Abstract
Soil bacterial population dynamics were examined in several crude-oil-contaminated soils to identify those organisms associated with alkane degradation and to assess patterns in microbial response across disparate soils. Seven soil types obtained from six geographically distinct areas of the United States (Arizona, Oregon, Indiana, Virginia, Oklahoma, and Montana) were used in controlled contamination experiments containing 2% (wt/wt) crude oil spiked with [1-14C]hexadecane. Microbial populations present during hydrocarbon degradation were analyzed using both 16S rRNA gene sequence analysis and by traditional methods for cultivating hydrocarbon-oxidizing bacteria. After a 50-day incubation, all seven soils showed comparable hydrocarbon depletion, where >80% of added crude oil was depleted and approximately 40 to 70% of added [14C]hexadecane was converted to 14CO2. However, the initial rates of hydrocarbon depletion differed up to 10-fold, and preferential utilization of shorter-chain-length n-alkanes relative to longer-chain-length n-alkanes was observed in some soils. Distinct microbial populations developed, concomitant with crude-oil depletion. Phylogenetically diverse bacterial populations were selected across different soils, many of which were identical to hydrocarbon-degrading isolates obtained from the same systems (e.g., Nocardioides albus, Collimonas sp., and Rhodococcus coprophilus). In several cases, soil type was shown to be an important determinant, defining specific microorganisms responding to hydrocarbon contamination. However, similar Rhodococcus erythropolis-like populations were observed in four of the seven soils and were the most common hydrocarbon-degrading organisms identified via cultivation.
Relatively little is known regarding the environmental determinants of microbial population selection in soil environments contaminated with complex hydrocarbon mixtures. The predominant factors influencing microbial community structure after contamination likely include (i) contaminant mixture type, (ii) soil type (i.e., physical, chemical, and biological characteristics of soils), and (iii) time. Complex petroleum hydrocarbon mixtures, including crude oil, diesel fuel, and creosote, consist of various concentrations of n- and branched alkanes, cycloalkanes, phenolics, aromatics, and polycyclic aromatic hydrocarbons. Although these mixtures contain similar constituents, the relative abundance of mixture components and toxic compounds (e.g., heterocyclics, chlorophenols) vary considerably, and these variations are potentially important in determining which microbial populations are involved in biodegradation. The physical/chemical/biological properties of soils (e.g., temperature, pH, conductivity, nutrient status, texture, biota) are expected to further influence the selection of adapted microbial populations. In actual contaminated environments, temporal changes in mixture components occur as a result of abiotic processes, such as aging, sorption, and sequestration (14, 19, 29), as well as microbial processes, such as sequential degradation of mixture constituents (27, 42). These temporal changes have been shown to affect subsequent changes in microbial community structure, as assayed by cultivation-independent molecular methods (17, 21). For example, changes in the bioavailability of nonpolar contaminants due to sorption or addition of surfactant have been shown to affect the selection of different contaminant-degrading microbial populations (6, 10, 11).
Our overarching goal is to introduce a population-based understanding to bioremediation ecology by investigating patterns of microbial population response to hydrocarbon perturbation. The patterns we observe may then suggest hypotheses about their mechanistic basis which can be investigated in future studies. Here, we focus on the influence of a single complex hydrocarbon mixture (i.e., crude oil) on bacterial population response across several disparate soil types. The fate of individual constituents within complex hydrocarbon mixtures may vary across soil types, in part due to the selection of different microbial populations that are adapted to a specific set of physical-chemical properties. Conversely, the selection pressure imparted by contamination with a particular complex mixture, such as crude oil, may result in the selection of similar microbial populations across widely different soil environments. Although research has been conducted in many ecologically and geographically diverse contaminated soil environments (12, 17, 20, 23, 46), there are no comprehensive studies that focus systematically on the identification of microbial populations associated with hydrocarbon degradation across different geographical locations where the various soil types exhibit distinctly different physical-chemical and biological properties. To understand the impacts of hydrocarbon mixture perturbation on soil microbial communities, it is important to determine whether microbial population responses to a constant complex mixture vary across soil types or if consistent patterns in the populations selected are observed across a wide range of soil environments.
Specific objectives of this study were to (i) examine the microbial response to hydrocarbon mixture contamination in diverse soil types from geographically distinct locations and (ii) assess the importance of soil type as a determinant for selecting specific hydrocarbon-degrading microbial populations. A cultivation-independent molecular analysis targeting the bacterial 16S rRNA gene was used to monitor bacterial population changes following crude-oil contamination. In addition, hydrocarbon-degrading bacteria were cultivated by direct plating from serially diluted samples from each contaminated soil, and their relevance was confirmed by comparing 16S rRNA gene sequences of isolates to those observed in crude-oil-contaminated soils. Using this combined approach, specific microbial populations were identified as important contributors to the observed hydrocarbon degradation.
MATERIALS AND METHODS
Soil types.
The surface horizons (A horizons) of seven common soil series (the official soil series descriptions are available from the United States Department of Agriculture's Natural Resources Conservation Service [http://soils.usda.gov/technical/classification/osd]) from six distinct geographical locations across the United States, including the Northwest, Southwest, Midwest, and Southeast (Table 1), were examined in this study. In each case, the soil types chosen for this study represent common landscape units observed within these regions and were collected from native sites with no history of crude-oil contamination (confirmed by gas chromatography-mass spectrometry [GC-MS] analysis which showed no detectable hydrocarbons [data not shown]). Each soil was passed through a 2-mm sieve and stored field moist at 4°C. Total organic carbon was determined using a LECO furnace with correction for inorganic carbonate (24); other soil properties, including extractable nutrients (NO3-N, K, and P), pH, and gravimetric water content at 33 kPa, were determined as described previously (24, 26, 30).
TABLE 1.
Physical and chemical characteristics of soils used in this study
| Soil | Location | Series name | Texture | pH | CaCO3 equivalent (%) | % Organic C | H2O contenta (%) | Concn (mg/kg) ofb:
|
||
|---|---|---|---|---|---|---|---|---|---|---|
| NO3-N | K | P | ||||||||
| AZ | Arizona | Casa Grande | Sandy loam | 8.8 | 1.7 | 0.2 | 12.0 | 8.1 | 192.0 | 4.5 |
| OR | Oregon | Jory | Clay | 5.4 | <0.1 | 5.7 | 38.9 | 0.2 | 452.0 | 11.7 |
| IN | Indiana | Chalmers | Silty clay loam | 6.1 | <0.1 | 2.2 | 27.1 | 2.0 | 260.0 | 6.0 |
| VA | Virginia | Groseclose | Silty loam | 7.4 | <0.1 | 4.5 | 35.8 | 17.2 | 260.0 | 12.8 |
| OK | Oklahoma | Konowa | Sandy loam | 7.1 | <0.1 | 3.3 | 15.8 | 51.6 | 350.0 | 31.9 |
| MT-N | Montana | Beaverton | Loam | 7.6 | 0.6 | 3.9 | 29.9 | 61.4 | 1732.0 | 75.9 |
| MT-O | Montana | Brocko | Loam | 8.4 | 3.8 | 1.2 | 21.6 | 52.4 | 968.0 | 22.9 |
Water content was determined at a corresponding water potential of 33 kPa (0.33 bar).
NO3-N, K, and P were extracted with KCl, NH4 acetate, and NaHCO3, respectively.
Crude-oil contamination and hexadecane mineralization experiments.
Crude-oil contamination assays were conducted in triplicate using 150-ml serum bottles containing 30 g (dry weight) soil. Sterile soils, prepared by autoclaving three times for 1 h with 24-h intervals at room temperature, were used as abiotic controls. The crude oil used in this study (Conoco Corp., Billings, MT) consisted of n-alkanes with chain lengths of C10 to C31: 80% of the total n-alkanes ranged from C12 to C24 (30% C12 to C15, 26% C16 to C19, and 26% C20 to C24) (data not shown). Crude oil was physically weathered for 100 h under a stream of N2 gas to remove volatile components, mixed with 300,000 dpm [1-14C]hexadecane (>98% purity, specific activity = 2.6 mCi mmol−1; Sigma Chemical, St. Louis, MO), and then added dropwise to each bottle using a 1-ml microsyringe to achieve uniform distribution and a final concentration of 2% crude oil (wt/wt). The depletion of individual hydrocarbon components was assessed by monitoring changes in chemical composition by capillary GC-MS after solvent extraction. The degradation of [1-14C]hexadecane was determined simultaneously by measuring the evolution of 14CO2 as a function of time.
Concentrations of several nutrient components important for microbial growth varied substantially among soils, especially nitrate concentrations, which ranged from 0.2 mg/kg in the OR soil to 61.4 mg/kg in the MT-N soil (Table 1). To eliminate the possibility of nutrient limitation being an additional variable among soils, we supplemented soils with nutrient solutions during crude-oil contamination assays to normalize C/N/P ratios after addition of 2% crude oil. The total volume of sterile deionized water and nutrient solution [final nutrient concentrations: (NH4)2SO4, 1.1 mM; NH4NO3, 5.9 mM; KH2PO4, 0.7 mM; K2HPO4, 0.7 mM; KOH, 0.28 mM; H2SO4, 0.28 mM; MgCl2, 1.6 mM; CaCl2, 3.2 mM; FeCl2, 0.02 mM] (11) added to each soil was calculated to achieve an equivalent matric potential of 33 kPa (Table 1).
Each bottle was sealed with a Teflon-coated rubber septum and an aluminum crimp top. The bottles were incubated at room temperature (25 ± 2°C) in the dark without shaking. For GC-MS analysis, 1-g (dry weight) subsamples were removed from replicate soil vessels at several time points, extracted, and analyzed as described previously (12). The efficiency of the solvent extraction procedure varied among soil types (ranging from 60 to 90%). The loss of total n-alkane fractions from control treatments with autoclaved soils was 15.0% ± 8.6% after a 50-day incubation, likely due to enhanced volatilization in air-purged reaction vessels. Thus, the percentage of n-alkane depletion in nonsterile treatments was normalized to the extractable amounts and then calculated relative to the amount of each n-alkane fraction recovered from control treatments at corresponding time points. The initial rates of hydrocarbon degradation were calculated as mg of total petroleum hydrocarbons degraded per gram of soil per day in each reaction vessel. The contaminated soils were purged twice a week with humidified CO2-free air to trap 14CO2, which also served to keep the system aerobic. Evolved 14CO2 was measured as described previously (12). After completion of the experiment, the mass balance was determined based on the sum of evolved 14CO2 plus residual soil 14C measured using total combustion (12) and resulted in average recoveries of 97.3% ± 4.6%.
Molecular analysis.
Bacterial populations responding to crude-oil contamination were examined by denaturing gradient gel electrophoresis (DGGE) analysis of PCR-amplified 16S rRNA gene fragments from each soil. DNA was extracted from 0.5-g (dry weight) subsamples after bead beating, and 16S rRNA gene fragments were PCR amplified using Bacteria-specific primer 1070F and the universal primer 1392R containing a GC clamp. Then the PCR products were separated using DGGE as described previously (9). Nucleotide sequences of dominant bands identified using DGGE were determined as described previously (9). These molecular approaches have been used successfully to amplify and identify phylogenetically diverse organisms throughout the domain Bacteria (6, 9, 22, 25) and to monitor changes in microbial populations associated with environmental perturbation (6, 22, 28, 44).
Sequences were assembled using Sequencher 4.1 (Gene Codes Corporation, Ann Arbor, MI) and compared to the GenBank database using BLAST (1). Alignments were performed by ClustalX (version 1.81) using default values (38) and edited manually. Distance analysis was performed using the Jukes and Cantor correction (15), followed by phylogenetic tree construction using the neighbor-joining method (34) of PAUP*4.0 software (Sinauer Associates, Sunderland, MA). The nucleotide sequences reported in this paper have been deposited in the GenBank database (see below).
Isolation of alkane degraders.
Following crude-oil contamination experiments, alkane-degrading bacteria were isolated by plating serially diluted soil slurry, prepared by vigorous mixing of a 1-g subsample of contaminated soil with 10 ml of sterile pyrophosphate buffer (2 mM sodium pyrophosphate, 24 mM NaCl, 2 mM sodium phosphate buffer [pH 8.0]) on minimal (Xm) medium plates (13), with n-hexadecane supplied as the sole carbon source through the vapor phase. Colonies were randomly selected from plates inoculated with dilutions of 10−4 to 10−5 soil slurry and restreaked for isolation. The isolates obtained were grown in liquid Xm medium containing 1% (vol/vol) n-hexadecane as a sole carbon source. Cells were collected by centrifugation and subjected to DNA extraction, followed by 16S rRNA gene amplification and sequencing as described previously (12).
Nucleotide sequence accession numbers.
The nucleotide sequences reported in this paper have been deposited in the GenBank database under accession numbers DQ149088 to DQ149100.
RESULTS
Crude-oil contamination assay.
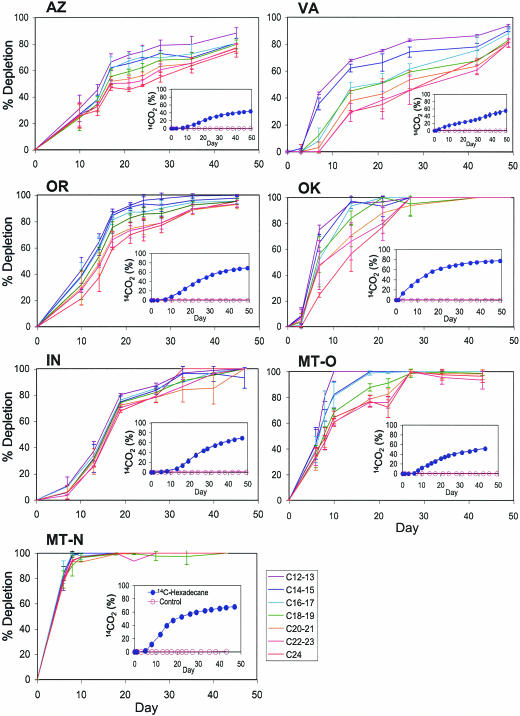
In all seven soils, greater than 80% of the added crude oil was depleted within the 50-day incubation (Fig. 1). Among the soils, differences in initial rates of hydrocarbon degradation (ranging from 0.19 to 2.00 mg hydrocarbons g soil−1 day−1 in VA and MT-N soils, respectively) and diverse patterns in utilization of various chain-length n-alkanes were observed. A pattern of sequential disappearance of shorter- before longer-chain-length n-alkanes was observed in five soils (AZ, OR, VA, OK, and MT-O). This pattern was most obvious in the VA soil, which showed initial loss of n-alkanes with chain lengths of 15 C or less followed by later loss of n-alkanes with chain lengths of at least 16 C. OK and MT-O soils showed greater chain-length-dependent depletion than OR and AZ soils. In contrast, all n-alkane components were depleted at approximately the same rate regardless of chain length in the IN and MT-N soils. The fastest depletion rate and shortest lag period were observed with the MT-N soil, where ∼81% of total n-alkanes disappeared by day 10. Only ∼40% depletion was observed by day 13 in the IN soil, likely due to a longer lag period (∼7 days).
FIG. 1.
Degradation of crude-oil components in different soils. Depletion of n-alkanes of C12 to C24 chain length is shown in main panels. Mineralization of [1-14C]hexadecane to 14CO2 is shown in inset panels with autoclaved soil as a control. Each point represents the average of results from triplicate bottles, and the error bars correspond to standard deviations. Where absent, error bars are smaller than symbol size.
Concomitant measurement of 14CO2 evolution from [1-14C]hexadecane confirmed the capability of indigenous microbial populations to mineralize an important crude-oil component in the soils (Fig. 1). Although the rates and extents of recovered 14CO2 varied among the soils, approximately 40 to 70% of added [14C]hexadecane was mineralized after the 50-day incubation. The total 14C recovered (14CO2 plus residual soil 14C) from the contaminated soil systems averaged 97.3% ± 4.6%. Control treatments with autoclaved soils showed no production of 14CO2, confirming the biological mineralization of hexadecane in these experiments. 14CO2 recovery ranged from 55 to 78% of the amount of hexadecane depletion measured using GC-MS among the soils, which likely reflects differences in the ratio of respiration versus incorporation of carbon into cellular mass by microbial populations.
Molecular analysis of bacterial community dynamics.
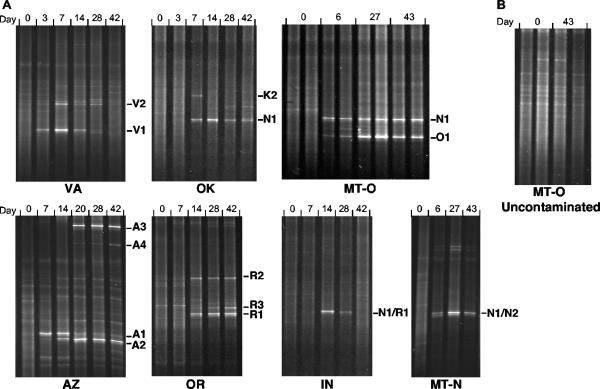
Bacterial populations associated with the depletion of hydrocarbon constituents in crude oil were examined by analysis of PCR-amplified 16S rRNA gene fragments using DGGE. The reproducibility of the techniques was confirmed using replicate samples from selected soils (a representative result from the MT-O soil is shown in Fig. 2). All seven soils showed the emergence of prominent DGGE bands during crude-oil depletion (Fig. 2A). Conversely, no obvious changes in DGGE banding patterns were observed in the uncontaminated control soils during the incubation period (Fig. 2B). Consequently, the populations corresponding to the prominent DGGE bands in treatments containing 2% crude oil were clearly due to hydrocarbon contaminations rather than other experimental factors such as nutrient addition or stimulation of microbial growth due to incubation at field-capacity (33 kPa) water content.
FIG. 2.
DGGE profiles of 16S rRNA gene fragments from crude-oil-contaminated soils (A) and uncontaminated control soil (B). The nucleotide sequences of the labeled bands were determined and are described further in Table 2 and Fig. 3.
Temporal changes in microbial populations also occurred during the degradation of crude oil in AZ, VA, and OK soils (Fig. 2). In the AZ soil, a prominent band A1 appeared on day 7, followed by emergence of band A2 on day 14, and two additional bands (A3 and A4) on day 20. Band A1 became less prominent after day 20, while bands A2 and A3 persisted throughout the incubation period. In the VA soil, band V1 emerged before (day 3) band V2 (day 7); both bands became less prominent upon further incubation. In the OK soil, two prominent bands (N1 and K2) appeared on day 7, but only one band (N1) persisted throughout the incubation period. In contrast, the other four soils (OR, IN, MT-N, and MT-O) showed no obvious successional patterns, where DGGE bands emerging early in the experiment remained relatively prominent throughout crude-oil degradation (Fig. 2). The prominent DGGE bands in soils from IN (N1 and R1) and MT-N (N1 and N2), which appear to constitute single bands, are actually two closely migrating bands that have significantly different 16S rRNA gene sequences (see below) (Table 2). It is noteworthy that the populations associated with DGGE bands selected in the IN and VA soils became less prominent after nearly complete depletion of crude oil by day 42, while in other soils, dominant bands persisted for many days after the complete depletion of crude oil (e.g., up to 30 days in MT-N soil).
TABLE 2.
Sequence analysis of 16S rRNA gene fragments detected in soils contaminated with crude oil
| DGGE banda | Soil(s) | Accession no. | Closest GenBank relativeb
|
% Similarity | |
|---|---|---|---|---|---|
| Phylogenetic groupc | Strain, species, or clone | ||||
| A1 | AZ | DQ149088 | Actinobacteria | Nocardioides jensenii | 99.7 |
| A2 | AZ | DQ149089 | Actinobacteria | Nocardioides albus | 100.0 |
| A3 | AZ | DQ149090 | Candidate division TM7 | Uncultured bacterial clone MAFB-C4-28 | 94.8 |
| A4 | AZ | DQ149091 | Uncultured bacterial clone 44a-U1-9 | 98.1 | |
| β-Proteobacteria | Chromobacterium sp. strain 70 | 91.6 | |||
| R1 | OR, IN | DQ149092 | Uncultured bacterial clone WD284 | 99.7 | |
| γ-Proteobacteria | Nevskia ramosa strain Soe1 | 99.7 | |||
| R2 | OR | DQ149093 | β-Proteobacteria | Collimonas sp. strain CTO 300 | 100.0 |
| R3 | OR | DQ149094 | γ-Proteobacteria | Rhodanobacter fulvus | 99.6 |
| V1 | VA | DQ149095 | Uncultured bacterial clone 156ds20 | 99.4 | |
| γ-Proteobacteria | Acinetobacter calcoaceticus | 94.1 | |||
| V2 | VA | DQ149096 | Uncultured bacterial clone WD2102 | 99.1 | |
| β-Proteobacteria | Leptothrix mobilis | 99.1 | |||
| K2 | OK | DQ149097 | γ-Proteobacteria | Alkanindiges hongkongensis | 97.5 |
| N1 | IN, OK, MT-N, MT-O | DQ149098 | Actinobacteria | Rhodococcus erythropolis | 100.0 |
| N2 | MT-N | DQ149099 | Actinobacteria | Rhodococcus coprophilus | 100.0 |
| O1 | MT-O | DQ149100 | Actinobacteria | Nocardia sp. strain DSM6249 | 100.0 |
The DNA sequences of prominent DGGE bands corresponding to bacterial populations selected during crude-oil degradation are reported along with their closest phylogenetic relatives in Table 2. Phylogenetically diverse populations related to β- and γ-Proteobacteria, Actinobacteria, and candidate division TM7 were identified across the set of contaminated soils (Fig. 3). However, the DNA sequences of bands N1 and R1 were detected in multiple soils; band N1 was detected in four diverse soils (IN, OK, MT-N, and MT-O), and band R1 was detected in two soils (OR and IN). The sequences corresponding to band N1 were 100% identical to the partial 16S rRNA gene sequence of Rhodococcus erythropolis NRRL B-16531. All 16S rRNA gene sequences obtained from the MT-N and MT-O soils were affiliated with gram-positive organisms, while all sequences obtained from the OR and VA soils were affiliated with gram-negative organisms. AZ, IN, and OK soils contained 16S rRNA gene sequences affiliated with both gram-positive and gram-negative organisms. All of the gram-positive-related sequences clustered within the Nocardia-Rhodococcus-Nocardioides complex, members of which are known to degrade hydrocarbons (41), while the gram-negative-related sequences were represented by phylogenetically diverse groups (Fig. 3).
FIG. 3.
Phylogenetic positions of 16S rRNA gene sequences from crude-oil-contaminated soils. The neighbor-joining tree was generated based on partial 16S rRNA gene sequences from the GenBank database, corresponding to nucleotide positions 677 to 1337 of the Escherichia coli 16S rRNA gene. The partial-length DGGE band sequences (<323 nucleotides) were used for the analysis, and their positions are indicated by dashed line segments. GenBank sequences from isolates are in italics, GenBank sequences from environmental gene clones are in plain type, and DGGE band sequences from this study are in boldface type. The tree was rooted with the 16S rRNA sequence of Sulfolobus acidocaldarius (accession no. D14876). Bootstrap values (per 100 trials) are shown. Bar, 0.01 substitutions per sequence position.
Isolation of alkane-degrading bacteria.
To link microbial populations detected by molecular methods with their function in crude-oil metabolism, alkane-degrading isolates were cultivated from each of the crude-oil-contaminated soils following the 50-day incubation (Table 3). Isolates that grew on minimal medium with n-hexadecane as a sole carbon source were examined for their relevance in contaminated soils. At least one of the prominent populations identified using DGGE analysis (above) was successfully cultivated from six of the seven soils. None of the isolates from the VA soil corresponded to the DGGE bands (V1 and V2) identified in the crude-oil-contaminated VA soil (data not shown). However, in many cases, the populations observed in contaminated soils using molecular methods were recovered most frequently during isolation of alkane-degrading microorganisms. Isolates include known alkane-degrading genera, Rhodococcus sp. and Nocardioides sp., as well as a Collimonas sp. We also obtained numerous isolates that did not correspond to prominent DGGE bands in contaminated soils (Table 3); many of these isolates were closely related to known hydrocarbon-degrading organisms, including Variovorax, Burkholderia, Ralstonia, Nocardia, Gordonia, Acinetobacter, and Pseudomonas (Table 3).
TABLE 3.
Alkane-degrading isolates obtained from each soil type and correlation with 16S rRNA gene sequences observed in crude-oil-contaminated soils
| Soil | No. of isolates | DGGE banda | Closest GenBank relative
|
% Similarity | |
|---|---|---|---|---|---|
| Phylogenetic group | Strain or species (accession no.) | ||||
| AZ | 6 | A2 | Actinobacteria | Nocardioides albus (AF005005) | 100.0 |
| 4 | Actinobacteria | Nocardioides alkalitolerans (AY633972) | 100.0 | ||
| 1 | Actinobacteria | Cellulomonas sp. strain X7 (AF060791) | 100.0 | ||
| 1 | β-Proteobacteria | Variovorax paradoxus (AF532868) | 100.0 | ||
| OR | 2 | R2 | β-Proteobacteria | Collimonas sp. strain CTO 300 (AY281145) | 100.0 |
| 3 | β-Proteobacteria | Burkholderia sp. strain CI6 (AY178099) | 100.0 | ||
| 2 | β-Proteobacteria | Burkholderia sp. strain Ch1-1 (AY367011) | 98.9 | ||
| 1 | β-Proteobacteria | Ralstonia sp. strain AU3369 (AF500587) | 100.0 | ||
| 1 | Actinobacteria | Streptomyces sp. strain EF-79 (AF112167) | 100.0 | ||
| IN | 7 | N1 | Actinobacteria | Rhodococcus erythropolis (X81929) | 100.0 |
| 4 | α-Proteobacteria | Sphingomonas asaccharolytica (AJ871435) | 98.9 | ||
| 2 | α-Proteobacteria | Mesorhizobium plurifarium (AF516883) | 100.0 | ||
| 1 | Actinobacteria | Nocardia otitidiscaviarum (X80599) | 100.0 | ||
| 1 | β-Proteobacteria | Burkholderia sp. strain Br3469 (AY773197) | 100.0 | ||
| VA | 9 | γ-Proteobacteria | Pseudomonas sp. strain K94.08 (AY456703) | 100.0 | |
| 3 | β-Proteobacteria | Hydrogenophaga taeniospiralis (AF078768) | 99.6 | ||
| 3 | α-Proteobacteria | Sphingopyxis alaskensis (AY337601) | 100.0 | ||
| 2 | Actinobacteria | Streptomyces sp. strain EF-79 (AF112167) | 99.2 | ||
| 2 | α-Proteobacteria | Sphingomonas agrestis (AY506539) | 100.0 | ||
| 1 | β-Proteobacteria | Phenanthrene-degrading strain 90 (AY177371) | 96.7 | ||
| OK | 8 | N1 | Actinobacteria | Rhodococcus erythropolis (X81929) | 100.0 |
| 3 | Actinobacteria | Microbacterium sp. strain SD-9 (AY336120) | 100.0 | ||
| 1 | Actinobacteria | Gordonia rubripertincta (X80632) | 100.0 | ||
| 1 | β-Proteobacteria | Variovorax paradoxus (AF532868) | 100.0 | ||
| 1 | γ-Proteobacteria | Pseudomonas sp. strain B65 (AF332541) | 100.0 | ||
| 1 | Actinobacteria | Streptomyces africanus (AY208912) | 100.0 | ||
| MT-N | 5 | N1 | Actinobacteria | Rhodococcus erythropolis (X81929) | 100.0 |
| 1 | N2 | Actinobacteria | Rhodococcus coprophilus (U93340) | 100.0 | |
| 2 | β-Proteobacteria | Burkholderia cepacia (AY268162) | 100.0 | ||
| 1 | Actinobacteria | Cellulomonas sp. strain UFZ-B529 (AF237956) | 100.0 | ||
| 1 | Actinobacteria | Streptomyces sp. strain An53 (AM039887) | 100.0 | ||
| MT-O | 1 | N1 | Actinobacteria | Rhodococcus erythropolis (X81929) | 100.0 |
| 5 | γ-Proteobacteria | Pseudomonas fluorescens (AY267197) | 100.0 | ||
| 1 | γ-Proteobacteria | Acinetobacter rhizosphaerae (AY364536) | 100.0 | ||
| 1 | Actinobacteria | Arthrobacter chlorophenolicus (AY167845) | 100.0 | ||
| 1 | Firmicutes | Bacillus silvestris (AJ006086) | 100.0 | ||
Identical to DGGE band sequences in Table 2.
DISCUSSION
This study demonstrates that crude-oil contamination results in the selection of diverse microbial populations across geographically and physiochemically distinct soils. Although previous observations using cultivation-dependent (3, 4, 35) and cultivation-independent (31, 40, 46) approaches have shown that hydrocarbon-degrading bacteria are ubiquitous in soil and water environments, this is the first study to systematically evaluate microbial population responses to a common contaminant (crude oil) across different soil types under conditions in which other parameters (e.g., water potential, nutrients, temperature, aeration) were held constant. Different soil properties reflect the complex set of climatic, biotic, geologic, and hydrologic factors responsible for soil formation. Consequently, although most of the sites we studied are geographically dispersed, any effects of geographic isolation on microbial population response are confounded with variation in soil properties. For example, pH values ranged from 5.4 to 8.8, and soil textures ranged from loam to clay. These properties influence microbial community composition and contribute to the observed variation in microbial populations responsible for crude-oil degradation. Conversely, any common patterns in the microorganisms selected across such a disparate group of soils would be useful for understanding responses to crude-oil contamination across numerous different locations. Previously, Bundy et al. (5) examined the effect of diesel contamination on the microbial communities in three soil types (cambisol, podsol, and fluvisol) with similar texture and pH using phospholipid fatty acid profiling and Biolog plate assays. They also showed that different community profiles developed in different soil types; however, the phylogeny of community members was not identified, thus no insights were obtained regarding the specific microbial populations selected. Our observations show that in situ biodegradation of alkane mixtures in different soil environments often involves different microbial population but that many soils demonstrate a consistent pattern in the types of microorganisms that respond to alkane perturbation.
The most consistent pattern observed in the current study was the emergence of Rhodococcus erythropolis-like microorganisms (Fig. 2, DGGE bands N1) in 4 of 7 contrasting soil types subjected to crude-oil contamination. Several previous studies showed that Pseudomonas spp. are commonly detected in various hydrocarbon-contaminated environments using culture-dependent (3, 4, 35) and culture-independent (17, 23, 31) approaches. However, our results indicate that this is not a universal trend, as Pseudomonas-like sequences were not detected using molecular approaches in any of the 7 crude-oil-contaminated soils examined. The absence of Pseudomonas-like sequences in this study is not an artifact of the molecular techniques employed, since the same PCR-DGGE approach (with same PCR primer set) has detected Pseudomonas-like organisms in hexadecane-contaminated soils (6) as well as in crude-oil-contaminated MT-N soil incubated at 10 and 4°C (N. Hamamura, D. M. Ward, and W. P. Inskeep, unpublished data).
Wide distribution of the R. erythropolis-like N1 population across geographically and physiochemically distinct soils and its association with crude-oil contamination indicate the importance of Rhodococcus spp. in hydrocarbon degradation in various soil environments. This observation is consistent with results from previous studies showing the prevalence of alkane-degrading Rhodococcus spp. in various hydrocarbon-contaminated soil environments using culture-dependent (2, 37) and culture-independent approaches (20, 23, 37, 46). Successful cultivation of R. erythropolis-like N1-type alkane-degrading isolates from four different soils (IN, OK, MT-N, and MT-O) further suggests that these organisms play a central role in hydrocarbon metabolism in situ. The partial 16S rRNA gene sequences obtained from R. erythropolis N1-type isolates from different soil types were confirmed to be identical (Table 3). However, these N1-type isolates may not be functionally identical, since various R. erythropolis strains with identical 16S rRNA gene sequences have been shown to contain three to five alkane hydroxylase gene homologues, whose functions may correlate with the oxidation of a different range of n-alkanes (39). Differences in crude-oil depletion patterns among the four soils (IN, OK, MT-N, and MT-O) with common R. erythropolis-like N1 populations (Fig. 1) may reflect functional variations within R. erythropolis strains, and this is the subject of further investigation.
The microbial population dynamics observed during crude-oil depletion may be associated with degradation patterns of specific n-alkane fractions. For instance, in the VA soil, the emergence of population V1 at day 3 correlated with preferential degradation of n-alkanes with chain lengths of ≤C15, which was followed by the emergence of an additional population V2 at day 7 corresponding to the degradation of n-alkanes with chain lengths of ≥C16 observed after day 7 (Fig. 1 and 2). This suggests that the VA soil may harbor specialist populations with different substrate ranges based on n-alkane chain lengths. In contrast, in soils where no population succession was observed during crude-oil depletion (OR, IN, MT-N, and MT-O) (Fig. 2), the dominant bacterial populations may function as generalists that possess alkane degradation pathways with a broad substrate range of n-alkane components of different chain lengths. For example, the detection of persistent and prominent N1 populations in IN, MT-N, and MT-O soils during the depletion of all n-alkane components in crude oil (Fig. 1 and 2) supports the hypothesis that R. erythropolis-like N1 populations function as generalists, and this is consistent with previous reports that R. erythropolis NRRL B-16531 is capable of degrading C6 to C36 n-alkanes (39).
The cultivation strategy employed in the current study was successful in retrieving a significant subset of the alkane-degrading bacteria in crude-oil-contaminated soils shown to be present using molecular methods. Included were Rhodococcus sp., Nocardioides sp., and Collimonas sp. Collimonas spp. have not been previously recognized as alkane degraders but rather for their chitinase activity and ability to grow on living fungal hyphae (7). It was shown that the 16S rRNA gene sequences of Collimonas strains were highly similar (98.9 to 99.9%) to that of a naphthalene-degrading strain, which was originally classified as Herbaspirillum sp. (7). This suggests that the ability to metabolize hydrocarbons may be extended to Collimonas spp.
It is clear, however, that additional strategies (e.g., media composition, incubation conditions and duration, analysis of larger number of colonies) will be necessary for isolating other organisms observed using molecular approaches. For instance, Ferrari et al. (8) recently described a novel microcultivation method for soil bacteria that mimics natural conditions. This approach successfully isolated previously “unculturable” organisms belonging to candidate division TM7, and one of the sequences observed using molecular approaches (e.g., band A3) (Fig. 2) was highly related to organisms from this division. Alternatively, it is possible that some populations detected using molecular methods were not directly responsible for alkane degradation but rather may have facilitated hydrocarbon bioavailability (e.g., production of surfactants) (16, 43), or utilized secondary products produced by alkane-degraders (10). For instance, band R3 was associated with Rhodanobacter spp., a genus that includes an isolate capable of stimulating the mineralization of benzo[a]pyrene (16). Populations indirectly associated with alkane degradation may not have been cultivated on n-alkanes as sole carbon sources.
We also obtained numerous alkane-degrading isolates that did not correspond to prominent bands in contaminated soils (Table 3). Traditional cultivation strategies are often selective and favor faster-growing populations, whose role in situ may not be relevant. Thus, it is possible that such isolates were obtained only because our cultivation techniques favored their selection. The cultivation of numerous hydrocarbon-degrading isolates from soil environments might overestimate their importance in situ without confirmation using culture-independent molecular analyses (6, 10). Alternatively, it is possible that these isolates were simply not involved in the degradation of crude-oil components under the conditions used in the current study. This would not rule out their potential importance as hydrocarbon-degrading populations under different environmental conditions with different niches. For instance, Colores et al. (6) showed that the addition of surfactant to a hydrocarbon-contaminated soil displaced dominant gram-positive Rhodococcus and Nocardia populations with gram-negative Pseudomonas and Alcaligenes populations. It is also possible that the PCR-DGGE approach employed in this study was unable to detect dominant hydrocarbon-degrading populations in the contaminated soils, although we believe this is highly unlikely considering the success in cultivating relevant hydrocarbon degraders from the crude-oil-contaminated soils and the success of molecular approaches in detecting 16S rRNA gene fragments from a wide range of Bacteria representatives (6, 9, 22, 25), as well as in revealing the presence and identity of dominant microorganisms in numerous habitats (9, 18, 25, 33, 36, 45). It is noteworthy that the dominant populations associated with prominent DGGE bands in some soils became less prominent after the depletion of crude oil concomitant with the increase in community diversity (IN and VA soils) (Fig. 2). A rapid recovery (in 26 days) of bacterial community diversity to near prepollution level has been reported in oil-contaminated beach sediment microcosms by Röling et al. (32), although there were considerable qualitative differences in the community structure before and after bioremediation.
It is clear from our studies that the properties of geographically and physiochemically distinct soils can influence the kinetics and patterns of hydrocarbon degradation as well as the specific microbial populations that respond to crude-oil contamination. Thus, the underlying mechanistic basis for population selection in soils and natural waters contaminated with complex hydrocarbon mixtures is of significant interest for predicting and managing in situ bioremediation. For example, different hydrocarbon-degrading populations likely occur in different soils in differing abundances and, thus, may not be equally available at the time of hydrocarbon contamination. Furthermore, physical-chemical properties of soils may control which organisms respond to the introduction of hydrocarbons. In such cases, further work will be necessary to determine why soils of different character exhibit unique population responses to hydrocarbon contamination. By studying a larger number and variety of soils, it might be possible to gain insight through statistical correlation of populations responding to hydrocarbon contamination and the soil properties to which they are adapted. However, R. erythropolis-like, N1-type populations were more frequently observed after crude-oil contamination than any other microorganisms, several of which were only detected in one soil. Observations across additional soil types will be necessary to confirm the importance of Rhodococcus-like and other gram-positive Actinobacteria in the degradation of alkanes in contaminated soils, however, the patterns observed in this study suggest that members of this lineage commonly respond to hydrocarbon perturbation and are important in the degradation of alkanes with various chain lengths, independent of soil type and geographic location.
Acknowledgments
We thank the following individuals for collecting soil samples from their respective states: John Baham, Oregon State University; Brian Carter, Oklahoma State University; Matt Eick, Virginia Polytechnic University; Bill McPhee and Linda Lee, Purdue University; and Jeff Silvertooth, University of Arizona. We also appreciate technical assistance from Mary Bateson, Katherine Schultz, and Rich Macur. We are indebted to the Conoco refinery (Billings, MT) for providing us with crude oil.
This work was supported by the USEPA (project no. 829357-01-0) and the Montana Agricultural Experiment Station (projects 911398 and 911352).
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Bej, A. K., D. Saul, and J. Aislabie. 2000. Cold-tolerant alkane-degrading Rhodococcus species from Antarctica. Polar Biol. 23:100-105. [Google Scholar]
- 3.Belhaj, A., N. Desnoues, and C. Elmerich. 2002. Alkane biodegradation in Pseudomonas aeruginosa strains isolated from polluted zone: identification of alkB and alkB-related genes. Res. Microbiol. 153:339-344. [DOI] [PubMed] [Google Scholar]
- 4.Bhattacharya, D., P. M. Sarma, S. Krishnan, S. Mishra, and B. Lal. 2003. Evaluation of genetic diversity among Pseudomonas citronellolis strains isolated from oily sludge-contaminated sites. Appl. Environ. Microbiol. 69:1435-1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bundy, J. G., G. I. Paton, and C. D. Campbell. 2002. Microbial communities in different soil types do not converge after diesel contamination. J. Appl. Microbiol. 92:276-288. [DOI] [PubMed] [Google Scholar]
- 6.Colores, G. M., R. E. Macur, D. M. Ward, and W. P. Inskeep. 2000. Molecular analysis of surfactant-driven microbial population shifts in hydrocarbon-contaminated soil. Appl. Environ. Microbiol. 66:2959-2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Boer, W., J. H. J. Leveau, G. A. Kowalchuk, P. J. A. K. Gunnewiek, E. C. A. Abeln, M. J. Figge, K. Sjollema, J. D. Janse, and J. A. van Veen. 2004. Collimonas fungivorans gen. nov., sp. nov., a chitinolytic soil bacterium with the ability to grow on living fungal hyphae. Int. J. Syst. Evol. Microbiol. 54:857-864. [DOI] [PubMed] [Google Scholar]
- 8.Ferrari, B. C., S. J. Binnerup, and M. Gillings. 2005. Microcolony cultivation on a soil substrate membrane system selects for previously uncultured soil bacteria. Appl. Environ. Microbiol. 71:8714-8720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferris, M. J., G. Muyzer, and D. M. Ward. 1996. Denaturing gradient gel electrophoresis profiles of 16S rRNA-defined populations inhabiting a hot spring microbial mat community. Appl. Environ. Microbiol. 62:340-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Friedrich, M., R. J. Grosser, E. A. Kern, W. P. Inskeep, and D. M. Ward. 2000. Effect of model sorptive phases on phenanthrene biodegradation: molecular analysis of enrichments and isolates suggests selection based on bioavailability. Appl. Environ. Microbiol. 66:2703-2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grosser, R. J., M. Friedrich, D. M. Ward, and W. P. Inskeep. 2000. Effect of model sorptive phase on phenanthrene biodegradation: different enrichment conditions influence bioavailability and selection of phenanthrene-degrading isolates. Appl. Environ. Microbiol. 66:2695-2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hamamura, N., S. H. Olson, D. M. Ward, and W. P. Inskeep. 2005. Diversity and functional analysis of bacterial communities associated with natural hydrocarbon seeps in acidic soils at Rainbow Springs, Yellowstone National Park. Appl. Environ. Microbiol. 71:5943-5950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hamamura, N., R. T. Storfa, L. Semprini, and D. J. Arp. 1999. Diversity in butane monooxygenases among butane-grown bacteria. Appl. Environ. Microbiol. 65:4586-4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hatzinger, P. B., and M. Alexander. 1995. Effect of aging of chemicals in soil on their biodegradability and extractability. Environ. Sci. Technol. 29:537-545. [DOI] [PubMed] [Google Scholar]
- 15.Jukes, T. H., and C. R. Cantor. 1969. Evolution of protein molecules, p. 21-132. In H. N. Munro (ed.), Mammalian protein metabolism. Academic Press, Inc., New York, N.Y.
- 16.Kanaly, R. A., S. Harayama, and K. Watanabe. 2002. Rhodanobacter sp. strain BPC1 in a benzo[a]pyrene-mineralizing bacterial consortium. Appl. Environ. Microbiol. 68:5826-5833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaplan, C. W., and C. L. Kitts. 2004. Bacterial succession in petroleum land treatment unit. Appl. Environ. Microbiol. 70:1777-1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koizumi, Y., H. Kojima, K. Oguri, H. Kitazato, and M. Fukui. 2004. Vertical and temporal shifts in microbial communities in the water column and sediment of saline meromictic Lake Kaiike (Japan), as determined by a 16S rDNA-based analysis, and related to physicochemical gradients. Environ. Microbiol. 6:622-637. [DOI] [PubMed] [Google Scholar]
- 19.Luthy, R. G., G. R. Aiken, M. L. Brusseau, S. D. Cunningham, P. M. Gschwend, J. J. Pignatello, M. Reinhard, S. J. Traina, W. J. Weber, Jr., and J. C. Westall. 1997. Sequestration of hydrophobic organic contaminants by geosorbents. Environ. Sci. Technol. 31:3341-3347. [Google Scholar]
- 20.Luz, A. P., V. H. Pellizari, L. G. Whyte, and C. W. Greer. 2004. A survey of indigenous microbial hydrocarbon degradation genes in soils from Antarctica and Brazil. Can. J. Microbiol. 50:323-333. [DOI] [PubMed] [Google Scholar]
- 21.MacNaughton, S. J., J. R. Stephen, A. D. Venosa, G. A. Davis, Y. Chang, and D. C. White. 1999. Microbial population changes during bioremediation of an experimental oil spill. Appl. Environ. Microbiol. 65:3566-3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Macur, R. E., C. R. Jackson, L. M. Botero, T. R. McDermott, and W. P. Inskeep. 2004. Bacterial populations associated with the oxidation and reduction of arsenic in an unsaturated soil. Environ. Sci. Technol. 38:104-111. [DOI] [PubMed] [Google Scholar]
- 23.Margesin, R., D. Labbé, F. Schinner, C. W. Greer, and L. G. Whyte. 2003. Characterization of hydrocarbon-degrading microbial populations in contaminated and pristine Alpine soils. Appl. Environ. Microbiol. 69:3085-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nelson, D. W., and L. E. Sommers. 1982. Total carbon, organic carbon and organic matter, p. 539-579. In A. L. Page (ed.), Methods of soil analysis. Part 2. Chemical and microbiological properties. American Society of Agronomy, Madison, Wis.
- 25.Norris, T. B., J. M. Wraith, R. W. Castenholz, and T. R. McDermott. 2002. Soil microbial community structure across a thermal gradient following a geothermal heating event. Appl. Environ. Microbiol. 68:6300-6309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olsen, S. R., and L. E. Sommers. 1982. Phosphorus, p. 403-430. In A. L. Page (ed.), Methods of soil analysis. Part 2. Chemical and microbiological properties. American society of Agronomy, Madiosn, Wis.
- 27.Olson, J. J., G. L. Mills, B. E. Herbert, and P. J. Morris. 1999. Biodegradation rates of separated diesel components. Environ. Toxicol. Chem. 18:2448-2453. [Google Scholar]
- 28.Ovreas, L., S. Jensen, F. L. Daae, and V. Torsvik. 1998. Microbial community changes in a perturbed agricultural soil investigated by molecular and physiological approaches. Appl. Environ. Microbiol. 64:2739-2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pignatello, J. J., and B. Xing. 1996. Mechanisms of slow sorption of organic chemicals to natural particles. Environ. Sci. Technol. 30:1-11. [Google Scholar]
- 30.Richards, L. A. 1965. Physical condition of water in soil, p. 128-152. In C. A. Black (ed.), Methods of soil analysis. Part 1. Physical and mineralogical properties. American Society of Agronomy, Madison, Wis.
- 31.Röling, W. F. M., M. G. Milner, D. M. Jones, F. Fratepietro, R. P. J. Swannell, F. Daniel, and I. M. Head. 2004. Bacterial community dynamics and hydrocarbon degradation during a field-scale evaluation of bioremediation on a mudflat beach contaminated with buried oil. Appl. Environ. Microbiol. 70:2603-2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Röling, W. F. M., M. G. Milner, D. M. Jones, K. Lee, F. Daniel, R. J. P. Swannell, and I. M. Head. 2002. Robust hydrocarbon degradation and dynamics of bacterial communities during nutrient-enhanced oil spill bioremediation. Appl. Environ. Microbiol. 68:5537-5548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rooney-Varga, J. N., R. T. Anderson, J. L. Fraga, D. Ringelberg, and D. R. Lovley. 1999. Microbial communities associated with anaerobic benzene degradation in a petroleum-contaminated aquifer. Appl. Environ. Microbiol. 65:3056-3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 35.Stapleton, R. D., N. G. Bright, and G. S. Sayler. 2000. Catabolic and genetic diversity of degradative bacteria from fuel-hydrocarbon contaminated aquifers. Microb. Ecol. 39:211-221. [DOI] [PubMed] [Google Scholar]
- 36.Sun, H. Y., S. P. Deng, and W. R. Raun. 2004. Bacterial community structure and diversity in a century-old manure-treated agroecosystem. Appl. Environ. Microbiol. 70:5868-5874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thomassin-Lacroix, E. J. M., Z. Yu, M. Eriksson, K. J. Reimer, and W. W. Mohn. 2001. DNA-based and culture-based characterization of a hydrocarbon-degrading consortium enriched from Arctic soil. Can. J. Microbiol. 47:1107-1115. [DOI] [PubMed] [Google Scholar]
- 38.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Beilen, J. B., T. H. M. Smits, L. G. Whyte, S. Schorcht, M. Röthlisberger, T. Plaggemeier, K.-H. Engesser, and B. Witholt. 2002. Alkane hydroxylase homologues in Gram-positive strains. Environ. Microbiol. 4:676-682. [DOI] [PubMed] [Google Scholar]
- 40.Van Hamme, J. D., A. Singh, and O. P. Ward. 2003. Recent advances in petroleum microbiology. Microbiol. Mol. Biol. Rev. 67:503-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vomberg, A., and U. Klinner. 2000. Distribution of alkB genes within n-alkane-degrading bacteria. J. Appl. Microbiol. 89:339-348. [DOI] [PubMed] [Google Scholar]
- 42.Ward, D. M., R. M. Atlas, P. D. Boehm, and J. A. Calder. 1980. Microbial biodegradation and chemical evolution of oil from the Amoco spill. AMBIO 9:277-283. [Google Scholar]
- 43.Ward, O., A. Singh, and J. Van Hamme. 2003. Accelerated biodegradation of petroleum hydrocarbon waste. J. Ind. Microbiol. Biotechnol. 30:260-270. [DOI] [PubMed] [Google Scholar]
- 44.Watanabe, K., M. Teramoto, H. Futamata, and S. Harayama. 1998. Molecular detection, isolation, and physiological characterization of functionally dominant phenol-degrading bacteria in activated sludge. Appl. Environ. Microbiol. 64:4396-4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.West, N. J., and D. J. Scanlan. 1999. Niche-partitioning of Prochlorococcus populations in a stratified water column in the eastern North Atlantic Ocean. Appl. Environ. Microbiol. 65:2585-2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Whyte, L. G., A. Schultz, J. B. van Beilen, A. P. Luz, V. Pellizari, D. Labbé, and C. W. Greer. 2002. Prevalence of alkane monooxygenase genes in Arctic and Antarctic hydrocarbon-contaminated and pristine soils. FEMS Microbiol. Ecol. 41:141-150. [DOI] [PubMed] [Google Scholar]