Abstract
Rhodococcus sp. RHA1 grows on a broad range of aromatic compounds and vigorously degrades polychlorinated biphenyls (PCBs). Previous work identified RHA1 genes encoding multiple isozymes for most of the seven steps of the biphenyl (BPH) pathway, provided evidence for coexpression of some of these isozymes, and indicated the involvement of some of these enzymes in the degradation of BPH, ethylbenzene (ETB), and PCBs. To investigate the expression of these isozymes and better understand how they contribute to the robust degradative capacity of RHA1, we comprehensively analyzed the 9.7-Mb genome of RHA1 for BPH pathway genes and characterized the transcriptome of RHA1 growing on benzoate (BEN), BPH, and ETB. Sequence analyses revealed 54 potential BPH pathway genes, including 28 not previously reported. Transcriptomic analysis with a DNA microarray containing 70-mer probes for 8,213 RHA1 genes revealed a suite of 320 genes of diverse functions that were upregulated during growth both on BPH and on ETB, relative to growth on the control substrate, pyruvate. By contrast, only 65 genes were upregulated during growth on BEN. Quantitative PCR assays confirmed microarray results for selected genes and indicated that some of the catabolic genes were upregulated over 10,000-fold. Our analysis suggests that up to 22 enzymes, including 8 newly identified ones, may function in the BPH pathway of RHA1. The relative expression levels of catabolic genes did not differ for BPH and ETB, suggesting a common regulatory mechanism. This study delineated a suite of catabolic enzymes for biphenyl and alkyl-benzenes in RHA1, which is larger than previously recognized and which may serve as a model for catabolism in other environmentally important bacteria having large genomes.
Rhodococcus sp. strain RHA1 was isolated from hexachlorocyclohexane-contaminated soil (30) and has remarkably broad catabolic diversity. In particular, this actinomycete can degrade a broad range of aromatic compounds, including a number of important pollutants. Published reports describe growth of RHA1 on biphenyl (BPH), ethylbenzene (ETB), benzoate (BEN), phthalate, and phenylacetate (14, 19, 24, 28, 32). In addition, we have observed growth on phenol, 4-hydroxybenzoate, toluene, o-xylene, benzene, terephthalate, dibenzothiophene, 2-ethoxyphenol, guaiacol, 3-hydroxyphenylpropionic acid, 3-(2-hydroxyphenyl) propionic acid, isopropylbenzene, 4-methoxybenzoic acid, phenylacetonitrile, protocatechuate, styrene, vanillate, and veratrol (unpublished data). Further, RHA1 degrades a very broad range of polychlorinated biphenyls (PCBs), which it cometabolizes concurrently with BPH (31) or ETB (30). The catabolic potential of RHA1 and its adaptation to the soil environment make it a promising organism for bioremediation of polluted soil.
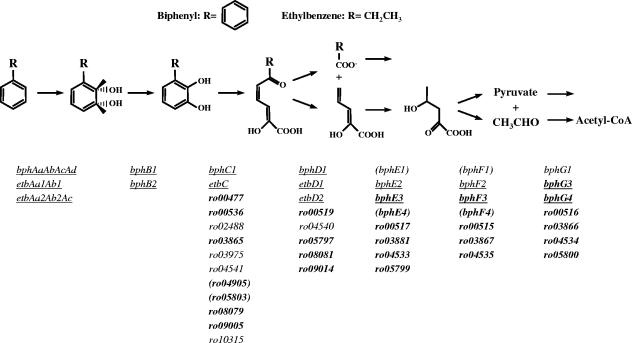
Fukuda and coworkers have characterized the BPH pathway of RHA1, which degrades biphenyl via ring dihydroxylation and oxygenolytic cleavage of a catecholic metabolite (Fig. 1). In RHA1, the same pathway also degrades other substituted benzenes, such as ETB and isopropylbenzene, as well as PCBs. A striking characteristic of RHA1 is the large number of genes potentially encoding multiple isozymes of the BPH pathway. Three gene clusters encoding ring-hydroxylating dioxygenase systems, bphAaAbAcAd (formerly bphA1A2A3A4), etbAa1Ab1C (formerly etbA1A2C), and etbAa2Ab2AcD2 (formerly ebdA1A2A3-etbD2), were detected on large linear plasmids (15, 19, 20, 30, 38). These clusters encode complete (four-component) or partial hydroxylating dioxygenase systems as well as associated enzymes for subsequent steps of the pathway. But none of the clusters encodes the complete BPH pathway. Additional genes potentially encoding other steps of the BPH pathway are distributed throughout the RHA1 genome. In total, published evidence exists for six homologues of bphC (27), three homologues of bphD (38), and two homologues each of bphE and bphF (28). The recently completed genome sequence for RHA1 (22) revealed additional homologues of BPH pathway enzymes as summarized in Fig. 1. It is proposed that this complex suite of enzymes contributes to the exceptional ability of RHA1 to degrade PCBs (27). Such multiplicity of catabolic genes appears to be typical of rhodococci (37).
FIG. 1.
The BPH pathway, a common pathway for degradation of biphenyl and ethylbenzene. Homologous genes potentially encoding each step are listed and classified as follows: bold, newly identified in this study; underlined, upregulated on BPH and ETB; in parentheses, constitutively expressed at high levels.
Evidence indicates that one ring-hydroxylating dioxygenase system, BphA, is important for BPH and PCB degradation but does not exclude a role for the other two isozymes, EtbA1 and EtbA2 (nearly identical to one another), in this process (31). PCB degradation by a bphAa mutant with ETB as a primary substrate suggests that multiple homologues of some or all of the bphA genes may be responsible for PCB degradation (31). One hydrolase, BphD, was shown to efficiently attack 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate (HOPDA), the ring-cleaved metabolite of BPH degradation, while two homologous hydrolases, EtbD1 and EtbD2, were shown to efficiently attack 2-hydroxy-6-oxohepta-2,4-dienoate (HOHDA), the metabolite of toluene degradation analogous to that of ETB degradation (38). Knockout analysis of bphE1, bphF1, and bphG1 suggests that these genes have a key, but not exclusive, role in BPH degradation and a lesser role in ETB degradation (28). Many questions about the functions of individual enzymes remain.
Genes encoding BEN biodegradation have also been identified in RHA1. A chromosomally located operon, benABCDK, was characterized (14). Proteomic analysis confirmed the upregulation of some products of the ben, cat (catechol degradation), and pca (protocatechuate degradation) genes, predicted to encode complete degradation of benzoate, during growth on that compound (25). The ben, cat, and pca genes are also predicted to function in growth on BPH, which yields BEN as an intermediate (Fig. 1).
Fragmentary information about the regulation of the above-described BPH pathway genes in RHA1 is available. Recent work suggests that the bphS1T1 genes, encoding a two-component regulatory system, mediate the induction of a BPH regulon consisting of operons involved in the degradation of BPH, ETB, and other aromatic compounds (32, 33). The bphS gene is essential for induction of the pathway by BPH, but it is possible that homologous regulatory genes permit induction by other compounds potentially degraded by the same pathway. Of the bphC homologues, bphC1, etbC, and ro04541 were shown by slot blot analysis to be inducibly expressed during growth on both BPH and ETB (27). The bphG1E1F1 genes were shown by reverse transcriptase (RT)-PCR analysis to be inducibly expressed during growth on both BPH and ETB (28). The limited evidence available does not indicate whether there is differential regulation of various isozymes of the BPH pathway with the different substrates of the pathway. This poorly understood regulatory system may have important consequences for the degradation potential of RHA1.
This study addressed important outstanding questions about the functions of the many genes encoding BPH, ETB, and BEN degradation by RHA1 and their regulation. To do so, we analyzed the complete genome sequence of RHA1 and developed and employed a microarray with probes for 8,313 genes (about 90% of those in the genome). We examined gene expression in RHA1 during exponential growth on the above-described three compounds. Quantitative PCR (Q-PCR) was used to verify results for selected genes.
MATERIALS AND METHODS
Cell culture and harvesting.
Rhodococcus sp. strain RHA1 was grown on W medium (3) plates with BPH vapor from crystals. Plates and liquid cultures were incubated at 30°C. Isolated colonies were transferred to 50-ml liquid precultures in 200-ml baffled flasks. Liquid medium consisted of W medium plus one of the following substrates: pyruvate (PYR; 20 mM), BEN (20 mM), BPH (10 mM), or ETB (vapors from 1 ml of ETB in a glass bulb in the flask). Liquid cultures were shaken at 200 rpm, and all flasks had foam stoppers permitting air exchange. The precultures were grown to stationary phase, and 5-ml aliquots were transferred to 500-ml liquid cultures in 2-liter baffled flasks with the corresponding substrate. Cells were grown to mid-log phase. This corresponded to an optical density at 600 nm of 2.0 for BPH, ETB, and BEN and 1.0 for PYR. To preserve RNA, a 1/10 volume of “stop solution,” 10% phenol (pH 5.0) in ethanol (2), was added to the cultures. Cells were harvested by centrifugation at 7,400 × g for 10 min at 4°C. The cell pellet was suspended in 0.5 ml ice-cold TE buffer (10 mM Tris-HCl and 1 mM EDTA, pH 7.5), and 1.0 ml RNAprotect bacterial reagent (QIAGEN) was added according to the manufacturer's instructions. The cell mixture was incubated for 5 min at room temperature and centrifuged at 10,000 × g for 5 min. The supernatant was removed, and the cells were frozen in a dry ice bath and stored at −80°C.
Triplicate cultures on each substrate were used to estimate growth rates (± standard deviations) in numbers of doublings/hour, which were 0.04 ± 0.02 on BPH, 0.05 ± 0.04 on ETB, 0.05 ± 0.01 on PYR, and 0.13 ± 0.02 on BEN. Another set of triplicate cultures was used for transcriptomic analysis. A third set of triplicate cultures was used for Q-PCR analysis.
Total RNA isolation.
Total RNA was isolated from RHA1 cells as follows. Cell pellets from 250 ml of culture were suspended in 2 ml of cold diethyl pyrocarbonate-treated water with EDTA (5 mM). Sodium dodecyl sulfate (SDS) and acidified phenol (pH 5.0) were added at final concentrations of 1.25% and 0.25% (vol/vol), respectively, and in a 50-ml Falcon tube, glass beads (3-mm diameter) were added to make a final volume of 10 ml. The tubes were transferred to a 64°C water bath and subsequently vortexed 10 times for 1 min, alternating with 1-min intervals in the water bath to keep the solution hot. To each sample, 0.07 ml of 3.0 M sodium acetate (pH 5.4) plus 2.5 ml of acidified phenol-chloroform (1:1, vol/vol) was added and followed by further vortexing for 10 min as described above. The liquid phases were transferred to a new tube and purified by phenol-chloroform extraction (29). To precipitate the total RNA, a 1/10 volume of 3.0 M sodium acetate plus one volume of isopropanol was added. The RNA was treated twice with DNase according to the manufacturer's instructions (Invitrogen) and then purified using the RNeasy system (QIAGEN). RNA was quantified by measuring absorbance at 260 nm.
cDNA synthesis and labeling.
cDNA probes were indirectly labeled by reverse transcription in the presence of amino allyl dUTP (Amersham-Pharmacia). Six micrograms of total RNA and 2.5 μg random hexamers (Invitrogen) were mixed, and the volume was brought to 13.3 μl with diethyl pyrocarbonate-treated water. The RNA was denatured for 10 min at 65°C and cooled on ice for 5 min. Then, the following were added: 3 mM each of dATP, dCTP, and dGTP; 1.2 mM dTTP; 1.8 mM amino allyl dUTP (Ambion); 0.01 mM dithiothreitol; 40 U cloned RNase inhibitor (Ambion); and 6 μl 5× RT buffer plus 380 U Superscript II reverse transcriptase (Invitrogen). The samples were incubated at 42°C for 2 h. The RNA template was hydrolyzed by adding 10 μl 1.0 M NaOH plus 10 μl 0.5 M EDTA and incubated at 65°C for 30 min. The sample was neutralized with 25 μl of 1 M HEPES (pH 7.5), purified using a Microcon YM30 column (Eppendorf), and dried in a vacuum. The coupling of either Cy3 or Cy5 dye to the amino allyl dUTP in the cDNA was done according to instructions from Ambion. The labeled probe was purified using the QIAquick PCR purification system (QIAGEN) and concentrated using a Microcon YM30 column. To quantify the labeled probes, samples of labeled cDNA were aliquoted onto a glass plate and scanned using a Typhoon scanner (Amersham Pharmacia). The signal was quantified using ImageQuant 5.2 (Molecular Dynamics).
Microarray preparation.
An array of 70-mer oligonucleotides was designed based on the RHA1 genome sequence assembly available in April 2005. Sequences from 8,213 putative genes were used to design oligonucleotides. As negative controls, genes were selected from Burkholderia xenovorans LB400 and Pseudomonas aeruginosa PAO1, which have a G+C content similar to that of RHA1. The LB400 and PAO1 genes were initially selected by blasting them against the RHA1 genome sequence. Their suitability as negative controls was verified by hybridizing Cy-labeled genomic DNA of RHA1 to spotted arrays of LB400 and PA01 (kindly supplied by J. Park, Michigan State University, and R. E. W. Hancock, University of British Columbia, Canada, respectively). Six genes, i.e., two from PAO1 and four from LB400 were selected as negative controls. The oligonucleotides were designed and synthesized by Operon/QIAGEN and arrayed on Superamine slides at the Genome BC Microarray Platform (Vancouver, Canada) according to the manufacturer's protocol (ArrayIt). All the probes were printed in duplicate, side by side. The controls were randomly distributed on the array in three sets of duplicate spots.
Microarray hybridization and data analysis.
The microarray slides were prehybridized using 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) containing 0.1% SDS and 0.2% bovine serum albumin for 45 min at 48°C and used immediately for hybridization in a GeneTac HybStation (Genomic Solution). The hybridization was carried out at 42°C for 18 h with mixing by using 120 μl per slide of SlideHyb#1 hybridization solution (Ambion). The posthybridization washing consisted of three cycles of 20-second incubations with each of the following solutions: 2× SSC plus 0.1% SDS (medium stringency) at 42°C, 0.1× SSC plus 0.05% SDS (high stringency) at 25°C, and 0.1× SSC (low stringency) at 25°C. After being washed, the slides were dried by centrifugation for 5 min at 350 × g at room temperature and scanned with a GenePix 4000B scanner (Axon Instruments). Nine hybridizations were conducted, one for each of the triplicate cultures grown on each of the three aromatic substrates. For each aromatic substrate, two triplicate cDNA samples were labeled with Cy5 and one was labeled with Cy3. Respectively, these were hybridized in competition with cDNA samples from the triplicate PYR control cultures, two labeled with Cy3 and one labeled with Cy5. Equal amounts of Cy3 and Cy5 were used in all hybridizations.
The spot intensities were quantified using Imagene 5.6 (BioDiscovery, Inc.). To correct for nonspecific (background) signal for each channel (each dye), the mean signal for 10% of the probes in each subgrid with the lowest intensity was subtracted from that for all probes in the corresponding subgrid. Using GeneSpring version 6.0 (Silicon Genetics), expression ratios were normalized using the LOWESS method. Average normalized expression ratios (treatment/control) were calculated for each gene and tested for significant variation between treatments (analysis of variance [ANOVA], P < 0.05). Treatments were further screened for difference from the control, which was defined as having an expression ratio of either >2.0 or <0.5. A heat map showing expression patterns for selected genes was generated using MeV 3.1 (The Institute for Genomic Research).
Quantitative PCR.
To validate the microarray data, transcripts from five genes from four different operons were quantified by real-time PCR analysis. TaqMan probes and primers (Table 1) were designed using the default parameters for the software Primer Express 2.0 (Applied Biosystems). As an internal standard, multiplex reactions additionally quantified the gene encoding DNA polymerase IV. This gene was selected because it showed high and constant expression levels on all substrates, including the PYR control. All reactions were performed using the following probe combination: 6FAM (5′ reporter) for genes of interest and VIC (5′ reporter) for the internal control. TAMRA (6-carboxytetramethylrhodamine) was used as a quencher for both probes in the same tube.
TABLE 1.
Primers and probes used for TaqMan quantitative PCR assays
| Target gene | Primer or probe |
|---|---|
| bphAa | Sense primer 5′TCGGATGGTGTTGATGCC3′ |
| Antisense primer 5′TTGGTGAGTGTGCCTTTCG3′ | |
| Probe (6FAM)TGTGGGGAGCGATCCTGACTGGAC (TAMRA) | |
| etbC | Sense primer 5′CCAGCCCAGCAGGAACACT3′ |
| Antisense primer 5′GCCATAGCCTTCAACCTCGTT3′ | |
| Probe (6FAM)CCTACGCGACATCTTCGGTCACGA (TAMRA) | |
| etbAc | Sense primer 5′ACGCACAGCTACTCGTTGCTT3′ |
| Antisense primer 5′CCGTGCACTTCACATTCGAT3′ | |
| Probe (6FAM)CCGACGGGTTTCAAGAGGGCG (TAMRA) | |
| benA | Sense primer 5′CCCGAATGTCGGCGACTA3′ |
| Antisense primer 5′TGTTGCGGGAGATCACGAT3′ | |
| Probe (6FAM)TTCACCACGTACATGGGCCGCC (TAMRA) | |
| benB | Sense primer 5′CCGCAGTAGCCGAAAGCA3′ |
| Antisense primer 5′GCGGGCCTCACGGTAGA3′ | |
| Probe (6FAM)TGTCACCCAGCACGACATCGAACA (TAMRA) | |
| DNApol IV | Sense primer 5′GACAACAAGTTACGAGCCAAGATC3′ |
| Antisense primer 5′CCTCCGTCAGCCGGTAGAT3′ | |
| Probe (VIC)CGACGGACTTCGGCAAACCGC(TAMRA) |
cDNA was synthesized using the ThermoScript RT-PCR system and random hexamers according to the manufacturer's instructions (Invitrogen Life Technology) and 1 μg of total RNA in a total volume of 20 μl. The cDNA was diluted, and samples corresponding to 0.05 μl from the original tube were used for the real-time PCR. All reactions were performed with an MJ Chromo4 real-time PCR system with the following conditions: 10 min at 95°C followed by 40 cycles of 15 seconds at 95°C and 1 min at 60°C for extension. All reactions were performed in triplicate, and the data were normalized using the average for the internal standard. Standards for the assays were prepared by cloning PCR amplicons containing the target genes into the TOPO-TA vector (Invitrogen). A standard curve was constructed by comparing the copy numbers of 10-fold dilutions of each standard to their respective threshold cycles.
Sequence analyses.
Amino acid sequence alignments and distance matrices were calculated using CLUSTALX version 1.83 (35). Trees were calculated by applying the neighbor-joining method (26) to the distance matrix and were displayed using GeneDoc. The sequences for potential BphAa, BphB, and BphC homologues were searched for the corresponding PROSITE signatures (10). To facilitate the phylogenetic analyses of RHA1 pathway enzymes, reference enzymes of experimentally verified substrate preference were included in these analyses.
Accession numbers.
The RHA1 genome was submitted to NCBI (accession numbers NC8268, NC8269, NC8270, and NC8271). Additional data, including files for whole-genome visualization (in Artemis and GBrowse formats), are available at http://www.rhodococcus.ca/. Details of the microarray design, transcriptomic experimental design, and transcriptomic data have been deposited in the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO Series accession number GSE5280.
RESULTS
Analysis of RHA1 genome.
The RHA1 genome sequence (http://www.rhodococcus.ca) confirmed the existence of the previously reported bph, etb, and ben gene clusters and identified additional homologues of most of those genes. Genes that potentially encode enzymes involved in the catabolism of BPH, ETB, and BEN are summarized in Fig. 1. Analyses of the genome sequence predicted the existence of 21 genes encoding Rieske nonheme iron oxygenases (ROs) (22). In phylogenetic analyses, many of these did not cluster with enzymes classified by Gibson and Parales (7), and their physiological roles in RHA1 are unclear. Of the 21 predicted RHA1 ROs, 9 were predicted to be part of ring-hydroxylating dioxygenases, including those previously identified to be encoded by the bph, etb, pad, and ben genes. Phylogenetic analyses indicated that only the three previously identified BphA, EtbA1, and EtbA2 dioxygenases likely transform PBH or ETB. The oxygenases of the six other predicted ring-hydroxylating ROs were each predicted to transform aromatic acids. We are currently investigating the function of the 12 predicted RHA1 ROs that could not be classified as ring-hydroxylating enzymes based on the phylogenetic analyses. Similarly, the genome encodes two BphB-like dehydrogenases of the short-chain dehydrogenase/reductase family that likely transform cis-dihydrodiols derived from BPH or ETB. Sequence analyses identified genes encoding 13 type I extradiol dioxygenases. A large number of extradiol dioxygenases reported in the literature are poorly characterized, confounding sequence-based prediction of enzyme specificity. Thus, all 13 bphC homologues are listed in Fig. 1. Genes for eight α/β-fold C-C bond hydrolases were identified, of which, based on phylogenetic analyses, six were predicted to possibly transform 2-hydroxypentadienoates originating from BPH or ETB. These six BphD homologues share five key active site residues and a minimum of 23% amino acid sequence identity. We identified 22 genes potentially encoding degradation of 2-hydroxy-2,4-pentadieneoate (HPD) to pyruvate plus acetyl-coenzyme A (CoA) in eight clusters of two or three genes. These genes include eight that potentially encode BphE-type hydratases, sharing a minimum of 26% amino acid sequence identity, some of which may alternately encode 4-oxalocrotonate decarboxylases; seven BphF-type aldolases, potentially able to transform 4-hydroxy-2-oxovalerate and sharing a minimum of 41% sequence identity; and seven BphG-type acetaldehyde dehydrogenases, sharing a minimum of 40% amino acid sequence identity. Importantly, each of the predicted enzymes for the first four steps of the pathway contained conserved amino acids known to be critical to catalytic function or structural stability.
Gene expression trends on different substrates.
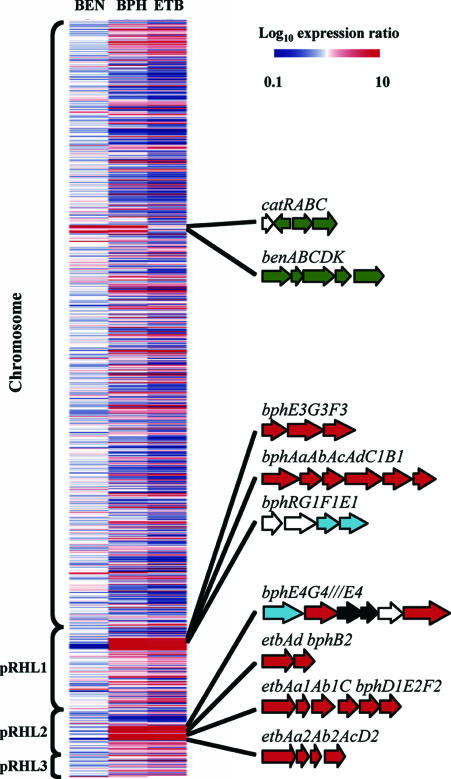
The expression of 8,313 genes was measured during exponential growth of RHA1 on BPH, ETB, BEN, and the control substrate, PYR, as the sole organic substrates. The expression of 926 genes differed significantly on the three aromatic substrates. The patterns of gene expression were overwhelmingly similar on BPH and ETB, with a large number of genes upregulated relative to their regulation on the PYR control (Fig. 2). These genes are distributed among all the genomic elements, but there are some obvious regions with a high density of upregulated genes on pRHL1 and pRHL2 that contain the bph and etb catabolic genes.
FIG. 2.
Expression of 926 RHA1 genes whose expression differed significantly on three substrates plus additional genes in Table 2. Color scale indicates log10 values for average normalized expression ratio (treatment/control). Genes are ordered according to location on the four genomic elements. Clusters of genes discussed in the text are shown, with genes color coded as follows: red, upregulated on BPH and ETB; green, upregulated on BEN and BPH; blue, constitutively expressed at high levels; black, possible transposases; white, other.
By contrast, only 65 genes were upregulated on BEN. The ben and cat catabolic genes were part of a cluster of genes that was upregulated on BEN and BPH but downregulated on ETB, relative to their regulation on the PYR control. Expression of the ben genes on BPH was expected, as BEN is an intermediate of BPH degradation (Fig. 1).
BPH-ETB transcriptome.
A common set of 320 genes was upregulated on both BPH and ETB. Upregulated genes in the BPH-ETB transcriptome were in diverse functional categories (see Table S1 in the supplemental material) based on clusters of orthologous groups of proteins (34). The majority of these genes have unknown functions. The next-largest groups are distributed throughout the clusters of orthologous groups of proteins within the general group of metabolism, including the genes predicted to specify BPH and ETB degradation (see below) plus metabolic genes whose roles are not apparent. These include ro00423, ro02511, and ro02355 and/or ro04667 (the latter two are not distinguished by probes), which encode P450 monooxygenases. The metabolism genes also include many that may actually be involved in transport or environmental sensing. Another large group of genes in the BPH-ETB transcriptome is distributed throughout the general group of information storage and processing, including many genes encoding transcriptional regulators.
BPH pathway genes.
The BPH-ETB transcriptome includes a common suite of catabolic genes, similarly expressed on both substrates, encoding multiple isozymes for each step in the BPH pathway (Fig. 1 and Table 2). We did not observe the alternative possibility, differential regulation of these genes on the two substrates. The levels of expression (based on probe signal intensities) appear to be very high for many of the BPH pathway genes. Further, most of the BPH pathway genes were strongly downregulated on BEN relative to their regulation on the PYR control.
TABLE 2.
Identity and expression of genes discussed in the textaa
| Gene ID | Gene name | Gene product | Expression ratiob
|
Signal intensityc
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| BEN | BPH | ETB | PYR | BEN | BPH | ETB | |||
| ro00423 | Cytochrome P450 CYP254 | 0.96 | 3.3 | 4.4 | 390 | 270 | 1,300 | 2,200 | |
| ro00477d | Type I extradiol dioxygenase | 0.71 | 0.70 | 0.63 | 260 | 160 | 230 | 150 | |
| ro00515d | Probable 4-hydroxy-2-oxovalerate aldolase | 1.3 | 2.1 | 3.4 | 70 | 72 | 210 | 220 | |
| ro00516d | Acetaldehyde dehydrogenase | 1.2 | 1.0 | 1.4 | 86 | 100 | 90 | 130 | |
| ro00517d | Probable 2-oxopent-4-enoate hydratase | 0.87 | 1.6 | 1.3 | 180 | 12 | 350 | 240 | |
| ro00519d | ohpC | 2-Hydroxy-6-ketonona-2,4-dienedoic acid hydrolase | 0.84 | 0.95 | 0.87 | 1,000 | 590 | 1,100 | 970 |
| ro00536d | Type I extradiol dioxygenase | 1.1 | 1.2 | 2.4 | 16 | 23 | 20 | 33 | |
| ro01333d | pcaJ1 | 3-Oxoacid CoA-transferase | 5.8 | 3.2 | 1.1 | 71 | 400 | 270 | 75 |
| ro01334d | pcaI | Probable 3-oxoacid CoA-transferase alpha subunit | 4.1 | 3.5 | 4.5 | 110 | 360 | 440 | 510 |
| ro01336 | pcaG | Protocatechuate dioxygenase alpha subunit | 2.9 | 1.3 | 0.38 | 440 | 1,100 | 610 | 180 |
| ro01338d | pcaL | 3-Oxoadipate enol-lactone hydrolase/ 4-carboxymuconolactone decarboxylase | 3.2 | 2.4 | 1.3 | 140 | 410 | 380 | 160 |
| ro01509 | Possible NADPH: quinone reductase | 0.67 | 11 | 10 | 1,400 | 470 | 13,000 | 20,000 | |
| ro02371 | catC | Muconolactone delta-isomerase | 38 | 43 | 2.2 | 27 | 960 | 1,300 | 81 |
| ro02372 | catB | Muconate cycloisomerase | 40 | 23 | 1.7 | 62 | 2,200 | 1,800 | 90 |
| ro02373 | catA1 | Catechol 1,2-dioxygenase | 17 | 12 | 1.5 | 330 | 5,100 | 4,600 | 420 |
| ro02374d | catR | Transcriptional regulator, IclR family | 0.96 | 0.62 | 0.51 | 12,000 | 7,400 | 6,500 | 7,800 |
| ro02381d | benR | Transcriptional regulator, AraC family | 2.1 | 1.5 | 0.19 | 82 | 140 | 150 | 39 |
| ro02383 | Cytochrome P450, reductase | 2.0 | 5.2 | 5.5 | 61 | 99 | 350 | 380 | |
| ro02384 | benA | Benzoate 1,2-dioxygenase alpha subunit | 33 | 14 | 1.1 | 270 | 9,100 | 5,000 | 190 |
| ro02385d | benB | Benzoate 1,2-dioxygenase beta subunit | 3.3 | 1.2 | 0.52 | 3,800 | 8,200 | 5,100 | 2,300 |
| ro02386 | benC | Benzoate 1,2-dioxygenase reductase subunit | 32 | 10 | 0.70 | 470 | 13,000 | 6,600 | 260 |
| ro02387 | benD | cis-1,6-Dihydroxycyclohexa-3,5-diene-1-carboxylate dehydrogenase | 8.1 | 4.7 | 0.68 | 250 | 2,100 | 1,400 | 140 |
| ro02388d | benK | Benzoate transporter, MFS superfamily | 6.9 | 5.0 | 2.2 | 440 | 2,700 | 2,400 | 990 |
| ro02488d | Type I extradiol dioxygenase | 1.1 | 1.2 | 2.4 | 32 | 32 | 40 | 84 | |
| ro02511 | Cytochrome P450 CYP147 | 1.1 | 2.2 | 4.1 | 110 | 110 | 280 | 470 | |
| ro03865d | Type I extradiol dioxygenase | 1.0 | 1.0 | 1.4 | 120 | 110 | 140 | 160 | |
| ro03866d | Acetaldehyde dehydrogenase | 0.82 | 1.4 | 1.2 | 210 | 150 | 310 | 250 | |
| ro03867d | Possible 4-hydroxy-2-oxovalerate aldolase | 0.91 | 0.72 | 0.63 | 3,300 | 2,100 | 2,300 | 2,800 | |
| ro03881d | 2-Oxopent-4-enoate hydratase | 1.2 | 1.0 | 1.1 | 46 | 48 | 47 | 50 | |
| ro03975d | Type I extradiol dioxygenase | 1.3 | 0.85 | 1.3 | 450 | 520 | 400 | 650 | |
| ro04533d | Hydratase | 0.79 | 0.94 | 0.97 | 920 | 580 | 880 | 1,100 | |
| ro04534d | Acetaldehyde dehydrogenase | 1.1 | 2.2 | 2.7 | 66 | 63 | 150 | 200 | |
| ro04535d | 4-Hydroxy-2-oxovalerate aldolase | 0.95 | 0.86 | 0.84 | 41 | 29 | 37 | 36 | |
| ro04540d | Alpha/beta-fold C-C bond hydrolase | 1.2 | 2.7 | 2.8 | 58 | 66 | 170 | 170 | |
| ro04541 | Type I extradiol dioxygenase | 0.92 | 17 | 16 | 52 | 33 | 850 | 1,100 | |
| ro04667 or ro02355e | Cytochrome P450 CYP125 | 0.79 | 3.5 | 4.9 | 290 | 170 | 1,200 | 1,700 | |
| ro04905d | Type I extradiol dioxygenase | 0.79 | 1.1 | 1.0 | 2,200 | 1,200 | 3,000 | 2,800 | |
| ro05797d | Alpha/beta-fold C-C bond hydrolase | 1.2 | 1.0 | 1.1 | 350 | 340 | 390 | 480 | |
| ro05799d | 2-Keto-4-pentenoate hydratase | 1.1 | 0.88 | 0.91 | 830 | 660 | 910 | 820 | |
| ro05800 | Acetaldehyde dehydrogenase (acetylating) | 2.1 | 1.6 | 1.0 | 660 | 1,100 | 1,000 | 810 | |
| ro05803d | Type I extradiol dioxygenase | 1.1 | 0.70 | 0.71 | 6,200 | 4,600 | 4,600 | 6,900 | |
| ro08044 or ro10146e | etbD1/etbD2 | 2-Hydroxy-6-oxohepta-2,4-dienoate hydrolase | 0.54 | 42 | 36 | 130 | 36 | 6,200 | 6,200 |
| ro08051 or ro10121e | bphT1/bphT2 | Response regulator, two-component system | 0.68 | 1.4 | 1.4 | 1,200 | 570 | 1,600 | 2,200 |
| ro08052 | bphS1 | Sensor kinase, two-component system | 0.85 | 25 | 24 | 720 | 280 | 16,000 | 22,000 |
| ro08054 | bphB1 | cis-2,3-Dihydrobiphenyl-2,3-diol dehydrogenase | 0.16 | 29 | 14 | 2100 | 54 | 32,000 | 32,000 |
| ro08055 | bphC1 | 2,3-Dihydroxybiphenyl 1,2-dioxygenase | 0.15 | 21 | 8.7 | 2700 | 51 | 29,000 | 29,000 |
| ro08057 | bphAd | Biphenyl 2,3-dioxygenase, reductase | 0.34 | 33 | 21 | 750 | 51 | 18,000 | 21,000 |
| ro08058 | bphAc | Biphenyl 2,3-dioxygenase, ferredoxin component | 0.16 | 37 | 21 | 1400 | 49 | 37,000 | 39,000 |
| ro08059 | bphAb | Biphenyl 2,3-dioxygenase beta subunit | 0.17 | 22 | 14 | 2,100 | 70 | 39,000 | 35,000 |
| ro08060 | bphAa | Biphenyl 2,3-dioxygenase alpha subunit | 0.36 | 43 | 43 | 1,200 | 300 | 41,000 | 39,000 |
| ro08079d | Type I extradiol dioxygenase | 0.90 | 0.88 | 0.76 | 980 | 780 | 870 | 980 | |
| ro08081d | Alpha/beta-fold C-C bond hydrolase | 1.0 | 1.3 | 1.6 | 110 | 99 | 180 | 160 | |
| ro08083 | bphF3 | 4-Hydroxy-2-oxovalerate aldolase | 0.72 | 13 | 16 | 180 | 76 | 2,400 | 3,900 |
| ro08084 | bphG3 | Acetaldehyde dehydrogenase | 1.3 | 12 | 11 | 180 | 37 | 2,500 | 3,100 |
| ro08085d | bphE3 | 2-Oxopent-4-enoate hydratase | 0.98 | 3.8 | 3.7 | 340 | 260 | 1,500 | 1,400 |
| ro09005d | Type I extradiol dioxygenase | 0.91 | 0.64 | 1.1 | 90 | 71 | 76 | 82 | |
| ro09014d | Alpha/beta-fold C-C bond hydrolase | 0.79 | 1.3 | 1.7 | 310 | 190 | 430 | 600 | |
| ro09018d | bphG1 | Acetaldehyde dehydrogenase | 1.1 | 1.2 | 2.0 | 37 | 41 | 70 | 33 |
| ro09019d | bphF1 | 4-Hydroxy-2-oxovalerate aldolase | 0.88 | 0.59 | 0.49 | 1,600 | 1,000 | 1,100 | 860 |
| ro09021d | bphE1 | 2-Hydroxypenta-2,4-dienoate hydratase | 0.84 | 1.1 | 1.3 | 460 | 310 | 610 | 570 |
| ro10112d | bphF4 | 4-Hydroxy-2-oxovalerate aldolase | 0.81 | 1.5 | 1.8 | 590 | 320 | 870 | 1,400 |
| ro10116 | bphG4 | Acetaldehyde dehydrogenase | 0.79 | 25 | 22 | 170 | 61 | 4,800 | 5,100 |
| ro10117d | bphE4 | 2-Hydroxypenta-2,4-dienoate hydratase | 1.2 | 1.3 | 1.1 | 3,300 | 2,900 | 3,500 | 4,900 |
| ro10121 or ro08051e | bphT2/bphT1 | Response regulator, two-component system | 0.79 | 1.6 | 1.7 | 990 | 600 | 1,600 | 1,900 |
| ro10122 or ro08052e | bphS2/bphS1 | Sensor kinase, two-component system | 0.96 | 18 | 19 | 630 | 370 | 12,000 | 15,000 |
| ro10125 | etbAd | Ferredoxin reductase | 1.1 | 15 | 12 | 1,500 | 490 | 21,000 | 29,000 |
| ro10126 | bphB2 | cis-3-Phenylcyclohexa-3,5-diene-1,2-diol dehydrogenase | 0.47 | 13 | 6.4 | 2,700 | 100 | 30,000 | 31,000 |
| ro10135d | etbC | 2,3-Dihydroxybiphenyl 1,2-dioxygenase | 0.20 | 79 | 40 | 1,500 | 100 | 47,000 | 35,000 |
| ro10136 | bphD1 | 2-Hydroxy-6-oxo-6-phenylhexa-2,4-dienoate hydrolase | 0.71 | 44 | 24 | 980 | 110 | 37,000 | 32,000 |
| ro10137 | bphE2 | 2-Oxopent-4-enoate hydratase | 0.89 | 38 | 19 | 1,400 | 240 | 35,000 | 33,000 |
| ro10138 | bphF2 | 4-Hydroxy-2-oxovalerate aldolase | 1.1 | 41 | 28 | 260 | 70 | 12,000 | 10,000 |
| ro10143 or ro10133e | etbAa2/etbAa1 | Ethylbenzene dioxygenase alpha subunit | 0.31 | 68 | 79 | 2,000 | 210 | 45,000 | 46,000 |
| ro10144 or ro10134e | etbAb2/etbAa1 | Ethylbenzene dioxygenase beta subunit | 0.29 | 140 | 75 | 670 | 57 | 52,000 | 41,000 |
| ro10145 | etbAc | Ethylbenzene dioxygenase, ferredoxin component | 0.27 | 58 | 39 | 890 | 99 | 26,000 | 32,000 |
| ro10303 | Probable ferredoxin FdxD | 0.88 | 5.3 | 8.5 | 84 | 54 | 520 | 780 | |
| ro10315d | Type I extradiol dioxygenase | 0.92 | 1.1 | 1.6 | 560 | 410 | 590 | 1,100 | |
Expression of all genes differed significantly for the four substrates tested, unless otherwise noted.
Average normalized expression ratio (treatment/control; n = 3).
Uncorrected average probe signal intensity (n = 3).
Expression did not differ significantly among substrates (P ≥ 0.05 for ANOVA).
Probe lacks specificity due to high similarity of genes.
Genes encoding three complete or partial ring-hydroxylating dioxygenase systems were upregulated on BPH and ETB. These genes (underlined) are encoded in three clusters, bphAaAbAcAdC1B1, etbAa1Ab1C-bphD1E2F2, and etbAa2Ab2AcD2 (Fig. 2), which were all highly induced on the three substrates (Table 2). The highly coordinated regulation of genes within each of these clusters (Fig. 3) confirms that these clusters are operons. For some pairs of genes, the transcripts could not be discriminated by array probes, due to their high sequence similarity. In the above-described cases of etbAa1, etbAa2, etbAb1, and etbAb2, it is likely that these four genes had a shared pattern of expression, since other genes with specific probes in the two corresponding operons all had similar expression patterns. Only the bph operon encodes all four components of a dioxygenase system, while the etb1 operon lacks genes for the ferredoxin and reductase components, and the etb2 operon lacks a gene for the reductase component. We examined transcriptomic data for an additional 57 genes putatively encoding reductases or ferredoxins, which were not proximal to the above-described gene clusters. Most of those genes were not upregulated on BPH or ETB. One ferredoxin gene (ro10303) and two reductase genes (ro01509 and ro02383) were upregulated on BPH and ETB, but there is no further evidence to suggest whether these components can function with the above-described dioxygenases. Both genes predicted to encode cis-dihydrodiol dehydrogenases homologous to bphB were upregulated on BPH and ETB.
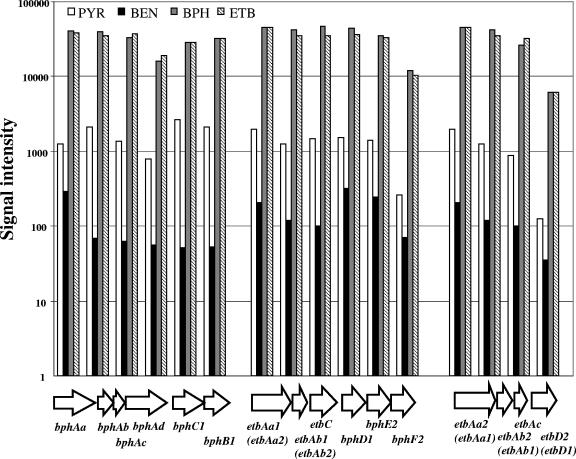
FIG. 3.
Uncorrected microarray signal intensities for genes in three putative operons in RHA1 grown on four substrates.
Of the 13 identified bphC homologues encoding type I extradiol dioxygenases (Fig. 1), bphC1 and ro04541 appeared upregulated on BPH and ETB (Table 2). The etbC gene did not meet the ANOVA criterion for significantly different expression on the three aromatic substrates (P = 0.062), but its pattern of expression and signal intensities were consistent with those of the other genes in the etbAa1Ab1C-bphD1E2F2 operon. Both ro04905 and ro05080 appeared to be constitutively expressed at high levels on all four substrates, including PYR. The remaining eight homologues were not upregulated or highly expressed in our experiments.
Of the eight bphD homologues encoding α/β-fold serine hydrolases (Fig. 1), the previously identified bphD1 was highly upregulated during growth of cells on BPH and ETB (Table 2). The etbD1 and etbD2 genes are 97% identical, so their expression could not be distinguished by a microarray probe. The probe representing both genes indicated upregulation on BPH and ETB. Given its location in the etbAa2Ab2AcD2 operon, etbD2 was presumably upregulated, but it is unclear whether etbD1 was also upregulated. The remaining five bphD homologues were not upregulated on BPH or ETB.
Of the genes potentially encoding the three steps of HPD degradation (Fig. 1), the bphE2F2 genes in the etbAa1Ab1C-bphD1E2F2 operon (Fig. 3) were highly upregulated, and those in the bphF3G3E3 cluster on pRHL1 were also upregulated (4- to 16-fold) on BPH and ETB (Table 2). The bphE1F1G1 cluster localized on plasmid pRHL1 was not upregulated on any of the tested substrates. However, based on the signal intensity of the three corresponding probes on all substrates, including PYR, the bphE1F1 genes appeared to be constitutively expressed at a relatively high level. Finally, an additional cluster on pRHL2, bphE4G4F4, was partially upregulated on the three substrates and has several unusual features. The ancestral bphG4 gene is interrupted by two possible transposases (ro10114 and ro10115), which split that gene into two open reading frames, ro10113 and bphG4. The latter gene appears to encode an intact, potentially functional acylating acetaldehyde dehydrogenase domain and was upregulated (22- to 25-fold) on BPH and ETB. By contrast, bphE4 and bphF4 appeared to be constitutively expressed at high levels on all substrates. The remaining 11 bphEFG homologues, in four clusters, were not upregulated on any substrate tested.
Quantitative PCR analyses.
Quantitative reverse transcriptase PCR was used as an alternative to measure the expression levels of genes. This was done to confirm the accuracy of the microarray analyses, to check microarray results that appeared doubtful in light of inconsistent results within putative operons, and to quantify upregulation of genes whose signals appeared to be beyond the dynamic range of the microarray assay.
The expression of three genes, bphAa, etbC, and etbAc, representing the highly expressed operons bphAaAbAcAdC1, etbAa1Ab1C-bphD1E2F2, and etbAa2Ab2AcD2, was analyzed by Q-PCR (Table 3). These genes were selected because they are unique in the genome and their expression levels appeared similar to those of the other genes in their respective operons (Fig. 3). When the expression ratios (treatment/control) of the three genes were compared using the two methods, the difference between methods was consistently less than fivefold, which is considered to indicate good agreement between the methods (39). Further, the trends for the expression of the three genes on the different substrates measured by the two methods agreed very well. This agreement confirms the accuracy of the microarray analyses. The results also confirm that despite the microarray results for etbC not meeting the ANOVA criterion for statistically different expression, that gene was clearly upregulated on BPH and ETB.
TABLE 3.
Numbers of gene transcripts measured by quantitative PCR (n = 9)
| Substrate | No. of gene transcripts (nmol/μg RNA)
|
||||
|---|---|---|---|---|---|
| bphAa | EtbC | etbAc | benA | benB | |
| Pyruvate | 160 | 26 | 143 | 1.4 | 1.2 |
| Benzoate | 18 | 22 | 52 | 14,200 | 15,900 |
| Biphenyl | 29,700 | 20,200 | 25,100 | 2,150 | 1,810 |
| Ethylbenzene | 31,400 | 17,400 | 27,500 | 1.4 | 1.5 |
The benABCDK gene cluster was highly upregulated on BEN. However, the benB microarray signal intensity was anomalously high on PYR (Table 2). In contrast to the microarray results, the Q-PCR assay indicated that benA and benB are expressed at similar levels on PYR (Table 3). The microarray analysis indicated that the maximum upregulation of ben genes on BEN was 64-fold, while Q-PCR analysis showed that the maximum upregulation was ca. 10,000-fold. These results indicate that the ben genes do comprise a coordinately expressed operon, which is upregulated on BEN and BPH but not on PYR. The extent of upregulation is less on BPH than on BEN, which is consistent with the lower growth rate on BPH than on BEN. The results also indicate that the microarray analysis underestimated the expression of this operon, probably due to the limited dynamic range of the technique (i.e., saturation of the probes).
DISCUSSION
Multiplicity of homologues for BPH pathway genes.
The analysis of the RHA1 genome revealed a considerable multiplicity of the genes potentially involved in BPH and alkyl benzene catabolism, including at least two homologues potentially encoding each step of the BPH pathway (Fig. 1). The multiplicity of these genes seems to be common in rhodococci (16, 37) and has been reported for other genera as well (8). In RHA1, previous reports indicate multiplicities of bphA (11, 14, 22, 27), bphB (32), bphC (1, 9, 15, 27), bphD (27, 38), bphE, and bphF (19). Here, we report 7 new bphC homologues (for a total of 13 in the genome), 3 new bphD homologues (for a total of 8), and 17 new homologues of genes encoding HPD degradation, bphEFG. The HPD degradation genes are located in eight clusters of two or three genes. Nearly all of the BPH pathway genes are located on the two largest plasmids, pRHL1 and pRHL2 (Fig. 2). Surrounding many of these genes are probable transposase genes, suggesting recombination processes involving these genes and their possible lateral transfer. Recombination and duplication are also indicated by the fact that pRHL1 and pRHL2 share a large cluster of phthalate and putative terephthalate degradation genes (25). Further, lateral transfer is indicated by a 330-kb plasmid in Rhodococcus sp. DK17, pDK2, which shares near-complete nucleotide sequence identity with pRHL2. The sequences for pRHL2 and pDK2 (GenBank accession number AY502075) are 100% identical, except for one 0.9-kb insertion in pRHL2. This insertion corresponds to the two probable transposase genes that disrupted the ancestral bphG4, yielding ro10113 and the existing bphG4. Transposon-mediated bph gene transfer was previously described for pseudomonads (23). Rhodococci appear capable of genetic exchange with distantly related organisms. For example, the catabolic gene dhaA, encoding a haloalkane halohydrolase, is believed to have been transferred between Pseudomonas and Rhodococcus (12).
Identification of BEN pathway genes.
Our transcriptomic analysis confirmed the roles of the ben and cat genes and their enzyme products in the degradation of BEN (Table 2). The pcaLIJF genes are also predicted to be necessary for complete BEN degradation to acetyl-CoA plus succinyl-CoA. Our results suggest that these genes may be somewhat upregulated on BEN and BPH above a moderate basal level of expression on PYR, but the data do not meet our criteria for significant upregulation. All of the corresponding proteins were previously found to be upregulated on BEN relative to their regulation on PYR by proteomic analysis (25). The available evidence is consistent with the predicted involvement of the pca genes in BEN and BPH degradation. The BEN pathway was regulated independently of the BPH pathway and was not part of the common BPH-ETB transcriptome. The extent of BEN gene upregulation appears to be much greater during growth on BEN than on BPH (Table 3). The BEN pathway could be induced during BPH degradation by the accumulation of BEN or one of its degradation intermediates. The expression pattern of benR is consistent with a role as a positive regulator.
Identification of BPH pathway genes.
Previous reports of BPH pathway gene regulation in RHA1 are fragmentary, and some are qualitative. Our transcriptomic analyses showed that multiple homologous genes encoding every step of the BPH pathway (Fig. 1) are simultaneously induced by BPH or ETB (Fig. 2 and 3 and Table 2). We confirmed the expression and probable role of a number of genes in the degradation of these two substrates, including genes encoding three biphenyl dioxygenase systems (bphAaAbAcAd, etbAa1Ab1Ad, and etbAa2Ab2Ac), two dihydrodiol dehydrogenases (bphB1 and bphB2), two dihydroxybiphenyl dioxygenases (bphC1 and etbC), two hydrolases (bphD and etbD2), and two HPD pathway enzymes (bphE2 and bphF2). The results confirmed our prediction that no additional ring-hydroxylating dioxygenase systems are involved in BPH or ETB degradation.
In contrast to previous findings (28), we found no evidence for induction of the bphE1F1G1 operon on BPH or ETB. Rather, probe signal intensity suggested that bphE1F1 genes were constitutively expressed at relatively high levels on all substrates (Table 2). The difference between results could be due to the use of a rich substrate, LB medium, for the control in the previous study versus mineral medium plus pyruvate in ours. Knockout analysis indicates that bphE1F1G1 genes are involved in biodegradation of biphenyl and alkyl benzenes (28), but these genes may also be involved in the biodegradation of additional compounds which yield HPD and its derivatives as intermediates, such as catechol and 3-(2-hydroxyphenyl) propionic acid.
The roles of four genes in the BPH pathway are questionable. The expression level of etbD1 is impossible to distinguish from that of the very similar etbD2, and the genomic context of etbD1 does not provide further insight, as it does for etbD2. The ro04541 gene was previously reported to be induced during growth on BPH or ETB and was designated bphC5 (27). Our results suggested that ro04541 is much less upregulated and expressed at much lower levels than were other confirmed BPH pathway genes (Table 2). Because of the similarity of ro04541 to other bphC homologues, we cannot completely rule out the possibility that its probe cross-hybridized (despite the probe meeting design criteria for specificity). The extreme expression levels of bphC1 and etbC greatly increase the potential for detectable cross-hybridization. Further, ro04541 clearly appears to be cotranscribed with the gene immediately downstream, ro04540, a bphD homologue, which overlaps with ro04540 by one nucleotide. Since ro04540 was not upregulated on BPH or ETB, the apparent upregulation of ro04541 in both studies was likely due to cross-hybridization, and it appears doubtful that this gene has a role in BPH or ETB degradation. Finally, ro04905 and ro05803 appeared to be constitutively expressed at high levels. Without evidence regarding the expression and specificity of the encoded extradiol dioxygenases, it is impossible to know whether ro04905 or ro05803 contributes to BPH or ETB degradation.
We identified several novel genes in the BPH-ETB transcriptome, which are likely involved in the degradation of those compounds. These include three HPD pathway genes in an apparent operon, bphF3G3E3 (Fig. 2). We identified an additional HPD operon, bphE4G4F4, which appears to have been disrupted by a transposon. The bphG4 gene is truncated but encodes a complete catalytic domain and is upregulated on BPH and ETB, so it may contribute to their biodegradation. The bphE4 and bphF4 genes were not upregulated on BPH or ETB but appear to be expressed constitutively at high levels (Table 2), particularly the latter gene, suggesting that one or both genes may contribute to BPH and ETB biodegradation. Two additional HPD pathway genes, ro04533 and ro03867, had high signal intensities on all substrates. However, both of these genes are clustered in apparent operons with other HPD pathway genes that did not have high signal intensities, suggesting that the former high signal intensities may have been due to cross-hybridization. Further evidence of protein expression and enzyme activity are required to confirm the roles of these novel genes.
We ruled out the involvement of 26 genes potentially encoding BPH pathway enzymes, because these genes were not upregulated or, on the basis of probe signal intensity, highly expressed on BPH or ETB (Table 2). These genes include eight bphC homologues, five bphD homologues, and nine HPD pathway gene homologues (Fig. 1).
High expression levels.
When induced, genes for the catabolism of BPH, ETB, and BEN by RHA1 appear to be very highly upregulated and expressed at extremely high levels (Table 2). These genes generally had the highest signal intensities on the microarray, except for rRNA genes. Q-PCR assays, with a dynamic range much greater than that of microarray analysis (13), were necessary to quantify the upregulation of these genes. These assays indicated >120-fold increases in the expression of the bph genes and a 10,000-fold increase in the expression of the ben genes (Table 3). We estimate that when induced, transcripts from these individual genes range from 6.75 pg to 1.32 ng per μg of total RNA. There are few comparable results published. In Burkholderia xenovorans LB400, a ∼1,000-fold increase in the expression of bph genes was observed during growth on BPH compared to that observed during growth on BEN (3). Another study reported a ∼400-fold increase in the expression of tfd genes responsible for 2,4-dichlorophenoxyacetate degradation following induction with that compound (17). Thus, such high levels of upregulation and of expression may be typical for catabolic genes for such aromatic substrates.
A single catabolic system for BPH and alkyl benzenes.
Importantly, expression levels for the various bph homologues were very similar on both BPH and ETB. For each gene, microarray hybridization signal intensities were similar on the two substrates (Table 2 and Fig. 3). The Q-PCR assay further confirmed the similar expression levels of bphAa, etbC, and etbAc on the two substrates (Table 3). With a previous prototype microarray, we found very similar expression levels for many of the catabolic genes on BPH and ETB as well as isopropyl benzene, indicating that the latter compound also induces and is degraded by the same suite of enzymes. Thus, RHA1 does not differentially express particular enzymes that are most efficient for the degradation of these individual substrates. Rather, there appears to be a common BPH-alkyl benzene catabolic system, including one suite of catabolic enzymes similarly employed for a broad group of structurally similar substrates. In part, this uniform response may reflect the requirement of genes from at least two gene clusters to encode a complete BPH pathway, but this general response could additionally be the result of adaptation to mixtures of substrates that typically occur in natural environments. Alternatively, this general response might reflect a lack of optimization of the regulation of genes likely obtained via horizontal transfer.
Multiple isozymes with different catalytic characteristics can be advantageous to bacteria in the catabolism of mixtures of related compounds (1). Further, for bioremediation applications, the spectrum of PCBs degraded by a microorganism is directly related to the specificity of the biphenyl dioxygenase (21), and it was also suggested that the presence of multiple bphC homologues with different activities can avoid the accumulation of inhibitory metabolites produced during the biodegradation of aromatic compounds, including PCBs (9). Clearly, the simultaneous expression of a multiplicity of BPH pathway isozymes in RHA1, as shown in this study, may contribute to its exceptional ability to degrade PCBs. The simultaneous expression of three biphenyl dioxygenases also creates the intriguing possibility that hybrid enzyme systems (combinations of subunits encoded in different gene clusters) might assemble, yielding additional systems with distinct catalytic properties (5, 6, 18). Despite the above-described potential benefits of expressing multiple dioxygenase systems, it is also known that ring-hydroxylating and extradiol dioxygenases can be inactivated during the transformation of suboptimal substrates (11, 36). Therefore, to evaluate the effects of expressing multiple dioxygenase systems, it will be important to determine the substrate specificities of those enzymes.
BPH-ETB transcriptome.
A striking trend in the transcriptomic data was the large set of genes induced by both BPH and ETB versus the much smaller group of genes induced by BEN (Fig. 2). Since nearly half of the 320 genes upregulated on both BPH and ETB have unknown functions, it is difficult to deduce the physiological role of this large suite of genes, but that role clearly extends beyond the catabolism of the growth substrates. One possibility is that the hydrophobicity of biphenyl and alkyl benzenes causes a solvent stress response, which could account for the observed induction of genes of many functional categories located throughout the genome (Fig. 2; also see Table S1 in the supplemental material). The induction on BPH and ETB of 26 putative regulatory protein genes and 11 putative transposase genes is consistent with a stress response. However, there is very little similarity between the BPH-ETB transcriptome and transcriptomes observed during osmotic, desiccation, and starvation stresses (unpublished data). Also, beyond the BPH-ETB transcriptomes of similarly regulated genes, there are differences in gene expression on the two substrates. In particular, ETB stands out as causing significant upregulation of 75 more genes than BPH. This difference may explain why RHA1 was found to more extensively degrade PCBs when growing on ETB than when growing on BPH (4).
Supplementary Material
Acknowledgments
We thank Masao Fukuda for sharing unpublished data and helpful discussions, Christine Florizone for technical assistance, and Clinton Fernandes for bioinformatic assistance.
This work was supported by a grant from Genome Canada/ Genome BC.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Asturias, J. A., L. D. Eltis, M. Prucha, and K. N. Timmis. 1994. Analysis of three 2,3-dihydroxybiphenyl 1,2-dioxygenases found in Rhodococcus globerulus P6. Identification of a new family of extradiol dioxygenases. J. Biol. Chem. 269:7807-7815. [PubMed] [Google Scholar]
- 2.Bernstein, J. A., A. B. Khodursky, P. H. Lin, S. Lin-Chao, and S. N. Cohen. 2002. Global analysis of mRNA decay and abundance in Escherichia coli at single-gene resolution using two-color fluorescent DNA microarrays. Proc. Natl. Acad. Sci. USA 99:9697-9702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Denef, V. J., J. Park, T. V. Tsoi, J. M. Rouillard, H. Zhang, J. A. Wibbenmeyer, W. Verstraete, E. Gulari, S. A. Hashsham, and J. M. Tiedje. 2004. Biphenyl and benzoate metabolism in a genomic context: outlining genome-wide metabolic networks in Burkholderia xenovorans LB400. Appl. Environ. Microbiol. 70:4961-4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fukuda, M., S. Shimizu, N. Okita, M. Seto, and E. Masai. 1998. Structural alteration of linear plasmids encoding the genes for polychlorinated biphenyl degradation in Rhodococcus strain RHA1. Antonie Leeuwenhoek 74:169-173. [DOI] [PubMed] [Google Scholar]
- 5.Furukawa, K., J. Hirose, S. Hayashida, and K. Nakamura. 1994. Efficient degradation of trichloroethylene by a hybrid aromatic ring dioxygenase. J. Bacteriol. 176:2121-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ge, Y., and L. D. Eltis. 2003. Characterization of hybrid toluate and benzoate dioxygenases. J. Bacteriol. 185:5333-5341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gibson, D. T., and R. E. Parales. 2000. Aromatic hydrocarbon dioxygenases in environmental biotechnology. Curr. Opin. Biotechnol. 11:236-243. [DOI] [PubMed] [Google Scholar]
- 8.Gonzalez, V., R. I. Santamaria, P. Bustos, I. Hernandez-Gonzalez, A. Medrano-Soto, G. Moreno-Hagelsieb, S. C. Janga, M. A. Ramirez, V. Jimenez-Jacinto, J. Collado-Vides, and G. Davila. 2006. The partitioned Rhizobium etli genome: genetic and metabolic redundancy in seven interacting replicons. Proc. Natl. Acad. Sci. USA 103:3834-3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hauschild, J. E., E. Masai, K. Sugiyama, T. Hatta, K. Kimbara, M. Fukuda, and K. Yano. 1996. Identification of an alternative 2,3-dihydroxybiphenyl 1,2-dioxygenase in Rhodococcus sp. strain RHA1 and cloning of the gene. Appl. Environ. Microbiol. 62:2940-2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hulo, N., C. J. A. Sigrist, V. Le Saux, P. S. Langendijk-Genevaux, L. Bordoli, A. Gattiker, E. De Castro, P. Bucher, and A. Bairoch. 2004. Recent improvements to the PROSITE database. Nucleic Acids Res. 32:D134-D137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Imbeault, N. Y. R., J. B. Powlowski, C. L. Colbert, J. T. Bolin, and L. D. Eltis. 2000. Steady-state kinetic characterization and crystallization of a polychlorinated biphenyl-transforming dioxygenase. J. Biol. Chem. 275:12430-12437. [DOI] [PubMed] [Google Scholar]
- 12.Irvine, V. A., L. A. Kulakov, and M. J. Larkin. 2000. The diversity of extradiol dioxygenase (edo) genes in cresol degrading rhodococci from a creosote-contaminated site that express a wide range of degradative abilities. Antonie Leeuwenhoek 78:341-352. [DOI] [PubMed] [Google Scholar]
- 13.Johnson, M. R., K. Wang, J. B. Smith, M. J. Heslin, and R. B. Diasio. 2000. Quantitation of dihydropyrimidine dehydrogenase expression by real-time reverse transcription polymerase chain reaction. Anal. Biochem. 278:175-184. [DOI] [PubMed] [Google Scholar]
- 14.Kitagawa, W., K. Miyauchi, E. Masai, and M. Fukuda. 2001. Cloning and characterization of benzoate catabolic genes in the gram-positive polychlorinated biphenyl degrader Rhodococcus sp. strain RHA1. J. Bacteriol. 183:6598-6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kitagawa, W., A. Suzuki, T. Hoaki, E. Masai, and M. Fukuda. 2001. Multiplicity of aromatic ring hydroxylation dioxygenase genes in a strong PCB degrader, Rhodococcus sp. strain RHA1 demonstrated by denaturing gradient gel electrophoresis. Biosci. Biotechnol. Biochem. 65:1907-1911. [DOI] [PubMed] [Google Scholar]
- 16.Kulakov, L. A., and M. J. Larkin. 2002. Genetic organization of Rhodococcus, p. 15-46. In A. Danchin (ed.), Genomics of GC-rich gram-positive bacteria. Caister Academic Press, Wymondham, United Kingdom.
- 17.Leveau, J. H. J., F. König, H. Füchslin, C. Werlen, and J. R. van der Meer. 1999. Dynamics of multigene expression during catabolic adaptation of Ralstonia eutropha JMP134 (pJP4) to the herbicide 2,4-dichlorophenoxyacetate. Mol. Microbiol. 33:396-406. [DOI] [PubMed] [Google Scholar]
- 18.Maeda, T., Y. Takahashi, H. Suenage, A. Suyama, M. Goto, and K. Furukawa. 2001. Functional analyses of Bph-Tod hybrid dioxygenase, which exhibits high degradation activity toward trichloroethylene. J. Biol. Chem. 276:29833-29838. [DOI] [PubMed] [Google Scholar]
- 19.Masai, E., K. Sugiyama, N. Iwashita, S. Shimizu, J. E. Hauschild, T. Hatta, K. Kimbara, K. Yano, and M. Fukuda. 1997. The bphDEF meta-cleavage pathway genes involved in biphenyl/polychlorinated biphenyl degradation are located on a linear plasmid and separated from the initial bphACB genes in Rhodococcus sp. strain RHA1. Gene 187:141-149. [DOI] [PubMed] [Google Scholar]
- 20.Masai, E., A. Yamada, J. M. Healy, T. Hatta, K. Kimbara, M. Fukuda, and K. Yano. 1995. Characterization of biphenyl catabolic genes of gram-positive polychlorinated biphenyl degrader Rhodococcus sp. strain RHA1. Appl. Environ. Microbiol. 61:2079-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McKay, D. B., M. Prucha, W. Reineke, K. N. Timmis, and D. H. Pieper. 2003. Substrate specificity and expression of three 2,3-dihydroxybiphenyl 1,2-dioxygenases from Rhodococcus globerulus strain P6. J. Bacteriol. 185:2944-2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McLeod, M. P., R. L. Warren, N. Araki, W. W. L. Hsiao, M. Myhre, C. Fernandes, D. Miyazawa, W. Wong, A. L. Lillquist, D. Wang, M. Dosanjh, H. Hara, A. Petrescu, R. D. Morin, G. Yang, J. M. Stott, J. E. Schein, H. Shin, D. Smailus, A. S. Siddiqui, M. A. Marra, S. J. M. Jones, R. Holt, F. S. L. Brinkman, K. Miyauchi, M. Fukuda, J. E. Davies, W. W. Mohn, and L. D. Eltis. Submitted for publication.
- 23.Merlin, C., D. Springael, and A. Toussaint. 1999. Tn4371: A modular structure encoding a phage-like integrase, a Pseudomonas-like catabolic pathway, and RP4/Ti-like transfer functions. Plasmid 41:40-54. [DOI] [PubMed] [Google Scholar]
- 24.Navarro-Llorens, J., M. Patrauchan, G. Stewart, J. Davies, L. Eltis, and W. Mohn. 2005. Phenylacetate catabolism in Rhodococcus sp. strain RHA1: a central pathway for degradation of aromatic compounds. J. Bacteriol. 187:4497-4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patrauchan, M. A., C. Florizone, M. Dosanjh, W. W. Mohn, J. Davies, and L. D. Eltis. 2005. Catabolism of benzoate and phthalate in Rhodococcus sp. strain RHA1: redundancies and convergence. J. Bacteriol. 187:4050-4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 27.Sakai, M., E. Masai, H. Asami, K. Sugiyama, K. Kimbara, and M. Fukuda. 2002. Diversity of 2,3-dihydroxybiphenyl dioxygenase genes in a strong PCB degrader, Rhodococcus sp strain RHA1. J. Biosci. Bioeng. 93:421-427. [DOI] [PubMed] [Google Scholar]
- 28.Sakai, M., K. Miyauchi, N. Kato, E. Masai, and M. Fukuda. 2003. 2-Hydroxypenta-2,4-dienoate metabolic pathway genes in a strong polychlorinated biphenyl degrader, Rhodococcus sp. strain RHA1. Appl. Environ. Microbiol. 69:427-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sambrook, J., E. F. Fritisch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed., vol. 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 30.Seto, M., E. Masai, M. Ida, T. Hatta, K. Kimbara, M. Fukuda, and K. Yano. 1995. Multiple polychlorinated biphenyl transformation systems in the gram-positive bacterium Rhodococcus sp. strain Rha1. Appl. Environ. Microbiol. 61:4510-4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seto, M., K. Kimbara, M. Shimura, T. Hatta, M. Fukuda, and K. Yano. 1995. A novel transformation of polychorinated biphenyls by Rhodococcus sp. strain RHA1. Appl. Environ. Microbiol. 61:3353-3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takeda, H., N. Hara, M. Sakai, A. Yamada, K. Miyauchi, E. Masai, and M. Fukuda. 2004. Biphenyl-inducible promoters in a polychlorinated biphenyl-degrading bacterium, Rhodococcus sp. RHA1. Biosci. Biotechnol. Biochem. 68:1249-1258. [DOI] [PubMed] [Google Scholar]
- 33.Takeda, H., A. Yamada, K. Miyauchi, E. Masai, and M. Fukuda. 2004. Characterization of transcriptional regulatory genes for biphenyl degradation in Rhodococcus sp. strain RHA1. J. Bacteriol. 186:2134-2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tatusov, R. L., N. D. Fedorova, J. D. Jackson, A. R. Jacobs, B. Kiryutin, E. V. Koonin, D. M. Krylov, R. Mazumder, S. L. Mekhedov, A. N. Nikolskaya, B. S. Rao, S. Smirnov, A. V. Sverdlov, S. Vasudevan, Y. I. Wolf, J. J. Yin, and D. A. Natale. 2003. The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vaillancourt, F. H., G. Labbe, N. M. Drouin, P. D. Fortin, and L. D. Eltis. 2002. The mechanism-based inactivation of 2,3-dihydroxybiphenyl 1,2-dioxygenase by catecholic substrates. J. Biol. Chem. 277:2019-2027. [DOI] [PubMed] [Google Scholar]
- 37.van der Geize, R., and L. Dijkhuizen. 2004. Harnessing the catabolic diversity of rhodococci for environmental and biotechnological applications. Curr. Opin. Microbiol. 7:255-261. [DOI] [PubMed] [Google Scholar]
- 38.Yamada, A., H. Kishi, K. Sugiyama, T. Hatta, K. Nakamura, E. Masai, and M. Fukuda. 1998. Two nearly identical aromatic compound hydrolase genes in a strong polychlorinated biphenyl degrader, Rhodococcus sp. strain RHA1. Appl. Environ. Microbiol. 64:2006-2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yuen, T., E. Wurmbach, R. L. Pfeffer, B. J. Ebersole, and S. C. Sealfon. 2002. Accuracy and calibration of commercial oligonucleotide and custom cDNA microarrays. Nucleic Acids Res. 30:e48. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.