Abstract
Two methods for accurate poly(3-hydroxybutyrate) (PHB) depolymerase activity determination and quantitative and qualitative hydrolysis product determination are described. The first method is based on online determination of NaOH consumption rates necessary to neutralize 3-hydroxybutyric acid (3HB) and/or 3HB oligomers produced during the hydrolysis reaction and requires a pH-stat apparatus equipped with a software-controlled microliter pump for rapid and accurate titration. The method is universally suitable for hydrolysis of any type of polyhydroxyalkanoate or other molecules with hydrolyzable ester bonds, allows the determination of hydrolysis rates of as low as 1 nmol/min, and has a dynamic capacity of at least 6 orders of magnitude. By applying this method, specific hydrolysis rates of native PHB granules isolated from Ralstonia eutropha H16 were determined for the first time. The second method was developed for hydrolysis product identification and is based on the derivatization of 3HB oligomers into bromophenacyl derivates and separation by high-performance liquid chromatography. The method allows the separation and quantification of 3HB and 3HB oligomers up to the octamer. The two methods were applied to investigate the hydrolysis of different types of PHB by selected PHB depolymerases.
Polyhydroxyalkanoates (PHAs) are typical storage compounds of carbon and energy and are widely found in prokaryotes. The most common PHA is poly(3-hydroxybutyrate) (PHB), and this polymer can be accumulated at up to 90% of the cellular dry weight during unbalanced growth in some bacteria (2a, 37, 49). PHAs are thermoplasts, and despite the relatively high production costs, PHB and copolymers of 3-hydroxybutyrate (3HB) and 3-hydroxyvalerate are industrially produced on a scale of a few hundred tons per year.
One reason for the interest in biologically produced PHAs lies in the environmentally friendly production from renewable resources and in the biodegradability of PHAs. The ester bonds of the PHAs are the Achilles' heel of the polymer that can be hydrolyzed by a large variety of hydrolytic enzymes (PHA depolymerases). In vivo, PHAs can be hydrolyzed by the accumulating strain itself during periods of starvation (intracellular PHA hydrolysis by intracellular PHA depolymerases) (41). PHAs that were produced by humans or that were released from PHA-accumulating microorganisms (e.g., after death) can be cleaved by extracellular PHB depolymerases, and many examples of extracellular PHA depolymerases were described during the last two decades (14, 21). A differentiation of extracellular degradation from intracellular degradation is necessary because PHA exists in two biophysical conformations: in vivo, PHB is completely amorphous (native) and is covered by a surface layer that is about half the size of a cytoplasmic membrane (4, 5, 31) but apparently consists mainly of proteins, so-called phasins (48). After the release of the polymer from the cell (e.g., after cell lysis or by solvent extraction) or after damage of the surface layer, the polymer denatures (33) and becomes more or less crystalline (paracrystalline). The terms “native” and “denatured” have been introduced previously by Merrick et al. (32). For the sake of clarity, we used the same definitions in this contribution: PHB in its amorphous form with an intact surface layer is called native PHB (nPHB), and extracellular, partially crystalline PHB without or with a damaged surface layer is called denatured PHB (dPHB). Artificial PHB (aPHB) represents another form of amorphous PHB and can be prepared by dissolving PHB in a solvent (trichloromethane) and by emulsifying this solution with an aqueous solution of a surfactant (sodium dodecyl sulfate, cholate, oleate, and cetyltrimethylammonium bromide) according to a method described previously by Horowitz and Sanders (20; also see references 30 and 34). After evaporation of the solvent, a suspension of amorphous aPHB granules was obtained. The amorphous state of aPHB granules is stable for a long time if aggregation of the granules and close packing (e.g., by centrifugation) are avoided (7, 20).
Enzymes that hydrolyze PHB are often specific for one of the two forms (nPHB/aPHB or dPHB). For example, extracellular PHB depolymerases hydrolyze dPHB, while intracellular PHB depolymerases are specific for nPHB and possibly for aPHB. As far as we know, intracellular PHB depolymerases do not hydrolyze dPHB. More than 20 extracellular PHA depolymerases (PhaZs) were characterized during the last two decades, and the function of extracellular PHA depolymerases is well understood (23); recently, the three-dimensional structure of the first PHB depolymerase has been solved (19). However, knowledge of the structure and molecular mechanism of intracellular depolymerases (iPhaZs) was poor until recently; iPhaZs have been postulated for many bacteria, and some bacteria such as Rhodospirillum rubrum and Ralstonia eutropha H16 (other names: Alcaligenes entrophus, Wautersia eutropha, Cupriavidus necator) apparently contain more than one iPhaZ (1, 14, 15, 56). The number of working groups and publications investigating the intracellular hydrolysis of PHAs has increased considerably since 2000 (1, 12, 14-16, 26, 28, 53, 56). Unfortunately, the contributions from different working groups and sometimes even contributions from the same group are almost impossible to compare because different methods for PHB granule isolation and/or preparation and different methods for intracellular PHB depolymerase activity and product determination were used. We therefore suggest that all researchers interested in PHB depolymerases should use at least one common standard method in the future. In this study, we investigated a combination of nPHB isolation by glycerol density gradient centrifugation with an optimized pH-stat assay and a high-performance liquid chromatography (HPLC)-based method for characterization of PHB hydrolysis products. To show proof of concept, we investigated the (rapid) hydrolysis of nPHB and dPHB by PhaZ7 and PhaZ5, respectively, of Pseudomonas lemoignei and the (slow) endogenous hydrolysis of isolated nPHB granules.
MATERIALS AND METHODS
Isolation of different types of PHB granules.
dPHB granules without any surface proteins were isolated from PHB-rich cells of R. eutropha by sodium hypochlorite treatment as described previously (36).
nPHB granules were isolated from R. eutropha H16 cell extracts obtained by passage (three times) through a French press and subsequent glycerol density gradient centrifugation (first gradient, 5 ml of 87% glycerol and 10 ml 50% glycerol; second gradient, 5 ml each of 87, 80, 60, and 40% glycerol) in an SW28 swing-out rotor (Beckmann) at 20,000 rpm (4°C, 40 min) as described in detail previously (13, 55). Isolated nPHB granules were stored at −20°C. If desired, PHB granules can be dialyzed to remove glycerol. A sample of each batch of isolated nPHB granules was incubated in sodium hypochlorite (6%, wt/vol) at room temperature overnight to oxidize and solubilize any organic material except PHB. The protein-free PHB granules obtained were centrifuged and dried to constant weight, and the mass of PHB was determined gravimetrically.
Artificial cholate-coated PHB (aPHB) granules were prepared from crystalline PHB that had been isolated by sodium hypochlorite digestion of PHB-rich R. eutropha cells according to procedures described previously (20, 30, 34). In brief, a 15% (wt/vol) solution of crystalline PHB in trichloromethane was stirred at 60°C until PHB was dissolved. Ten volumes of 50 mM sodium cholate were added. The mixture was sonicated for 2 min until the solution became milky. Trichloromethane was evaporated. The amorphous state of the artificial PHB granule preparation was confirmed by hydrolysis with PhaZ7, which is specific for amorphous PHB and cannot hydrolyze dPHB (13).
Purification of PHB depolymerases.
PHB depolymerases PhaZ5 (specific for dPHB) and PhaZ7 (specific for nPHB) were purified from succinate-grown cells of P. lemoignei as described previously (13, 36, 51). Care was taken that none of the two purified depolymerases contained traces of the other respective isoenzyme. Both PHB depolymerases were homogeneous, as revealed by denaturing sodium dodecyl sulfate-polyacrylamide gel electrophoresis and subsequent silver staining (data not shown).
Assay of PHB depolymerase by pH-stat.
PHB depolymerase activity was assayed by the determination of the NaOH consumption rates necessary to keep the pH of a low-buffered suspension of nPHB granules constant at pH 8.5, unless otherwise indicated. A pH-stat apparatus equipped with Tiamo 1.1 software was used (Titrando 842, Metrohm, Switzerland). The reaction temperature was 37°C, and 0.001 to 1 N NaOH was used for titration. The volume of the reaction mixture varied from 2 to 50 ml (0.5 to 1.0 mM Tris-HCl, pH 8.5). Depending on the type of reaction vessel, and by use of a micro-pH electrode, reaction mixture volumes of as low as 1 ml are possible. Routinely, a reaction mixture volume of 15 ml was used in this study. The minimum and maximum rates of NaOH addition were adjusted to the expected hydrolysis activity and ranged from 5 μl/min to 2 ml/min. It was necessary to optimize this range for different experiments. Acidification of buffer by solubilization of CO2 was significantly above pH 7.5 and had to be taken into account for low activities. The resulting endogenous NaOH consumption rate necessary to maintain the pH at the desired value was recorded for 5 to 10 min before substrate (PHB granules) was added, and the increased NaOH consumption rate was measured for at least an additional 20 min. Depending on the type of experiment, PHB depolymerase PhaZ5 or PhaZ7 purified from P. lemoignei was added (13, 22, 36). For dPHB and nPHB depolymerase activity determination, 25 mg of purified dPHB and 25 mg of nPHB granules, respectively, were used (15-ml reaction mixture volume), unless otherwise stated.
Isolation and derivatization of hydrolysis products.
After measuring the NaOH consumption rate in a PHB depolymerase assay, the reaction mixture was centrifuged (30 min at 20,000 rpm and 4°C with an SW28 rotor). The supernatant containing the reaction products was concentrated at least 10-fold (rotary evaporator) and used for product determination. Alternatively, the reaction mixture was acidified with HCl to pH 3.5 to 4 to stop any enzymatic activity and to protonate 3HB oligomers. After centrifugation for 30 min at 20,000 rpm (SW28 rotor) and 4°C, the supernatant was extracted with ethylacetate. 3HB oligomers solubilized in the organic phase were isolated by evaporation of the solvent. Note that monomeric 3HB is only partially recovered by ethylacetate extraction. Solvent extraction should therefore be omitted if quantitative determination of all hydrolysis products is necessary. The method used for the derivatization of PHB hydrolysis products is based on a procedure described previously by Durst et al. for fatty acids (10). One hundred microliters of sample containing 1 mM to a maximal 5 mM 3HB or 3HB oligomers was alkalized by the addition of 100 μl triethylamine (0.1 M in acetone). The solvents were evaporated under nitrogen gas. One hundred fifty microliters of bromophenacyl bromide (BPB) (10 mM in acetonitrile) and 150 μl crown ether (decyl-18-crown-6) (2 mM in acetonitrile) were added, and the reaction mixture was incubated with a sealed lid at 80°C for 90 min. Note that reaction times shorter than 90 min result in an incomplete derivatization of higher 3HB oligomers. The reaction tubes were vortexed several times during the incubation and were cooled to room temperature at the end of the incubation time.
Detection and quantification of PHB hydrolysis products.
One to five microliters of the derivatization mixture was loaded onto a reverse-phase C18 HPLC column (5 μm, 4.6 by 150 mm) (Eclipse XDB-C18; Agilent). Samples were eluted at a flow rate of 0.8 ml/min. Buffers were 0.01 M ammonium formate with 2% methanol, pH 4 (solution A), and pure methanol (solution B). Elution conditions were as follows: gradient of 60% solution A-40% solution B to 100% solution B within 28 min, isocratic run at 100% solution B for 5 min (33 min), gradient to starting conditions (100% solution B to 60% solution A-40% solution B) within 1 min (34 min), and an isocratic run at 60% solution A-40% solution B for 5 min (39 min). The absorbance of BPB derivates was detected at 254 nm. A standard of BPB derivates with defined numbers of monomers was used as a reference. Defined 3HB oligomers were a gift from D. Seebach (ETH Zürich). A peak at 16.1 min corresponded to unreacted bromophenacyl bromide. Note that if the peak at 16.1 min is missing in a chromatogram, the molar amount of the 3HB oligomer exceeded the molar amount of bromophenacyl bromide, and accurate quantification of 3HB oligomers is not possible. The molar absorption coefficient of bromophenacyl fatty acid ester is 18,700 M−1 cm−1 (10).
RESULTS
Hydrolysis of dPHB and nPHB granules by purified PHB depolymerases.
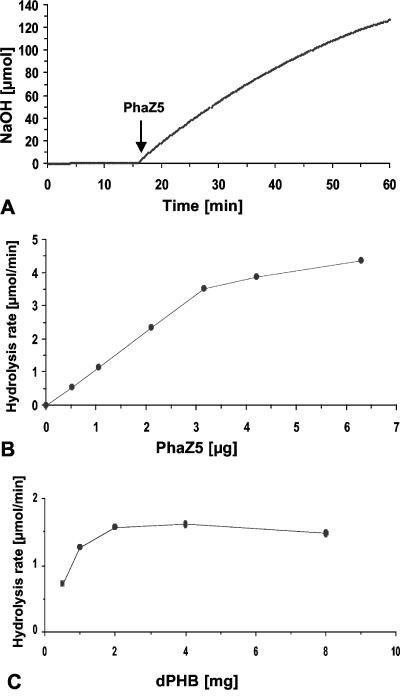
Purified PHB depolymerase PhaZ5 was used to hydrolyze dPHB granules at pH 8.5 in the pH-stat experiment. An almost constant rate of NaOH consumption (3.5 μmol/min) was obtained for about 30 min using the conditions described in the legend of Fig. 1A and confirmed that the enzyme is active in low-buffered water. The rate of NaOH consumption necessary to keep the pH at 8.5 corresponded to the activity of PHB depolymerase. When the amount of PhaZ5 was increased from 0.5 μg to 6.3 μg depolymerase protein (determined by the Bradford method [5a]), a linear dependence of the observed NaOH consumption rate on the amount of enzyme was obtained at up to 3 μg under the conditions applied (Fig. 1B) and confirmed that the enzyme is the rate-limiting step. A specific activity of 1,100 U mg−1 (μmol acid released min−1 mg−1 PhaZ5) was calculated for purified PhaZ5 under these conditions. The hydrolysis rates increased only slowly above 3 μg and indicated that the ratio of depolymerase to substrate was too high. A variation of the amount of substrate (dPHB) at a constant concentration of depolymerase resulted in a typical saturation kinetic above 2 mg of dPHB granules (15-ml reaction mixture volume) (Fig. 1C). Routinely, 25 mg dPHB granules was used for assays with a 15-ml volume.
FIG. 1.
pH-stat-controlled hydrolysis of dPHB granules by purified PHB depolymerase PhaZ5. (A) Twenty-five milligrams of hypochlorite-purified dPHB granules was incubated in 15 ml of 1 mM Tris-HCl, pH 8.5, at 37°C. Three micrograms of purified PhaZ5 was added, as indicated by the arrow. The pH-stat unit was kept constant at pH 8.5, and the amount of NaOH necessary to keep the pH at 8.5 was recorded. (B) Dependence of (initial) hydrolysis rates (μmol NaOH consumed min−1) on the amount of PhaZ5. Conditions were the same as described above for A, except that 5 mg dPHB granules was used. (C) Dependence of (initial) hydrolysis rates (μmol NaOH consumed min−1) on the amount of PHB. Conditions were the same as those described above for A, except that the amount of PhaZ5 was 1 μg and the amount of dPHB granules was variable.
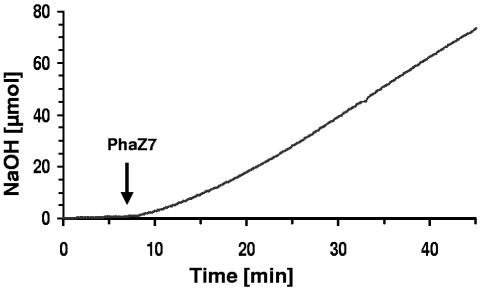
When purified PhaZ5 was used for hydrolysis of nPHB granules (instead of dPHB), almost no NaOH was consumed. The addition of trypsin to the assay mixture to partially remove the phasin surface layer of nPHB granules, as was necessary for PHB depolymerase of Rhodospirillum rubrum (15-17), also did not result in a significant hydrolysis of nPHB. This confirmed that PhaZ5 is not able to hydrolyze nPHB granules. No significant consumption of NaOH was observed upon the addition of different amounts of purified PHB depolymerase PhaZ7 to dPHB granules and confirmed previous findings that PhaZ7 is not able to hydrolyze dPHB. When PhaZ7 was added to nPHB granules in pH-stat experiments, a lag phase of several minutes was observed before a constant NaOH consumption rate of 2.2 μmol/min−1 was recorded (Fig. 2). A specific hydrolysis activity of 1,000 μmol/min−1 mg−1 was calculated for PhaZ7 at 37°C in 1 mM Tris-HCl, pH 8.5.
FIG. 2.
pH-stat-controlled hydrolysis of nPHB granules by purified PHB depolymerase PhaZ7. Twenty-five milligrams of glycerol density gradient-purified nPHB granules was incubated in 15 ml of 1 mM Tris-HCl, pH 8.5, at 37°C. Three micrograms of purified PhaZ7 was added, as indicated by the arrow. The pH-stat unit was kept constant at pH 8.5, and the amount of NaOH necessary to keep the pH at 8.5 was recorded.
Endogenous hydrolysis of nPHB granules.
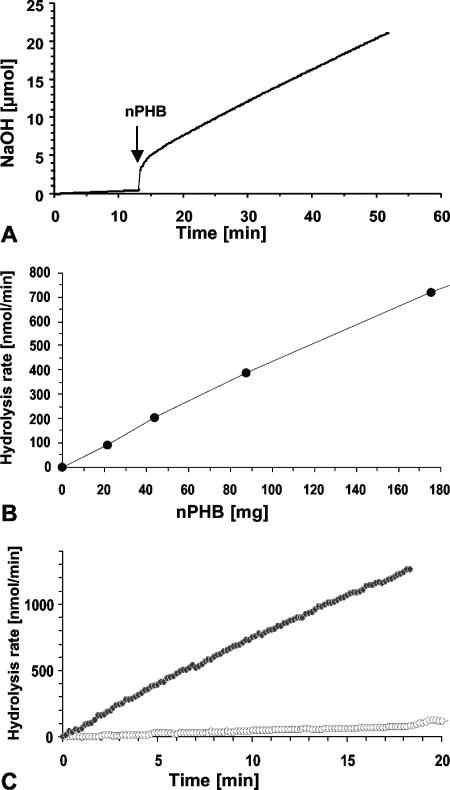
From the experiments described above, we conclude that the determination of the NaOH consumption rate is a good tool for the determination of dPHB and nPHB depolymerase activities of the highly active depolymerases PhaZ5 and PhaZ7. To find out whether this method is also suited for assays of very low hydrolysis activities, we determined the endogenous hydrolysis activity of isolated nPHB granules. Endogenous hydrolysis of isolated nPHB granules by intracellular PHB depolymerases attached to the surface layer of the granules has been postulated previously (14, 32) but has not yet been measured quantitatively. We incubated different amounts of isolated nPHB granules at 37°C and kept the pH constant at pH 8.5. As shown in Fig. 3A and B, a linear dependence of the NaOH consumption rate on the amount of nPHB granules was determined, and a specific hydrolysis rate of 4.3 nmol released acid min−1 mg−1 PHB was calculated for isolated nPHB granules. Depending on the batch of nPHB granules, the absolute value varied by a factor of up to 5. This result confirmed that endogenous PHB depolymerase activity was associated with nPHB granules. To find evidence that this activity was caused by an enzymatic reaction and to exclude that this activity could be caused by spontaneous chemical hydrolysis, the experiment was repeated using aPHB (Fig. 3C). aPHB does not contain any proteins. A poor hydrolysis rate (0.19 nmol acid min−1 mg−1 aPHB) was determined. Apparently, chemical hydrolysis of amorphous PHB is significant but contributed only very little to the activity observed with nPHB granules. Purified PhaZ7 was added as a control at the end of the experiment and confirmed that the aPHB granules were amorphous and could be hydrolyzed principally by nPHB depolymerases (data not shown).
FIG. 3.
pH-stat-controlled self-hydrolysis of nPHB granules. (A) Eighty-eight milligrams of purified nPHB granules was added (arrow) to 15 ml of 1 mM Tris-HCl, pH 8.5, and incubated at a constant pH at 37°C. (B) Dependence of (initial) hydrolysis rates (nmol NaOH consumed min−1) on the amount of nPHB granules. Conditions were the same as those described above for A. (C) Comparison of hydrolysis rates of nPHB (black symbols) with those of aPHB (white symbols). PHB granules (25 mg/15 ml) were incubated in 1 mM Tris-HCl, pH 8, at 37°C, and the amount of NaOH necessary to keep the pH constant was recorded.
Incubation of isolated nPHB granules in a boiling water bath for 15 min before the start of the pH-stat experiment resulted in an almost complete reduction of the NaOH consumption rate (data not shown), confirming that the endogenous hydrolysis rate is caused by heat-labile compounds (proteins) attached to isolated nPHB granules. The addition of PhaZ7 at the end of the experiment and the subsequent rapid hydrolysis of PHB confirmed that the sample still contained amorphous PHB. Taken together, the results described above strongly suggest that the major portion of the hydrolysis rate of isolated PHB granules is caused by an enzymatic activity.
Product analysis of the PHB depolymerase reaction.
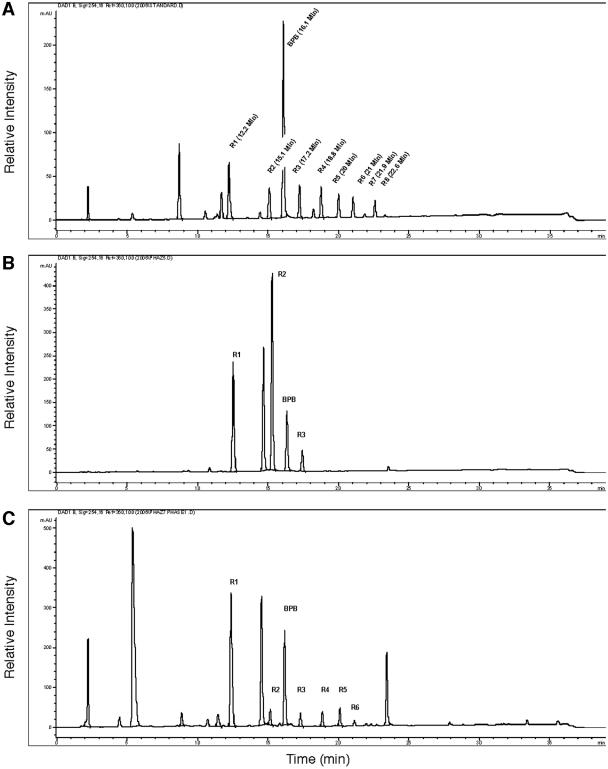
Alkylations of fatty acids with BPB as a chromophore and separation by HPLC have been described previously by Durst et al. (10). We were able to modify and apply this method for the derivatization of 3HB and 3HB oligomers (see Materials and Methods). Figure 4A shows an HPLC chromatogram of a mixture of bromophenacyl derivates of 3HB and oligomers of up to eight 3HB units. The identity of the 3HB oligomer peaks indicated in Fig. 4A with the respective 3HB oligomers was confirmed by detection of the expected respective masses (m/z) of the quasimolecular ions ([M + H]+, [M + NH4]+) by HPLC-coupled electrospray ionization-mass spectrometry (MS). Each peak was present in double form with a mass increment of 2 because bromine is present in two stable isotope forms (79Br and 81Br) (data not shown). To assess the suitability of the method for product analysis of enzymatic hydrolyzed nPHB granules, we analyzed the PhaZ5- and PhaZ7-derived hydrolysis products of dPHB and nPHB granules. Figure 4B shows the product analysis of the PhaZ5-catalyzed hydrolysis of dPHB. A mixture of monomer, dimer, and trimer in relative proportions of 5.9:9.4:1 was determined. No evidence of 3HB oligomers higher than the trimer was found. The same method was applied for PhaZ7-derived hydrolysis products of nPHB granules: a mixture of 3HB oligomers with one to six 3HB units was obtained (Fig. 4C). The relative composition varied with the incubation time and shifted to lower oligomers during prolonged incubation times (data not shown).
FIG. 4.
Separation of bromophenacyl derivates by HPLC. (A) 3-Hydroxybutyrate (R1) and 3HB oligomers ranging from two to eight monomer units (R2 to R8) were reacted with bromophenacyl bromide as described in Materials and Methods and separated on a reverse-phase C18 column. Detection was performed at 254 nm. The retention times of each of the individual oligomers were tested in separate experiments and by HPLC-MS (data not shown). The peak at 16.1 min corresponded to unreacted bromophenacyl bromide. (B) Products derived from PhaZ5-catalyzed hydrolysis of dPHB. (C) Products derived from PhaZ7-catalyzed hydrolysis of nPHB. Note that the appearance of peaks at 14.7 min (B and D) coincided with the presence of Tris-HCl during the hydrolysis reaction (1 mM). The nature of the peaks at 8.8 min (A), 5.5 min (C), and 23.5 min (C) is unknown. mAU, milliabsorption units.
DISCUSSION
PHB granules in native, pure denatured, or artificial form cannot be purchased and have to be isolated from bacteria and prepared by biochemical and chemical methods. dPHB is commercially available but only with relatively low purity in comparison to the hypochlorite-purified PHB used in this study. The methods for nPHB and dPHB granule preparation differ highly among PHA research laboratories: while Merrick et al. and our laboratory isolated nPHB granules by glycerol density gradient centrifugation (32, 34), others centrifuged crude cell extracts without glycerol and incubated the obtained polymer fraction with hydrolytic enzymes (25). The Saito laboratory used artificial PHB granules prepared according to the procedure described previously by Horowitz and Sanders (20) and used different surfactants such as oleate or phosphatidylglycerol in early investigations of intracellular PHB degradation (40, 42, 43). In recent studies, they used native PHB granules obtained by density gradient centrifugation (glycerol and sucrose) as the PHB depolymerase substrate (1, 26-28). Since close packing of amorphous PHB granules during centrifugation and other physical or chemical stresses are sufficient to decrease the susceptibility of the granules to enzymatic hydrolysis by Rhodospirillum rubrum PHB depolymerase (13, 32, 33), data from different laboratories cannot be compared.
A high level of variation in methods of PHB depolymerase activity determination also exists: many laboratories performed weight loss measurements (6, 8, 11, 39, 52, 54). This method is practicable when the degradation of macroscopically visible specimens of polymer samples (e.g., PHB films) is determined over a time scale of many hours. For short assays of PHB depolymerase activity, most laboratories determine the rate of clearing of a turbid suspension of nPHB or dPHB granules for activity determination (1, 13, 22, 24, 28, 32, 45, 46). However, this method is limited principally to very diluted suspensions of PHB granules. As a consequence of the low substrate concentration, the relative change in substrate concentration (Δ surface area) during the reaction is very high, and high PHB depolymerase activities may be underestimated (Fig. 1B). Some laboratories perform a combination of turbidity assay and pH-stat assay, depending on the type of reaction and substrate (1, 13, 32). The detection of 3HB at 210 nm after the removal of the remaining nonhydrolyzed substrate, as performed by the Doi laboratory, has relatively low sensitivity, does not allow online detection of activity (PHB granules), or is limited to relatively low PHB surface areas (PHB films) (2, 9). The quantification of 3HB by NAD-dependent 3HB dehydrogenase is possible but is also time-consuming, allows the detection of monomers only, and gives no online information for the hydrolysis reaction. 3HB oligomers that are major products of most PHB depolymerases can only be indirectly taken into account either by chemical cleavage of 3HB oligomers to monomers by alkali treatment (29) or by enzymatic hydrolysis by 3HB oligomer hydrolase (40). Some exotic methods for recording the degradation of polyesters include electron microscopy (35, 38), 1H-nuclear magnetic resonance (47), or tracking of radiolabeled polymers (18, 50), but apparently, they are not suitable for routine assays. In contrast to turbidity measurements, the initial substrate concentration (nPHB, dPHB, or aPHB granules) can be very high in a pH-stat apparatus. Moreover, a combination of a pH-stat with a sensitive Micro pH electrode and an intelligently regulated pump for pH control allows a very sensitive and accurate determination of PHB depolymerase activity. By using nearly unbuffered suspensions of PHB granules and pH-stat pump pulses of diluted (or concentrated) NaOH solutions in the low (or high) μl/min range, PHB depolymerase activity can be determined at very low or at very high levels of activity in a dynamic range from nmol/min up to mmol/min. We therefore suggest that future work on enzymatic PHB hydrolysis should always include pH-stat data.
The high sensitivity of the method allowed us to determine the endogenous PHB depolymerase activity of isolated nPHB granules for the first time. In addition, the previously observed striking differences in substrate specificities of PHB depolymerases PhaZ5 (specific for dPHB) (22) and PhaZ7 (specific for nPHB) (13) of P. lemoignei and PHB depolymerase of Rhodospirillum rubrum (specific for nPHB after trypsin treatment) (15-17) were confirmed by the pH-stat method.
Despite the high sensitivity and accuracy of the pH-stat assay method for determination of PHB depolymerase activity, the rate of acid production does not provide any information on the type of products (monomer versus oligomers). However, PHB depolymerases significantly differ in the composition of end products. Some investigators previously used NADdependent 3HB dehydrogenase to quantify hydrolysis products. Since 3HB dehydrogenase cannot utilize 3HB oligomers as a substrate, product determination with 3HB dehydrogenase is not useful. 3HB and its oligomeric esters have only poor absorption at 210 nm, and therefore, direct UV detection of the oligomers is possible only for high concentrations of products. Bachmann and Seebach previously described an HPLC-based method for the separation and detection of 3HB oligomers after derivatization with diazomethane to the corresponding methylesters (3). This method is suitable to separate the 3HB oligomers with one to eight 3HB units and has been used to investigate the substrate specificity and reaction products of dPHB depolymerase from R. pickettii. However, the derivatization method requires the handling of volatile and very toxic diazomethane and therefore appears to be less suitable as a routine method. The derivatization method with bromophenacyl bromide can be performed in about 2 h for many samples simultaneously, and most of this time is only the incubation period. The method is easy to perform and omits the handling of volatile toxic chemicals. As shown in Fig. 4, separation of 3HB and 3HB oligomers up to octamers is very efficient, and the peak broadening at the high oligomers, as was found for the diazomethane-derivatized 3HB octamer, is significantly less for the bromophenacyl derivate of the octamer.
Acknowledgments
We thank W. Armbruster for HPLC-MS measurements. We also thank B. Neubert (Metrohm, Germany) for continuous support and helpful discussion and D. Seebach for providing 3HB oligomers with definite numbers of 3HB units.
This work was supported by a grant of the Deutsche Forschungsgemeinschaft.
REFERENCES
- 1.Abe, T., T. Kobayashi, and T. Saito. 2005. Properties of a novel intracellular poly(3-hydroxybutyrate) depolymerase with high specific activity (PhaZd) in Wautersia eutropha H16. J. Bacteriol. 187:6982-6990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abe, T., I. Madsubara, Y. Doi, Y. Hori, and A. Yamaguchi. 1994. Physical properties and enzymatic degradability of poly(3-hydroxybutyrate) stereoisomers with different stereoregularities. Macromolecules 27:6018-6025. [Google Scholar]
- 2a.Babel, W., and A. Steinbüchel (ed.). 2001. Advances in biochemical engineering/biotechnology, vol. 71. Biopolyesters. Springer, Berlin, Germany.
- 3.Bachmann, B., and D. Seebach. 1999. Investigation of the enzymatic cleavage of diastereomeric oligo(3-hydroxybutanoates) containing two to eight HB units. A model for the stereoselectivity of PHB depolymerase from Alcaligenes faecalis T1. Macromolecules 32:1777-1784. [Google Scholar]
- 4.Barnard, G. N., and J. K. Sanders. 1989. The poly-beta-hydroxybutyrate granule in vivo. A new insight based on NMR spectroscopy of whole cells. J. Biol. Chem. 264:3286-3291. [PubMed] [Google Scholar]
- 5.Boatman, E. S. 1964. Observations on the fine structure of spheroplasts of Rhodospirillum rubrum. J. Cell Biol. 20:297-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5a.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 6.Briese, B. H., D. Jendrossek, and H. G. Schlegel. 1994. Degradation of poly(3-hydroxybutyrate-co-3-hydroxyvalerate) by aerobic sewage sludge. FEMS Microbiol. Lett. 117:107-111. [DOI] [PubMed] [Google Scholar]
- 7.de Koning, G. J. M., and P. J. Lemstra. 1992. The amorphous state of poly[(R)-3-hydroxyalkanoate] in vivo. Polymer 33:3292-3294. [Google Scholar]
- 8.de Koning, G. J. M., H. M. M. van Bilsen, P. J. Lemstra, W. Hazenberg, B. Witholt, H. Preusting, J. G. van der Galien, A. Schirmer, and D. Jendrossek. 1994. A biodegradable rubber by cross-linking poly(hydroxyalkanoate) from Pseudomonas fluorescens. Polymer 35:2090-2097. [Google Scholar]
- 9.Doi, Y., M. Katsuyuki, K. Kasuya, and K. Yamada. 1994. Biodegradation of biosynthetic and chemosynthetic polyhydroxyalkanoates, p. 39-51. In Y. Doi and K. Fukuda (ed.), Biodegradable plastics and polymers. Elsevier Science B.V., Amsterdam, The Netherlands.
- 10.Durst, H., M. Milano, E. Kikta, S. Conelly, and E. Grushka. 1975. Phenacyl esters of fatty acids via crown ether catalysts for enhanced ultraviolet detection in liquid chromatography. Anal. Chem. 47:1797-1801. [DOI] [PubMed] [Google Scholar]
- 11.Foster, L. J. R., E. S. Stuart, A. Tehrani, R. W. Lenz, and R. C. Fuller. 1996. Intracellular depolymerase and polyhydroxyoctanoate granule integrity in Pseudomonas oleovorans. Int. J. Biol. Macromol. 19:177-183. [DOI] [PubMed] [Google Scholar]
- 12.Gao, D., A. Maehara, T. Yamane, and S. Ueda. 2001. Identification of the intracellular polyhydroxyalkanoate depolymerase gene of Paracoccus denitrificans and some properties of the gene product. FEMS Microbiol. Lett. 196:159-164. [DOI] [PubMed] [Google Scholar]
- 13.Handrick, R., S. Reinhardt, M. L. Focarete, M. Scandola, G. Adamus, M. Kowalczuk, and D. Jendrossek. 2001. A new type of thermoalkalophilic hydrolase of Paucimonas lemoignei with high specificity for amorphous polyesters of short chain-length hydroxyalkanoic acids. J. Biol. Chem. 276:36215-36224. [DOI] [PubMed] [Google Scholar]
- 14.Handrick, R., S. Reinhardt, and D. Jendrossek. 2000. Mobilization of poly(3-hydroxybutyrate) in Ralstonia eutropha. J. Bacteriol. 182:5916-5918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Handrick, R., S. Reinhardt, P. Kimmig, and D. Jendrossek. 2004. The “intracellular” poly(3-hydroxybutyrate) (PHB) depolymerase of Rhodospirillum rubrum is a periplasm-located protein with specificity for native PHB and with structural similarity to extracellular PHB depolymerases. J. Bacteriol. 186:7243-7253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Handrick, R., S. Reinhardt, D. Schultheiss, T. Reichart, D. Schüler, V. Jendrossek, and D. Jendrossek. 2004. Unraveling the function of the Rhodospirillum rubrum activator of polyhydroxybutyrate (PHB) degradation: the activator is a PHB-granule-bound protein (phasin). J. Bacteriol. 186:2466-2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Handrick, R., U. Technow, T. Reichart, S. Reinhardt, T. Sander, and D. Jendrossek. 2004. The activator of the Rhodospirillum rubrum PHB depolymerase is a polypeptide that is extremely resistant to high temperature (121 degrees C) and other physical or chemical stresses. FEMS Microbiol. Lett. 230:265-274. [DOI] [PubMed] [Google Scholar]
- 18.Hippe, H., and H. G. Schlegel. 1967. Hydrolyse von PHBS durch intracelluläre Depolymerase von Hydrogenomonas H16. Arch. Mikrobiol. 56:278-299. [PubMed] [Google Scholar]
- 19.Hisano, T., K. Kasuya, Y. Tezuka, N. Ishii, T. Kobayashi, M. Shiraki, E. Oroudjev, H. Hansma, T. Iwata, Y. Doi, T. Saito, and K. Miki. 2006. The crystal structure of polyhydroxybutyrate depolymerase from Penicillium funiculosum provides insights into the recognition and degradation of biopolyesters. J. Mol. Biol. 356:993-1004. [DOI] [PubMed] [Google Scholar]
- 20.Horowitz, D. M., and K. M. Sanders. 1994. Amorphous, biomimetic granules of polyhydroxybutyrate: preparation, characterization, and biological implications. J. Am. Chem. Soc. 116:2695-2702. [Google Scholar]
- 21.Jendrossek, D. 2001. Microbial degradation of polyesters. Adv. Biochem. Eng. Biotechnol. 71:293-325. [DOI] [PubMed] [Google Scholar]
- 22.Jendrossek, D., A. Frisse, A. Behrends, M. Andermann, H. D. Kratzin, T. Stanislawski, and H. G. Schlegel. 1995. Biochemical and molecular characterization of the Pseudomonas lemoignei polyhydroxyalkanoate depolymerase system. J. Bacteriol. 177:596-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jendrossek, D., and R. Handrick. 2002. Microbial degradation of polyhydroxyalkanoates. Annu. Rev. Microbiol. 56:403-432. [DOI] [PubMed] [Google Scholar]
- 24.Jendrossek, D., I. Knoke, R. B. Habibian, A. Steinbüchel, and H. G. Schlegel. 1993. Degradation of poly(3-hydroxybutyrate), PHB, by bacteria and purification of a novel PHB depolymerase from Comamonas sp. J. Environ. Polym. Degrad. 1:53-63. [Google Scholar]
- 25.Kawaguchi, Y., and Y. Doi. 1990. Structure of native poly(3-hydroxybutyrate) granules characterized by X-ray diffraction. FEMS Microbiol. Lett. 70:151-156. [Google Scholar]
- 26.Kobayashi, T., K. Nishikori, and T. Saito. 2004. Properties of an intracellular poly(3-hydroxybutyrate) depolymerase (PhaZ1) from Rhodobacter spheroides. Curr. Microbiol. 49:199-202. [DOI] [PubMed] [Google Scholar]
- 27.Kobayashi, T., M. Shiraki, T. Abe, A. Sugiyama, and T. Saito. 2003. Purification and properties of an intracellular 3-hydroxybutyrate-oligomer hydrolase (PhaZ2) in Ralstonia eutropha H16 and its identification as a novel intracellular poly(3-hydroxybutyrate) depolymerase. J. Bacteriol. 185:3485-3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kobayashi, T., K. Uchino, T. Abe, Y. Yamazaki, and T. Saito. 2005. Novel intracellular 3-hydroxybutyrate-oligomer hydrolase in Wautersia eutropha H16. J. Bacteriol. 187:5129-5135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee, S. Y., and Y. Lee. 2003. Metabolic engineering of Escherichia coli for production of enantiomerically pure (R)-(−)-hydroxycarboxylic acids. Appl. Environ. Microbiol. 69:3421-3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marchessault, R. H., and I. Saracovan. 1995. Artificial granules suspension of long side chain poly(3-hydroxyalkanoate). Can. J. Microbiol. 41(Suppl. 1):138-142. [Google Scholar]
- 31.Mayer, F. 1992. Structural aspects of poly-β-hydroxybutyrate granules. FEMS Microbiol. Rev. 103:265-268. [Google Scholar]
- 32.Merrick, J. M., and M. Doudoroff. 1964. Depolymerization of poly-β-hydroxybutyrate by intracellular enzyme system. J. Bacteriol. 88:60-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Merrick, J. M., D. G. Lundgren, and R. M. Pfister. 1965. Morphological changes in poly-β-hydroxybutyrate granules associated with decreased susceptibility to enzymatic hydrolysis. J. Bacteriol. 89:234-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Merrick, J. M., R. Steger, and D. Dombroski. 1999. Hydrolysis of native poly(hydroxybutyrate) granules (PHB), crystalline PHB, and artificial amorphous PHB granules by intracellular and extracellular depolymerases. Int. J. Biol. Macromol. 25:129-134. [DOI] [PubMed] [Google Scholar]
- 35.Molitoris, H. P., S. T. Moss, G. J. M. de Koning, and D. Jendrossek. 1996. Scanning electron microscopy of polyhydroxyalkanoate degradation by bacteria. Appl. Microbiol. Biotechnol. 46:570-579. [Google Scholar]
- 36.Müller, B., and D. Jendrossek. 1993. Purification and properties of a poly(3-hydroxyvalerate) depolymerase from Pseudomonas lemoignei. Appl. Microbiol. Biotechnol. 38:487-492. [Google Scholar]
- 37.Müller, H.-M., and D. Seebach. 1993. Poly(hydroxyalkanoates): a fifth class of physiologically important organic biopolymers? Angewandte Chemie 32:477-502. [Google Scholar]
- 38.Nobes, G. A. R., R. H. Marchessault, H. Chanzy, B. H. Briese, and D. Jendrossek. 1996. Splintering of poly(3-hydroxybutyrate) single crystals by PHB-depolymerase A from Pseudomonas lemoignei. Macromolecules 29:8330-8333. [Google Scholar]
- 39.Ohura, T., Y. Aoyagi, K. Takagi, Y. Yoshida, K. Kasuya, and Y. Doi. 1999. Biodegradation of poly(3-hydroxyalkanoic acids) fibers and isolation of poly(3-hydroxybutyric acid)-degrading microorganisms under aquatic environments. Polym. Degrad. Stab. 63:23-29. [Google Scholar]
- 40.Saegusa, H., M. Shiraki, C. Kanai, and T. Saito. 2001. Cloning of an intracellular poly[d(−)-3-hydroxybutyrate] depolymerase gene from Ralstonia eutropha H16 and characterization of the gene product. J. Bacteriol. 183:94-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saito, T., and T. Kobayashi. 2002. Intracellular degradation of PHAs, p. 23-40 In Y. Doi and A. Steinbüchel (ed.), Biopolymers, vol. 3b. Polyesters II. Wiley-VCH, Weinheim, Germany. [Google Scholar]
- 42.Saito, T., H. Saegusa, Y. Miyata, and T. Fukui. 1992. Intracellular degradation of poly(3-hydroxybutyrate) granules of Zoogloea ramigera I-16-M. FEMS Microbiol. Rev. 9:333-338. [DOI] [PubMed] [Google Scholar]
- 43.Saito, T., K. Takizawa, and H. Saegusa. 1995. Intracellular poly(3-hydroxybutyrate) depolymerase in Alcaligenes eutrophus. Can. J Microbiol. 41(Suppl. 1):187-191. [Google Scholar]
- 44.Reference deleted.
- 45.Schirmer, A., C. Matz, and D. Jendrossek. 1995. Substrate specificities of poly(hydroxyalkanoate)-degrading bacteria and active site studies on the extracellular poly(3-hydroxyoctanoic acid) depolymerase of Pseudomonas fluorescens GK13. Can. J. Microbiol. 41(Suppl. 1):170-179. [DOI] [PubMed] [Google Scholar]
- 46.Schöber, U., C. Thiel, and D. Jendrossek. 2000. Poly(3-hydroxyvalerate) depolymerase of Pseudomonas lemoignei. Appl. Environ. Microbiol. 66:1385-1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spyros, A., R. Kimmich, B. H. Briese, and D. Jendrossek. 1997. 1H NMR imaging study of enzymatic degradation of poly(3-hydroxybutyrate) and poly(3-hydroxyvalerate). Macromolecules 30:8218-8225. [Google Scholar]
- 48.Steinbüchel, A., K. Aerts, W. Babel, C. Follner, M. Liebergesell, M. H. Madkour, F. Mayer, U. Pieper-Fürst, A. Pries, H. E. Valentin, et al. 1995. Considerations on the structure and biochemistry of bacterial polyhydroxyalkanoic acid inclusions. Can. J. Microbiol. 41(Suppl. 1):94-105. [DOI] [PubMed] [Google Scholar]
- 49.Steinbüchel, A., and S. Hein. 2001. Biochemical and molecular basis of microbial synthesis of polyhydroxyalkanoates in microorganisms. Adv. Biochem. Eng. Biotechnol. 71:81-123. [DOI] [PubMed] [Google Scholar]
- 50.Taidi, B., D. Mansfield, and A. J. Anderson. 1995. Turnover of poly(3-hydroxybutyrate) (PHB) and its influence on the molecular mass of the polymer accumulated by Alcaligenes eutrophus during batch culture. FEMS Microbiol. Lett. 129:201-206. [Google Scholar]
- 51.Terpe, K., K. Kerkhoff, E. Pluta, and D. Jendrossek. 1999. Relationship between succinate transport and production of extracellular poly(3-hydroxybutyrate) depolymerase in Pseudomonas lemoignei. Appl. Environ. Microbiol. 65:1703-1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tomasi, G., M. Scandola, B. H. Briese, and D. Jendrossek. 1996. Enzymatic degradation of bacterial poly(3-hydroxybutyrate) by a depolymerase from Pseudomonas lemoignei. Macromolecules 29:507-513. [Google Scholar]
- 53.Uchino, K., and T. Saito. 2006. Thiolysis of poly(3-hydroxybutyrate) with polyhydroxyalkanoate synthase from Ralstonia eutropha. J. Biochem. (Tokyo) 139:615-621. [DOI] [PubMed] [Google Scholar]
- 54.Uefuji, M., K. Ken-ichi, and Y. Doi. 1997. Enzymatic degradation of poly[(R)-3-hydroxybutyrate]: secretion and properties of PHB depolymerase from Pseudomonas stutzeri. Polym. Degrad. Stab. 58:275-281. [Google Scholar]
- 55.Wieczorek, R., A. Steinbuchel, and B. Schmidt. 1996. Occurrence of polyhydroxyalkanoic acid granule-associated proteins related to the Alcaligenes eutrophus H16 GA24 protein in other bacteria. FEMS Microbiol. Lett. 135:23-30. [DOI] [PubMed] [Google Scholar]
- 56.York, G. M., J. Lupberger, J. Tian, A. G. Lawrence, J. Stubbe, and A. J. Sinskey. 2003. Ralstonia eutropha H16 encodes two and possibly three intracellular poly[d-(−)-3-hydroxybutyrate] depolymerase genes. J. Bacteriol. 185:3788-3794. [DOI] [PMC free article] [PubMed] [Google Scholar]