative staining. The two bands in lane 1 correspond to the S. cerevisiae catalases Ctt1 and Cta1. (B) Native polyacrylamide gel electrophoresis of protein extracts from C. neoformans cat1, cat2, cat3, and cat4 mutant strains and wild-type strain H99 grown at 30°C in YPD medium. A single activity band was observed for lysates from all strains except the cat1 mutant strain. (C) Native polyacrylamide gel electrophoresis was performed with protein extracts and cell culture supernatants of C. neoformans wild-type strain H99 cells that were either treated with hydrogen peroxide (0.5 mM or 1.0 mM), grown at an elevated temperature (37°C), or grown in yeast nitrogen base (YNB) medium with 2% glucose. Each lane was loaded with 20 μg of total protein. Each gel represents one of at least three independent experiments.
Although Cat2, Cat3, and Cat4 lacked detectable catalase activity in vitro, we were able to detect a transcript for each of the catalases by real-time quantitative PCR. However, none of the transcripts were elevated in the cat1 mutant strain relative to that in wild-type strain H99, suggesting that compensatory transcriptional activation of CAT2, CAT3, or CAT4 did not occur in the cat1 mutant strain (data not shown). We hypothesized that given the presence of transcript, Cat2, Cat3, or Cat4 might exhibit activities under appropriate growth conditions. To assess this possibility, we grew wild-type strain H99 in the presence of oxidative stress (hydrogen peroxide concentrations of 0.25 mM and 0.5 mM) and at an elevated temperature (37°C). Similar to the previous results, we were unable to detect Cat2, Cat3, or Cat4 enzyme activity under these conditions (Fig. 4C). Densitometry analysis of Cat1 bands revealed no major differences in the magnitude of catalase activity under any of these conditions, suggesting that Cat1 activity is not substantially regulated in response to exogenous oxidative stress or elevated temperature (Fig. 4C).
Catalase mutant strains do not exhibit an oxidative-stress phenotype.
C. neoformans possesses several well-characterized phenotypes that contribute to the virulence composite, including capsule synthesis, melanin production, and the ability to grow at 37°C. The cat1, cat2, cat3, cat4, and cat1 cat2 cat3 cat4 mutant strains were assessed to determine if these phenotypes were affected by the catalase gene mutations. We found no phenotypic differences between any of the catalase mutant strains and wild-type strain H99 with respect to these phenotypes (data not shown).
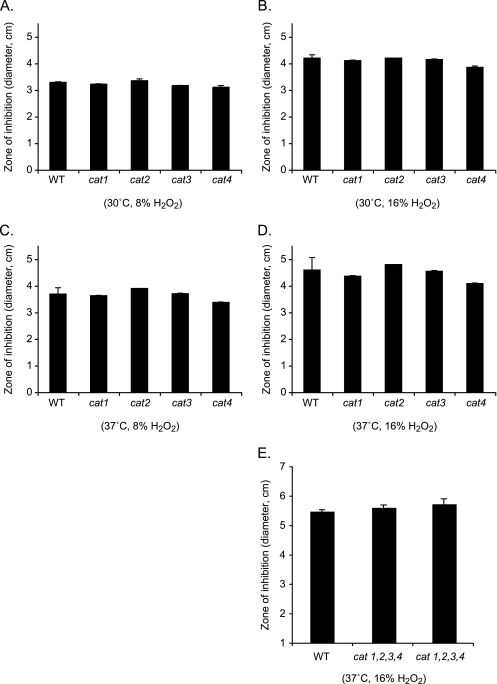
Catalases are an important component of the antioxidant defense systems of many bacteria, providing protection against the oxidative stress that results from exogenous and endogenous reactive oxygen species. To determine if this was also the case for C. neoformans, disc diffusion assays were performed to assess the susceptibilities of the cat1, cat2, cat3, cat4, and cat1 cat2 cat3 cat4 mutant strains and wild-type strain H99 to exogenous oxidative stress. None of the C. neoformans catalase mutant strains exhibited an increase in sensitivity to exogenous oxidative stress compared to that of wild-type strain H99 at 30°C or 37°C (Fig. 5). The same results were observed independent of whether strains were treated with 8 or 16% hydrogen peroxide (Fig. 5). In addition, none of the catalase double or triple mutant strains exhibited an oxidative-stress phenotype. These results were confirmed using liquid growth assays.
FIG. 5.
Catalase mutant strains do not exhibit an oxidative-stress phenotype. Sensitivity to oxidative stress was assessed by a disc diffusion assay. Sterile discs were saturated with 10 μl of 8% (A and C) or 16% (B, D, and E) hydrogen peroxide. Plates were incubated at either 30°C (A and B) or 37°C (B, D, and E). No differences in the diameters of zones of inhibition were observed among any of the catalase mutant strains compared to that of the wild-type (WT) strain. Results represent mean values ± the standard errors of the means for three or more experiments.
Although none of the individual catalase mutant strains exhibited increased sensitivity to exogenous oxidative stress, we could not rule out the possibility that catalase activity contributed to protection against endogenous oxidative stress. Reactive oxygen species are produced during beta-oxidation of short-, medium-, and long-chain fatty acids in the mitochondria and during beta-oxidation chain shortening of long-chain fatty acids in the peroxisome. We compared the abilities of the cat1 mutant and wild-type strain H99 to grow on a variety of carbon sources. Under these conditions, the dependency on beta-oxidation and respiration for energy production would be expected to result in elevated levels of endogenous reactive oxygen species. We observed that the growth levels of wild-type strain H99 and the cat1 mutant were the same when Tween 20, Tween 40, Tween 60, Tween 80, oleic acid 735, oleic acid 73, and lignoceric acid were provided as the sole carbon sources (data not shown). Both strains utilized lignoceric acid and oleic acid 73 well as the sole carbon sources. However, Tween 20, Tween 40, Tween 60, Tween 80, and oleic acid 735 served as poor carbon sources for both strains. These results suggested that the loss of catalase activity does not impair the ability of C. neoformans to respond to changes in the steady-state concentration of endogenous reactive oxygen species.
Cat1 and Cat3 are not essential for mating.
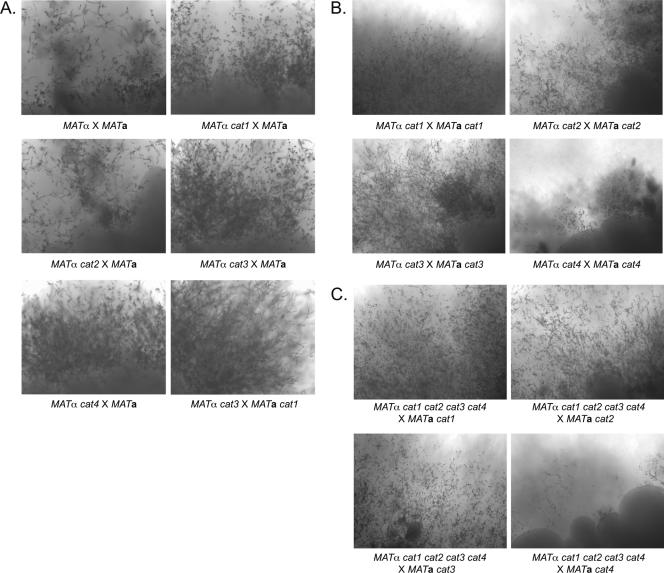
The assignment of CAT1 and CAT3 to the same clade (clade A) as the conidium-specific euascomycete catalase genes (43) suggested that they may participate in processes required for mating. We performed mating reactions to assess the contributions of Cat1 and Cat3 to mating and observed that the MATαcat1 × MATa wild-type, MATα cat3 × MATa wild-type, and MATα cat3 × MATa cat1 strains were just as able to form mating structures and produce basidiospore chains as the MATα × MATa wild-type strain (Fig. 6A). Unilateral crosses between either the cat2 or cat4 mutants and a wild-type mating partner resulted in normal mating structures and basidiospore chains (Fig. 6A). In addition, there was no impact on mating in either the cat1, cat2, or cat3 mutant bilateral cross (Fig. 6B). We did, however, observe a striking mating defect when both the mating partners were cat4 mutants. The majority of the hyphae resulting from the cross were embedded within the agar, in contrast to the result with wild-type crosses, in which abundant aerial hyphae produced basidia and basidiospores (Fig. 6B). Although the amount of filament production was dramatically decreased in the cat4 mutant bilateral cross, a limited number of well-formed basidia and basidiospore chains were eventually produced. A decrease in mating was also observed when the MATα cat1 cat2 cat3 cat4 mutant strain was crossed with the MATa cat4 mutant strain but not for crosses with the MATa cat1, MATa cat2, or MATa cat3 mutant strains (Fig. 6C). These results suggest that Cat4, but not Cat1, Cat2, or Cat3, contributes to C. neoformans sexual differentiation.
FIG. 6.
Role of catalases in C. neoformans sexual differentiation. (A) Cat1 and Cat3 are not required for sexual differentiation in C. neoformans. Wild-type MATα and cat1α and cat3α mutant strains were each cocultured with a MATa wild-type tester strain on V8 mating medium. In addition, the cat3α mutant strain was cocultured with the cat1a mutant strain. (B) Cat4 appears to contribute to the sexual differentiation of C. neoformans. MATα and MATa pairs of each catalase mutant strain were cocultured on V8 mating medium. (C) A quadruple catalase mutant is not impaired for sexual differentiation. The MATα quadruple strain was cocultured with each MATa catalase mutant strain on V8 mating medium. Plates were incubated for 2 weeks. Representative crosses were photographed at a magnification of ×90.
The cat1 and cat1 cat2 cat3 cat4 mutant strains are virulent in a murine model of cryptococcosis.
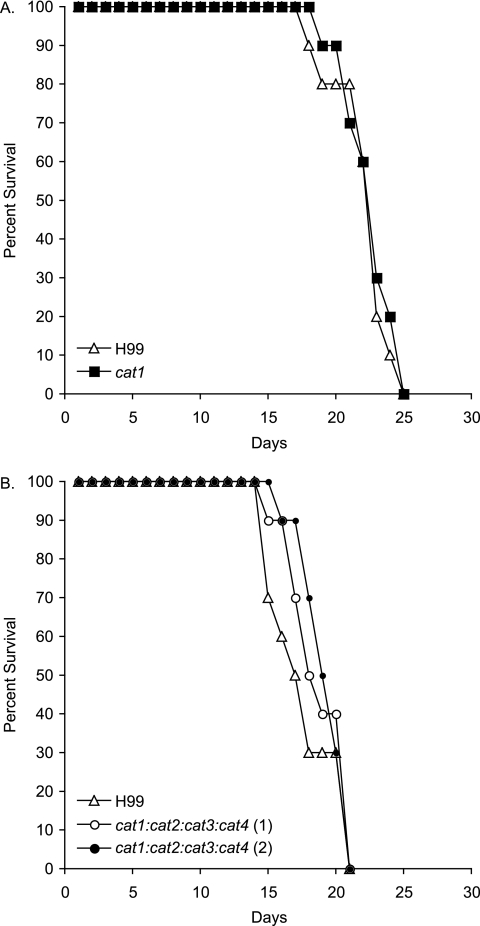
Given that the cat1 and cat1 cat2 cat3 cat4 mutant strains did not exhibit oxidative-stress phenotypes and completely lacked detectable catalase activity in vitro, we predicted that they would not exhibit virulence defects in mice. To test this hypothesis, 10 A/Jcr mice were infected by intranasal inoculation with 5 × 105 CFU of cat1 or cat1 cat2 cat3 cat4 (two independent mutant strains) mutant strains or wild-type strain H99. By day 25 of infection, all infected mice succumbed to infection, with no significant differences in the mean times to mortality (P > 0.05) between any of the groups of mice (Fig. 7).
FIG. 7.
Cat1 activity is not required for virulence. Groups of 10 A/Jcr mice were anesthetized by intraperitoneal injection of pentobarbital and infected with 5 × 105 CFU of cat1 (A) and cat1 cat2 cat3 cat4 (B) mutant strains (two independent mutants) and wild-type strain H99. All mice infected with the catalase mutant strains and wild-type strain H99 succumbed to infection and died by day 25. The absence of differences in the mean times to death demonstrates that catalase does not contribute to the virulence composite.
DISCUSSION
Numerous attributes collectively contribute to the success of pathogenic microorganisms in the host environment. One such attribute is the ability to resist damage by ROS elicited by host effector cells that contribute to the innate immune response. This resistance can be accomplished in a number of different ways, including the production of factors that prevent host effector cells from eliciting an oxidative burst, the possession of nonenzymatic antioxidants (such as melanin and mannitol), and the utilization of enzymatic antioxidants (such as catalases, superoxide dismutases, and various peroxidases) to degrade ROS. The complexity of the antioxidant defense system is illustrated by the retained ability of many C. neoformans mutants with antioxidant defense defects to cause morbidity and mortality in a manner undistinguishable from that of wild-type strains (20, 36). C. neoformans can colonize the lung, which suggests that it possesses adequate antioxidant defenses to overcome the oxygen-dependent killing mechanisms of alveolar macrophages (14, 15, 29, 30, 42, 50, 52) and other host phagocytes. Indeed, a number of studies have reported correlations between resistance to oxidative stress in vitro and virulence in a murine cryptococcosis model (2, 6, 56).
In the present study, we utilized a bioinformatics approach to identify and characterize all four members of the C. neoformans catalase gene family, which is the largest antioxidant gene family thus far identified for C. neoformans. We hypothesized that the catalases might contribute individually or collectively to antioxidant defense against endogenous or exogenous sources of ROS. To definitively test this hypothesis, we deleted the entire catalase family of genes, in addition to individual catalase genes. This is the first study that has assessed the contribution of an entire C. neoformans gene family to antioxidant defense and virulence. We have demonstrated that the loss of the entire catalase gene family does not alter the in vitro resistance to intracellular or extracellular oxidative stress. Additionally, the mutation of all four catalase genes does not diminish the virulence potential of C. neoformans. This observation is in agreement with studies that have assessed the contribution of catalase to the virulence composite of A. fumigatus and A. nidulans (5, 44). Our results suggest that C. neoformans possesses a robust and redundant antioxidant defense system.
Our phylogenetic analysis provides insight into the history of the fungal catalases. As shown in Fig. 1, it appears that there are four distinct clades of fungal catalases and that C. neoformans possesses catalases in three of the four clades. The peroxisomal catalases form clade P, cytoplasmic catalases form clade C, spore-specific catalases form clade A, and clade B is made up primarily of secreted catalases. We interpret the catalase gene tree by first studying the species phylogeny of the three major fungal groups as shown in Fig. 1: basidiomycetes (C. neoformans, C. cinereus, and P. chrysosporium), euascomycetes (A. fumigatus, A. nidulans, N. crassa, F. graminearum, M. grisea, and P. anserina), and hemiascomycetes (S. cerevisiae, Kluyveromyces lactis, A. gossypii, and C. albicans). We observe both basidiomycetes and euascomycetes in clades P, C, and A of the catalase gene tree, suggesting that these three catalase clades were present at least as recently as the fungal ancestor. This interpretation further suggests that ancestral fungi possessed spore-specific, peroxisomal, and cytoplasmic catalases. The observation that clade B catalases have members only from the euascomycetes suggests either that the secreted catalases arose by duplication and divergence from the spore-specific catalase or the less parsimonious possibility that this form of catalase was lost independently from the hemiascomycetes, basidiomycetes, and Schizosaccharomyces pombe. Evaluation of the clade A members suggests that the spore-specific catalase was lost twice, once from the S. pombe lineage and once from the hemiascomycete ancestor.
Using this approach to evaluate the phylogenetic relationships of the genes across the fungi allows us to hypothesize a function for each of the four C. neoformans catalases. This approach, often-dubbed “phylogenomics” (13), is more robust than assigning a function based on the most similar gene identified through BLAST analysis. As shown in Fig. 1, clade P contains several peroxisomal catalase genes, such as the A. nidulans CATC gene (26) and the S. cerevisiae CTA1 gene (48). We can assign a putative peroxisomal function to C. neoformans CAT2 since it also was found in clade P. Similarly, the C. neoformans CAT4 gene is likely a cytosolic catalase gene due to its presence in clade C, which contains the S. cerevisiae cytosolic catalase gene CTT1. The presence of CAT1 and CAT3 in clade A, which contains the conidium-specific A. fumigatus and A. nidulans euascomycete catalase genes (43), suggested that they might participate in spore-related processes, such as germination. However, a preliminary analysis of the CAT1 and CAT3 promoter regions did not indicate the presence of significant shared motifs. Without observable phenotypic data for CAT3, related to Cat3 function, it is difficult to define unique functions for Cat3 compared to those of Cat1, the only C. neoformans catalase with detectable in vitro activity. These findings are surprising given that these paralogs are 85% similar at the protein level. Clade B is composed of the secreted CATB catalase genes from fungi such as A. fumigatus and Histoplasma capsulatum (24, 25). The absence of a C. neoformans catalase gene from this clade is consistent with our in vitro results, since we were unable to detect any secreted catalase activity for C. neoformans culture supernatants. Furthermore, our phylogenetic analysis suggests that only the euascomycete fungi will possess secreted catalases.
Based on our phylogenetic analysis, we predicted that the cat1 or cat3 mutant might exhibit a mating defect. Although we did not detect mating defects or diminished spore viability in strains resulting from unilateral or bilateral crosses between the cat1 or cat3 mutant strain, we did observe decreased filament production in a strain resulting from the cat4 mutant bilateral cross. Our phylogenetic analysis predicts that Cat4 is likely a cytosolic catalase. These observations suggest that some catalase-related event, such as compartmentalized oxidative stress, might contribute to C. neoformans sexual differentiation. The absence of mating defects in strains resulting from cat1 mutant unilateral and bilateral crosses, along with the observation that Cat1 appears to be constitutively active, suggests that the function of Cat1 may have become specialized in C. neoformans compared to that in related fungi. Although C. neoformans can undergo sexual reproduction, C. neoformans populations in the environment are largely clonal, consisting predominantly of MATα strains, which suggests that mating in the environment is probably a rare event (31). It is possible that Cat1 initially provided specialized antioxidant defense during processes involved in mating, such as spore production or germination, but that an evolutionary shift away from sexual reproduction resulted in Cat1 being co-opted to provide antioxidant defense during vegetative growth. It is equally plausible that the presence of several other gene families that encode antioxidants, including the glutathione peroxidases, thioredoxin peroxidases, and the cytochrome c peroxidase, provide redundant and compensatory antioxidant defenses against hydrogen peroxide.
The absence of detectable oxidative-stress phenotypes for any of the C. neoformans catalase mutants is consistent with the results of similar studies of Saccharomyces and Aspergillus. Saccharomyces cta1, ctt1, and cta1 ctt1 mutant strains exhibited growth rates and susceptibilities to hydrogen peroxide under exponential growth conditions that were similar to those of the wild-type strain (23). Furthermore, A. fumigatus mutant strains lacking the conidial catalase (CatA) or the mycelial catalases (Cat1 and Cat2) exhibited slight susceptibilities to oxidative stress in vitro compared to that of the wild-type strain but exhibited no significant virulence defect in vivo (44). Similar results were also observed for A. nidulans: catA, catB, and catA catB mutant strains were just as virulent as the wild-type strain in a murine model of chronic granulomatous disease (5). In contrast to these studies, it was reported that a C. albicans cat1 mutant strain exhibited an oxidative-stress phenotype in vitro and a virulence defect (39, 55). However, reconstitution of the mutant strains with CAT1 did not restore resistance against oxidative stress, so the role that the catalase plays in the antioxidant defense of C. albicans remains to be clarified.
Indeed, the role of catalase in antioxidant defense for many of these fungi is enigmatic, given that the loss of catalase activity does not correlate with oxidative stress or developmental defects. One interpretation of these results is that the catalases function interchangeably with other constituents of the antioxidant defense systems as part of a robust and multipronged response to oxidative stress. For example, in Saccharomyces, it has been reported that the mitochondrial cytochrome c peroxidases (Ccp1) and the cytoplasmic catalase (Ctt1) exhibit interchangeable and compensatory antioxidant activities (35). Furthermore, glutathione has been reported to exhibit an antioxidant defense that overlaps with that of catalases in S. cerevisiae (21). S. cerevisiae mutant strains lacking glutathione (gsh1) or glutathione reductase (glr1) exhibited increased sensitivity to hydrogen peroxide (21). Glutathione and glutathione reductase mutant strains that lacked catalases (cta1 ctt1 glr1 and cta1 ctt1 gsh1 strains) exhibited even more severe oxidative-stress defects (21).
The presence of redundancy in the antioxidant defense system provides a plausible explanation as to why the loss of many of the individual components of the C. neoformans antioxidant defense system does not result in reduced cell viability or developmental defects. Potential elements that would create redundant layers of antioxidant defense include cytochrome c peroxidase (CCP1) (20), the catalases (CAT1, CAT2, CAT3, and CAT4), the thiol peroxidases (TSA1, TSA3, and TSA4) (37), the glutathione peroxidases (GPX1 and GPX2) (36), alternative oxidase (AOX1) (2), Cu,Zn superoxide dismutase (SOD1) (6, 40), and Mn superoxide dismutase (SOD2) (19, 41). Among these potential antioxidant defense proteins, only mutations of the AOX1, CCP1, SOD1, SOD2, TSA1, GPX1, and GPX2 genes are associated with increased susceptibility to extracellular oxidant stress (2, 7, 19, 20, 36, 37, 40, 41). Additionally, only the AOX1, SOD1, SOD2, and TSA1 mutants are attenuated for virulence in animal models (2, 6, 19, 37). That the catalase gene family is not required for the virulence of C. neoformans reinforces the concept that these varied proteins create a complex and partially redundant system for antioxidant defense of this human fungal pathogen.
. .
Acknowledgments
This work was supported by Public Health Service grants AI028388 (J.R.P.) and AI50128 (J.A.A.) from NIAID. S.S.G. was supported by a National Institutes of Health RSUM grant (AI028388). J.E.S. was supported by an NSF predoctoral fellowship, and C.N. was supported by an Interdisciplinary Research Training Grant in AIDS (T32 AI07392).
We thank Gary Cox for suggesting the idea of disrupting the catalase genes, and we are grateful to Irwin Fridovich for assistance with the catalase activity assay.
REFERENCES
- 1.Adachi, J., and M. Hasegawa. 1996. MOLPHY version 2.3, programs for molecular phylogenetics based on maximum likelihood. The Institute of Statistical Mathematics, Tokyo, Japan.
- 2.Akhter, S., H. C. McDade, J. M. Gorlach, G. Heinrich, G. M. Cox, and J. R. Perfect. 2003. Role of alternative oxidase gene in pathogenesis of Cryptococcus neoformans. Infect. Immun. 71:5794-5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 4.Bateman, A., L. Coin, R. Durbin, R. D. Finn, V. Hollich, S. Griffiths-Jones, A. Khanna, M. Marshall, S. Moxon, E. L. Sonnhammer, D. J. Studholme, C. Yeats, and S. R. Eddy. 2004. The Pfam protein families database. Nucleic Acids Res. 32:D138-D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang, Y. C., B. H. Segal, S. M. Holland, G. F. Miller, and K. J. Kwon-Chung. 1998. Virulence of catalase-deficient Aspergillus nidulans in p47phox−/− mice. Implications for fungal pathogenicity and host defense in chronic granulomatous disease. J. Clin. Investig. 101:1843-1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cox, G. M., T. S. Harrison, H. C. McDade, C. P. Taborda, G. Heinrich, A. Casadevall, and J. R. Perfect. 2003. Superoxide dismutase influences the virulence of Cryptococcus neoformans by affecting growth within macrophages. Infect. Immun. 71:173-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cox, G. M., J. Mukherjee, G. T. Cole, A. Casadevall, and J. R. Perfect. 2000. Urease as a virulence factor in experimental cryptococcosis. Infect. Immun. 68:443-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davidson, R. C., J. R. Blankenship, P. R. Kraus, M. de Jesus Berrios, C. M. Hull, C. D'Souza, P. Wang, and J. Heitman. 2002. A PCR-based strategy to generate integrative targeting alleles with large regions of homology. Microbiology 148:2607-2615. [DOI] [PubMed] [Google Scholar]
- 9.Day, W. A., Jr., J. L. Sajecki, T. M. Pitts, and L. A. Joens. 2000. Role of catalase in Campylobacter jejuni intracellular survival. Infect. Immun. 68:6337-6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diamond, R. D., R. K. Root, and J. E. Bennett. 1972. Factors influencing killing of Cryptococcus neoformans by human leukocytes in vitro. J. Infect. Dis. 125:367-376. [DOI] [PubMed] [Google Scholar]
- 11.Eddy, S. R. 1998. Profile hidden Markov models. Bioinformatics 14:755-763. [DOI] [PubMed] [Google Scholar]
- 12.Edgar, R. C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eisen, J. A., and C. M. Fraser. 2003. Phylogenomics: intersection of evolution and genomics. Science 300:1706-1707. [DOI] [PubMed] [Google Scholar]
- 14.Feldmesser, M., Y. Kress, P. Novikoff, and A. Casadevall. 2000. Cryptococcus neoformans is a facultative intracellular pathogen in murine pulmonary infection. Infect. Immun. 68:4225-4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feldmesser, M., S. Tucker, and A. Casadevall. 2001. Intracellular parasitism of macrophages by Cryptococcus neoformans. Trends Microbiol. 9:273-278. [DOI] [PubMed] [Google Scholar]
- 16.Flesch, I. E., G. Schwamberger, and S. H. Kaufmann. 1989. Fungicidal activity of IFN-gamma-activated macrophages. Extracellular killing of Cryptococcus neoformans. J. Immunol. 142:3219-3224. [PubMed] [Google Scholar]
- 17.Fraser, J. A., S. S. Giles, E. C. Wenink, S. G. Geunes-Boyer, J. R. Wright, S. Diezmann, A. Allen, J. E. Stajich, F. S. Dietrich, J. R. Perfect, and J. Heitman. 2005. Same-sex mating and the origin of the Vancouver Island Cryptococcus gattii outbreak. Nature 437:1360-1364. [DOI] [PubMed] [Google Scholar]
- 18.Galagan, J. E., S. E. Calvo, K. A. Borkovich, E. U. Selker, N. D. Read, D. Jaffe, W. FitzHugh, L. J. Ma, S. Smirnov, S. Purcell, B. Rehman, T. Elkins, R. Engels, S. Wang, C. B. Nielsen, J. Butler, M. Endrizzi, D. Qui, P. Ianakiev, D. Bell-Pedersen, M. A. Nelson, M. Werner-Washburne, C. P. Selitrennikoff, J. A. Kinsey, E. L. Braun, A. Zelter, U. Schulte, G. O. Kothe, G. Jedd, W. Mewes, C. Staben, E. Marcotte, D. Greenberg, A. Roy, K. Foley, J. Naylor, N. Stange-Thomann, R. Barrett, S. Gnerre, M. Kamal, M. Kamvysselis, E. Mauceli, C. Bielke, S. Rudd, D. Frishman, S. Krystofova, C. Rasmussen, R. L. Metzenberg, D. D. Perkins, S. Kroken, C. Cogoni, G. Macino, D. Catcheside, W. Li, R. J. Pratt, S. A. Osmani, C. P. DeSouza, L. Glass, M. J. Orbach, J. A. Berglund, R. Voelker, O. Yarden, M. Plamann, S. Seiler, J. Dunlap, A. Radford, R. Aramayo, D. O. Natvig, L. A. Alex, G. Mannhaupt, D. J. Ebbole, M. Freitag, I. Paulsen, M. S. Sachs, E. S. Lander, C. Nusbaum, and B. Birren. 2003. The genome sequence of the filamentous fungus Neurospora crassa. Nature 422:859-868. [DOI] [PubMed] [Google Scholar]
- 19.Giles, S. S., I. Batinic-Haberle, J. R. Perfect, and G. M. Cox. 2005. Cryptococcus neoformans mitochondrial superoxide dismutase: an essential link between antioxidant function and high-temperature growth. Eukaryot. Cell 4:46-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giles, S. S., J. R. Perfect, and G. M. Cox. 2005. Cytochrome c peroxidase contributes to the antioxidant defense of Cryptococcus neoformans. Fungal Genet. Biol. 42:20-29. [DOI] [PubMed] [Google Scholar]
- 21.Grant, C. M., G. Perrone, and I. W. Dawes. 1998. Glutathione and catalase provide overlapping defenses for protection against hydrogen peroxide in the yeast Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 253:893-898. [DOI] [PubMed] [Google Scholar]
- 22.Idnurm, A., J. L. Reedy, J. C. Nussbaum, and J. Heitman. 2004. Cryptococcus neoformans virulence gene discovery through insertional mutagenesis. Eukaryot. Cell 3:420-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Izawa, S., Y. Inoue, and A. Kimura. 1996. Importance of catalase in the adaptive response to hydrogen peroxide: analysis of acatalasaemic Saccharomyces cerevisiae. J. Biochem. 320:61-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson, C. H., M. G. Klotz, J. L. York, V. Kruft, and J. E. McEwen. 2002. Redundancy, phylogeny and differential expression of Histoplasma capsulatum catalases. Microbiology 148:1129-1142. [DOI] [PubMed] [Google Scholar]
- 25.Johnson, H., J. R. Whiteford, S. E. Eckert, and P. D. Spanu. 2003. Production and secretion of Aspergillus nidulans catalase B in filamentous fungi driven by the promoter and signal peptide of the Cladosporium fulvum hydrophobin gene hcf-1. Curr. Genet. 44:155-163. [DOI] [PubMed] [Google Scholar]
- 26.Kawasaki, L., and J. Aguirre. 2001. Multiple catalase genes are differentially regulated in Aspergillus nidulans. J. Bacteriol. 183:1434-1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klotz, M. G., Y. C. Kim, J. Katsuwon, and A. J. Anderson. 1995. Cloning, characterization and phenotypic expression in Escherichia coli of catF, which encodes the catalytic subunit of catalase isozyme CatF of Pseudomonas syringae. Appl. Microbiol. Biotechnol. 43:656-666. [DOI] [PubMed] [Google Scholar]
- 28.Kwon-Chung, K. J., A. Varma, J. C. Edman, and J. E. Bennett. 1992. Selection of ura5 and ura3 mutants from the two varieties of Cryptococcus neoformans on 5-fluoroorotic acid medium. J. Med. Vet. Mycol. 30:61-69. [PubMed] [Google Scholar]
- 29.Levitz, S. M. 2002. Receptor-mediated recognition of Cryptococcus neoformans. Nippon Ishinkin Gakkai Zasshi 43:133-136. [DOI] [PubMed] [Google Scholar]
- 30.Levitz, S. M., S.-H. Nong, K. F. Seetoo, T. S. Harrison, R. A. Speizer, and E. R. Simons. 1999. Cryptococcus neoformans resides in an acidic phagolysosome of human macrophages. Infect. Immun. 67:885-890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Litvintseva, A. P., L. Kestenbaum, R. Vilgalys, and T. G. Mitchell. 2005. Comparative analysis of environmental and clinical populations of Cryptococcus neoformans. J. Clin. Microbiol. 43:556-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manca, C., S. Paul, C. E. Barry, III, V. H. Freedman, and G. Kaplan. 1999. Mycobacterium tuberculosis catalase and peroxidase activities and resistance to oxidative killing in human monocytes in vitro. Infect. Immun. 67:74-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martinez, D., L. F. Larrondo, N. Putnam, M. D. Gelpke, K. Huang, J. Chapman, K. G. Helfenbein, P. Ramaiya, J. C. Detter, F. Larimer, P. M. Coutinho, B. Henrissat, R. Berka, D. Cullen, and D. Rokhsar. 2004. Genome sequence of the lignocellulose degrading fungus Phanerochaete chrysosporium strain RP78. Nat. Biotechnol. 22:695-700. [DOI] [PubMed] [Google Scholar]
- 34.Miller, G. P., and S. Kohl. 1983. Antibody-dependent leukocyte killing of Cryptococcus neoformans. J. Immunol. 131:1455-1459. [PubMed] [Google Scholar]
- 35.Minard, K. I., and L. McAlister-Henn. 2001. Antioxidant function of cytosolic sources of NADPH in yeast. Free Radic. Biol. Med. 31:832-843. [DOI] [PubMed] [Google Scholar]
- 36.Missall, T. A., J. F. Cherry-Harris, and J. K. Lodge. 2005. Two glutathione peroxidases in the fungal pathogen Cryptococcus neoformans are expressed in the presence of specific substrates. Microbiology 151:2573-2581. [DOI] [PubMed] [Google Scholar]
- 37.Missall, T. A., M. E. Pusateri, and J. K. Lodge. 2004. Thiol peroxidase is critical for virulence and resistance to nitric oxide and peroxide in the fungal pathogen, Cryptococcus neoformans. Mol. Microbiol. 51:1447-1458. [DOI] [PubMed] [Google Scholar]
- 38.Mitchell, T. G., and J. R. Perfect. 1995. Cryptococcosis in the era of AIDS—100 years after the discovery of Cryptococcus neoformans. Clin. Microbiol. Rev. 8:515-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakagawa, Y., T. Kanbe, and I. Mizuguchi. 2003. Disruption of the human pathogenic yeast Candida albicans catalase gene decreases survival in mouse-model infection and elevates susceptibility to higher temperature and to detergents. Microbiol. Immunol. 47:395-403. [DOI] [PubMed] [Google Scholar]
- 40.Narasipura, S. D., J. G. Ault, M. J. Behr, V. Chaturvedi, and S. Chaturvedi. 2003. Characterization of Cu,Zn superoxide dismutase (SOD1) gene knock-out mutant of Cryptococcus neoformans var. gattii: role in biology and virulence. Mol. Microbiol. 47:1681-1694. [DOI] [PubMed] [Google Scholar]
- 41.Narasipura, S. D., V. Chaturvedi, and S. Chaturvedi. 2005. Characterization of Cryptococcus neoformans variety gattii SOD2 reveals distinct roles of the two superoxide dismutases in fungal biology and virulence. Mol. Microbiol. 55:1782-1800. [DOI] [PubMed] [Google Scholar]
- 42.Naslund, P. K., W. C. Miller, and D. L. Granger. 1995. Cryptococcus neoformans fails to induce nitric oxide synthase in primed murine macrophage-like cells. Infect. Immun. 63:1298-1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Navarro, R. E., M. A. Stringer, W. Hansberg, W. E. Timberlake, and J. Aguirre. 1996. catA, a new Aspergillus nidulans gene encoding a developmentally regulated catalase. Curr. Genet. 29:352-359. [PubMed] [Google Scholar]
- 43a.Nichols, C. B., J. A. Fraser, and J. Heitman. 2004.. PAK kinases Ste20 and Pak1 govern cell polarity at different stages of mating in Cryptococcus neoformans. Mol. Biol. Cell 15:4476-4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43b.Nielsen, K., G. M. Cox, P. Wang, D. L. Toffaletti, J. R. Perfect, and J. Heitman. 2003. Sexual cycle of Cryptococcus neoformans var. grubii and virulence of congenic a and α isolates. Infect. Immun. 71:4831-4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paris, S., D. Wysong, J.-P. Debeaupuis, K. Shibuya, B. Philippe, R. D. Diamond, and J.-P. Latgé. 2003. Catalases of Aspergillus fumigatus. Infect. Immun. 71:3551-3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pearson, W. R. 1990. Rapid and sensitive sequence comparison with FASTP and FASTA. Methods Enzymol. 183:63-98. [DOI] [PubMed] [Google Scholar]
- 45a.Perfect, J. R., S. D. R. Lang, and D. T. Durack. 1980. Chronic cryptococcal meningitis: a new experimental model. Am. J. Pathol. 101:177-193. [PMC free article] [PubMed] [Google Scholar]
- 46.Rice, P., I. Longden, and A. Bleasby. 2000. EMBOSS: the European Molecular Biology Open Software suite. Trends Genet. 16:276-277. [DOI] [PubMed] [Google Scholar]
- 47.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 48.Simon, M., G. Adam, W. Rapatz, W. Spevak, and H. Ruis. 1991. The Saccharomyces cerevisiae ADR1 gene is a positive regulator of transcription of genes encoding peroxisomal proteins. Mol. Cell. Biol. 11:699-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stajich, J. E., D. Block, K. Boulez, S. E. Brenner, S. A. Chervitz, C. Dagdigian, G. Fuellen, J. G. Gilbert, I. Korf, H. Lapp, H. Lehvaslaiho, C. Matsalla, C. J. Mungall, B. I. Osborne, M. R. Pocock, P. Schattner, M. Senger, L. D. Stein, E. Stupka, M. D. Wilkinson, and E. Birney. 2002. The Bioperl toolkit: Perl modules for the life sciences. Genome Res. 12:1611-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Steenbergen, J. N., H. A. Shuman, and A. Casadevall. 2001. Cryptococcus neoformans interactions with amoebae suggest an explanation for its virulence and intracellular pathogenic strategy in macrophages. Proc. Natl. Acad. Sci. USA 98:15245-15250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Toffaletti, D. L., T. H. Rude, S. A. Johnston, D. T. Durack, and J. R. Perfect. 1993. Gene transfer in Cryptococcus neoformans by use of biolistic delivery of DNA. J. Bacteriol. 175:1405-1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tucker, S. C., and A. Casadevall. 2002. Replication of Cryptococcus neoformans in macrophages is accompanied by phagosomal permeabilization and accumulation of vesicles containing polysaccharide in the cytoplasm. Proc. Natl. Acad. Sci. USA 99:3165-3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Varma, A., J. C. Edman, and K. J. Kwon-Chung. 1992. Molecular and genetic analysis of URA5 transformants of Cryptococcus neoformans. Infect. Immun. 60:1101-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wayne, L. G., and G. A. Diaz. 1986. A double staining method for differentiating between two classes of mycobacterial catalase in polyacrylamide electrophoresis gels. Anal. Biochem. 157:89-92. [DOI] [PubMed] [Google Scholar]
- 55.Wysong, D. R., L. Christin, A. M. Sugar, P. W. Robbins, and R. D. Diamond. 1998. Cloning and sequencing of a Candida albicans catalase gene and effects of disruption of this gene. Infect. Immun. 66:1953-1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xie, Q., K. Kawakami, N. Kudeken, T. Zhang, M. H. Qureshi, and A. Saito. 1997. Different susceptibility of three clinically isolated strains of Cryptococcus neoformans to the fungicidal effects of reactive nitrogen and oxygen intermediates: possible relationships with virulence. Microbiol. Immunol. 41:725-731. [DOI] [PubMed] [Google Scholar]
- 57.Xu, X. Q., and S. Q. Pan. 2000. An Agrobacterium catalase is a virulence factor involved in tumorigenesis. Mol. Microbiol. 35:407-414. [DOI] [PubMed] [Google Scholar]