Abstract
Detection of plasmid DNA uptake in river bacteria at the single-cell level was carried out by rolling-circle amplification (RCA). Uptake of a plasmid containing the green fluorescent protein gene (gfp) by indigenous bacteria from two rivers in Osaka, Japan, was monitored for 506 h using this in situ gene amplification technique with optimized cell permeabilization conditions. Plasmid uptake determined by in situ RCA was compared to direct counts of cells expressing gfp under fluorescence microscopy to examine differences in detection sensitivities between the two methods. Detection of DNA uptake as monitored by in situ RCA was 20 times higher at maximum than that by direct counting of gfp-expressing cells. In situ RCA could detect bacteria taking up the plasmid in several samples in which no gfp-expressing cells were apparent, indicating that in situ gene amplification techniques can be used to determine accurate rates of extracellular DNA uptake by indigenous bacteria in aquatic environments.
Genetic transformation, a principal mechanism of lateral gene transfer, is characterized by the uptake of free DNA by a recipient bacterium, its chromosomal integration or extrachromosomal stabilization, and its expression, which leads to a new phenotype (12, 24). About 90 bacterial species have been known to be transformable so far (34). Some species are naturally competent, and some species require manipulation to become competent (11). Ten percent of marine isolates have been reported to be transformable by plasmids, and 14% are transformable by chromosome fragments (13). One study suggested that Pseudomonas fluorescens is transformable in its native soil environment but not in vitro (10). Many more species may therefore be competent for transformation, but the conditions under which they develop competence are still unknown (6).
Uptake of free DNA by a recipient bacterium is the important first step of genetic transformation, and the machinery of uptake has been studied extensively in a limited number of bacterial species (5, 12). The frequency of DNA uptake by naturally occurring bacteria as determined by conventional culture methods has been underestimated and biased primarily because only a small fraction of metabolically active bacteria present in the environment is culturable (9, 31). Recently, a culture-independent technique based on the expression of the green fluorescent protein gene (gfp) has been used to monitor plasmid transformation of an Acinetobacter species monoculture (15). Using expression of gfp, Acinetobacter sp. transformation was detectable at a DNA concentration that was 106-fold lower than that previously reported. This technique is, however, limited by differences in cell physiology and the availability of organism-specific active promoters (34). Thus, measurements of natural competence in a diverse population of bacteria based on DNA sequence targeting are less likely to be biased by physiological or genetic (i.e., marker expression) limitations and may be required for accurate measurements of DNA uptake (35).
Detection of single-copy genes in individual cells is necessary to determine the frequency of DNA uptake in environmental bacteria, which should be possible with in situ gene amplification techniques (27). Although several kinds of these techniques have been reported previously (16, 25, 27, 39), only a few studies have applied in situ PCR to the quantitative monitoring of bacteria in natural environments (22, 36, 37). The main barrier for the application of these techniques to environmental samples is the optimization of cell permeabilization conditions for a broad spectrum of bacteria that would allow reagents to penetrate without detachment from an anchoring substrate or destruction of the cell (19, 47). To examine cell permeability, these techniques need to include all of the bacteria present in a sample by using universally conserved DNA sequences as a validation of the method before specific bacteria are targeted. Fluorescence in situ hybridization (FISH) targeting of 16S rRNA genes usually uses an 18-bp oligonucleotide DNA probe as a universal bacterial probe, which, however, requires another two probes to detect all bacterial species (8). In situ PCR requires two primers, each with a length of around 20 bp, which means that a target sequence of at least 40 bp within a specific region is required. However, the requirement for a longer primer makes it more difficult to amplify target sequences from all types of bacteria, since it is difficult to find a universal sequence long enough to satisfy technical requirements.
In this study, a specific gene amplification method, rolling-circle amplification (RCA), was used to monitor the number of river bacteria that take up a specific gene sequence at the single-cell level (27). We examined the optimization of cell permeabilization with three enzymes and heat treatment using river water bacteria and laboratory strains as test populations by fluorescently labeled protein staining. The accuracy of the in situ RCA method was confirmed by detecting bacteria carrying gfp that had been added to river water samples. Once the method was validated, plasmid DNA carrying gfp was added to two river water samples, and the uptake of the plasmid by river bacteria was monitored for 506 h by using in situ RCA. Frequencies of plasmid uptake determined by in situ RCA were compared with direct counts of cells expressing the gfp gene under fluorescence microscopy. The plasmid was also quantified by real-time PCR to validate the observed frequencies.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
Twenty-two bacterial strains from phylogenetically diverse groups were used to examine cell permeability: α-Proteobacteria (Agrobacterium tumefaciens and Brevundimonas diminuta), β-Proteobacteria (Alcaligenes xylosoxidans, Alcaligenes faecalis, Burkholderia cepacia, Comamonas testosteroni, and Ralstonia eutropha), γ-Proteobacteria (Acinetobacter calcoaceticus, Aeromonas hydrophila, Aeromonas sobria, Enterobacter gergoviae, Escherichia coli W3110, Moraxella nonliquefaciens, and Pseudomonas putida), the Bacteroides-Flavobacterium-Cytophaga phylum (Chryseobacterium meningosepticum, Cytophaga xantha, Empedobacter brevis, and Sphingobacterium thalpophilum), Firmicutes (Bacillus megaterium, Bacillus subtilis, and Staphylococcus epidermidis), and Actinobacteria (Streptomyces clavuligerus).
The applicability of in situ RCA for detecting bacteria in river water and selective cell loss by permeabilization were investigated using E. coli O157:H7 ATCC 43888 carrying plasmid pQE70-gfp, which was constructed from a ColE1-based plasmid, pQE70 (QIAGEN), and the gfp gene from plasmid pGFPuv (Clontech) as described elsewhere previously (2). E. coli O157:H7 cells were added to river water samples to a concentration approximately equal to 20% of the number of river bacteria and incubated for 2 h at 19°C before fixation as described below. E. coli JM109 was used for maintenance and amplification of plasmid pQE70-gfp for the DNA uptake experiment. Plasmid DNA was extracted and purified with a HiSpeed Plasmid Maxi kit according to the manufacturer's instructions (QIAGEN).
E. coli, S. epidermidis, and Streptomyces clavuligerus cells were grown aerobically at 37°C in Luria-Bertani (LB) medium (1% tryptone, 0.5% yeast extract, 0.5% NaCl [pH 7.0]). All other bacterial species were grown at 30°C. Media were modified for Flavobacterium (no NaCl) and Vibrio species (3% NaCl). E. coli O157:H7 or JM109 cells carrying plasmid pQE70-gfp were grown at 37°C in LB medium supplemented with 50 μg ml−1 ampicillin.
Cultures grown overnight were harvested by centrifugation, washed twice with phosphate-buffered saline (PBS) (137 mM NaCl, 2.7 mM KCl, 8.1 mM Na2HPO4, 1.5 mM KH2PO4 [pH 7.2]), and suspended in filtered 2% formaldehyde in PBS for 3 h. After fixation, cells were washed twice with PBS and suspended in 50% ethanol with PBS.
River samples.
Surface river water samples were collected at four locations (Juhachijo [industrial area], on the Kanzakigawa river; Kitahashi [commercial area], on the Neyagawa river; Takiue [forested area], on the Minohgawa river; and Kurumatsukuri [agricultural area], on the Inagawa river) from the northern part of Osaka, Japan (26, 44). The Kanzakigawa and Neyagawa rivers are considered to be polluted, and the Minohgawa and Inagawa rivers are relatively unpolluted by organic carbon (17, 45). All river samples were collected in 2-liter sterilized polycarbonate bottles and transported to the laboratory on ice. The densities (cells per milliliter) of bacterioplankton in the river waters were 2.0 × 107 (Juhachijo), 8.2 × 106 (Kitahashi), 4.0 × 105 (Takiue), and 6.8 × 105 (Kurumatsukuri), determined by 4′,6-diamidino-2-phenylindole (DAPI) staining at a final concentration of 1 μg ml−1. Twenty milliliters of the samples was fixed with 2% formaldehyde at 4°C for 3 h, and 0.5-ml to 10-ml portions were used for further experiments.
Sample preparation.
Fixed cells were filtered onto gelatin [0.1% gelatin, 0.01% CrK(SO4)2]-coated white polycarbonate membrane filters (25 mm [pore size, 0.2 μm]; Advantec, Tokyo, Japan), washed twice with sterile distilled, deionized water, dehydrated with 100% ethanol for 3 min, and dried thoroughly. Bacterial cells were then embedded in Metaphor agarose (0.2% [wt/vol] in sterilized distilled, deionized water; FMC Bioproducts, Rockland, ME), dried on glass slides, and dehydrated with 100% ethanol as previously described (29).
Cell permeabilization.
Cells immobilized on the membrane filters were permeabilized under various conditions using lysozyme, achromopeptidase, or proteinase K as well as heat treatment. Treatments included (i) no enzymatic digestion or heating as a control; (ii) incubation with 0.5 mg ml−1 lysozyme (100 mM Tris-HCl [pH 8.2], 50 mM EDTA) (Nacalai Tesque Inc., Kyoto, Japan) at 37°C for 15 min followed by 10 μg ml−1 proteinase K (100 mM Tris-HCl [pH 8.2], 50 mM EDTA) (Roche Diagnostics) at 37°C for 5 min, with or without heating at 94°C for 10 min in 1× Ampligase buffer (Epicenter, Madison, WI); (iii) incubation with 10 mg ml−1 lysozyme at 37°C for 60 min, with or without heat treatment; (iv) incubation with 65 U ml−1 achromopeptidase (Sigma) at 37°C for 30 min, with or without heat treatment; and (v) incubation with 65 U ml−1 achromopeptidase at 37°C for 30 min followed by 10 μg ml−1 proteinase K at 37°C for 45 min, with or without heat treatment (Table 1). Following treatment, the filters were washed twice in sterilized distilled, deionized water, dehydrated twice in 100% ethanol for 1 min, and dried thoroughly.
TABLE 1.
Percentages of permeabilized river bacteria and cells remaining on membrane filters after various cell permeabilization treatments for in situ RCAa
| Sample | Treatment
|
Mean % of cells (SD)b
|
|||||
|---|---|---|---|---|---|---|---|
| Lysozyme | Achromopeptidase | Proteinase K | Heat | Totalc | Permeabilizedd | Remaininge | |
| 1 | Not treated | Not treated | Not treated | Not treated | 100 (2) | 2 (0.3) | 100 (7) |
| 2 | Not treated | Not treated | Not treated | 94°C, 10 min | 97 (4) | 49 (6) | 96 (4) |
| 3 | 0.5 mg/ml, 15 min | Not treated | 10 μg/ml, 5 min | Not treated | 90 (8) | 2 (0.6) | 28 (4) |
| 4 | 0.5 mg/ml, 15 min | Not treated | 10 μg/ml, 5 min | 94°C, 10 min | 78 (7) | 39 (12) | 32 (10) |
| 5 | 10 mg/ml, 60 min | Not treated | Not treated | Not treated | 102 (10) | 5 (2) | 98 (7) |
| 6 | 10 mg/ml, 60 min | Not treated | Not treated | 94°C, 10 min | 99 (8) | 95 (3) | 100 (8) |
| 7 | 10 mg/ml, 60 min | Not treated | 10 μg/ml, 45 min | Not treated | 102 (10) | 4 (0.8) | 68 (8) |
| 8 | 10 mg/ml, 60 min | Not treated | 10 μg/ml, 45 min | 94°C, 10 min | 77 (6) | 96 (8) | 24 (3) |
| 9 | Not treated | 65 U/ml, 30 min | Not treated | Not treated | 82 (10) | 3 (0.7) | 19 (3) |
| 10 | Not treated | 65 U/ml, 30 min | Not treated | 94°C, 10 min | 47 (5) | 3 (1) | 10 (1) |
| 11 | Not treated | 65 U/ml, 30 min | 10 μg/ml, 45 min | Not treated | 81 (12) | 1 (0.3) | 11 (1) |
| 12 | Not treated | 65 U/ml, 30 min | 10 μg/ml, 45 min | 94°C, 10 min | 43 (7) | 13 (6) | 5 (1) |
River water samples were collected at Juhachijo on the Kanzakigawa river (9 July 2004). All enzymatic reactions were carried out at 37°C.
Means and standard deviations for triplicate determinations.
Percentage of total bacteria remaining on membrane filter.
Percentage of bacteria stained with Cy3-labeled rabbit phosphorylase used for examination of cell permeability.
Percentage of E. coli O157:H7 cells added to the river water remaining on the membrane filter to investigate the possibility of a selective loss of specific bacterial species.
Permeability assay.
Rabbit phosphorylase (97 kDa; Sigma) was labeled with cyanine Cy3 monofunctional dye (Amersham) according to the manufacturer's instructions. After cell permeabilization treatment, the bacterium-containing filters were incubated in a blocking solution (3% bovine serum albumin [BSA]) (Wako, Osaka, Japan) in PBS at 4°C for 15 min. Cy3-labeled phosphorylase in 3% BSA in PBS was added to the blocking solution at a final concentration of 10 ng ml−1. Two methods were used to stain the permeabilized cells. In the first method, the mixture was incubated at 4°C for 30 min followed by washing with sterile distilled, deionized water and counterstaining with 1 μg ml−1 DAPI for 10 min. In the second method, the mixture was incubated at 4°C for 20 min, and the filters were then transferred into 100 μl of 1× Ampligase buffer to prevent the heat denaturation of BSA. The filters were washed with sterile distilled, deionized water, subjected to the same conditions as those for the cell permeabilization by heat treatment described above, and counterstained with 1 μg ml−1 DAPI for 10 min.
Detection of cells with the gfp gene by in situ RCA.
RCA detection of gfp inside single cells (in situ RCA) was carried out basically as described previously (27). In brief, filters coated with agarose were incubated with 10 mg ml−1 lysozyme solution for 1 h at 37°C, rinsed twice with sterile water, dehydrated with 100% ethanol, and dried thoroughly. The filters were immersed in ligation buffer containing 0.8 μM gfp-RCA (27), 1× Ampligase buffer, and 25 U Ampligase. The ligation reaction was carried out at 94°C for 10 min, followed by 60°C for 80 min. The probe was labeled with a phosphate group at the 5′ end and circularized by ligation when hybridized to the target sequences. The filters were then rinsed twice with PBS containing 0.01% Nonidet P-40 (Sigma) at 50°C for 10 min, followed by rinsing with sterile water at room temperature for 5 min.
After ligation of the circularizable probe, the RCA reaction was carried out with 1 μM gfpf (27), 1 M Betain (Sigma), 1× ThermoPol buffer, and Bst DNA polymerase (New England Biolabs) at 63°C for 90 min. The RCA primer allowed the amplification of the complementary sequence of the circularized probe by hybridization to a specific region of the probe. The resulting amplicons were single-stranded DNA tandem repeats of the circularized probe sequence. After the RCA reaction, filters were rinsed with PBS containing 0.01% Nonidet P-40 at 50°C for 15 min and then washed twice with sterile water at room temperature for 5 min.
FISH was used to detect RCA amplicons inside cells. Filters were immersed in prewarmed hybridization solution (900 mM NaCl, 5 mM EDTA, 20 mM Tris-HCl [pH 7.5], 0.01% sodium dodecyl sulfate [SDS], 30% formamide) containing 1 ng μl−1 of gfpstx-d (27) at 46°C for 16 h. The detector probe consisted of sequences complementary to a specific region of the RCA amplicon and was labeled with Cy3 at the 5′ end. After hybridization, the filters were washed twice with PBS containing 0.01% Nonidet P-40 at 48°C for 15 min, followed by counterstaining with 1 μg ml−1 of DAPI for 10 min.
When E. coli O157:H7 with plasmid pQE70-gfp was used as target bacteria to test the specificity of in situ RCA, the filters were stained with 2 μg ml−1 of fluorescein isothiocyanate (FITC)-labeled anti E. coli O157:H7 antibody (goat polyclonal antibody; Kirkegaard and Perry Laboratories Inc., MD) in PBS with 30 mg ml−1 of BSA at 37°C for 30 min before counterstaining.
DNA uptake in river water.
Two surface river water samples collected at Juhachijo (polluted) on 11 August 2004 and at Takiue (unpolluted) on 25 June 2005 were used. River water was incubated in 5-liter round-bottomed flasks at 19°C (mean river water temperature) in the dark with shaking at 80 rpm for 506 h. Plasmid pQE70-gfp was added to the flasks at a final concentration of 1.5 × 1010 copies ml−1 for the river water from Juhachijo and 1.3 × 109 copies ml−1 for the river water from Takiue. Duplicate samples from each flask were collected at 0, 1, 4, 7, 13, 25, 75, 170, and 506 h. River water without the addition of the plasmid and autoclaved river water with the addition of the plasmid served as negative controls for DNA uptake by river bacteria.
For each sample, DNA uptake was detected as cells expressing gfp (green) under UV and blue excitation by fluorescence microscopy. In addition, cells carrying gfp were detected by in situ RCA as described above.
Microscopic examination.
Cells were observed by using an epifluorescence microscope (E-400; Nikon, Tokyo, Japan) with the UV-2A (30-350, DM400, and BA420), B-2A (Ex450/490, DM505, and BA520), and HQ-Cy3 (G535/50, FT565, and BP610/75) filter sets (Nikon) for UV, blue, and green excitation, respectively. Cooled charge-coupled-device cameras were used to obtain monochrome (Sensys 1401; Photometrics, Tucson, AZ) and colored (Cool Snap; Roper Photometrics) digital images. More than 1,000 DAPI-stained cells from 20 different fields were counted per sample in duplicate or triplicate.
Cells carrying plasmid pQE70-gfp were detected by two methods, based on either the expression of the gfp gene or in situ RCA targeting of the gfp gene. Both UV and blue excitation were used for the expression of gfp, and green excitation was used for the detection of cells by in situ RCA. When cells expressing gfp were counted, propidium iodide (2.5 μg ml−1 for 10 min at room temperature) was used to detect all bacteria (44). A total of 200 to 300 microscopic fields were examined on each filter in duplicate for quantitative evaluation as described previously by Dahlberg et al. (7). Composite images were constructed with Adobe Photoshop 8.0.1 (Adobe Systems, Inc.).
Quantification of gfp by real-time PCR.
Extraction of bacterial DNA from the water samples was carried out basically according to the method described previously by Tsai and Olson (40). Water samples (10 to 50 ml) were filtered through 0.2-μm polycarbonate membrane filters (25-mm diameter; Advantec) to collect bacteria, followed by two washes with 2 ml of sterile distilled, deionized water. Filters were aseptically cut into eight sections, placed into a sterile disposable 15-ml polypropylene centrifuge tube with 1 ml of lysis solution (0.15 M NaCl, 0.1 M Na2EDTA [pH 8.0]) containing 15 mg ml−1 of lysozyme, and incubated at 37°C for 2 h. One milliliter of extraction buffer (0.1 M NaCl, 0.5 M Tris-HCl [pH 8.0], 10% SDS) was added, and the sample was subjected to three cycles of freezing with liquid nitrogen and thawing at 65°C. DNA was then harvested and prepared using standard methods (32).
gfp copy numbers in river water samples were determined by real-time PCR amplification using a LightCycler (Roche Diagnostics) with a primer pair that yields a 282-bp gfp fragment (gfpuv436f [5′-AAC TCA CAC AAT GTA TAC ATC ACG-3′] and gfpuv717r [5′-TTA TTT GTA GAG CTC ATC CAT GC-3′]). For hybridization, amplification parameters were optimized using two oligomer probes for fluorescent resonance energy transfer to monitor the quantity of PCR amplicons during real-time PCR. One probe was labeled with FITC at the 3′ end (640F [5′-AAG CGT GAC CAC ATG GTC CTT CT-3′]), and another probe was labeled with LC Red640 at the 5′ end and phosphorylated at the 3′ end (690R [5′-GAG TTT GTA ACT GCT GCT GGG ATT ACA-3′]). The sensitivity and linear range of the assay were determined by using known amounts of purified template DNA. The PCR mixture (total volume of 20 μl) consisted of LightCycler DNA Master hybridization probes (Roche Diagnostics) with 4 mM Mg2+, 0.5 μM each forward primer (gfpuv436f) and reverse primer (gfpuv717r), 0.2 μM each hybridization probe, and 5 μl of DNA. After a hot start for 10 min at 95°C, 50 cycles were run with denaturation for 10 s at 95°C, annealing for 10 s at 60°C, and extension for 15 s at 72°C. About 5 × 100 to 5 × 107 copies of the gfp gene per reaction were used to establish a standard curve in each sample. Fluorescence acquisition occurred within the F2/F1 channel after the annealing stage of each cycle, and all subsequent analyses were carried out using the second-derivative-maximum option of the LightCycler software (version 3.01).
RESULTS
Optimization of cell permeabilization.
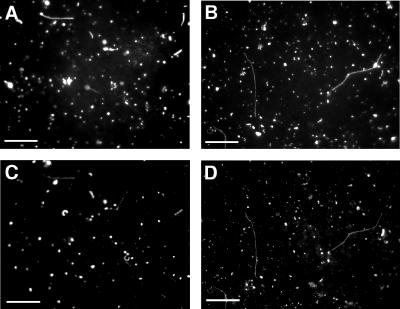
Three enzymes (lysozyme, achromopeptidase, and proteinase K) and heat treatment were used to optimize the cell permeabilization of bacteria in the river water samples for in situ RCA. Cy3-labeled phosphorylase was used to stain permeabilized cells, which were counted before and after treatment to monitor cell detachment from the membrane filter, and anti-E. coli O157:H7 antibody staining for selective cell loss was used to evaluate the applicability of each cell permeabilization treatment (Table 1). Two percent of cells were stained with Cy3-labeled phosphorylase, but 49% of cells were stained by the Cy3-labeled protein after heat treatment without enzymatic digestion (Fig. 1A and C). Both achromopeptidase and proteinase K or a combination of lysozyme and proteinase K resulted in a decrease in cell attachment to the membrane filters. Lysozyme treatment combined with heating stained 95% of cells without reducing the number of cells on the membrane filters (Fig. 1B and D), and 99% of E. coli cells were stained without the heat treatment (data not shown). All subsequent experiments were carried out using the lysozyme permeabilization with heat treatment.
FIG. 1.
Permeabilized cells stained with Cy3-labeled phosphorylase. River water samples taken at Juhachijo on the Inagawa river were used. Under UV excitation, all DAPI-stained bacterial cells were visualized (A and B). Under green excitation, permeabilized cells emitted red fluorescence of the Cy3-labeled phosphorylase (C and D). Forty-nine percent of bacteria were permeabilized with heating only (A and C). Ninety-five percent of cells were permeabilized with 10 mg/ml lysozyme for 60 min and heating. Scale bar, 5 μm.
The applicability of this treatment was examined using the 22 bacterial species listed in Materials and Methods. The treatment did not affect cell number or visibly alter cell morphology (data not shown). Twenty-one of the bacterial species stained with Cy3-labeled phosphorylase, but there was no apparent staining of S. epidermidis.
Modification of in situ RCA for aquatic bacteria.
The in situ RCA protocol was optimized for the detection of aquatic bacteria using the modifications listed in Table 2. Modifications were compared to the in situ RCA method for E. coli described in a previous study (27). Formaldehyde replaced paraformaldehyde to shorten the fixation time to 3 h from 16 h. Fixation times shorter than 3 h caused alterations of cell morphology and detachment from membrane filters after in situ RCA (data not shown).
TABLE 2.
Optimized experimental steps for in situ RCA targeting of the gfp gene in river water samples
| Stage | Procedure(s) |
|---|---|
| 1 (sample fixation) | 2% formaldehyde in PBS at 4°C for 3 h |
| 2 (sample preparation) | Filtrate samples onto gelatin coated membrane filters; wash with 5 ml sterile water twice; dehydrate with 100% ethanol for 1 min twice; vacuum dry |
| 3 (immobilization) | Soak filters in Metaphor agarose (0.2% in sterile water) and dry on a glass slide; dehydrate with 100% ethanol for 1 min twice and vacuum dry |
| 4 (cell permeabilization) | Incubate in 10 mg/ml lysozyme (100 mM Tris-HCl [pH 8.2], 50 mM EDTA) solution at 37°C for 60 min; wash with sterile water twice; dehydrate with 100% ethanol for 1 min twice; vacuum dry |
| 5 (ligation) | Incubate with ligation buffer containing 0.80 μM circularizable probe and 25 U Ampligase at 4°C for 15 min, followed by 94°C for 10 min and 60°C for 80 min for ligation of the probe; wash twice with PBS containing 0.01% Nonidet P-40 at 50°C for 10 min and with sterile water for 5 min, dehydrate with 100% ethanol for 1 min twice; vacuum dry |
| 8 (amplification) | Incubate with reaction buffer containing 1 μM RCA primer and 40 U Bst DNA polymerase at 4°C for 15 min, followed by 63°C for 90 min; wash twice with PBS containing 0.01% Nonidet P-40 at 50°C for 10 min and sterile water for 5 min; dehydrate with 100% ethanol for 1 min twice; vacuum dry |
| 9 (detection by FISH) | Immerse in prewarmed hybridization solution (900 mM NaCl, 5 mM EDTA, 20 mM Tris-HCl [pH 7.5], 0.01% SDS, 30% formamide) containing 1 ng μl−1 of Cy3-labeled detector probe at 46°C for 16 h; wash twice with PBS containing 0.01% Nonidet P-40 at 48°C for 15 min and with sterile water for 5 min |
| 10 (counterstaining) | Incubate with 1 μg ml−1 of DAPI for 10 min; wash with sterile water; vacuum dry |
In this work, samples were filtered onto a gelatin-coated membrane. This step replaces centrifugation, which may introduce a bias for environmental samples, because many of these cells are smaller than cells cultured in the laboratory, which could result in selective cell sedimentation. Incubation of 10 mg ml−1 lysozyme solution for 60 min was used rather than 0.5 mg ml−1 lysozyme solution for 30 min and 0.1 μg ml−1 proteinase K solution for 5 min for E. coli cell permeabilization (27), because the lysozyme treatment used in this study increased the number of river bacteria stained by Cy3-labeled protein. These modifications make in situ RCA more applicable to river water samples.
Recovery of added cells carrying a specific gene.
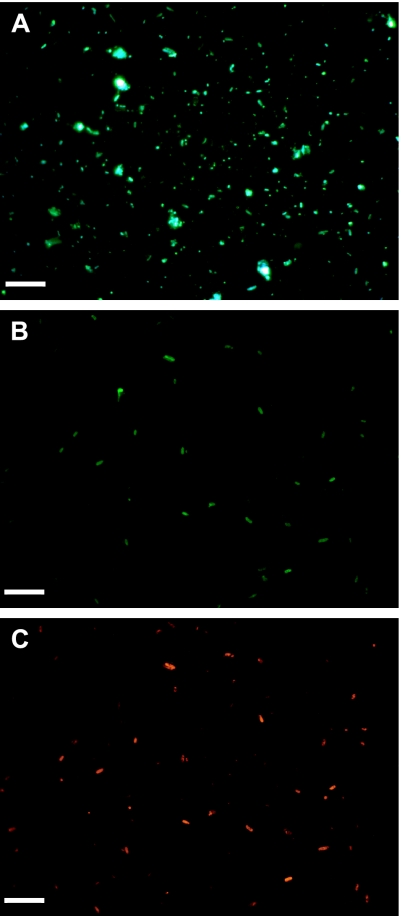
E. coli O157:H7 cells carrying gfp were added to four different river samples and subjected to modified in situ RCA. The applicability of permeabilization was examined with Cy3-labeled protein staining, total direct counting before and after the in situ RCA experimental steps, and anti-E. coli O157:H7 antibody staining after in situ RCA for examination of the specificity of the reaction (Table 3). At least 85% of bacteria remained on the filter after in situ RCA of all of the river samples. More than 83% of cells were permeabilized by the optimized treatment, and no decrease was observed in the numbers of Cy3-labeled E. coli O157:H7 cells after in situ RCA. The number of cells detected by in situ RCA targeting gfp was equal to the input number. The specificity of in situ RCA could be confirmed because cells were located at the same positions under both blue and green excitation (Fig. 2).
TABLE 3.
Applicability of optimized cell permeabilization conditions for in situ RCA using four different river water samplesa
| Sample siteb | Mean % of cells (SD)g
|
|||
|---|---|---|---|---|
| Totalc | Stainedd | Addede | In situ RCA detectedf | |
| Juhachijo (polluted) | 85 (10) | 85 (6) | 99 (6) | 100 (6) |
| Kitahashi (polluted) | 99 (16) | 96 (3) | 99 (6) | 99 (12) |
| Kurumatsukuri (unpolluted) | 99 (4) | 83 (4) | 99 (2) | 99 (7) |
| Takiue (unpolluted) | 100 (8) | 104 (4) | 102 (4) | 104 (5) |
Treatment consisted of 10 mg/ml lysozyme for 60 min at 37°C followed by heating at 94°C for 10 min.
River samples collected 21 June 2005 (Osaka, Japan).
Percentage of total bacteria remaining on the membrane filter after in situ RCA experiment.
Percentage of bacteria stained with Cy3-labeled rabbit phosphorylase used for examination of cell permeability.
Percentage of E. coli O157:H7 cells added to the river water samples remaining on the membrane filter detected by fluorescent antibody staining to investigate the possibility of a selective loss of specific bacterial species.
Percentage of E. coli O157:H7 cells carrying gfp gene added to the river water samples detected by in situ RCA targeting of the gene to validate the absence of the inhibitory effect of the RCA reaction.
Means and standard deviations for triplicate determinations.
FIG. 2.
In situ RCA of E. coli O157:H7 cells carrying the gfp gene added to river water samples. Total bacteria (A), E. coli O157:H7 cells (B), and cells carrying gfp (C) were visualized under UV, blue, and green excitation, respectively. E. coli O157:H7 cells carrying the gfp gene were fixed with 2% formaldehyde after 2 h of incubation in river water taken at Juhachijo, Osaka, Japan. E. coli O157:H7 cells were stained with FITC-labeled anti-E. coli O157:H7 antibody. Cells carrying gfp were detected by in situ RCA targeting of the gene. Scale bars, 5 μm.
Quantification of bacterial competence.
Frequencies of uptake of plasmid pQE70-gfp in water samples from the Kanzakigawa river at Juhachijo and the Minoh river at Takiue were determined by direct counting of cells expressing gfp and by in situ RCA (Table 4). In addition, the copy number of the gfp gene in the DNA extracted from bacterial cells on membrane filters was quantified by real-time PCR. No DNA uptake by bacteria was observed in the negative controls by counting cells expressing gfp or by in situ RCA targeting gfp.
TABLE 4.
Monitoring of DNA uptake frequencies and abundance of donor plasmid in >0.2 μm size fractions from models using two river water samplesg
| Location | Sampling time (h) | Frequencyexpressiona | Frequencyuptakeb | FrequencyPCRc | Frequencyuptaked/ Frequencyexpression | FrequencyPCRe/ Frequencyuptake |
|---|---|---|---|---|---|---|
| Juhachijo | 0 | <2.1 × 10−6 | <1.4 × 10−6 | NAf | NA | NA |
| 1 | <2.1 × 10−6 | <1.4 × 10−6 | 2 × 101 | NA | >1 × 107 | |
| 4 | <2.1 × 10−6 | <1.4 × 10−6 | 2 × 101 | NA | >1 × 107 | |
| 7 | <2.1 × 10−6 | 1.3 × 10−5 | 7 × 100 | >6 × 100 | 6 × 105 | |
| 13 | <1.9 × 10−6 | 3.4 × 10−5 | 8 × 100 | >2 × 101 | 2 × 105 | |
| 25 | <1.3 × 10−6 | 1.4 × 10−4 | 6 × 100 | >1 × 102 | 4 × 104 | |
| 75 | 9.2 × 10−6 | 1.3 × 10−4 | 1 × 100 | 1 × 101 | 9 × 103 | |
| 170 | 1.0 × 10−4 | 5.3 × 10−4 | 1 × 10−1 | 5 × 100 | 2 × 102 | |
| 506 | 2.5 × 10−5 | 2.0 × 10−4 | 5 × 10−3 | 8 × 100 | 2 × 101 | |
| Takiue | 0 | <3.1 × 10−5 | <2.1 × 10−5 | NA | NA | NA |
| 1 | <3.1 × 10−5 | 4.6 × 10−4 | 3 × 101 | >2 × 101 | 7 × 104 | |
| 4 | <3.0 × 10−5 | 7.0 × 10−4 | 2 × 100 | >2 × 101 | 3 × 103 | |
| 7 | <3.1 × 10−5 | 5.3 × 10−4 | 3 × 101 | >2 × 101 | 6 × 104 | |
| 13 | <2.9 × 10−5 | 1.6 × 10−3 | 4 × 100 | >6 × 101 | 2 × 103 | |
| 25 | <2.1 × 10−5 | 2.3 × 10−4 | 9 × 10−1 | >1 × 101 | 4 × 103 | |
| 75 | 1.5 × 10−4 | 3.1 × 10−3 | 2 × 10−1 | 2 × 101 | 7 × 101 | |
| 170 | 9.1 × 10−5 | 1.6 × 10−3 | 3 × 10−1 | 2 × 101 | 2 × 102 | |
| 506 | <2.0 × 10−5 | 1.4 × 10−4 | 5 × 10−1 | >7 × 100 | 4 × 103 |
Frequencyexpression, number of cells expressing the gfp gene divided by the total number of cells.
Frequencyuptake, number of cells taking up the gfp gene, which was detected by in situ RCA, divided by the total number of cells.
FrequencyPCR, number of donor plasmids, which was determined by real-time PCR targeting the gfp gene, divided by the total number of cells.
The value of Frequencyuptake was divided by that of Frequencyexpression.
The value of FrequencyPCR was divided by that of Frequencyuptake.
NA, not applicable.
Values indicate means for duplicate determinations.
Cells expressing gfp were observed after 75 h of incubation following the addition of plasmid. No cells were observed expressing gfp in the Takiue (unpolluted) river water sample up to 506 h after the addition of plasmid. Cells containing plasmid were detected by in situ RCA, however, after 1 to 7 h of incubation.
DNA uptake frequencies ranged from 3.4 × 10−5 to 3.1 × 10−3 when determined by in situ RCA and 9.2 × 10−6 to 1.5 × 10−4 when determined by direct cell counts in samples from both Juhachijo and Takiue (Table 4). In situ RCA always gave frequencies 5 to 20 times higher than direct counting of cells expressing gfp. For reasons of convenience, in situ RCA frequencies were estimated to be 6 to 100 times greater than those obtained by direct counts when there were no cells expressing gfp.
Either the direct counting of cells expressing gfp or in situ RCA can determine the number of cells carrying the gene. The total number of gfp copies in the bacterial DNA extracted from the membrane filter can be determined by real-time PCR. The gfp copy number detected by real-time PCR was greater than the number of cells detected by in situ RCA, which was also greater than the number of cells expressing gfp. Dividing each of these detection results by the number of total cells gives a good approximation of the relative sensitivities of each method. Plasmid stability and replication should also be considered when these methods are compared. The introduced plasmid is a high-copy-number plasmid and can be maintained at up to 200 copies only in E. coli and closely related enteric bacteria, depending on the physiological state. The plasmid is a narrow-host-range vector, and thus, it will not be able to replicate in the majority of indigenous bacteria in the water samples after being taken up by a cell. Thus, only in plasmids whose replication is strictly regulated, and whose copy number is close to 1 copy per cell, would the mean obtained by dividing the gfp copy number by the total number of bacteria be closely reflective of the maximum possible frequency of exogenous DNA uptake by bacteria, but it would not reflect the frequency of natural transformation in the sample. The frequencies determined by real-time PCR copy number were 5 × 10−3 to 2 × 101 gfp copies cell−1 (Juhachijo) and 2 × 10−1 to 3 × 101 gfp copies cell−1 (Takiue). The frequencies determined by real-time PCR divided by the frequencies determined by in situ RCA were 2 × 101 to 1 × 107 (Juhachijo) and 7 × 101 to 7 × 104 (Takiue) (Table 4).
Cells taking up the plasmid were observed by fluorescence microscopy (Fig. 3). Composite images showed that cells expressing gfp were attached to suspended particulate matter (Fig. 3A and D), while cells detected by in situ RCA were observed both in suspended particulate matter (Fig. 3B and E) and in a free planktonic state (Fig. 3C and F). Although two rivers were sampled for DNA uptake, all of the cells expressing gfp were found in aggregates, and cells detected by in situ RCA were observed both in aggregate and in a free planktonic state.
FIG. 3.
Bacteria taking up added plasmid DNA appeared during incubation of two river water samples taken at polluted (Juhachijo) and unpolluted (Kurumatsukuri) locations. Bacteria taking up the plasmid were detected based on gfp expression (A and D) and by in situ RCA targeting of gfp (B, C, E, and F). Samples from Juhachijo, in the upper row (A, B, and C), and Kurumatsukuri locations, in the lower row (D, E, and F), were taken 75 h after the addition of plasmid to the river water samples. The total population of bacteria emitted red with propidium iodide under green excitation, and bacteria taking up plasmid and expressing gfp emitted green under both UV and blue excitation (A and D). Three images obtained under UV, blue, and green excitation were used to generate the composite images shown (A and D). The total population of bacteria emitted blue by DAPI staining under UV excitation, and bacteria taking up plasmid detected by in situ RCA emitted red under green excitation (B, C, E, and F). Cells expressing the gfp gene were located in suspended particulate matter. Cells detected by in situ RCA were located as single cells (C and F), in addition to their association with suspended particulate matter (B and E). Two images obtained under UV and green excitation were used to generate the composite images shown (B, C, E, and F). Scale bars, 5 μm.
DISCUSSION
Examination of permeability with fluorescently labeled protein as a size marker.
Optimization of cell permeabilization conditions for in situ RCA was carried out by Cy3-labeled rabbit phosphorylase staining, because this protein (97 kDa) (21) was larger than any of the reagents used; thus, its entry into the cell indicated sufficient permeability to allow smaller molecules to enter. Ampligase and Bst DNA polymerase used for RCA are 80 kDa and 67 kDa (1), respectively. In this study, at least 83% of the aquatic bacteria were permeabilized with lysozyme and heat treatment (Table 3), indicating that a majority of bacteria in river water can be successfully monitored for competence using these methods.
For in situ PCR, the applicability of cell permeabilization was validated using EUB primers that amplified a 761-bp fragment from the rRNA gene of eubacteria (36). This method, however, may not confirm the applicability of cell permeabilization because of the inability to design truly universal primers from a limited amount of conserved sequence. Cell permeabilization was therefore examined by fluorescently labeled proteins as the method of choice in this study. By this approach, cell permeability could be evaluated without depending on a specific nucleotide sequence.
In situ RCA in river water samples.
Cell permeabilization was examined using lysozyme, achromopeptidase, proteinase K, and heat, which have been reported to be effective for bacteria in natural environments (29, 30, 33) and for archaeal species (38). Although more than 83% of the total bacterial population was permeabilized with the optimized conditions (Table 3), there were still impermeable cells, which did not change when the lysozyme treatment was extended to 90 min (data not shown). The refractory cells may be resistant to permeabilization because lysozyme sensitivity is dependent on the physiological state of bacteria (39). Alternatively, lysozyme resistance may be a characteristic of certain Archaea. Certain acid-fast or gram-positive species also have lysozyme resistance, although the gram-positive species Streptomyces clavuligerus, B. megaterium, and B. subtilis were permeabilized in this study.
Less than 1% of cells were gram-positive bacteria with a high G+C content, and Archaea represented 1.1% to 3.1% of species found in the same rivers (3, 18). Thus, the 17% of lysozyme-resistant cells may be accounted for by a combination of the physiological state and bacterial cells that are inherently resistant to lysozyme digestion.
In situ PCR requires thermal cycling, which causes significant detachment losses from the sample glass slide, making the application of the method to quantitative analysis difficult (26, 36). The isothermal gene amplification characteristic of RCA allowed 96% of cells, on average, to be retained on the membrane filter after in situ RCA with optimized cell permeabilization. As shown in Table 1, different permeabilization treatments greatly affected the number of cells retained, indicating the importance of the optimization of cell permeabilization conditions for the sample type.
Species-selective cell loss was examined by adding E. coli O157:H7 cells, which are rarely found in rivers (22). This isolate was chosen because all of the permeabilization treatments used caused cell destruction and/or cell detachment from the membrane filter when the cells were prepared without being embedded in agarose (data not shown) and because it is representative of bacteria that are easily destroyed by cell permeabilization. The percentages of total and E. coli O157:H7 cells retained sometimes varied with different cell permeabilization treatments (Table 1). This difference indicates that the loss could be species selective, and optimization of cell permeabilization should take the possibility that target cells could be destroyed by the chosen treatment into consideration. Thus, a method that does not bias cell remains based on species or genus level characteristics, even if the number of total cells is decreased slightly, should be chosen.
Differences in detection levels between culture-independent methods.
In genetic transformation studies, transformants have usually been detected by culture-dependent methods. These methods cause bias or an underestimation of the frequency and the extent of DNA uptake because many naturally occurring bacteria are not detected by conventional culture-based methods (31, 41, 42). In addition, the presence of naturally occurring bacteria that are resistant to ampicillin hampered the use of this selectable marker in this study (data not shown). DNA uptake by indigenous bacteria was therefore detected by the expression and amplification of gfp at the single-cell level.
Recent application of the introduction of fluorescent protein genes to gene transfer studies has revealed that gene transfer is more frequent in the environment than has been estimated by results using conventional culture-dependent methods (14). Fluorescent proteins, however, have limitations for DNA uptake studies resulting from being dependent on the expression of a phenotype rather than the presence of an introduced genotype. The result is that detection is affected by physiology and by the suitability of heterologous promoters in a wide range of bacteria found in the environment. DNA uptake frequencies determined by in situ RCA may thus be 5 to 20 times higher than frequencies determined by the direct counting of cells expressing gfp, and in situ RCA could detect bacteria taking up DNA even when cells do not express gfp. No cell was observed to take up plasmid in the autoclaved water control by in situ RCA, indicating that the uptake of plasmid in this experimental system is active and requires live cells.
A comparison of the frequencies obtained by in situ RCA and real-time PCR suggested that more copies of gfp were detected in the extracted DNA from bacterial cells on the membrane filters than expected from the in situ RCA results because the frequency determined by PCR divided by the frequency determined by DNA uptake was 2 × 101 to 1 × 107. Plasmid pQE70 used in this study is a narrow-host-range ColE1-based replicon and is not able to replicate in the majority of indigenous bacteria and archaea in a river. Therefore, only an E. coli isolate and its relatives can maintain up to 200 copies of a high-copy-number plasmid if the host is appropriately grown in a laboratory. This seems to be rare, and because of its overall negative charge, most of the plasmid DNA is likely to be adsorbed to clay and other soil particles and to cell surfaces by cation bridges in addition to uptake into live bacteria.
DNA uptake by river bacteria.
The frequencies determined by in situ RCA targeting of a specific gene sequence are indicative of the maximum possible frequencies of genetic transformation, and the data obtained in this study indicate that the frequencies could be higher in the environment than previously estimated (9, 24). A fluorescent protein gene such as gfp requires expression to be detected; thus, the actual genetic transformation frequency is between the values obtained by in situ RCA and those obtained by direct counting of cells expressing gfp if all the cells were permeabilized without a decrease in cell number.
In this study, all of the cells expressing gfp were found in the suspended particulate matter, which is ubiquitous in aquatic environments, and are the foci of microbial activity compared to surrounding water (4, 46). This might be because bacterial species expressing the gfp gene favor the attachment lifestyle or because the higher metabolic activity that accompanies attachment to organic material makes it possible to express the acquired gene. Cells detected by in situ RCA were found in both aggregate and planktonic states. This may be because DNA uptake by bacteria occurs in various physiological states (12, 24) and may be stimulated by nutrient limitation (23, 31). Because the suspended particulate matter could be self-organized during incubation (20), the river water samples incubated with aeration in a flask in this study may not accurately simulate the natural environment. Further studies are therefore required to determine the level of DNA uptake by indigenous bacteria using better model systems or in situ experiments (43). This study, however, is the first study in which DNA uptake by river bacteria was determined quantitatively based on the gene sequence without depending on cultivation or expression of a marker gene.
Conclusions and future perspectives.
In this study, uptake of plasmid DNA by indigenous bacteria from two rivers was quantitatively monitored at the single-cell level by in situ RCA with optimized cell permeabilization conditions. In situ RCA gave us the ability to detect the uptake of plasmid DNA with better accuracy than the direct counting of cells expressing the gfp gene under fluorescence microscopy. By optimizing cell permeabilization conditions for each sample type and confirming the applicability of the treatment by fluorescently labeled protein staining, in situ RCA may allow an accurate determination of the rates of free-DNA uptake by indigenous bacteria in other aquatic environments.
Future studies will include environmental factors that affect free-DNA uptake by indigenous bacteria in their natural environment or the spatial distribution of bacteria taking up free DNA in a biofilm, which is thought to be a hot spot of gene transfer (28).
Acknowledgments
The work was supported by the Development of Monitoring Methods for Microorganisms in Environment Project of the New Energy and Industrial Technology Development Organization (NEDO), Tokyo, Japan.
REFERENCES
- 1.Aliotta, J. M., J. J. Pelletier, J. L. Ware, L. S. Moran, J. S. Benner, and H. Kong. 1996. Thermostable Bst DNA polymerase I lacks a 3′→5′ proofreading exonuclease activity. Genet. Anal. 12:185-195. [PubMed] [Google Scholar]
- 2.Andersen, J. B., C. Sternberg, L. K. Poulsen, S. P. Bjørn, M. Givskov, and S. Molin. 1998. New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl. Environ. Microbiol. 64:2240-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Araya, R., K. Tani, T. Takagi, N. Yamaguchi, and M. Nasu. 2003. Bacterial activity and community composition in stream water and biofilm from an urban river determined by fluorescent in situ hybridization and DGGE analysis. FEMS Microbiol. Ecol. 43:111-119. [DOI] [PubMed] [Google Scholar]
- 4.Böckelmann, U., W. Manz, T. R. Neu, and U. Szewzyk. 2000. Characterization of the microbial community of lotic organic aggregates (′river snow') in the Elbe river of Germany by cultivation and molecular methods. FEMS Microbiol. Ecol. 33:157-170. [DOI] [PubMed] [Google Scholar]
- 5.Chen, I., and D. Dubnau. 2004. DNA uptake during bacterial transformation. Nat. Rev. Microbiol. 2:241-249. [DOI] [PubMed] [Google Scholar]
- 6.Claverys, J.-P., and B. Martin. 2003. Bacterial ‘competence’ genes: signatures of active transformation, or only remnants? Trends Microbiol. 11:161-165. [DOI] [PubMed] [Google Scholar]
- 7.Dahlberg, C., M. Bergström, M. Andreasen, B. B. Christensen, S. Molin, and M. Hermansson. 1998. Interspecies bacterial conjugation by plasmids from marine environments visualized by gfp expression. Mol. Biol. Evol. 15:385-390. [Google Scholar]
- 8.Daims, H., A. Bruhl, R. Amann, K. H. Schleifer, and M. Wagner. 1999. The domain-specific probe EUB338 is insufficient for the detection of all Bacteria: development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22:434-444. [DOI] [PubMed] [Google Scholar]
- 9.Day, M. J. 2004. Transformation, p. 158-172. In R. V. Miller and M. J. Day (ed.), Microbial evolution. ASM Press, Washington, D.C.
- 10.Demanèche, S., E. Kay, F. Gourbière, and P. Simonet. 2001. Natural transformation of Pseudomonas fluorescens and Agrobacterium tumefaciens in soil. Appl. Environ. Microbiol. 67:2617-2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Vries, J., and M. Wackernagel. 2004. Microbial horizontal gene transfer and the DNA release from transgenic crop plants. Plant Soil. 266:91-104. [Google Scholar]
- 12.Dubnau, D. 1999. DNA uptake in bacteria. Annu. Rev. Microbiol. 53:217-244. [DOI] [PubMed] [Google Scholar]
- 13.Frischer, M. E., G. J. Stewart, and J. H. Paul. 1994. Plasmid transfer to indigenous marine bacterial populations by natural transformation. FEMS Microbiol. Ecol. 15:127-135. [Google Scholar]
- 14.Hausner, M., and S. Wuertz. 1999. High rates of conjugation in bacterial biofilms as determined by quantitative in situ analysis. Appl. Environ. Microbiol. 65:3710-3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hendrickx, L., M. Hausner, and S. Wuertz. 2003. Natural genetic transformation in monoculture Acinetobacter sp. strain BD413 biofilms. Appl. Environ. Microbiol. 69:1721-1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jacobs, D., M. L. Angels, A. E. Goodman, and B. A. Neilan. 1997. Improved methods for in situ enzymatic amplification and detection of low copy number genes in bacteria. FEMS Microbiol. Lett. 152:65-73. [DOI] [PubMed] [Google Scholar]
- 17.Kenzaka, T., N. Yamaguchi, K. Tani, and M. Nasu. 1998. rRNA-targeted fluorescent in situ hybridization analysis of bacterial community structure in river water. Microbiology 144:2085-2093. [DOI] [PubMed] [Google Scholar]
- 18.Kenzaka, T., N. Yamaguchi, B. Prapagdee, E. Mikami, and M. Nasu. 2001. Bacterial community composition and activity in urban rivers in Thailand and Malaysia. J. Health Sci. 47:353-361. [Google Scholar]
- 19.Kenzaka, T., S. Tamaki, N. Yamaguchi, K. Tani, and M. Nasu. 2005. Recognition of individual genes in diverse microorganisms by cycling primed in situ amplification. Appl. Environ. Microbiol. 71:7236-7244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kerner, M., H. Hohenberg, S. Ertl, M. Reckermann, and A. Spitzy. 2003. Self-organization of dissolved organic matter to micelle-like microparticles in river water. Nature 422:150-154. [DOI] [PubMed] [Google Scholar]
- 21.Kurganov, B. I., B. A. Kornilaev, N. A. Chebotareva, V. P. Malikov, V. N. Orlov, A. E. Lyubarev, and N. B. Livanova. 2000. Dissociative mechanism of thermal denaturation of rabbit skeletal muscle glycogen phosphorylase b. Biochemistry 39:13144-13152. [DOI] [PubMed] [Google Scholar]
- 22.Kurokawa, K., K. Tani, M. Ogawa, and M. Nasu. 1999. Abundance and distribution of bacteria carrying sltII gene in natural river water. Let. Appl. Microbiol. 28:405-410. [DOI] [PubMed] [Google Scholar]
- 23.Lorenz, M. G., and W. Wackernagel. 1991. High frequency of natural genetic transformation of Pseudomonas stutzeri in soil extract supplemented with a carbon/energy and phosphorus source. Appl. Environ. Microbiol. 57:1246-1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lorenz, M. G., and W. Wackernagel. 1994. Bacterial gene transfer by natural genetic transformation in the environment. Microbiol. Rev. 58:563-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maruyama, F., T. Kenzaka, N. Yamaguchi, K. Tani, and M. Nasu. 2003. Detection of bacteria carrying the stx2 gene by in situ loop-mediated isothermal amplification. Appl. Environ. Microbiol. 69:5023-5028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maruyama, F., N. Yamaguchi, T. Kenzaka, K. Tani, and M. Nasu. 2004. Simplified sample preparation using frame spotting method for direct counting of total bacteria by fluorescence microscopy. J. Microbiol. Methods 59:427-431. [DOI] [PubMed] [Google Scholar]
- 27.Maruyama, F., T. Kenzaka, N. Yamaguchi, K. Tani, and M. Nasu. 2005. Visualization and enumeration of bacteria carrying a specific gene sequence by in situ rolling circle amplification. Appl. Environ. Microbiol. 71:7933-7940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Molin, S., and T. Tolker-Nielsen. 2003. Gene transfer occurs with enhanced efficiency in biofilms and induces enhanced stabilisation of the biofilm structure. Curr. Opin. Biotechnol. 14:255-261. [DOI] [PubMed] [Google Scholar]
- 29.Pernthaler, A., J. Pernthaler, and R. Amann. 2002. Fluorescence in situ hybridization and catalyzed reporter deposition for the identification of marine bacteria. Appl. Environ. Microbiol. 68:3094-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pernthaler, A., and R. Amann. 2004. Simultaneous fluorescence in situ hybridization of mRNA and rRNA in environmental bacteria. Appl. Environ. Microbiol. 70:5426-5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Redfield, R. J. 2001. Do bacteria have sex? Nat. Rev. Microbiol. 2:634-639. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 33.Sekar, R., A. Pernthaler, J. Pernthaler, F. Warnecke, T. Posch, and R. Amann. 2003. An improved protocol for quantification of freshwater Actinobacteria by fluorescence in situ hybridization. Appl. Environ. Microbiol. 69:2928-2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sørensen, S. J., M. Bailey, L. H. Hansen, N. Kroer, and S. Wuertz. 2005. Studying plasmid horizontal transfer in situ: a critical review. Nat. Rev. Microbiol. 3:700-710. [DOI] [PubMed] [Google Scholar]
- 35.Strätz, M., M. Mau, and K. N. Timmis. 1996. System to study horizontal gene exchange among microorganisms without cultivation of recipients. Mol. Microbiol. 22:207-215. [DOI] [PubMed] [Google Scholar]
- 36.Tani, K., K. Kurokawa, and M. Nasu. 1998. Development of a direct in situ PCR method for detection of specific bacteria in natural environments. Appl. Environ. Microbiol. 64:1536-1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tani, K., M. Muneta, K. Nakamura, K. Shibuya, and M. Nasu. 2002. Monitoring of Ralstonia eutropha KT1 in groundwater in an experimental bioaugmentation field by in situ PCR. Appl. Environ. Microbiol. 68:412-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Teira, E., T. Reinthaler, A. Pernthaler, J. Pernthaler, and G. J. Herndl. 2004. Combining catalyzed reporter deposition-fluorescence in situ hybridization and microautoradiography to detect substrate utilization by bacteria and archaea in the deep ocean. Appl. Environ. Microbiol. 70:4411-4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tolker-Nielsen, T., K. Holmstrøm, and S. Molin. 1997. Visualization of specific gene expression in individual Salmonella typhimurium cells by in situ PCR. Appl. Environ. Microbiol. 63:4196-4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsai, Y.-L., and B. H. Olson. 1992. Rapid method for separation of bacterial DNA from humic substances in sediments for polymerase chain reaction. Appl. Environ. Microbiol. 58:2292-2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Elsas, J. D., and M. J. Bailey. 2002. The ecology of transfer of mobile genetic elements. FEMS Microbiol. Ecol. 42:187-197. [DOI] [PubMed] [Google Scholar]
- 42.van Elsas, J. D., S. Turner, and M. J. Bailey. 2003. Horizontal gene transfer in the phytosphere. New Phytol. 157:525-537. [DOI] [PubMed] [Google Scholar]
- 43.Williams, H. G., M. J. Day, J. C. Fry, and G. J. Stewart. 1996. Natural transformation in river epilithon. Appl. Environ. Microbiol. 62:2994-2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamaguchi, N., and M. Nasu. 1997. Flow cytometric analysis of bacterial respiratory and enzymatic activity in the natural aquatic environment. J. Appl. Microbiol. 83:43-52. [Google Scholar]
- 45.Yokomaku, D., N. Yamaguchi, and M. Nasu. 2000. Improved direct viable count procedure for quantitative estimation of bacterial viability in freshwater environments. Appl. Environ. Microbiol. 66:5544-5548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zimmermann-Timm, H. 2002. Characteristics, dynamics and importance of aggregates in rivers—an invited review. Int. Rev. Hydrobiol. 87:197-240. [Google Scholar]
- 47.Zwirglmaier, K., W. Ludwig, and K.-H. Schleifer. 2004. Recognition of individual genes in a single bacterial cell by fluorescence in situ hybridization—RING-FISH. Mol. Microbiol. 51:89-96. [DOI] [PubMed] [Google Scholar]