Abstract
Trypanosomatids of the order Kinetoplastida are major contributors to global disease and morbidity, and understanding their basic biology coupled with the development of new drug targets represents a critical need. Additionally, trypanosomes are among the more accessible divergent eukaryote experimental systems. The genome of Trypanosoma brucei contains 8,131 predicted open reading frames (ORFs), of which over half have no known homologues beyond the Kinetoplastida and a substantial number of others are poorly defined by in silico analysis. Thus, a major challenge following completion of the T. brucei genome sequence is to obtain functional data for all trypanosome ORFs. As T. brucei is more experimentally tractable than the related Trypanosoma cruzi and Leishmania spp. and shares >75% of their genes, functional analysis of T. brucei has the potential to inform a range of parasite biology. Here, we report methods for systematic mRNA ablation by RNA interference (RNAi) and for phenotypic analysis, together with online data dissemination. This represents the first systematic analysis of gene function in a parasitic organism. In total, 210 genes have been targeted in the bloodstream form parasite, representing an essentially complete phenotypic catalogue of chromosome I together with a validation set. Over 30% of the chromosome I genes generated a phenotype when targeted by RNAi; most commonly, this affected cell growth, viability, and/or cell cycle progression. RNAi against approximately 12% of ORFs was lethal, and an additional 11% had growth defects but retained short-term viability in culture. Although we found no evidence for clustering or a bias towards widely evolutionarily conserved genes within the essential ORF cohort, the putative chromosome I centromere is adjacent to a domain containing genes with no associated phenotype. Involvement of such a large proportion of genes in robust growth in vitro indicates that a high proportion of the expressed trypanosome genome is required for efficient propagation; many of these gene products represent potential drug targets.
Protozoan parasites are responsible for a significant proportion of global morbidity, mortality, and economic hardship, and in most countries current control and treatment regimes are either failing or under serious threat (6). African trypanosomiasis, caused by Trypanosoma brucei spp., is responsible for tens to hundreds of thousands of human infections annually in areas of sub-Saharan Africa where the infection is endemic and is compounded by the impact of the disease on animal welfare and productivity. Without treatment trypanosomiasis is invariably fatal, and the need for new treatments is urgent. Mechanisms for combating pathogenic protozoa rely on development and exploitation of new drug targets and/or vaccine candidates as well as efforts at vector control, all of which require detailed understanding of the biology of the pathogen and its relationship with the host and vector. The presence of efficient antigenic variation in T. brucei makes development of a vaccine highly unlikely (7).
Recently, the genomes of several major protozoan pathogens have been decoded, including the three principal trypanosomatid pathogenic species, T. brucei, Trypanosoma cruzi, and Leishmania major (8, 16, 23). The availability of complete genome data for these organisms provides an opportunity to develop new methods and approaches to analyze gene function and identify and validate therapeutic targets, as well as to identify novel aspects of exploitable biology.
A major feature experimentally differentiating T. brucei from T. cruzi or Leishmania is the availability of a robust RNA interference (RNAi) system in the former (12, 36); recent data indicate that T. cruzi and Leishmania lack components required by the RNAi pathway (13, 32), and T. brucei has emerged as a pathogenic trypanosomatid model system. Comparison of the genomes of T. brucei, T. cruzi, and Leishmania indicates that, excluding pathogen-specific systems such as antigenic variation, abundant plasma membrane components, and mechanisms of cell invasion, there is a remarkable level of conservation of predicted gene function and genomic architecture (17). Specifically, 76% of T. brucei genes are shared between all three sequenced kinetoplastids, and a further 6% are shared with T. cruzi and 1% with Leishmania (17). Hence, understanding gene function in T. brucei is anticipated as contributing to the functional assignment of T. cruzi and Leishmania. Further, the position of T. brucei within the Excavata provides a valuable experimental system for the analysis of biology in eukaryotic taxa distant to the classical model systems, specifically metazoa and fungi (34).
In silico analysis of the T. brucei genome presents several significant challenges. In particular over half of trypanosome open reading frames (ORFs) have no identifiable counterpart in higher eukaryotes and can lack identifiable domain or motif features, posing difficulties for annotation and functional assignment (16, 17). Both the great evolutionary divergence and novel functions likely contribute to this aspect; the molecular systems mediating antigenic variation, kinetoplast/basal body assembly, chromosomal segregation, and intracellular signaling may all require the activity of novel protein factors that are not conserved across the eukaryota. Major noncoding features of the trypanosome genome also remain cryptic, including centromeres (29) and RNA maturation and turnover motifs (23). Experimental confirmation of in silico predicted gene models is thus highly important for defining the functional potential within the trypanosome genome. The T. brucei genome consists of 11 pairs of diploid megabase chromosomes together with a large number of mini and intermediate-sized chromosomes; the major repository of coding potential appears to reside on the megabase chromosomes (20).
We report an RNAi analysis of the ORFs on chromosome I of T. brucei. We address potential clustering of essential ORFs within predicted polycistrons, biases in the DNA strand location or position within a polycistron, over- or underrepresentation of conserved ORFs, and the functionality of ORFs of <150 codons. We find that a large proportion of ORFs are important for robust growth in vitro, with clear implications for the complexity of cellular homeostasis and potentially virulence, evolutionary fitness, and selection of drug targets. Except for an absence of phenotype-associated ORFs adjacent to a putative centromeric region, there are no detectable biases to ORF location correlating with phenotype.
MATERIALS AND METHODS
Target selection and construction of RNAi strains.
The nucleotide sequence of T. brucei chromosome I is predicted to contain 369 ORFs. For selection of the target set for RNAi analysis, these ORFs were parsed initially for membership of chromosome I-specific clusters, membership of widely dispersed but highly similar gene families, annotation as pseudogenes, or membership of the variant surface glycoprotein (VSG) gene family. We excluded the subtelomeric regions at either end as these contain repetitive sequence, VSG genes, and expression site-associated genes that are widely dispersed through the genome; this included ∼200 kbp at the left-hand end and ∼100 kbp at the right-hand end of the chromosome. For chromosome I clusters, we elected to design pan-specific RNAi constructs predicted to knock down the message from all members of the clusters. ORFs that were part of a multigene family represented on additional chromosomes were excluded from the analysis, as were ORFs clearly annotated as pseudogenes and/or VSGs. Further, the remaining set of ORFs was then subjected to a primer prediction algorithm, RNAit, designed to minimize the potential for cross talk and, hence, off-target effects (30); ORFs that failed either the cross talk or the primer prediction routine were also excluded. It should be noted that the microRNA complex may be absent from the trypanosome system, while the use of large portions of sequence minimizes the possibility of off-target effects which can be frequent when small oligonucleotide-based small interfering RNAs are used (9); hence only a single construct was used to target each ORF. This resulted in the selection of 197 knockdowns for inclusion and is 57% of the total number of ORFs and ∼70% of the ORFs within the core region of the chromosome. We also selected a small number of additional ORFs as a validation set (see Table S3 in the supplemental material), bringing the total to 210; this latter set was chosen based on an expectation that its ablation would be detectable by one or more of the assays used in the analysis.
All target ORFs were amplified from purified genomic DNA obtained from in vitro cultures of T. brucei 427 (26). All PCR primers were of ∼23 bases and were designed to amplify fragments of 400 to 600 bp for convenience in downstream manipulations. PCRs were analyzed by agarose gel electrophoresis and compared with the predicted product size. Positive reaction products were ligated into the RNAi vector p2T7TAblue, a derivative of p2T7 optimized for direct cloning of PCR products (1). Following transformation into Escherichia coli DH5α, plasmids were isolated using QIAGEN kits and screened by restriction enzyme digestion. All RNAi vectors were subsequently verified by dye deoxy sequencing prior to trypanosome transformation. Of ∼100 kbp of sequence produced in toto (ca. 500 bp times 200), we observed ∼5 nucleotide differences per kilobase, the vast majority of which were single base changes, likely reflecting a combination of single nucleotide polymorphisms between the 927 genome strain and the 427 experimental strain and errors introduced during amplification by Taq polymerase. Only two of the PCRs failed to generate a product (<0.9%).
A total of 10 μg of each RNAi vector was linearized with NotI, checked for linearization by agarose gel electrophoresis, and introduced into the SMB (T7RNAP::TETR::NEO) cell line (40) by electroporation. Stable transformants were selected with hygromycin (2.5 μg/ml) in HMI-11 medium. Note that the SMB line is routinely maintained with 2 μg/ml G418 to retain the T7 RNA polymerase and Tet repressor.
We elected to do our analysis in T. brucei strain 427, even though this is not the genome reference strain (T. brucei TREU 927/4) (8), for a number of reasons. First, T. brucei 427 enjoys multiple systems for expression and has been in use in many laboratories for decades, with the result that it is exceptionally well characterized. Second, our analysis indicates minimal differences between strains 427 and TREU 927/4 at the level of the primary structure of individual ORFs, and there is no published evidence suggesting a significant biological difference between 427 and TREU 927/4 with the exception of its ability to readily transmit through tsetse flies (see reference 37 for analysis of this line). Third, inducible RNAi systems are in place for Lister 427, and many of the standard phenotypic assays were developed and validated in that strain.
We elected to work with cell lines rather than clonal populations. Statistically significant clonal analysis would require the study of three or more clones, and while we are aware that this strategy may result in increased false negatives due to the presence of heterogeneous populations in some of the transfected lines, we considered the advantage in terms of throughput to be persuasive.
Assay design and protocols.
All assays were carried out using a standard operating procedure that was validated using wild-type 427 trypanosomes for simplicity, reproducibility, and sensitivity. Full details of experimental procedures are available at http://trypanofan.path.cam.ac.uk. For analysis of growth rates, transformed parasite cultures were split, and one subculture was induced with 1 μg ml−1 tetracycline. Growth of the induced and uninduced culture was then followed for up to 8 days, with cell number determined daily (except for days 5 and 6) using a Z2 Coulter counter (Beckman Coulter Inc.). Cultures were diluted daily and were counted in triplicate.
The copy number of the nucleus and kinetoplast, and hence cell cycle position, typically in 200 individual cells of the cultures, was determined by 4′,6-diamidino-2-phenylindole (DAPI) staining of fixed cells (31) followed by analysis on a Nikon Eclipse E600 microscope equipped with a CoolSNAP-fx charge-coupled-device camera (Roper Scientific) driven by Metamorph Software (Molecular Devices Inc.).
The endocytic and protein processing systems were probed using two well-characterized lectins, Canavalia ensiformis concanavalin A (ConA), which binds mainly oligomannose-containing N-glycans and the glycosylphosphatidylinositol (GPI) core glycan, and Lycopersicon esculentum (tomato) lectin, which interacts preferentially with polylactosamine ([Galβ1-4GlcNAcβ1-]n)-containing glycans (5). For tomato lectin, cells were incubated in the presence of fluorescein isothiocyanate-tomato lectin (Sigma) in serum-free medium-1% bovine serum albumin (BSA) for 30 min at 4°C prior to washing, fixing, and mounting for analysis. For ConA, cells were incubated at 4°C, 12°C, and 37°C for 30 min with 5 μg ml−1 fluorescein isothiocyanate-ConA (Molecular Probes Inc.) in serum-free medium-1% BSA prior to washing, fixing, and mounting to preferentially label the flagellar pocket, early endosomes, and lysosome, respectively (18). The structure and function of the mitochondrion and Golgi complex were monitored by staining cells with MitoTracker (Molecular Probes) or BODIPY-ceramide (Molecular Probes), respectively. Cells were incubated with 100 nM MitoTracker Red CMXROS in serum-free medium-1% BSA for 30 min prior to fixation or 0.5 mM Texas Red BODIPY-ceramide for 1 h in serum-free medium-1% BSA (18). Images were acquired as above and processed in Photoshop (Adobe Inc.) for presentation.
Morphology and motility were assessed by phase-contrast microscopy. For morphology, cells were fixed with 4% paraformaldehyde and mounted, and images were collected. For analysis of motility, live cells were spotted onto a microscope slide, and the sample was immediately analyzed on a Nikon E600. Images were accumulated in Metamorph at 15 frames per s, and the images were exported as a stack. Image stacks were converted to Quicktime movies using Quicktime Pro (Apple Computer Inc.) at 15 frames per s.
Data analysis and scoring criteria.
All primary phenotypic data were collected blind; geneDB accession numbers for individual ORFs were recoded with trypanoFAN designations (e.g., TFNX.XXX) for the RNAi plasmids, and the code was retained by the workers producing the DNA and not communicated to the phenotyping centers until analysis was complete (see Table S1 in the supplemental material). The numerical data obtained for growth were parsed according to the following criteria: a defect was recorded if, during the 4 days following induction, cell number in the induced culture was <75% (mild) or <50% (severe) of the uninduced culture for two consecutive days. DAPI analysis for wild-type cells indicated ∼85% one nucleus/one kinetoplast (1N:1K), ∼10% 1N:2K, and ∼5% 2N:2K. The numerical data obtained for DAPI analysis were parsed according to the following criteria. A defect was recorded if the following conditions were met: <4% or >17% 1N:2K; >12% 2N; >4.5% 0N, 0K, N > 2, or K > 2. If any two of these criteria were met, the phenotype was scored severe. This procedure was blinded; i.e., no information concerning the ORF targeted or the effect on other assays was provided. In all cases, a severe phenotype was recorded as red, mild as amber, and no phenotype or no discernible effect as green.
Data validation.
Assays and data quality were validated by several methods. The primary concerns were elimination of systematic and nonsystematic error and bias from the analysis and generation of false-positive and false-negative annotations.
To assess nonsystematic error we resequenced a random 5% of the RNAi plasmid inserts following completion of the whole collection. All of these were correct, suggesting low error rates in plasmid handling and archiving. Observational bias was eliminated by using a blind code, preventing the experimenter from predicting the outcome based on the sequence of the RNAi construct (see above). Further, all data were subjected to rescoring by two additional investigators.
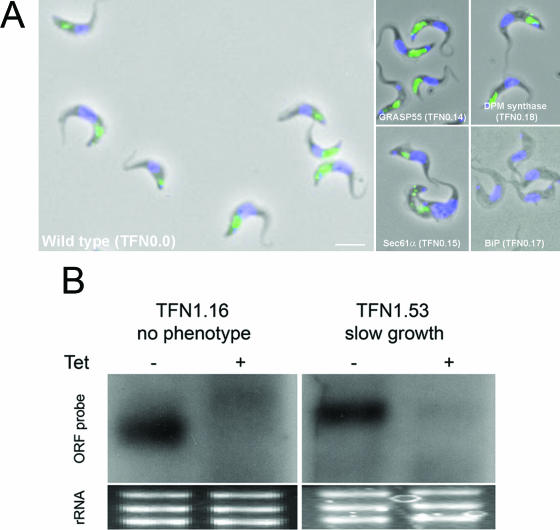
False negatives, i.e., cases where no phenotype is observed but one should be detected, can arise by several mechanisms, including an inappropriate time chosen for analysis, a broken RNAi system (11), mistargeting (27), and inappropriate/insensitive assays; false negatives can also arise because the gene product is stage specific and the ORF is not expressed in the mammalian infective form. First, due to the scale of the project, the time of analysis was selected as optimum for the majority of RNAi knockdowns and was adjusted following growth analysis if appropriate; typically, analysis was performed on day 4. Second, the effect on growth of an RNAi plasmid that contained a fragment of the Aequorea victoria green fluorescent protein (GFP) gene (TFN0. 1) or just the empty p2T7TAblue vector (TFN0.0) was analyzed. Both were predicted to have no impact on the replication time of transgenic trypanosomes, and, indeed, no significant growth defects were obtained. These data also served to standardize the threshold for a significant growth defect. Further, variability between growth curve replicates was typically ≪10%. Third, Northern blot analysis was used to demonstrate that RNAi knockdown had resulted in loss of mRNA for a small subset of RNAi inductions; cell lines exhibited mRNA suppression whether or not there was a resulting phenotype (see Fig. 5). Fourth, batch-to-batch variability in analysis was monitored by the inclusion of the T. brucei clathrin heavy chain (TbCLH) knockdown, a strong positive control (p2T7 · TbCLH) (2) in each cohort of plasmids analyzed; p2T7 · TbCLH always behaved as expected (>20 independent transfections resulted in inducible death in each case, without exception), with significant cell death following 24 h of induction (not shown).
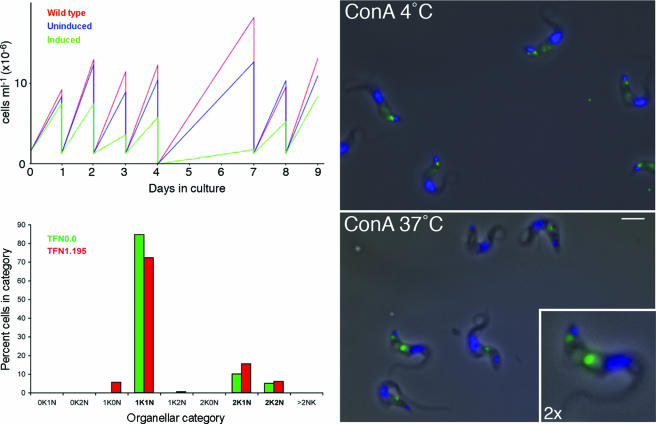
FIG. 5.
Example of phenotypic analysis of a pleomorphic defect. Data for TFN 1.195 (geneDB Tb927.1.1340) are shown (reproduced using data from the trypanoFAN website [http://trypanofan.path.cam.ac.uk]). The protein is weakly predicted to contain an FG-GAP motif (Pfam PF01839; a β-propeller), commonly observed in members of the integrin cell adhesion molecule family. (Left) Growth curves for wild-type (parental strain; red), uninduced p2T7 · TFN1.195 (blue), and induced p2T7 · TFN1.195 (green) cells are shown for a period of 9 days (top graph). An analysis of nucleus and kinetoplast copy number in TFN0.0 (empty vector; green) and TFN1.195 (red) is shown in the bottom graph. Categories that are part of the normal cell cycle progression are in bold. (Right) ConA uptake at 4°C and 37°C. ConA fluorescence is shown in green; DAPI (DNA) stain is shown in blue merged with the corresponding phase-contrast image. Scale bar, 5 μm. Inset shows a digital ×2 magnification of a cell at 37°C to illustrate the phase-light vacuole.
The level of false positives is less easy to quantitate, and while potentially arising from a number of sources, the principal mechanism is likely to be mistargeting of the RNAi construct, affecting expression of loci at other positions within the genome (27). Such an outcome, which is also low in frequency, requires considerable detailed additional work to characterize which could not be applied systematically to the set of transformants generated here. Given the comparatively high fidelity of homologous recombination in trypanosomes, we considered this to be a minor concern, and it was not addressed experimentally. Off-target effects due to RNAi we consider to be of very low frequency for the reasons discussed above.
To probe vital systems and cell cycle progression in trypanosomes well-characterized and simple assays were chosen; this selection also helped to remove some nonsystematic errors from the study. Specifically, use of antibodies and GFP chimeras was avoided due to a finite reagent supply (generating potential variability) or a lack of validation in the system at the time. Lethality was measured by growth analysis in liquid culture, together with analysis of the cell cycle, by virtue of DAPI staining. It should be noted that the threshold for detection of a phenotype by DAPI is significantly lower than for the growth analysis. For example, if 10% of cells arrest at cytokinesis, this would result in a highly significant increase in cells having two kinetoplasts and two nuclei, but such an effect may only result in a 10% decrease in growth, which is at the limit of reliable detection. Further, in many instances, cell cycle blocks are incomplete, such that while cells may be delayed in completing mitosis, the vast majority still manage to complete cell division; such a phenotype will also manifest as a strong DAPI phenotype with a weak growth defect.
Data archiving and dissemination.
Data were loaded into a MySQL (version 4.0.16) relational database using a custom PERL script. Web pages were designed and generated with Web Objects (Apple Computer Inc.) to receive image data, and to process and display graphically the growth and DAPI data, graphs were dynamically created from the data contained within the mySQL database and served from a Mac OSX (version 10.2.8) server. A Boolean search engine is also incorporated into the website to allow rapid location of specific data, as well as hyperlinks to geneDB at the Sanger Institute.
RESULTS
Statistics of the ORF cohort and data set.
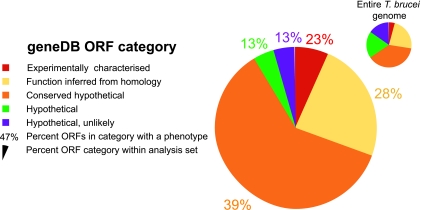
The chromosome I RNAi ORF set represented 197 individual knockdowns (see Material and Methods for details of selection and validation set). The targeted ORFs can be subdivided into five categories depending on their geneDB annotation; these categories are, specifically, 13 experimentally characterized ORFs, 47 ORFs with a potential function inferred from homology with other entries in the nonredundant database, 120 conserved hypothetical ORFs (mainly found in one or more additional kinetoplastid genomes), 8 hypothetical ORFs, and 9 ORFs designated as “unlikely,” on account of a prediction that they encode a protein of less than 150 amino acids (Table 1) (http://www.genedb.org/genedb/glossary.jsp [Status Code Descriptions]). A large proportion (∼73%) of trypanosome ORFs have unknown, poorly predictable, or unpredictable function (Fig. 1, inset).
TABLE 1.
Summary of phenotype data characterized by ORF categorya
| ORF category | Total no. of ORFs analyzed | No. of ORFs in category
|
% of ORFs with detected phenotype | ||||
|---|---|---|---|---|---|---|---|
| Growth defect | DAPI stainingb | Growth and DAPI stainingb | Pleiotropic phenotype | Total no. with any phenotype | |||
| Red (experimentally characterized) | 13 | 3 | 0 | 0 | 0 | 3 | 23 |
| No. with Pfam annotation | 13 | 3 | 23 | ||||
| No. with predicted signal sequence | 2 | 0 | 0 | ||||
| Yellow (function inferred from homology) | 47 | 13 | 3 | 3 | 0 | 13 | 28 |
| No. with Pfam annotation | 47 | 13 | 28 | ||||
| No. with predicted signal sequence | 8 | 1 | 13 | ||||
| Orange (conserved hypothetical) | 120 | 28 | 27 | 8 | 1 | 47 | 39 |
| No. with Pfam annotation | 24 | 9 | 38 | ||||
| No. with predicted signal sequence | 25 | 6 | 24 | ||||
| Green (hypothetical) | 8 | 1 | 0 | 0 | 0 | 1 | 13 |
| No. with Pfam annotation | 2 | 0 | 0 | ||||
| No. with predicted signal sequence | 1 | 0 | 0 | ||||
| Magenta (hypothetical, unlikely) | 9 | 0 | 1 | 0 | 0 | 1 | 13 |
| No. with Pfam annotation | 0 | NA | |||||
| No. with predicted signal sequence | 5 | 1 | 20 | ||||
| Total in all categories | 197 | 45 | 31 | 11 | 1 | 65 | 38 |
| No. with Pfam annotation | 86 | 25 | 29 | ||||
| No. with predicted signal sequence | 41 | 8 | 20 | ||||
RNAi analysis was performed on ORFs from chromosome I by transfection of trypanosomes and scored as described in Materials and Methods. Data are taken from the full data set (see Table S1 in the supplemental material). Annotation categories are as defined at geneDB. Boldface data are values for the whole data set taken as a single category. NA, not available.
Nucleus/kinetoplast copy number was determined by DAPI staining.
FIG. 1.
Summary of the frequency of phenotypes obtained for T. brucei. The main pie chart shows genes that were analyzed as part of this study and that were assigned categories following the annotation at geneDB and color-coded accordingly. The percentage values refer to the fraction of genes in each category that generated any detectable phenotype that could be scored. Data are summarized from Table S1 in the supplemental material. The small pie chart (inset) shows the distribution of predicted genes in each category for the entire T. brucei genome (8).
A high frequency of growth, cell cycle, and viability defects.
The collated data for all knockdowns are available in Table S1 in the supplemental material, in a summary in Table 1, and in the original data at http://trypanofan.path.cam.ac.uk. Overall, 65 knockdowns displayed a significant phenotype (33%). A high frequency of growth and cell cycle defects was obtained; 45 knockdowns (23%) had a growth defect, while 31 (16%) had abnormal cell cycle progression. Eleven knockdowns (6%) displayed a defect in both growth and cell cycle parameters, while one knockdown (0.5%) (TFN1.195) displayed a pleiotrophic defect; i.e., a number of cellular systems appear to be affected. A small number also displayed defects in morphology (cellular and/or organellar) but were almost invariably associated with a growth or cell cycle defect.
Bias in the incidence of knockdown-associated phenotypes was observed when the ORFs were considered by geneDB annotation (Table 1 and Fig. 1). Specifically, the three categories of ORF expected to give rise to functional protein products (experimentally characterized, function inferred from homology, and conserved hypothetical) all produced similar frequencies of phenotype (23%, 28%, and 39%, respectively). Given the sample sizes, there was no bias observed when these categories were further subdivided based on Pfam (protein family database) or signal sequence annotations (Table 1). By contrast, the two ORF categories considered less likely to produce functional gene products (hypothetical and “hypothetical, unlikely”) indeed produced a lower frequency of knockdown-associated phenotypes (both 13%). Of the “hypothetical, unlikely” class of knockdowns (predicted at <150 codons), only one produced a phenotype, but as this was at the limit of detection, the data should be treated with caution (TFN1.202 in Table S1 in the supplemental material). None of the small ORFs analyzed is significantly expressed based on microarray studies (S. Melville, K. Matthews, C. M. R. Turner, A. Tait, and A. Ivens, personal communication).
Detectable phenotypes are not preferentially associated with widely conserved genes.
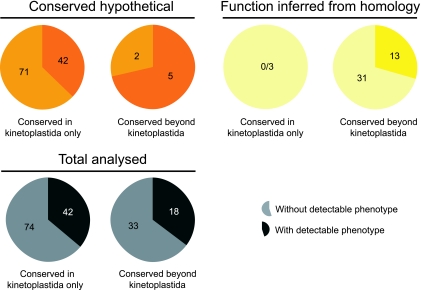
We considered the possibility that phenotypes are more likely to be associated with core functions than with proteins specifically required for specific life cycle stages, especially as our analysis was performed exclusively on the bloodstream form parasite in vitro (14). The ORFs of the former type are more likely to have arisen early in eukaryote evolution and, hence, to be widely distributed among multiple taxa, while those of the latter are more likely to be kinetoplastid or even species specific. We considered the conserved ORF categories (function inferred from homology and conserved hypothetical), which comprise 85% of the data set; no significant bias was present in the phenotype frequency associated with ORFs that are kinetoplastid specific or widely distributed among the eukaryotes (Fig. 2 and see Table S2 in the supplemental material). The presence of a significant number of nonconserved ORFs in the essential cohort indicates that there is a large proportion of kinetoplastid-specific ORFs required for basic functions in in vitro culture. This set of ORFs potentially contains excellent drug targets that could be assessed by in vitro methods as they are, by definition, not found in the host genome.
FIG. 2.
Frequency of phenotypes among genes annotated as conserved hypothetical and with function inferred from homology. Orange pie charts show the “conserved hypothetical” gene class, subdivided into those genes restricted to the kinteoplastida and a smaller category with some level of similarity to genes in nonkinetoplastid organisms. Yellow pies show data subdivided in the same manner but for the “function inferred from homology” gene class. The gray pie charts show merged data comprising both classes. Raw data are available in Table S1 and summarized in Table S2 in the supplemental material. By a chi-square test there is no significant difference between the distributions in the kinetoplastida only or widely conserved cohorts for the function inferred from homology or total analyzed categories, while the significance for these cohorts in the conserved hypothetical category is a P value of >0.07. Given the small sample size for the conserved hypothetical, widely conserved cohort, this is not considered significant.
Large-scale genomic features of functionality.
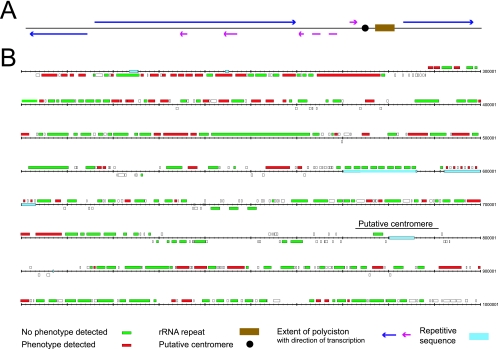
Trypanosome coding sequences are organized into large polycistronic transcriptional units. Such an arrangement has the potential to include biases in the position of particular classes of genes, and some evidence suggests functional clustering may be present (23). There appear to be three large polycistronic transcription units within the core region of chromosome I, one on the bottom strand and two on the top, together with four smaller polycistrons, three of which are on the bottom strand (20) (Fig. 3).
FIG. 3.
Locations of ORFs studied on chromosome I of T. brucei. (A) Overview of organization of the core of chromosome I from T. brucei TREU 927/4. Shown are the positions of both large (dark blue) and small (magenta) polycistronic transcription units and the position of an rRNA gene cluster. Also shown is the putative location of a centromere based on synteny with T. cruzi (29). (B) The ORF map is redrawn from reference 20 with permission of the publisher. The central region of chromosome I of the genome reference strain (T. brucei TREU 927/4) is shown, together with annotation of the ORFs according to phenotype. Green, no phenotype detected; red, phenotype detected; white, ORF not analyzed. Regions of repetitive sequence are highlighted in light blue, and a region that may correspond to a centromere is highlighted. Numbers refer to nucleotide position, counting from the left-hand end of the chromosome. Note that ORF numbers have been removed for clarity, but this figure corresponds exactly to the annotation used by Hall et al. (20).
The large transcription units do not display strong evidence of clustering, strand bias, or positional effects that correlate with a knockdown phenotype. For example, a relationship between a high proportion of phenotype-associated genes and short intergenic distances was examined in the large polycistronic unit on the bottom strand (Fig. 3) (between kbp 200 and 280). Such a relationship was not supported, as determined by comparison of the frequency of phenotype-associated ORFs between the 5′ third and the central or 3′ third of the cistron (P = 0.43 and P = 0.17, respectively, by a chi-square test). There was also no evidence of bias in the incidence of phenotype-associated ORFs between the three large cistrons (data not shown). Hence, phenotype-associated ORFs are not strongly clustered on chromosome I.
Two of the smaller polycistrons located between kbp 730 and 810 fall within a region displaying synteny with a potential centromeric region identified in T. cruzi (29). None of the eight ORFs sampled from these polycistrons had a phenotype, and microarray analysis did not reveal detectable transcription in this region (S. Melville et al., personal communication), suggesting that this region of chromosome I is unlikely to encode protein products. The remaining two small polycistrons were sampled with a single ORF each, and no phenotype was obtained. The first, located around kbp 520, contains only one ORF of >450 nucleotides in length, a 1.4-kbp ESAG2 (where ESAG is expression site-associated gene) gene. The final small polycistron around kbp 645 contains only three significant ORFs (geneDB accession no. Tb927.1.2820-80), all encoding a putative pteridine transporter.
Non-growth phenotypes.
A small proportion of genes are associated with nuclear, kinetoplast, and cytokinesis defects (3.3, 1.4, and 1.9%, respectively). Among seven genes associated with a nuclear defect (0N > 4.5%) were histone H3 (TFN1.218), cyclin 6 (TFN0.8), a (conserved hypothetical) protein with homology to the PRP38 pre-mRNA splicing factor (TFN1.212), and a probable arginine N-methyltransferase (TFN1.25). Topoisomerase II (TFN0.13) knockdown, as expected (35), and two additional conserved hypothetical proteins (TFN1.55 and TFN1.161) generated a kinetoplast defect (0K > 4.5%). BLAST analysis of the four conserved hypothetical genes associated with a block in cytokinesis (2N > 12%) revealed similarity to a histone methyltransferase (TFN1.24), intermediate filament proteins (TFN1.20), and a scaffold-type, growth control protein (TFN1.21). The small number and identity of the genes in these three classes suggest accurate annotation of these nuclear, kinetoplast-associated and cell cycle defects.
Several accessible cellular systems were monitored, specifically the morphology of the mitochondrion, the Golgi apparatus, and the endosomal system, using fluorescent lectin conjugates and validated vital stains (Fig. 4A). The ability of these assays to provide a meaningful readout was evaluated by construction of a set of 12 RNAi knockdown plasmids expected to affect targeted biological subsystems, specifically, glycosylation, Golgi structure, cytoskeletal integrity, protein secretion, and cell cycle progression. The majority of these knockdowns behaved as expected (Fig. 4A; see Tables S1 and S3 in the supplemental material). For example, ConA reacts with glycans present mainly on VSG as components of N-glycans and the core GPI-glycan; staining of cells with ConA at 37°C results in extensive labeling of the entire endocytic apparatus, including the flagellar pocket, endosomes, and the lysosome (Fig. 4A). Knockdown of GRASP55 and dolichoylphosphorylmannose (DPM) synthase, part of the Golgi matrix system and responsible for generation of dolichol-P-mannose, a mannosyl donor for GPI and N-glycosylation, respectively, resulted in comparatively minor alterations to the lectin stain, with a decrease in intensity but, on the other hand, did generate a severe growth defect. The absence of a strong effect on the lectin stain for these gene products is possibly due to incomplete suppression or partial redundancy; for example, in Saccharomyces cerevisiae, knockout of the Golgi reassembly stacking protein (GRASP) homologue is viable, and unless knockdown of DPM synthase approached 100%, even residual activity may facilitate continual glycosylation. For example, the level of the VSG GPI-anchor precursor P2 is approximately 10-fold greater than that required for GPI addition to newly synthesized VSG (25). Sec61α and BiP knockdowns, components of the endoplasmic reticulum (ER) membrane translocon and ER lumenal chaperone system, respectively, resulted in rather stronger effects on lectin staining, with BiP RNAi leading to a complete loss of ConA reactivity. Significantly, both of these gene products are also essential in S. cerevisiae, consistent with the potent phenotypes observed here. Further, we adopted a conservative scoring system for the lectin, BODIPY-ceramide, and lysotracker staining. Some knockdowns are scored as normal for ConA in the data set, for example, N-ethyl maleimide-sensitive factor (TFN1.40); in this particular example, there are some cells with abnormal staining, but the persistence of cells within the population retaining a wild-type stain is scored as normal. This regime increases the likelihood of false negatives but significantly reduces false-positive assignments. Hence, while it is still not possible to ascribe a level of statistical significance to the morphological assays, the data are consistent with a low error rate from nonpenetrance, sample handling, and biases and indicate that the assays are capable of detecting the systems targeted.
FIG. 4.
Validation of lectin assays, morphology, and RNA knockdown. A selected set of genes was analyzed for evaluation of the phenotyping assays (full data available in Tables S1 and S3 in the supplemental material). (A) Verification of four components of the secretory and glycosylation system as analyzed using the ConA uptake assay at 37°C. Trypanosomes harboring p2T7 RNAi constructs directed toward Sec61 (TFN0.15, geneDB Tb11.02.4100), a component of the ER translocase, BiP (TFN0.17, geneDB Tb11.02.5450/5500), the major ER chaperone, GRASP55 (TFN0.14, geneDB 11.02.0280), a Golgin responsible for maintenance of Golgi complex stacked cisterna, DPM synthase (TFN0.18, geneDB Tb10.70.2610), and the empty vector control (TFN0.0) were induced and analyzed by pulsed ConA uptake, followed by fluorescence microscopy as described in Materials and Methods. Green fluorescence is ConA, blue is DNA (DAPI); both channels are shown merged over a phase-contrast image. Scale bar, 5 μm. (B) RNAi knockdown in two cells lines, one with and one without a growth phenotype. Following induction, cells harboring RNAi constructs for TFN1.16 and TFN1.53 (geneDB Tb927.1.4300 and Tb927.1.3250, respectively) were extracted for total RNA, which was then separated on an agarose gel and transferred to nylon membranes. Each construct was probed using a digoxigenin-labeled riboprobe corresponding to the targeted gene. In both cases significant loss of hybridization was observed. Controls for loading (rRNA), visualized with ethidium bromide, are also shown.
One knockdown, TFN1.195 (geneDB accession no. Tb927.1.1340), produced a severe growth defect, perturbation of the cell cycle, and also defective accumulation of ConA. There was also a phase-light vacuole in the posterior region of the cell, likely associated with the endosomal system (Fig. 5). The associated predicted protein product indicates similarity to the Pfam FG-GAP integrin domain and the presence of a β-propeller structure, a common motif for proteins involved in membrane trafficking, for example, clathrin and nuclear pore proteins (15). The absence of an ER membrane-targeting domain or an extensive region of hydrophobicity that could encode a transmembrane domain suggests that the product is cytoplasmic. Given both the phenotype and the predicted organization of the Tb927.1.1340 gene product, it is tempting to speculate that Tb927.1.1340 may encode a novel vesicle coat system; however, additional experimental evidence is required to confirm this assignment.
No evidence for positional effects in RNAi in trypanosomes.
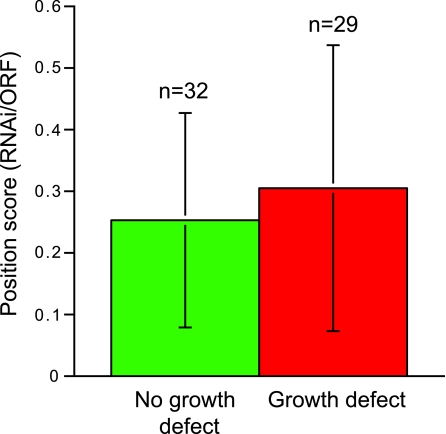
We analyzed a subset of the knockdown collection for evidence of position effects with respect to the RNAi construct sequence relative to the ORF start site. Our analysis of 61 RNAi knockdowns (52% with a growth defect and 48% without) indicated that there was no significant difference in the positions of the RNAi constructs between these two sets (Fig. 6). Hence, we suggest that target selection based on specificity is by far the more important parameter, and position within the ORF is unlikely to have a major influence on phenotype.
FIG. 6.
Position of RNAi target within the ORF has no significant effect on phenotype frequency. A total of 61 ORFs were selected at random from the target set, and the position of the RNAi construct was calculated with reference to the overall ORF size (distance from start codon ATG to 5′ end of RNAi construct/distance between start and stop codon). Data are shown for those with and without a growth defect; values are the means ± standard deviations.
DISCUSSION
Here, we report the first systematic study of gene function in a parasitic organism of major public health importance. Use of RNAi for genome-wide functional annotation has been demonstrated in yeast (14), nematode (24), insect (10), and human systems (38) and is now the method of choice for similar analyses in T. brucei. We chose the bloodstream form trypanosome as the life stage that is responsible for disease in mammals and that displays a range of metabolic, cellular, and genetic characteristics that are relevant to understanding and controlling infectivity and morbidity. A suite of protocols was established for rapid and systematic phenotypic analysis and applied to a large cohort of knockdowns.
The major findings of the study are as follows. First, approximately 12% of knockdowns were lethal, with a further ∼11% exhibiting impaired growth. Extrapolation to the whole genome suggests that at least 1,500 ORFs are needed for robust growth in vitro; this number is likely an underestimate for a combination of reasons; specifically, the data cover only a single life stage, and we are aware of a number of probable false negatives within the data set. Second, there is no higher-order genome organization associated with the presence of a knockdown phenotype, specifically, clustering or strand bias within the polycistronic transcription units. Third, there is no evidence to support an increased frequency of knockdown phenotypes associated with widely conserved genes.
As analysis was performed exclusively in one life cycle stage in in vitro culture and within a limited time frame, a significant number of apparent nonessential genes may, in fact, be essential under distinct conditions. Hence, the number of ORFs required to complete the life cycle is likely to be substantially greater than predicted from the present data set. Gene products required solely for completion of the many insect stage portions of the life cycle or needed specifically for survival in the mammalian host, e.g., those required for immune evasion, are likely to have been missed by this study. In addition, many of the experimentally characterized gene products are highly abundant proteins, which may be difficult to efficiently suppress by RNAi; however, a recent study indicates that VSG, which represents ∼10% of total mRNA, is efficiently targeted by RNAi (33). Protein products with enzymatic activity may be more difficult to completely silence, however.
A total of 180 of the knockdowns targeted ORFs that, based on in silico analysis (e.g., presence of orthologues in other organisms) and/or prior experimental study, are likely to be expressed. Of this set, 33% displayed growth or cell cycle phenotypes, with 23% exhibiting growth defects alone; 12% scored as severe, and 11% were considered mild. This is similar to findings in S. cerevisiae (19) and Schizosaccharomyces pombe (14), where approximately 18% of genes appear to be essential for growth in rich medium, with a further 15% associated with a growth defect. These latter studies concluded that essential genes are generally conserved among eukaryotes, which would support a model whereby core metabolic processes are essential and more specialized lineage-specific functions are less so, at least in in vitro cultures in rich medium (14, 19). However, this does not appear to hold true when analysis is extended to trypanosomatids, where no statistical difference was seen in the frequency of essential genes in the widely conserved and kinetoplastid-specific cohorts. Over 25% of T. brucei annotated ORFs are hypothetical and lack homology with any other genomes, and around half of these are small (<150 amino acids). These ORFs are significantly underrepresented among phenotype-associated knockdowns, although we note that some small ORFs are indeed functional (21, 22). Hence, the majority of these ORFs are either not expressed, encode dispensable T. brucei specific functions, or do not represent true genes.
The lack of a relatively increased frequency of essential genes within the widely conserved ORF cohort may reflect technical differences between the various studies (14, 19). However, an alternative explanation is that the evolutionary distance between the organisms is substantially greater here than in the earlier work, which was restricted to taxa within the Opisthokonta, i.e., metazoa and fungi, rather than between members of the Excavata and Opisthokonta. Thus, the high degree of divergence between the Opisthokonta and trypanosomes may explain the failure to identify homologues and the apparent lack of evolutionary conservation of the trypanosome essential gene cohort. It is unlikely that trypanosomatids contain a radically different core metabolome, despite a number of significant unique features, and therefore it is a possibility that the apparent emphasis on conserved genes being essential is a reflection of a relatively close evolutionary relationship. More extensive analysis of the trypanosome genome and also documentation of essential genes in additional divergent taxa will be required to determine which of these possibilities applies. The majority of T. brucei genes with associated growth defects (62%) are hypothetical proteins conserved among trypanosomatids but with no homology with any other sequences in the public databases. This finding highlights the evolutionary divergence of these organisms, and, since there is little information available from model organisms for most T. brucei genes, also underscores the importance of functional annotation of the T. brucei genome for full comprehension of eukaryote biology.
The dense coverage of chromosome I allowed examination of clustering or strand bias associated with function. Since no general association was identified, the forces maintaining the remarkable degree of synteny among trypanosomatids, i.e., the conserved order of genes along the chromosome, remain elusive (17). However, two of the smaller polycistrons fall within a region located between kbp 730 and 810; all eight ORFs sampled in this region gave no phenotype. This region is syntenic with a putative centromere recently described for T. cruzi (29), suggesting that the centromeric region of chromosome I of T. brucei resides here. As the analysis is limited to chromosome I, any potential functional clustering within the other megabase chromosomes of T. brucei is not addressed by the present study and must await further work.
RNAi is a powerful method for functional study of the T. brucei genome. Quantitative growth analysis has the advantage of permitting detection of subtle phenotypes or those associated with partially redundant functions. The strongest indicator of function is a growth or cell cycle phenotype, indicating that these parameters alone could be used for rapid screening. The data also imply that any defect in the biological systems that could be scored and analyzed here is likely of sufficient severity to have a negative impact on growth also. The small number of phenotypes obtained beyond the growth and cell cycle category opened the possibility that the assays are insufficiently sensitive to detect minor defects, while the validation set clearly indicates that the assays are capable of detecting significant defects. The observation that essentially all organellar system phenotypes were indeed associated with a growth/cell cycle phenotype suggests that significant perturbation of the structure and/or function of organelles is not well tolerated by the trypanosome.
Three RNAi knockdowns produced very mild morphological abnormalities that were not associated with a growth defect. TFN1.132 (β-COP) exhibited cells that were wider latitudinally across the center of the cell than the controls. TFN1.194 (phosphatidylinositol kinase) cells appear to contain phase-dense granules or vacuoles and TFN1.117 (developmentally regulated phosphoprotein [39]) exhibited phase-light vacuoles in the posterior of the cell. In each case the morphology was detected mainly in cells fixed for lectin stains, and due to the subtlety of the defect, these knockdowns are scored as normal in our data set. Additional studies are required to validate these defects; but while suggesting that additional phenotypes are present, an experienced investigator is essential to detect these abnormalities, and hence such phenotypes are unlikely to be easily observable in high throughput. We also observe in a small minority of cases that a phenotype was not observed where a potent effect was expected (28), perhaps most surprisingly for α-tubulin. Retesting of the α-tubulin and RPN6 RNAi constructs in a double-inducible cell line, with tighter control over expression of the double-stranded RNAi (3), did result in the expected phenotypes, suggesting that the inability to detect a phenotype is likely a result of lethality due to leakiness in the SMB cell background. While this indicates that a number of phenotypes may have been lost, the ability to reproduce the clathrin RNAi phenotype suggests that, in the main, leakiness of the p2T7 system is not a major concern.
Many otherwise-uncharacterized trypanosome genes, where informatics is not informative, are now known to be required for growth in culture. A significant new challenge is posed by these data; despite knowledge that the ORF encodes a protein required for growth, the absence of informatics data provides no clues for subsequent analysis. Individual functional analysis of genes lacking annotation is a slow and expensive process and clearly is not scalable to whole genomes. In addition to a need for more sensitive phenotype detection, the location of a gene product is a highly informative piece of data that can aid in the design of more detailed experimental approaches, and rapid methods for achieving this are clearly needed.
This report represents a robust validation of RNAi as an approach for systematic functional annotation of the T. brucei genome and provides experimental confirmation of many in silico predicted gene models. Extensive gene conservation among the trypanosomatids means that the present study also represents an advance in annotation of the T. cruzi and Leishmania genomes. The challenge is now to extend analysis to the remainder of the trypanosome genome and to establish more powerful tools and methodologies for in-depth phenotype detection and/or selection. For example, barcode RNAi selective screening, pioneered for use in mammalian cells (38) and tested in T. brucei (4), could facilitate analysis of a large numbers of ORFs, while creation of live-cell reporter strains is likely to improve throughput and sensitivity; this aspect is particularly critical as the availability of live-cell organellar markers would facilitate multiple interrogation of morphology of subcellular compartments without the use of staining protocols which are inherently variable and which likely underpin much of the low frequency of such phenotypes detected in the present work. Further functional screening will greatly enhance both our understanding of the basic biology of the trypanosomatids and the identification of new drug targets.
Supplementary Material
Acknowledgments
We are indebted to many in the trypanosome community for advice and support during this project and in particular to Dave Barry (Glasgow), Christine Clayton (Heidelberg), and Mike Turner (Glasgow) during planning phases and to Michael Boshart (Munich), Christiana Hertz-Fowler (Hinxton), Paul McKean (Lancaster), Sara Melville (Cambridge), and Derek Nolan (Dublin) for discussions and/or sharing of data. We also thank Margaret MacKinnon and Angela Pinot de Moira (Cambridge) for assistance with statistical analysis.
T. brucei sequence data were obtained from The Sanger Institute at www.sanger.ac.uk/Projects/T_brucei/. Sequencing of the T. brucei genome was accomplished as part of the Trypanosome Genome Network.
This work was funded by The Wellcome Trust (grant 064563 to M.C.F., K.G., K.M., and D.H.).
REFERENCES
- 1.Alibu, V. P., L. Storm, S. Haile, C. Clayton, and D. Horn. 2005. A doubly inducible system for RNA interference and rapid RNAi plasmid construction in Trypanosoma brucei. Mol. Biochem. Parasitol. 139:75-82. [DOI] [PubMed] [Google Scholar]
- 2.Allen, C. L., D. Goulding, and M. C. Field. 2003. Clathrin-mediated endocytosis is essential in Trypanosoma brucei. EMBO J. 22:4991-5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alsford, S., T. Kawahara, L. Glover, and D. Horn. 2005. Tagging a T. brucei RRNA locus improves stable transfection efficiency and circumvents inducible expression position effects. Mol. Biochem. Parasitol. 144:142-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alsford, S., L. Glover, and D. Horn. 2005. Multiplex analysis of RNA interference defects in Trypanosoma brucei. Mol. Biochem. Parasitol. 139:129-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Atrih, A., J. M. Richardson, A. R. Prescott, and M. A. Ferguson. 2005. Trypanosoma brucei glycoproteins contain novel giant poly-N-acetyllactosamine carbohydrate chains. J. Biol. Chem. 280:865-871. [DOI] [PubMed] [Google Scholar]
- 6.Barrett, M. P., R. J. Burchmore, A. Stich, J. O. Lazzari, A. C. Frasch, et al. 2003. The trypanosomiases. Lancet 362:1469-1480. [DOI] [PubMed] [Google Scholar]
- 7.Barry, J. D., and R. McCulloch. 2001. Antigenic variation in trypanosomes: enhanced phenotypic variation in a eukaryotic parasite. Adv. Parasitol. 49:1-70. [DOI] [PubMed] [Google Scholar]
- 8.Berriman, M., E. Ghedin, C. Hertz-Fowler, G. Blandin, H. Renauld, et al. 2005. The genome of the African trypanosome Trypanosoma brucei. Science 309:416-422. [DOI] [PubMed] [Google Scholar]
- 9.Birmingham, A., E. M. Anderson, A. Reynolds, D. Ilsley-Tyree, D. Leake, Y. Fedorov, S. Baskerville, E. Maksimova, K. Robinson, J. Karpilow, W. S. Marshall, and A. Khvorova. 2006. 3′ UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat. Methods 3:199-204. [DOI] [PubMed] [Google Scholar]
- 10.Boutros, M., A. A. Kiger, S. Armknecht, K. Kerr, M. Hild, B. Koch, S. A. Haas, H. F. Consortium, R. Paro, and N. Perrimon. 2004. Genome-wide RNAi analysis of growth and viability in Drosophila cells. Science 303:832-835. [DOI] [PubMed] [Google Scholar]
- 11.Chen, Y., C.-H. Hung, T. Burderer, and G.-S. M. Lee. 2003. Development of RNA interference revertants in Trypanosoma brucei cell lines generated with a double-stranded RNA expression construct driven by two opposing promoters. Mol. Biochem. Parasitol. 126:275-279. [DOI] [PubMed] [Google Scholar]
- 12.Clayton, C. E., A. M. Esteacutevez, C. Hartmann, V. P. Alibu, M. C. Field, et al. 2005. Down-regulating gene expression by RNA interference in Trypanosoma brucei. Methods Mol. Biol. 309:39-60. [DOI] [PubMed] [Google Scholar]
- 13.DaRocha, W. D., K. Otsu, S. M. Teixeira, and J. E. Donelson. 2004.. Tests of cytoplasmic RNA interference (RNAi) and construction of a tetracycline-inducible T7 promoter system in Trypanosoma cruzi. Mol. Biochem. Parasitol. 133:175-186. [DOI] [PubMed] [Google Scholar]
- 14.Decottignies, A., I. Sanchez-Perez, and P. Nurse. 2003. Schizosaccharomyces pombe essential genes: a pilot study. Genome Res. 13:399-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Devos, D., S. Dokudovskaya, F. Alber, R. Williams, B. T. Chait, A. Sali, and M. P. Rout. 2004. Components of coated vesicles and nuclear pore complexes share a common molecular architecture. PLoS Biol. 2:e380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.El-Sayed, N. M., P. J. Myler, D. C. Bartholomeu, D. Nilsson, G. Aggarwal, et al. 2005. The genome sequence of Trypanosoma cruzi, etiologic agent of Chagas disease. Science 309:409-415. [DOI] [PubMed] [Google Scholar]
- 17.El-Sayed, N. M., P. J. Myler, G. Blandin, M. Berriman, J. Crabtree, et al. 2005. Comparative genomics of trypanosomatid parasitic protozoa. Science 309:404-409. [DOI] [PubMed] [Google Scholar]
- 18.Field, M. C., C. L. Allen, V. Dhir, D. Goulding, B. S. Hall, G. W. Morgan, P. Veazey, and M. Engstler. 2004. New approaches to the microscopic imaging of Trypanosoma brucei. Microsc. Microanal. 10:621-636. [DOI] [PubMed] [Google Scholar]
- 19.Giaever, G., A. M. Chu, L. Ni, C. Connelly, L. Riles, S. Veronneau, et al. 2004. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418:387-391. [DOI] [PubMed] [Google Scholar]
- 20.Hall, N., M. Berriman, N. J. Lennard, B. R. Harris, C. Hertz-Fowler, et al. 2003. The DNA sequence of chromosome I of an African trypanosome: gene content, chromosome organisation, recombination and polymorphism. Nucleic Acids Res. 31:4864-4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hendriks, E. F., D. R. Robinson, M. Hinkins, and K. R. Matthews. 2001. A novel CCCH protein which modulates differentiation of Trypanosoma brucei to its procyclic form. EMBO J. 20:6700-6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hendriks, E. F., and K. R. Matthews. 2005. Disruption of the developmental programme of Trypanosoma brucei by genetic ablation of TbZFP1, a differentiation-enriched CCCH protein. Mol. Microbiol. 57:706-716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ivens, A. C., C. S. Peacock, E. A. Worthey, L. Murphy, G. Aggarwal, et al. 2005. The genome of the kinetoplastid parasite, Leishmania major. Science 309:436-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kamath, R. S., A. G. Fraser, Y. Dong, G. Poulin, R. Durbin, M. Gotta, A. Kanapin, N. Le Bot, S. Moreno, M. Sohrmann, D. P. Welchman, P. Zipperlen, and J. Ahringer. 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421:231-237. [DOI] [PubMed] [Google Scholar]
- 25.Mayor, S., A. K. Menon, and G. A. Cross. 1991. Transfer of glycosyl-phosphatidylinositol membrane anchors to polypeptide acceptors in a cell-free system. J. Cell Biol. 114:61-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Medina-Acosta, E., and G. A. Cross. 1993. Rapid isolation of DNA from trypanosomatid protozoa using a simple “mini-prep” procedure. Mol. Biochem. Parasitol. 59:327-329. [DOI] [PubMed] [Google Scholar]
- 27.Motyka, S. A., Z. Zhao, K. Gull, and P. T. Englund. 2004. Integration of pZJM library plasmids into unexpected locations in the Trypanosoma brucei genome. Mol. Biochem. Parasitol. 134:163-167. [DOI] [PubMed] [Google Scholar]
- 28.Ngo, H., C. Tschudi, K. Gull, and E. Ullu. 1998. Double-stranded RNA induces mRNA degradation in Trypanosoma brucei. Proc. Natl. Acad. Sci. USA 95:14687-14692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Obado, S. O., M. C. Taylor, S. R. Wilkinson, E. V. Bromley, and J. M. Kelly. 2005. Functional mapping of a trypanosome centromere by chromosome fragmentation identifies a 16-kb GC-rich transcriptional “strand-switch” domain as a major feature. Genome Res. 15:36-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Redmond, S., J. Vadivelu, and M. C. Field. 2003. RNAit: an automated web-based tool for the selection of RNAi targets in Trypanosoma brucei. Mol. Biochem. Parasitol. 128:115-118. [DOI] [PubMed] [Google Scholar]
- 31.Robinson, D. R., and K. Gull. 1991. Basal body movements as a mechanism for mitochondrial genome segregation in the trypanosome cell cycle. Nature 352:731-733. [DOI] [PubMed] [Google Scholar]
- 32.Robinson, K. A., and S. M. Beverley. 2003. Improvements in transfection efficiency and tests of RNA interference (RNAi) approaches in the protozoan parasite Leishmania. Mol. Biochem. Parasitol. 128:217-228. [DOI] [PubMed] [Google Scholar]
- 33.Sheader, K., S. Vaughan, J. Minchin, K. Hughes, K. Gull, and G. Rudenko. 2005. Variant surface glycoprotein RNA interference triggers a precytokinesis cell cycle arrest in African trypanosomes. Proc. Natl. Acad. Sci. USA 102:8716-8721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simpson, A. G., and A. J. Roger. 2004. The real “kingdoms” of eukaryotes. Curr. Biol. 14:R693-R696. [DOI] [PubMed] [Google Scholar]
- 35.Timms, M. W., F. J. van Deursen, E. F. Hendriks, and K. R. Matthews. 2002. Mitochondrial development during life cycle differentiation of African trypanosomes: evidence for a kinetoplast-dependent differentiation control point. Mol. Biol. Cell 13:3747-3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ullu, E., C. Tschudi, and T. Chakraborty. 2004. RNA interference in protozoan parasites. Cell. Microbiol. 6:509-519. [DOI] [PubMed] [Google Scholar]
- 37.van Deursen, F. J., S. K. Shahi, C. M. Turner, C. Hartmann, C. Guerra-Giraldez, K. R. Matthews, and C. E. Clayton. 2001. Characterisation of the growth and differentiation in vivo and in vitro of bloodstream-form Trypanosoma brucei strain TREU 927. Mol. Biochem. Parasitol. 112:163-171. [DOI] [PubMed] [Google Scholar]
- 38.Westbrook, T. F., E. S. Martin, M. R. Schlabach, Y. Leng, A. C. Liang, et al. 2005. A genetic screen for candidate tumor suppressors identifies REST. Cell 121:837-848. [DOI] [PubMed] [Google Scholar]
- 39.Wirtz, E., D. Sylvester, and G. C. Hill. 1991. Characterization of a novel developmentally regulated gene from Trypanosoma brucei encoding a potential phosphoprotein. Mol. Biochem. Parasitol. 47:119-128. [DOI] [PubMed] [Google Scholar]
- 40.Wirtz, E., M. Hoek, and G. A. Cross. 1998. Regulated processive transcription of chromatin by T7 RNA polymerase in Trypanosoma brucei. Nucleic Acids Res. 26:4626-4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.