Abstract
Made of more than 40 subunits, the rotenone-sensitive NADH:ubiquinone oxidoreductase (complex I) is the most intricate membrane-bound enzyme of the mitochondrial respiratory chain. In vascular plants, fungi, and animals, at least seven complex I subunits (ND1, -2, -3, -4, -4L, -5, and -6; ND is NADH dehydrogenase) are coded by mitochondrial genes. The role of these highly hydrophobic subunits in the enzyme activity and assembly is still poorly understood. In the unicellular green alga Chlamydomonas reinhardtii, the ND3 and ND4L subunits are encoded in the nuclear genome, and we show here that the corresponding genes, called NUO3 and NUO11, respectively, display features that facilitate their expression and allow the proper import of the corresponding proteins into mitochondria. In particular, both polypeptides show lower hydrophobicity compared to their mitochondrion-encoded counterparts. The expression of the NUO3 and NUO11 genes has been suppressed by RNA interference. We demonstrate that the absence of ND3 or ND4L polypeptides prevents the assembly of the 950-kDa whole complex I and suppresses the enzyme activity. The putative role of hydrophobic ND subunits is discussed in relation to the structure of the complex I enzyme. A model for the assembly pathway of the Chlamydomonas enzyme is proposed.
Complex I (NADH:ubiquinone oxidoreductase; EC 1.6.5.3) is the largest and the most complicated enzyme of the mitochondrial respiratory chain. It is a membrane-bound assembly of approximately 1,000 kDa composed of 42 subunits in the land plants Arabidopsis thaliana and Oryza sativa (36), 42 or 43 subunits in the green alga Chlamydomonas reinhardtii (7, 8), 46 subunits in the mammal Bos taurus (9, 38), and 39 subunits in the fungus Neurospora crassa (52). The complex I enzyme shows a general L shape with a peripheral arm protruding in the matrix and another arm embedded in the inner membrane of the mitochondrion (18, 21, 32). Among the 33 components common to all of these eukaryotes, 14 are homologues of the constituents of the bacterial type I NADH dehydrogenase (ND) and are considered to represent the core of the enzyme (8). Seven subunits of the bacterial enzyme bind the FMN and nine iron-sulfur clusters (37). In contrast, the role of the seven other subunits, which are counterparts of the ND1, -2, -3, -4, -4L, -5, and -6 hydrophobic polypeptides of eukaryotes and are encoded in the mitochondrial DNA (mtDNA) of plants, mammals, and fungi, is still unclear.
In land plants, the ND7 and ND9 polypeptides are also encoded in the organelle genome. In maize, the NCS2 mitochondrial mutation is a deletion of the 3′ end of the nd4 gene, which creates a fused nd4-nd7 chimeric gene and prevents the assembly of the whole complex (43, 51). NCS2 mutant plants, which are maintained at the heteroplasmic state (a mixture of normal and mutated mitochondrial genomes), display poor growth, pale striping of leaves, and decreased yield of kernels. Cytoplasmic male-sterile mutants I and II of Nicotiana sylvestris, with a deletion of the nd7 mitochondrial gene, show reduced respiration and breakdown of complex I activity and assembly (34, 63). Mutations affecting mitochondrion-encoded subunits are also known to be responsible for many hereditary diseases in humans (12, 74). However, due to the heteroplasmic state of the cells that contain both normal and mutant mitochondria and to the presence of allelic variants, it is often difficult to establish the real impact of these mutations on complex I assembly and activity in patients. In this respect, the unicellular green alga C. reinhardtii constitutes an invaluable tool to study the role of the ND subunits since homoplasmic mutations that affect mitochondrial genes encoding respiratory chain components are not lethal in this organism (53, 69). Moreover, genetic transformation allowing the insertion of point mutations into these genes has been very recently achieved (70). Previously, characterization of mutations altering the nd1, nd4, and nd6 mitochondrial genes allowed us to determine the impact of the loss of the corresponding subunits on the activity and assembly of the enzyme (6). The mtDNA of Chlamydomonadaceae algae has the particularity that it codes for only five complex I subunits (ND1, -2, -4, -5, and -6) and lacks the genes that encode the ND3 and ND4L subunits (4, 57). This observation led to the suggestion that the corresponding genes have been transferred to the nucleus (60). Thanks to the data of the Chlamydomonas nuclear genome sequencing (33) (data available at the Joint Genome Institute website, http://genome.jgi-psf.org/Chlre3/Chlre3.home.html), we have identified the NUO3 and NUO11 nuclear genes as homologs of the ND3 and ND4L coding sequences, respectively (7, 8).
In this work, we investigate the modifications that have occurred following the transfer of these two genes to the nucleus, which allows their proper expression, and the sorting and import of their polypeptidic products to the mitochondria. Moreover, to analyze the role of ND3 and ND4L subunits, we here describe the suppression of NUO3 and NUO11 gene expression and determine the impact of the loss of the corresponding subunits on the activity and assembly of complex I and other respiratory chain complexes.
MATERIALS AND METHODS
Strains and culture conditions.
The C. reinhardtii strain used in this study is the cw15 arg7-8 mt+ mutant. This strain is deficient for the cell wall and is auxotroph for arginine because of a mutation in the ARG7 gene coding for argininosuccinate lyase (16). Cells were grown in liquid or on solid agar medium under continuous illumination (75 μE m−2 s−1) at 25°C. The routinely used medium was Tris-acetate-phosphate medium (30) in which 7.5 mM NH4Cl is replaced by 4 mM NaNO3, and supplemented with 100 μg of l-arginine per ml when necessary.
Escherichia coli DH5α was used for cloning gene and cDNA sequences, and E. coli transformants were grown in Luria medium in the presence of ampicillin (50 μg/ml) at 37°C.
Construction of plasmids for dsRNA expression.
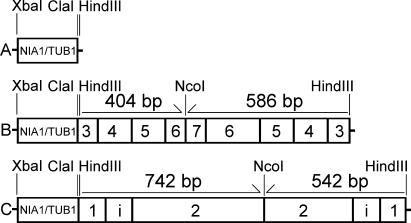
The pNB1 plasmid (2,895 bp) used to express double-stranded RNA (dsRNA) contains a 229-bp chimeric strong promoter comprising 84 bp of the C. reinhardtii nitrate reductase (NIA1 gene) promoter and 98 bp of the C. reinhardtii β-tubulin (TUB1 gene) minimal promoter. This NIA1/TUB1 promoter is inserted in the XbaI and HindIII sites of the pUC19 vector (50) (Fig. 1A).
FIG. 1.
Schematic representation of dsRNAi constructs. XbaI/HindIII fragments of pNB1 (A), pND3-RNAi (B), and pND4L-RNAi (C). NIA1/TUB1, 229-bp chimeric promoter. Numbers within rectangles correspond to the selected exons of the NUO3 and NUO11 genes (i, intron).
Construction of plasmid pND3-RNAi (3,849 bp).
Two fragments of the NUO3 cDNA (GenBank accession number AY351265) (586 and 404 bp) were amplified by PCR using as forward primer ND3-3F (5′-ATCGATAAGCTTCAGCAGTACGTGCGCGAGCA-3′) and as reverse primers ND3-2R (5′-AAGCTTCCATGGCGTGCGTCATGGCGTAGGGG-3′) or ND3-5R (5′-AAGCTTCCATGGACAATGAAGACGGTGGAGATGGC-3′). The oligonucleotides contain ClaI, HindIII, or NcoI restriction sites at their 5′ ends for further constructions. These PCR fragments were cloned into pGEM-T Easy Vector (Promega) to obtain pND3-1 (ND3-1F/ND3-2R) and pND3-2 (ND3-1F/ND3-5R), respectively. The excised HindIII fragment of pND3-1 was inserted into the pNB1 plasmid, and the construct with inverse orientation of ND3-1F/ND3-2R fragment was selected by a PCR analysis to obtain pND3-AS. The ClaI-NcoI fragment of pND3-2 was then inserted into the ClaI-NcoI sites of pND3-AS, giving the plasmid pND3-RNAi (where RNAi is RNA interference) to be used for RNA inactivation of NUO3 (Fig. 1B).
Construction of plasmid pND4L-RNAi (4,190 bp).
Two NUO11 gene fragments containing a 90-bp intron (541 bp and 742 bp) (GenBank accession number AY216718) were amplified by PCR with the primer ND4L-1F (5′-ATCGATAAGCTTTAGAGTCACAAGAATGTCGCGGA-3′) in association with ND4L-3R (5′-AAGCTTCCATGGGATACCGGCCCAGAACATCATGT-3′) and ND4L-2R (5′-AAGCTTCCATGGGTGGAAGTACGCCACGCACAG-3′), respectively. The cloning strategy into pNB1 was the same as for the pND3-RNAi construct. It gave rise to the pND4L-RNAi plasmid to be used for RNA inactivation of NUO11 (Fig. 1C).
Transformation of C. reinhardtii and selection of RNAi clones.
Transformation of the Chlamydomonas cw15 arg7-8 mt+ strain was carried out using the glass bead method (46) with 5 μg of plasmid pRNAi (linearized with ScaI for pND3-RNAi or SspI for pND4L-RNAi) and 1 μg of pASL, linearized with BamHI. This pASL plasmid bears the Chlamydomonas ARG7 gene that codes for the argininosuccinate lyase (16) and is used as a selectable marker. Prototroph transformants were selected on Tris-acetate-phosphate agar plates. The presence of sequences belonging to the right and to the left part of the RNAi plasmids in the transformants was checked by PCR with primers hybridizing in the NUO3 or NUO11 sequences and in the vector (universal primers 5′-GTAAAACGACGGCCAG-3′ and 5′-CAGGAAACAGCTATGAC-3′) on a DNA extract prepared as described elsewhere (58). The stability of the phenotype observed for the transformants mentioned in this study was checked 1 year after their isolation.
RNA analyses.
Total RNA was extracted from 5 × 108 cells as previously described (49). For RNA blot analyses, RNA (15 μg) was separated on 0.8% agarose-formaldehyde gels and transferred onto Hybond N+ membrane (Amersham Pharmacia Biotech). Digoxigenin-labeled PCR products of cDNA fragments were used as gene probes and detected with anti-digoxigenin-AP conjugates and CDP-Star as substrate (Roche). ND4L-1F and ND4L-3R primers were used to synthesize the probe for detection of NUO11 transcripts; ND3-3F and ND3-2R primers were used for NUO3; ND9-3F (5′-CAGGAGCCCACGATATACACCACG-3′) and ND9-2R (5′-GCGGTGGTTGAACACCTTGCAGA-3′) were used for NUO9 (GenBank accession number AY351261); ND7-1F (5′-GAACAACTTCACGCTGAACTTCGG-3′) and ND7-9R (5′-TGAACTGCATCTTGCCGTACGC-3′) were used for NUO7 (GenBank accession number AY347483); ESSS-3F (5′-CTCAGACAGGCGGAGGCA-3′) and ESSS-6R (5′-TGCGCAACGAGTACAAGG-5′) were used for NUO17 (GenBank accession number AY538676); NUOP1-1F (5′-ATGGGCTGGGCGTACTCTG-3′) and NUOP1-3R (5′-CTCCCCAGTGCACATTACCTC-3′) were used for NUOP1 (GenBank accession number AY549572).
Protein complex analyses.
The starting material was purified mitochondria (6) or crude membrane fractions (68). The protein content was determined by the Bradford method (2).
Activity of complexes I, IV, and NADH:ferricyanide oxidoreductase was measured as previously described (6, 68).
To conduct blue native polyacrylamide gel electrophoresis (BN-PAGE) analyses, protein complexes were first solubilized in the presence of 2.5% (wt/vol) dodecylmaltoside, 375 mM 6-aminohexanoic acid, 250 mM EDTA, and 25 mM Bis-Tris, pH 7.0, and centrifuged for 20 min at 15,000 × g at 4°C to remove insoluble matters. One percent (wt/vol) sodium taurodeoxycholate was then added to the supernatant prior to separation by electrophoresis on a 4 to 12% acrylamide gradient BN gel (73). NADH/nitroblue tetrazolium (NBT) staining and Coomassie blue staining of the gels were performed as previously published (7).
BN gels were electroblotted (20) according to standard protocols. The following antisera raised in rabbits were used: polyclonal antiserum against whole purified complex I from N. crassa (provided by H. Weiss and U. Schulte) or monoclonal antiserum against the PSST subunit from Yarrowia lipolytica (provided by V. Zickermann and U. Brandt). Detection was performed using a BM Chemiluminescence Western blotting kit (Roche) with anti-rabbit peroxidase-conjugated antibodies.
In silico analyses.
Frequency of codon utilization was compared with data available for C. reinhardtii nuclear genes (http://www.kazusa.or.jp/codon/). Alpha-helix predictions were computed with the Deleage-Roux and Levitt scales with the Protscale program on the Expasy server (http://us.expasy.org/). The positions of residues along alpha helixes (3.6 residues per turn) were determined with the Pepwheel program (http://emboss.sourceforge.net/apps/pepwheel.html). Hydropathy profiles were determined by using the scale of Kyte and Doolittle with a window of seven residues (48). Local hydrophobicity (<H>) and average regional hydrophobicity (termed mesohydrophobicity) were calculated using scanning windows of 17 and 60 to 80 residues, respectively, with the MITOPROT, version 2.0, program (http://ihg.gsf.de/ihg/mitoprot.html) (11).
RESULTS AND DISCUSSION
NUO3 and NUO11 genes possess characteristics of Chlamydomonas nuclear genes, and their polypeptide products are adapted for sorting and import into mitochondria.
The endosymbiotic event that gave rise to mitochondria was followed by a massive migration of genes to the nucleus (31). Like other highly hydrophobic components of complex I, ND3 and ND4L subunits from vascular plants, animals, and fungi stay encoded in the mitochondrial genome. In contrast, the nd3 and nd4l genes are missing from the mitochondrial genome of C. reinhardtii, Chlamydomonas eugametos, and Polytomella parva, three members of the “reinhardtii” clade (65). In the course of proteomic analysis of the complex I components of C. reinhardtii, we have identified the homolog of the ND3 subunit, whereas other small or hydrophobic polypeptides, including the ND4L subunit, escaped detection using our experimental approach (7). We have also identified the NUO3 and NUO11 nuclear genes of C. reinhardtii as homologs of the ND3 and ND4L coding sequences, respectively (7, 8). However, no bioinformatic or expression analysis of these two genes has ever been undertaken.
To be efficiently expressed, mitochondrial genes that have been transferred to the nucleus have to acquire several distinct traits typical of nuclear genes and absent in mitochondrial genes (3, 29). Moreover, this transfer of genes from the mitochondrial genome to the nucleus also raises the question of the import of their hydrophobic polypeptidic products. We thus investigated the expression of the NUO3 and NUO11 nuclear genes and the presence of modifications in their sequences.
(i) Gene expression.
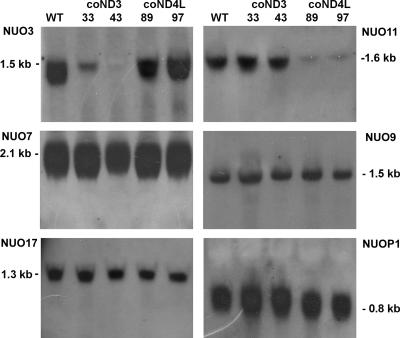
To analyze NUO3 and NUO11 gene expression, RNA blots from wild-type cells were hybridized with cDNA probes. Strong single signals were detected at 1.5 and 1.6 kb, respectively (Fig. 2, WT lanes in top blots), indicating that both genes are highly expressed. The sizes of the transcripts were in good agreement with the sizes predicted from the GenBank database (1,585 bp for NUO3, accession number AY351265; 1,616 bp for NUO11, accession number AY216718). Several methionine codons were found in the 5′ ends of the NUO3 and NUO11 cDNA sequences. For each sequence, the first ATG was chosen as the putative initiation codon because its neighborhood was the closest to the (A/C)A(A/C)(A/C)ATG(G/C)C(C/G) consensus sequence defined for initiation codons of Chlamydomonas nuclear genes (76). The ND3 and ND4L polypeptides should thus be 279 and 227 amino acids long, respectively (GenBank accession numbers AAQ55461 and AAO61142).
FIG. 2.
Transcriptional analysis of several complex I genes in wild-type and coND3 and coND4L mutant strains. Hybridization patterns were obtained with NUO3, NUO7, NUO9, NUO11, NUO17, and NUOP1 probes on RNA blots from wild-type, coND3, and coND4L mutants.
(ii) Changes in codon usage.
Since the codon usage strongly differs between nuclear and mitochondrial genes in Chlamydomonas (76), the pattern of codon utilization in NUO3 and NUO11 genes was determined. Data obtained showed that codon usage in both genes was typical of nuclear genes. For example, we found that among the 25 leucine codons present in the NUO3 coding sequence, 17 are CTG, 5 are CTC, 2 are CTA, and 1 is TTG. The first two codons are preferentially found in nuclear sequences, whereas the latter two are more frequently used in mitochondrial genes. Moreover, the GC content of both genes is about 65% and fits well with that of the nuclear genome including the protein coding sequences (more than 60%) (35). In contrast, the GC content is only 45% for mtDNA (GenBank accession number U03843).
(iii) Acquisition of polyadenylation signal.
The polyadenylation signal of C. reinhardtii nuclear genes (TGTAA) (76) is found at the end of both cDNA sequences.
(iv) Acquisition of introns.
Intronic sequences with orthodox splicing sites (76) were identified; six were found in the NUO3 gene, and one was found in the NUO11 gene (Joint Genome Institute accession numbers 24195 and 58686).
(v) Presence of a putative mitochondrial targeting sequence.
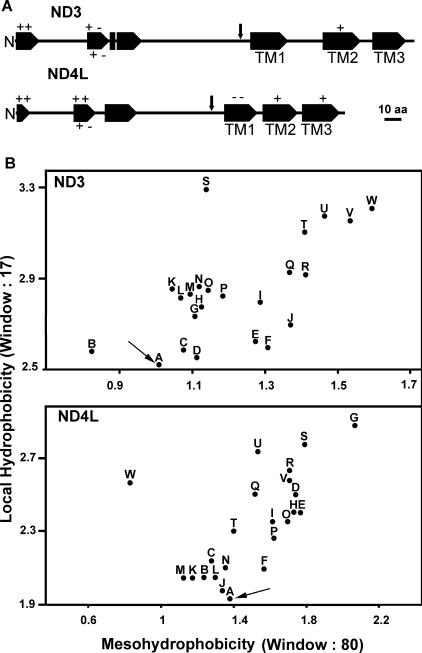
The large majority of nucleus-encoded mitochondrial proteins contain an N-terminal targeting sequence (MTS, or mitochondrial targeting sequence) that drives the import to the mitochondria (28). When the deduced full-length ND3 and ND4L sequences were compared to the polypeptidic sequences of other organisms (prokaryotic and eukaryotic), long N-terminal extensions of about 160 and 130 residues, respectively, were found (Fig. 3A). Such unusually long presequences (>100 amino acids) are also present in ATP6, COX IIA, and COX III proteins from Chlamydomonas (26, 61, 62) and may improve the efficiency of import of nucleus-encoded, highly hydrophobic proteins across mitochondrial membranes (10, 27). Both ND3 and ND4L N-terminal sequences possess the characteristics of an MTS. (a) Their amino acid composition is similar to the composition described for the MTS of land plants (28); they are rich in Arg (7.8%), Leu (10%), Ala (15%), and Ser (10%) and poor in Cys, His, Ile, Trp, and Asp (about 1% each). (b) The N-terminal residues (1 to 17) have the potential to form an amphiphilic α helix with one hydrophilic positively charged face and one apolar face (Fig. 3A). This amphiphilicity seems to be essential for the function of the MTS (23, 28). (c) Putative cleavage sites as defined by Glaser et al. (28) are found at positions that are consistent with the length of their protein homologues encoded by mtDNA (indicated by ↓), i.e., after residue 158 (RTFQ↓FT) for ND3 and after residue 123 (RSY↓YT) for ND4L. In particular, the deduced molecular mass of mature ND3 subunit (13,547 Da) is in good agreement with its identification by mass spectrometry as a 14-kDa band in a sodium dodecyl sulfate gel (7).
FIG. 3.
In silico analyses of C. reinhardtii ND3 and ND4L polypeptides. (A) Predicted alpha helixes are shown as large arrows, and positions of the charged amino acid residues within helixes are indicated. TM, transmembrane helix. Putative cleavage sites of the transit peptide are indicated by vertical arrows. (B) Maximal local hydrophobicity versus mesohydrophobicity plots for ND subunits 3 and 4L from different organisms using the Kyte and Doolittle scale (48). Arrows point to the spots representing the Chlamydomonas polypeptides. The GenBank accession numbers of the ND3 and ND4L sequences, respectively, for the identified points are as follows: A (C. reinhardtii), AAQ55461 and AAO61142; B (Beta vulgaris), BAA99467 and BAA99307; C (Danio rerio), AAF74304 and AAF74305; D (Rattus norvegicus), AAV31046 and AAV31047; E (Reclinomonas americana), AAD11900 and AAD11914; F (Chondrus crispus), CAA87616 and CAA87597; G (Cyanidioschyzon merolae), NP_059358 and BAA36543; H (Prototheca wickerhamii), Q37625 and Q37627; I (Homo sapiens), AAX55498 and AAX55499; J (Xenopus laevis), P03900 and P03904; K (A. thaliana), P92533 and CAA47479; L (O. sativa), BAC19871 and CAC79145; M (Gallus gallus), AAC83281 and BAE16034; N (Boa constrictor), YP_313688 and YP_313689; O (Marchantia polymorpha), P26847 and P26851; P (Equus asinus), P92482 and P92483; Q (Scenedesmus obliquus), CAB90363 and CAB90362; R (Aspergillus niger), ABA33725 and Q5YF91; S (Thalassiosira pseudonana), YP_316616 and YP_316614; T (Anopheles gambiae), NP_008075 and P34858; U (Drosophila melanogaster), NP_008283 and NP_008286; V (Y. lipolytica), Q9B6C7 and CAC28106; W (Caenorhabditis elegans), P24895 and P24886.
(vi) Diminished hydrophobicity.
It was proposed that the ND3 and ND4L subunits, which possess three transmembrane domains (24), are too hydrophobic to be imported across the mitochondrial membranes (66). However, the algal polypeptides also showed three hydrophobic segments predicted to be transmembrane alpha helixes (Fig. 3A). The likelihood that a polypeptide chain could be imported into the mitochondria can be appreciated on the basis of two parameters: the highest average hydrophobicity over 80 amino acids (termed mesohydrophobicity or mesoH) and the maximum hydrophobicity (<H>) over 17 amino acids (a range that is relevant to determine the hydrophobicity of putative transmembrane segments) (10, 14). Figure 3B shows an <H> versus mesoH plot using the Kyte and Doolittle scale (48) for ND3 and ND4L sequences of various origins. In comparison with mtDNA-encoded polypeptides, the ND3 and ND4L polypeptides from Chlamydomonas (Fig. 3B, dots A) exhibit the lowest <H> values while their mesoH values are low or close to the average, respectively. Similar results were obtained with the GES, GVH1, or ECS scales (11) (data not shown). Such reduced <H> and mesoH values compared to their mtDNA-encoded counterparts have also been reported for the nucleus-encoded COX IIA, COX IIB, COX III, and ATP6 proteins of Chlamydomonas. Reduced hydrophobicity of the most hydrophobic domains is considered to facilitate the import into mitochondria (29, 60, 61). Daley et al. have demonstrated that structural changes in COX2 polypeptide that diminish hydrophobicity can, indeed, allow import into mitochondria of a nucleus-encoded protein that is normally mtDNA encoded (14).
We thus show here that, following their transfer to the nucleus, both the NUO3 and NUO11 genes have acquired several features allowing their proper expression and the importation of their translation products into mitochondria: acquisition of an efficient promoter, as suggested by the abundance of the transcripts; modification of codon frequencies to the usage of the nucleus; acquisition of a polyadenylation signal and of one or several introns that are essential in the expression and subsequent maturation of the mRNA (67); addition of a mitochondrial targeting amino acid presequence; and hydrophobicity diminution of transmembrane stretches that probably promote sorting and import to mitochondria.
Transformants inactivated for the NUO3 and NUO11 nuclear genes can be isolated by RNAi.
In Chlamydomonas, efficient targeted gene disruption by homologous recombination is not yet available for nuclear genes (59). However, interference with the expression of specific gene by dsRNA is a powerful tool to investigate protein function in this organism (25, 75). To suppress the expression of NUO3 and NUO11 genes, a strain of Chlamydomonas lacking cell walls and auxotrophic for arginine was cotransformed with the plasmid pASL (bearing the ARG7 gene) and the plasmid designed for RNAi (pND3-RNAi or pND4L-RNAi) (Fig. 1) (see Materials and Methods). One hundred arg+ colonies of each transformation experiment were selected and tested by PCR for the presence of sequences belonging to the pRNAi plasmids, as described in Materials and Methods. A total of 26 and 28 positive colonies, called coND3 and coND4L clones, respectively, were obtained in transformation experiments with the pND3-RNAi and pND4L-RNAi constructs, respectively.
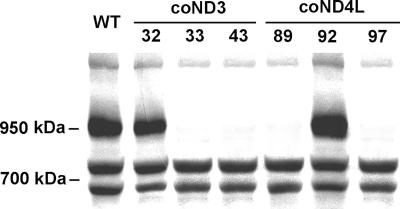
To investigate whether complex I enzyme activity was suppressed in the cotransformants, high-molecular-mass complexes from crude membrane fractions were separated by BN-PAGE. NADH dehydrogenase activity of the complex I matrical arm was then tested using NBT as an electron acceptor. Wild-type complex I activity was associated with a single band at 950 kDa (6, 7). Among the 28 coND4L clones analyzed, only two (89 and 97) showed no activity or very low activity (observed only after 2 h of incubation). Similarly, only two clones (33 and 43) of the 26 coND3 isolates showed no activity or very low staining activity, whereas the other ones exhibited no significant modification of complex I coloration compared to wild type (Fig. 4 and data not shown).
FIG. 4.
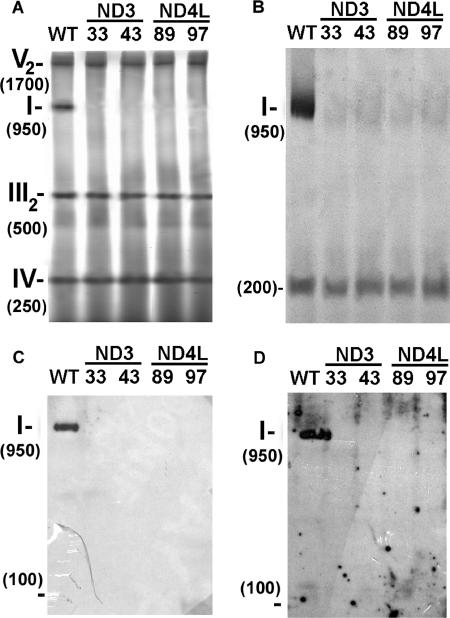
Screening of clones for complex I defect. Crude membrane extracts (120 μg of protein solubilized with 1.5% [wt/wt] dodecyl-β-d-maltoside) of Chlamydomonas strains were loaded on a BN gel. After electrophoresis, the gel was stained for NADH dehydrogenase activity of complex I using NBT as an electron acceptor. Mitochondrial complex I is detected at 950 kDa. The two lower-molecular-mass bands (about 750 and 700 kDa) are shown as loading controls and correspond to green bands of photosystem II and I, respectively, associated with light harvesting complexes (71).
To confirm the alteration of complex I in the four defective clones, enzyme activity was measured in crude membrane fractions compared to wild type and to transformants coND3-32 and coND4L-92 that behaved like wild type in the BN-PAGE experiment (Fig. 4). Cytochrome c oxidase (complex IV) enzyme activity was determined as a control, and no significant difference could be observed between strains (Table 1). In contrast, the NADH:duroquinone oxidoreductase activity sensitive to rotenone (considered to represent the activity of complex I) was found to be null or extremely low in the four mutant cell lines. The NADH:ferricyanide oxidoreductase activity was also measured. In the Chlamydomonas whole-membrane fraction, this reaction is mainly (70 to 85%) catalyzed by the peripheral arm of complex I (6). The four mutant strains exhibited an average activity of 15 to 30% of wild-type activity, again indicating the loss of the peripheral arm activity of complex I. These results fully confirm the complex I defect observed in BN gels by NADH/NBT staining (Fig. 4).
TABLE 1.
Activities of respiratory chain complexes in wild-type and transformant strains
| Strain | Specific activity (± SD)a
|
||
|---|---|---|---|
| Complex I | NADH:Fe(CN)63− | Complex IV | |
| Wild type (325.2) | 50 ± 22 | 2,489 ± 370 | 245 ± 29 |
| coND3-32 | 48 ± 9 | 2,234 ± 175 | 250 ± 12 |
| coND3-33 | 2 ± 1 | 808 ± 119 | 248 ± 18 |
| coND3-43 | 0 | 620 ± 74 | 244 ± 5 |
| coND4L-89 | 1 ± 1 | 739 ± 208 | 245 ± 12 |
| coND4L-92 | 53 ± 19 | 2,378 ± 89 | 212 ± 28 |
| coND4L-97 | 0 | 401 ± 219 | 229 ± 41 |
Values are specific activities (± SD) from three to six experiments. Specific activity was calculated as follows: for complex I, rotenone-sensitive NADH:duroquinone oxidoreductase, as nmol of NADH oxidized min−1 mg of protein−1; for NADH:Fe(CN)63−, NADH:ferricyanide oxidoreductase, as nmol of K3Fe(CN)63− reduced min−1 mg of protein−1; for complex IV, cytochrome c oxidase, as nmol of cytochrome c oxidized min−1 mg of protein−1.
To determine whether the loss of complex I activity detected in coND3-33, coND3-43, coND4L-89, and coND4L-97 strains was correlated to specific degradation of the corresponding transcripts, RNA blotting was performed using NUO3 and NUO11 cDNA sequences as probes (Fig. 2). A specific and drastic reduction of NUO3 transcript level was observed in coND3-33 (more than 95%) and coND3-43 (no transcript detected), while the amounts of NUO11 transcripts were below 2% in coND4L-89 and coND4L-97 clones. The transcription levels of the NUO7, NUO9, NUO17, and NUOP1 genes encoding other complex I subunits were also investigated as controls. The three first genes encode homologous subunits of the mammalian 49-kDa, 30-kDa, and ESSS complex I subunits, whereas NUOP1 codes for a 10-kDa protein typical of complex I from plants (7, 8). No signal difference was observed between wild-type and mutant strains. The transcript levels from coND3-32 and coND4L-92 strains were also analyzed as controls and were similar to those observed for wild-type cells (data not shown).
Taken together, these results demonstrate that the complex I enzyme activity defect detected in the four mutants is due to the absence of or to a drastic decrease in the ND3 and ND4L protein levels triggered by the expression of the NUO3 or NUO11 dsRNA. These results also indicate that no transcription downregulation of nuclear genes coding for other complex I subunits is engaged following inactivation of NUO3 and NUO11 gene expression. As observed in several other experiments designed for RNA interference in Chlamydomonas (75), the rates of clones altered in NUO3 or NUO11 gene expression were less than 10%. Such low frequencies of transformed clones for which suppression of gene expression was effective can be due to rearrangements of the transformant plasmid at the time of the insertion process or to the integration of the construction in transcriptionally inactive areas of the genome, as previously described in Chlamydomonas (46).
Loss of complex I assembly in ND3- and ND4L-deficient strains; function and localization of both proteins within the membrane domain of complex I.
In order to investigate the impact of the loss of ND3 or ND4L on complex I assembly, mitochondria were purified from wild-type and mutant cells, and the organelle extracts were subjected to BN-PAGE analyses. By staining the gel with Coomassie blue, four main bands of 250, 500, 950, and 1,700 kDa were observed over 100 kDa for wild type (Fig. 5A). They correspond to respiratory chain complexes IV, III (dimeric state), I, and V (ATP synthase under dimeric state), respectively (7, 78). No signal was observed at 950 kDa in the mutants, while the amount of complexes III, IV, and V was not altered. Moreover, no additional blue band that could correspond to a putative complex I subcomplex was found.
FIG. 5.
Complex I assembly in coND3 and coND4L mutants. For each strain, 150 μg of total mitochondrial protein was loaded on a BN gel. After electrophoresis, the gel was submitted to Coomassie blue staining (A) or NADH/NBT staining (B) or blotted and probed with antisera against the N. crassa whole complex I (C) or against the Y. lipolytica PSST subunit (D). I, III2, IV, and V2 correspond to respiratory chain complexes I, III (dimeric state), IV, and V (dimeric state), respectively.
To confirm these results, the protein complexes were investigated by immunological reactions with antisera against whole N. crassa complex I (Fig. 5C) or against the PSST subunit of Y. lipolytica (homologous of C. reinhardtii complex I subunit encoded by the NUO10 gene; GenBank accession number AAQ63698) (Fig. 5D). In wild type, the whole complex I is highlighted (950 kDa) by both antibodies, while no signal was detected in lanes showing results with mutants. Hence, the absence of the ND3 or ND4L subunit prevents the assembly of the whole complex I.
Finally, when the gel obtained from purified mitochondria was submitted to NADH/NBT staining, the 950-kDa complex I activity was observed solely in the wild-type mitochondria (as previously shown from the crude membrane fraction in Fig. 4), but an additional signal, in the range of 200 kDa, was also observed in both wild-type and mutant strains (Fig. 5B). We previously demonstrated that this 200-kDa signal corresponds to a free soluble enzyme present in Chlamydomonas mitochondria and displays NADH:ferricyanide and NADH:duroquinone oxydoreductase activities (6). Immunological studies, moreover, showed (6) that this enzyme comprises the 49-kDa (NUOD) and the 76-kDa (NUOG) polypeptides, two components of the complex I peripheral arm which are thought to be involved in the electron flow from NADH to ubiquinone. Hence, the 200-kDa soluble enzyme can be considered as a fragment originating from the peripheral arm of the complex I enzyme. Interestingly, a similar 200- to 300-kDa NADH dehydrogenase that contains the 76-kDa subunit has also been identified in mitochondria from potato and pea (5). In contrast to the 49- and 76-kDa subunits, the PSST subunit could be absent from the Chlamydomonas 200-kDa subcomplex, since no signal was obtained using an antibody directed against the Y. lipolytica PSST (Fig. 5D). Finally, it has to be mentioned that the 200-kDa subcomplex is much larger than the type II NADH dehydrogenases (55 to 65 kDa) found in land plant mitochondria (22) and also probably present in Chlamydomonas, as suggested from sequence alignment studies (8).
While the presence of ND3 and ND4L in mitochondrial complex I has been demonstrated for more than 20 years (24), very little is known about the role of these two subunits in the activity and assembly of the eukaryotic enzyme. Some data were recently obtained, mainly by studying their prokaryotic counterparts. Concerning the ND3 subunit, it has been shown in the bacterium Paracoccus denitrificans that the corresponding polypeptide (Nqo7) directly interacts with subunits Nqo6 (PSST) and Nqo4 (49 kDa) of the peripheral arm (17, 41). Both of these hydrophilic polypeptides are considered to bind the last iron-sulfur cluster (N2) that reduces the quinone at the interface between the two arms and to participate to a quinone binding site (15, 19, 37, 44, 64). In E. coli, evidence of the implication of ND3 in the enzyme activity is provided by the double mutant D79N/E81Q, which almost completely lacks the energy-transducing activity of the enzyme (40). Similarly, point mutations affecting the nd3 gene sequence in human cause specific defects in complex I activity (13, 56, 77). With regard to ND4L, this subunit could be involved in the proton translocation machinery of the enzyme. As a matter of fact, in E. coli, NuoK (ND4L) along with NuoL (ND5), NuoM (ND4), and NuoN (ND2) shows primary sequence similarity to antiporters of the prokaryotic Mrp class (54, 55). Moreover, mutations of nearly perfectly conserved Glu-36 in NuoK (ND4L) of E. coli lead to almost null activities of coupled electron transfer with a concomitant loss of generation of the electrochemical gradient (42, 45).
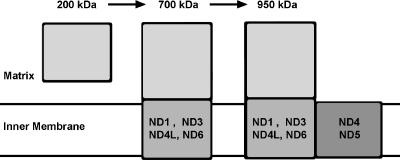
Not all hydrophobic subunits of the core enzyme play the same role in the assembly of complex I. As a matter of fact, in the course of the characterization of other Chlamydomonas complex I mutants (6), we showed that the loss of ND4 or ND4+ND5 subunits leads to the assembly of a 700-kDa membrane-bound subcomplex; however, as for ND3 and ND4L, the absence of intact ND1 or ND6 subunits totally prevents complex I assembly. This suggests that ND4 and ND5, on one hand, and ND1, ND3, ND4L, and ND6, on the other hand, are located in two different domains of the complex I membrane arm. This hypothesis is in good agreement with the structural models proposed for the localization of these subunits within the bovine complex I (9, 72) and the prokaryotic NDH-1 enzyme (39). Following this hypothesis, we propose a preliminary model for the assembly pathway of Chlamydomonas complex I (Fig. 6). In this model, the association of the 200-kDa soluble fragment displaying NADH dehydrogenase activity (see above) with other hydrophilic subunits and with hydrophobic proteins (notably ND1, ND3, ND4L, and ND6) would lead to the formation of a membrane-bound 700-kDa subcomplex. Then, the association of another membrane domain located at a distal position and comprising ND4, ND5, and other hydrophobic proteins would give the 950-kDa fully assembled complex I. This simple model is very close to the model that was proposed for the human complex I assembly from the study of various mutant cell lines producing different assembly intermediates (1). A divergent model was proposed for complex I assembly in the fungus N. crassa, based on the existence of a stable peripheral arm composed mainly of hydrophilic proteins and of a membrane arm composed of hydrophobic subunits (79). A priori, the existence of two different complex I assembly pathways is rather surprising. The discrepancy between the two models could be related to the absence or the nondetection of some assembly intermediates in one of the other organisms. Some factors specific to the organisms could also influence the relative stability of the subcomplexes: the rate of proteolysis of unassembled protein sets or the presence of specific chaperones. In this context, only two chaperones involved in complex I assembly have been identified to date: CIA30 and CIA84 (47). While CIA30 is widely found among eukaryotes, homologous sequences of CIA84 are found only in fungi (8). A more detailed analysis of the composition of the various subcomplexes is required to elucidate the assembly pathway of the huge multimeric complex I enzyme.
FIG. 6.
Schematic representation of the putative assembly pathway of C. reinhardtii complex I (reference 6 and the present study). The locations of six hydrophobic subunits within subcomplexes are also indicated.
Acknowledgments
This research was supported by grants from the Belgian FRFC (2.4587.04 and 2.4582.05) to C.R. P.C. and F.F. are postdoctoral researcher and research associate, respectively, of the Belgian FNRS. M.L. is a fellow of the Belgian FRIA.
We are grateful to D. González-Halphen for critical reading of the manuscript and to H. Weiss and U. Schulte (Institut fur Biochimie, Düsseldorf) and V. Zickermann and U. Brandt (Institut fur Biochimie, Frankfurt) for the generous gifts of antisera. We also thank M. Radoux and E. Schmetz for technical assistance.
The gene sequence data were produced by the U.S. Department of Energy Joint Genome Institute (http://www.jgi.doe.gov/) and are provided for use in this publication only.
REFERENCES
- 1.Antonicka, H., I. Ogilvie, T. Taivassalo, R. P. Anitori, R. G. Haller, J. Vissing, N. G. Kennaway, and E. A. Shoubridge. 2003. Identification and characterization of a common set of complex I assembly intermediates in mitochondria from patients with complex I deficiency. J. Biol. Chem. 278:43081-43088. [DOI] [PubMed] [Google Scholar]
- 2.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 3.Brennicke, A., L. Grohmann, R. Hiesel, V. Knoop, and W. Schuster. 1993. The mitochondrial genome on its way to the nucleus: different stages of gene transfer in higher plants. FEBS Lett. 325:140-145. [DOI] [PubMed] [Google Scholar]
- 4.Bullerwell, C. E., and M. W. Gray. 2004. Evolution of the mitochondrial genome: protist connections to animals, fungi and plants. Curr. Opin. Microbiol. 7:528-534. [DOI] [PubMed] [Google Scholar]
- 5.Bykova, N. V., A. G. Rasmusson, A. U. Igamberdiev, P. Gardestrom, and I. M. Moller. 1999. Two separate transhydrogenase activities are present in plant mitochondria. Biochem. Biophys. Res. Commun. 265:106-111. [DOI] [PubMed] [Google Scholar]
- 6.Cardol, P., R. F. Matagne, and C. Remacle. 2002. Impact of mutations affecting ND mitochondria-encoded subunits on the activity and assembly of complex I in Chlamydomonas. Implication for the structural organization of the enzyme. J. Mol. Biol. 319:1211-1221. [DOI] [PubMed] [Google Scholar]
- 7.Cardol, P., F. Vanrobaeys, B. Devreese, J. Van Beeumen, R. F. Matagne, and C. Remacle. 2004. Higher plant-like subunit composition of the mitochondrial complex I from Chlamydomonas reinhardtii: 31 conserved components among eukaryotes. Biochim. Biophys. Acta 1658:212-224. [DOI] [PubMed] [Google Scholar]
- 8.Cardol, P., D. González-Halphen, A. Reyes-Prieto, D. Baurain, R. F. Matagne, and C. Remacle. 2005. The mitochondrial oxidative phosphorylation proteome of Chlamydomonas reinhardtii deduced from the Genome Sequencing Project. Plant Physiol. 137:447-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carroll, J., I. M. Fearnley, R. J. Shannon, J. Hirst, and J. E. Walker. 2003. Analysis of the subunit composition of complex I from bovine heart mitochondria. Mol. Cell Proteomics 2:117-126. [DOI] [PubMed] [Google Scholar]
- 10.Claros, M. G., J. Perea, Y. Shu, F. A. Samatey, J. L. Popot, and C. Jacq. 1995. Limitations to in vivo import of hydrophobic proteins into yeast mitochondria. The case of a cytoplasmically synthesized apocytochrome b. Eur. J. Biochem. 228:762-771. [PubMed] [Google Scholar]
- 11.Claros, M. G., and P. Vincens. 1996. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur. J. Biochem. 241:779-786. [DOI] [PubMed] [Google Scholar]
- 12.Crimi, M., M. Sciacco, S. Galbiati, A. Bordoni, G. Malferrari, R. Del Bo, I. Biunno, N. Bresolin, and G. P. Comi. 2002. A collection of 33 novel human mtDNA homoplasmic variants. Hum. Mutat. 20:409. [DOI] [PubMed] [Google Scholar]
- 13.Crimi, M., A. Papadimitriou, S. Galbiati, P. Palamidou, F. Fortunato, A. Bordoni, U. Papandreou, D. Papadimitriou, G. M. Hadjigeorgiou, E. Drogari, N. Bresolin, and G. P. Comi. 2004. A new mitochondrial DNA mutation in ND3 gene causing severe Leigh syndrome with early lethality. Pediatr. Res. 55:842-846. [DOI] [PubMed] [Google Scholar]
- 14.Daley, D. O., R. Clifton, and J. Whelan. 2002. Intracellular gene transfer: reduced hydrophobicity facilitates gene transfer for subunit 2 of cytochrome c oxidase. Proc. Natl. Acad. Sci. USA 99:10510-10515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Darrouzet, E., J. P. Issartel, J. Lunardi, and A. Dupuis. 1998. The 49-kDa subunit of NADH-ubiquinone oxidoreductase (complex I) is involved in the binding of piericidin and rotenone, two quinone-related inhibitors. FEBS Lett. 431:34-38. [DOI] [PubMed] [Google Scholar]
- 16.Debuchy, R., S. Purton, and J. D. Rochaix. 1989. The argininosuccinate lyase gene of Chlamydomonas reinhardtii: an important tool for nuclear transformation and for correlating the genetic and molecular maps of the ARG7 locus. EMBO J. 8:2803-2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Di Bernardo, S., and T. Yagi. 2001. Direct interaction between a membrane domain subunit and a connector subunit in the H(+)-translocating NADH-quinone oxidoreductase. FEBS Lett. 508:385-388. [DOI] [PubMed] [Google Scholar]
- 18.Djafarzadeh, R., S. Kerscher, K. Zwicker, M. Radermacher, M. Lindahl, H. Schagger, and U. Brandt. 2000. Biophysical and structural characterization of proton-translocating NADH-dehydrogenase (complex I) from the strictly aerobic yeast Yarrowia lipolytica. Biochim. Biophys. Acta 1459:230-238. [DOI] [PubMed] [Google Scholar]
- 19.Duarte, M., H. Populo, A. Videira, T. Friedrich, and U. Schulte. 2002. Disruption of iron-sulphur cluster N2 from NADH: ubiquinone oxidoreductase by site-directed mutagenesis. Biochem. J. 364:833-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duby, F., P. Cardol, R. F. Matagne, and C. Remacle. 2001. Structure of the telomeric ends of mt DNA, transcriptional analysis and complex I assembly in the dum24 mitochondrial mutant of Chlamydomonas reinhardtii. Mol. Genet. Genomics 266:109-114. [DOI] [PubMed] [Google Scholar]
- 21.Dudkina, N. V., H. Eubel, W. Keegstra, E. J. Boekema, and H. P. Braun. 2005. Structure of a mitochondrial supercomplex formed by respiratory-chain complexes I and III. Proc. Natl. Acad. Sci. USA 102:3225-3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elhafez, D., M. W. Murcha, R. Clifton, K. L. Soole, D. A. Day, and J. Whelan. 2006. Characterization of mitochondrial alternative NAD(P)H dehydrogenases in Arabidopsis: intraorganelle location and expression. Plant Cell. Physiol. 47:43-54. [DOI] [PubMed] [Google Scholar]
- 23.Emanuelsson, O., H. Nielsen, S. Brunak, and G. von Heijne. 2000. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300:1005-1016. [DOI] [PubMed] [Google Scholar]
- 24.Fearnley, I. M., and J. E. Walker. 1992. Conservation of sequences of subunits of mitochondrial complex I and their relationships with other proteins. Biochim. Biophys. Acta 1140:105-134. [DOI] [PubMed] [Google Scholar]
- 25.Fuhrmann, M., A. Stahlberg, E. Govorunova, S. Rank, and P. Hegemann. 2001. The abundant retinal protein of the Chlamydomonas eye is not the photoreceptor for photoaxis and photophobic responses. J. Cell Sci. 114:3857-3863. [DOI] [PubMed] [Google Scholar]
- 26.Funes, S., E. Davidson, M. G. Claros, R. van Lis, X. Perez-Martinez, M. Vazquez-Acevedo, M. P. King, and D. González-Halphen. 2002. The typically mitochondrial DNA-encoded ATP6 subunit of the F1Fo-ATPase is encoded by a nuclear gene in Chlamydomonas reinhardtii. J. Biol. Chem. 277:6051-6058. [DOI] [PubMed] [Google Scholar]
- 27.Galanis, M., R. J. Devenish, and P. Nagley. 1991. Duplication of leader sequence for protein targeting to mitochondria leads to increased import efficiency. FEBS Lett. 282:425-430. [DOI] [PubMed] [Google Scholar]
- 28.Glaser, E., S. Sjoling, M. Tanudji, and J. Whelan. 1998. Mitochondrial protein import in plants. Signals, sorting, targeting, processing and regulation. Plant Mol. Biol. 38:331-338. [DOI] [PubMed] [Google Scholar]
- 29.González-Halphen, D., S. Funes, X. Perez-Martinez, A. Reyes-Prieto, M. G. Claros, E. Davidson, and M. P. King. 2004. Genetic correction of mitochondrial diseases: using the natural migration of mitochondrial genes to the nucleus in chlorophyte algae as a model system. Ann. N. Y. Acad. Sci. 1019:232-239. [DOI] [PubMed] [Google Scholar]
- 30.Gorman, D. S., and R. P. Levine. 1965. Cytochrome f and plastocyanin: their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 54:1665-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gray, M. W., G. Burger, and B. F. Lang. 1999. Mitochondrial evolution. Science 283:1476-1481. [DOI] [PubMed] [Google Scholar]
- 32.Grigorieff, N. 1998. Three-dimensional structure of bovine NADH:ubiquinone oxidoreductase (complex I) at 22 Å in ice. J. Mol. Biol. 277:1033-1046. [DOI] [PubMed] [Google Scholar]
- 33.Grossman, A. R., E. E. Harris, C. Hauser, P. A. Lefebvre, D. Martinez, D. Rokhsar, J. Shrager, C. D. Silflow, D. Stern, O. Vallon, and Z. Zhang. 2003. Chlamydomonas reinhardtii at the crossroads of genomics. Eukaryot. Cell 2:1137-1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gutierres, S., M. Sabar, C. Lelandais, P. Chetrit, P. Diolez, H. Degand, M. Boutry, F. Vedel, Y. de Kouchkovsky, and R. De Paepe. 1997. Lack of mitochondrial and nuclear-encoded subunits of complex I and alteration of the respiratory chain in Nicotiana sylvestris mitochondrial deletion mutants. Proc. Natl. Acad. Sci. USA 94:3436-3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harris, E. H. 1989. The Chlamydomonas sourcebook. Academic Press, San Diego, Calif. [DOI] [PubMed]
- 36.Heazlewood, J. L., K. A. Howell, and A. H. Millar. 2003. Mitochondrial complex I from Arabidopsis and rice: orthologs of mammalian and fungal components coupled with plant-specific subunits. Biochim. Biophys. Acta 1604:159-169. [DOI] [PubMed] [Google Scholar]
- 37.Hinchliffe, P., and L. A. Sazanov. 2005. Organization of iron-sulfur clusters in respiratory complex I. Science 309:771-774. [DOI] [PubMed] [Google Scholar]
- 38.Hirst, J., J. Carroll, I. M. Fearnley, R. J. Shannon, and J. E. Walker. 2003. The nuclear encoded subunits of complex I from bovine heart mitochondria. Biochim. Biophys. Acta 1604:135-150. [DOI] [PubMed] [Google Scholar]
- 39.Holt, P. J., D. J. Morgan, and L. A. Sazanov. 2003. The location of NuoL and NuoM subunits in the membrane domain of the Escherichia coli complex I: implications for the mechanism of proton pumping. J. Bio. Chem. 278:43114-43120. [DOI] [PubMed] [Google Scholar]
- 40.Kao, M. C., S. Di Bernardo, M. Perego, E. Nakamaru-Ogiso, A. Matsuno-Yagi, and T. Yagi. 2004. Functional roles of four conserved charged residues in the membrane domain subunit NuoA of the proton-translocating NADH-quinone oxidoreductase from Escherichia coli. J. Biol. Chem. 279:32360-32366. [DOI] [PubMed] [Google Scholar]
- 41.Kao, M. C., A. Matsuno-Yagi, and T. Yagi. 2004. Subunit proximity in the H+-translocating NADH-quinone oxidoreductase probed by zero-length cross-linking. Biochemistry 43:3750-3755. [DOI] [PubMed] [Google Scholar]
- 42.Kao, M. C., E. Nakamaru-Ogiso, A. Matsuno-Yagi, and T. Yagi. 2005. Characterization of the membrane domain subunit NuoK (ND4L) of the NADH-quinone oxidoreductase from Escherichia coli. Biochemistry 44:9545-9554. [DOI] [PubMed] [Google Scholar]
- 43.Karpova, O. V., and K. J. Newton. 1999. A partially assembled complex I in ND4-deficient mitochondria of maize. Plant J. 17:511-521. [Google Scholar]
- 44.Kashani-Poor, N., K. Zwicker, S. Kerscher, and U. Brandt. 2001. A central functional role for the 49-kDa subunit within the catalytic core of mitochondrial complex I. J. Biol. Chem. 276:24082-24087. [DOI] [PubMed] [Google Scholar]
- 45.Kervinen, M., J. Patsi, M. Finel, and I. E. Hassinen. 2004. A pair of membrane-embedded acidic residues in the NuoK subunit of Escherichia coli NDH-1, a counterpart of the ND4L subunit of the mitochondrial complex I, are required for high ubiquinone reductase activity. Biochemistry 43:773-781. [DOI] [PubMed] [Google Scholar]
- 46.Kindle, K. L. 1990. High frequency nuclear transformation of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 87:1228-1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kuffner, R., A. Rohr, A. Schmiede, C. Krull, and U. Schulte. 1998. Involvement of two novel chaperones in the assembly of mitochondrial NADH:ubiquinone oxidoreductase (complex I). J. Mol. Biol. 283:409-417. [DOI] [PubMed] [Google Scholar]
- 48.Kyte, J., and R. F. Doolittle. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105-132. [DOI] [PubMed] [Google Scholar]
- 49.Loppes, R., and M. Radoux. 2001. Identification of short promoter regions involved in the transcriptional expression of the nitrate reductase gene in Chlamydomonas reinhardtii. Plant Mol. Biol. 45:215-227. [DOI] [PubMed] [Google Scholar]
- 50.Loppes, R., and M. Radoux. 2002. Two short regions of the promoter are essential for activation and repression of the nitrate reductase gene in Chlamydomonas reinhardtii. Mol. Genet. Genomics 268:42-48. [DOI] [PubMed] [Google Scholar]
- 51.Marienfeld, J. R., and K. J. Newton. 1994. The maize NCS2 abnormal growth mutant has a chimeric nad4-nad7 mitochondrial gene and is associated with reduced complex I function. Genetics 138:855-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marques, I., M. Duarte, J. Assuncao, A. V. Ushakova, and A. Videira. 2005. Composition of complex I from Neurospora crassa and disruption of two “accessory” subunits. Biochim. Biophys. Acta 1707:211-220. [DOI] [PubMed] [Google Scholar]
- 53.Matagne, R. F., M. R. Michel-Wolwertz, C. Munaut, C. Duyckaerts, and F. Sluse. 1989. Induction and characterization of mitochondrial DNA mutants in Chlamydomonas reinhardtii. J. Cell Biol. 108:1221-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mathiesen, C., and C. Hagerhall. 2002. Transmembrane topology of the NuoL, M and N subunits of NADH:quinone oxidoreductase and their homologues among membrane-bound hydrogenases and bona fide antiporters. Biochim. Biophys. Acta 1556:121-132. [DOI] [PubMed] [Google Scholar]
- 55.Mathiesen, C., and C. Hagerhall. 2003. The “antiporter module” of respiratory chain complex I includes the MrpC/NuoK subunit—a revision of the modular evolution scheme. FEBS Lett. 549:7-13. [DOI] [PubMed] [Google Scholar]
- 56.McFarland, R., D. M. Kirby, K. J. Fowler, A. Ohtake, M. T. Ryan, D. J. Amor, J. M. Fletcher, J. W. Dixon, F. A. Collins, D. M. Turnbull, R. W. Taylor, and D. R. Thorburn. 2004. De novo mutations in the mitochondrial ND3 gene as a cause of infantile mitochondrial encephalopathy and complex I deficiency. Ann. Neurol. 55:58-64. [DOI] [PubMed] [Google Scholar]
- 57.Michaelis, G., C. Vahrenholz, and E. Pratje. 1990. Mitochondrial DNA of Chlamydomonas reinhardtii: the gene for apocytochrome b and the complete functional map of the 15.8 kb DNA. Mol. Gen. Genet. 223:211-216. [DOI] [PubMed] [Google Scholar]
- 58.Newman, S. M., J. E. Boynton, N. W. Gillham, B. L. Randolph-Anderson, A. M. Johnson, and E. H. Harris,. 1990. Transformation of chloroplast ribosomal RNA genes in Chlamydomonas: molecular and genetic characterization of integration events. Genetics 126:875-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pazour, G. J., and G. B. Witman. 2000. Forward and reverse genetic analysis of microtubule motors in Chlamydomonas. Methods 22:285-298. [DOI] [PubMed] [Google Scholar]
- 60.Perez-Martinez, X., M. Vazquez-Acevedo, E. Tolkunova, S. Funes, M. G. Claros, E. Davidson, M. P. King, and D. González-Halphen. 2000. Unusual location of a mitochondrial gene. Subunit III of cytochrome c oxidase is encoded in the nucleus of chlamydomonad algae. J. Biol. Chem. 275:30144-30152. [DOI] [PubMed] [Google Scholar]
- 61.Perez-Martinez, X., A. Antaramian, M. Vazquez-Acevedo, S. Funes, E. Tolkunova, J. d'Alayer, M. G. Claros, E. Davidson, M. P. King, and D. González-Halphen. 2001. Subunit II of cytochrome c oxidase in chlamydomonad algae is a heterodimer encoded by two independent nuclear genes. J. Biol. Chem. 276:11302-11309. [DOI] [PubMed] [Google Scholar]
- 62.Perez-Martinez, X., S. Funes, E. Tolkunova, E. Davidson, M. P. King, and D. González-Halphen. 2002. Structure of nuclear-localized cox3 genes in Chlamydomonas reinhardtii and in its colorless close relative Polytomella sp. Curr. Genet. 40:399-404. [DOI] [PubMed] [Google Scholar]
- 63.Pineau, B., C. Mathieu, C. Gerard-Hirne, R. De Paepe, and P. Chetrit. 2005. Targeting the NAD7 subunit to mitochondria restores a functional complex I and a wild type phenotype in the Nicotiana sylvestris CMS II mutant lacking nad7. J. Bio. Chem. 280:25994-26001. [DOI] [PubMed] [Google Scholar]
- 64.Prieur, I., J. Lunardi, and A. Dupuis. 2001. Evidence for a quinone binding site close to the interface between NUOD and NUOB subunits of complex I. Biochim. Biophys. Acta 1504:173-178. [DOI] [PubMed] [Google Scholar]
- 65.Pröschold, T., B. Marin, U. G. Schlosser, and M. Melkonian. 2001. Molecular phylogeny and taxonomic revision of Chlamydomonas (Chlorophyta). I. Emendation of Chlamydomonas Ehrenberg and Chloromonas Gobi, and description of Oogamochlamys gen. nov. and Lobochlamys gen. nov. Protist 152:265-300. [DOI] [PubMed] [Google Scholar]
- 66.Race, H. L., R. G. Herrmann, and W. Martin. 1999. Why have organelles retained genomes? Trends Genet. 15:364-370. [DOI] [PubMed] [Google Scholar]
- 67.Reddy, A. S. N. 2001. Nuclear pre-mRNA splicing in plants. Crit. Rev. Plant Sci. 20:523-571. [Google Scholar]
- 68.Remacle, C., D. Baurain, P. Cardol, and R. F. Matagne. 2001. Mutants of Chlamydomonas reinhardtii deficient in mitochondrial complex I. Characterization of two mutations affecting the nd1 coding sequence. Genetics 158:1051-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Remacle, C., F. Duby, P. Cardol, and R. F. Matagne. 2001. Mutations inactivating mitochondrial genes in Chlamydomonas reinhardtii. Biochem. Soc. Trans. 29:442-446. [DOI] [PubMed] [Google Scholar]
- 70.Remacle, C., P. Cardol, N. Coosemans, M. Gaisne, and N. Bonnefoy. 2006. High-efficiency biolistic transformation of Chlamydomonas mitochondria can be used to insert mutations in complex I genes. Proc. Natl. Acad. Sci. USA 103:4771-4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rexroth, S., J. M. Meyer zu Tittingdorf, F. Krause, N. A. Dencher, and H. Seelert. 2003. Thylakoid membrane at altered metabolic state: challenging the forgotten realms of the proteome. Electrophoresis 24:2814-2823. [DOI] [PubMed] [Google Scholar]
- 72.Sazanov, L. A., S. Y. Peak-Chew, I. M. Fearnley, and J. E. Walker. 2000. Resolution of the membrane domain of bovine complex I into subcomplexes: implications for the structural organization of the enzyme. Biochemistry 39:7229-7235. [DOI] [PubMed] [Google Scholar]
- 73.Schägger, H., and G. Von Jago. 1991. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 199:223-231. [DOI] [PubMed] [Google Scholar]
- 74.Schapira, A. H. 1998. Human complex I defects in neurodegenerative diseases. Biochim. Biophys. Acta 1364:261-270. [DOI] [PubMed] [Google Scholar]
- 75.Schroda, M. 2006. RNA silencing in Chlamydomonas: mechanisms and tools. Curr. Genet. 49:69-84. [DOI] [PubMed] [Google Scholar]
- 76.Silflow, C. D.1998. Organization of the nuclear genome, p. 25-40. In J. D. Rochaix, M. Goldschmidt-Clermont, and S. Merchant (ed.), The molecular biology of chloroplasts and mitochondria in Chlamydomonas. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 77.Taylor, R. W., R. Singh-Kler, C. M. Hayes, P. E. Smith, and D. M. Turnbull. 2001. Progressive mitochondrial disease resulting from a novel missense mutation in the mitochondrial DNA ND3 gene. Ann. Neurol. 50:104-107. [DOI] [PubMed] [Google Scholar]
- 78.van Lis, R., A. Atteia, G. Mendoza-Hernández, and D. González-Halphen. 2003. Identification of novel mitochondrial protein components of Chlamydomonas reinhardtii. A proteomic approach. Plant Physiol. 132:318-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Videira, A., and M. Duarte. 2002. From NADH to ubiquinone in Neurospora mitochondria. Biochim. Biophys. Acta 1555:187-191. [DOI] [PubMed] [Google Scholar]