Abstract
Winter wheats require several weeks at low temperature to flower. This process, vernalization, is controlled mainly by the VRN1 gene. Using 6,190 gametes, we found VRN1 to be completely linked to MADS-box genes AP1 and AGLG1 in a 0.03-centimorgan interval flanked by genes Cysteine and Cytochrome B5. No additional genes were found between the last two genes in the 324-kb Triticum monococcum sequence or in the colinear regions in rice and sorghum. Wheat AP1 and AGLG1 genes were similar to Arabidopsis meristem identity genes AP1 and AGL2, respectively. AP1 transcription was regulated by vernalization in both apices and leaves, and the progressive increase of AP1 transcription was consistent with the progressive effect of vernalization on flowering time. Vernalization was required for AP1 transcription in apices and leaves in winter wheat but not in spring wheat. AGLG1 transcripts were detected during spike differentiation but not in vernalized apices or leaves, suggesting that AP1 acts upstream of AGLG1. No differences were detected between genotypes with different VRN1 alleles in the AP1 and AGLG1 coding regions, but three independent deletions were found in the promoter region of AP1. These results suggest that AP1 is a better candidate for VRN1 than AGLG1. The epistatic interactions between vernalization genes VRN1 and VRN2 suggested a model in which VRN2 would repress directly or indirectly the expression of AP1. A mutation in the promoter region of AP1 would result in the lack of recognition of the repressor and in a dominant spring growth habit.
Winter wheats differ from spring wheats in their requirement for a long period at low temperatures to become competent to flower. This process, vernalization, prevents the damage of the cold-sensitive flowering meristem during the winter. VRN1 and VRN2 (unrelated to the genes with similar names in Arabidopsis) are the main genes involved in the vernalization response in diploid wheat Triticum monococcum (1, 2). However, most of the variation in vernalization requirement in the economically important polyploid species of wheat is controlled by the VRN1 locus (2, 3). This gene is critical in polyploid wheats for their adaptation to autumn sowing and divides wheat varieties into the winter and spring market classes.
The VRN1 gene has been mapped in colinear regions of the long arm of chromosomes 5A (1, 3, 4), 5B (5, 6), and 5D (3). This region of wheat chromosome 5 is colinear with a region from rice chromosome 3 that includes the HD-6 quantitative trait locus for heading date (7). However, it was recently demonstrated that VRN1 and HD-6 are different genes (8).
Despite the progress made in the elucidation of the vernalization pathway in Arabidopsis, little progress has been made in the characterization of wheat vernalization genes. The two main genes involved in the vernalization pathway in Arabidopsis, FRI and FLC (9–11), have no clear homologues in the complete draft sequences of the rice genome (12). This finding may not be surprising, considering that rice is a subtropical species that has no vernalization requirement. Because no clear orthologues of the Arabidopsis vernalization genes were found in rice or among the wheat or barley ESTs, a map-based cloning project for the wheat VRN1 gene was initiated in our laboratory.
Chromosome walking in wheat is not a trivial exercise because of the large size of its genomes (5,600 Mb per T. monococcum haploid genome) and the abundance of repetitive elements (13, 14). To minimize the probability that these repetitive elements would stop the chromosome walking, simultaneous efforts were initiated in the orthologous regions in rice, sorghum, and wheat. The initial sequencing of rice, sorghum, and wheat bacterial artificial chromosomes (BACs) selected with restriction fragment length polymorphism marker WG644 [0.1 centimorgan (cM) from VRN1] showed good microcolinearity among these genera (14–16). The low gene density observed in the wheat region and the large ratio of physical-to-genetic distances (14) suggested that large mapping populations and comparative physical maps would be necessary for a successful positional cloning of VRN1.
In this article we report the construction of detailed genetic and physical maps of VRN1 in diploid wheat and the comparison of the sequences from selected BACs from these contigs with the sequences of colinear regions in rice and sorghum. We also present the expression patterns of two MADS-box genes completely linked to VRN1 and discuss the evidence supporting one of these genes as a strong candidate gene for VRN1.
Materials and Methods
Mapping Population.
The high-density map was based on 3,095 F2 plants from the cross between T. monococcum ssp. aegilopoides accessions G2528 (spring, Vrn1) with G1777 (winter, vrn1). These two lines have the same dominant allele at the VRN2 locus and therefore, plants from this cross segregate only for VRN1 in a clear 3:1 ratio (1, 2).
Plants were grown in a greenhouse at 20–25°C without vernalization and under long photoperiod (16-h light). Under these conditions, winter plants flowered 1–2 months later than spring plants. All F2 plants were analyzed for molecular markers flanking VRN1, and progeny tests were performed for plants showing recombination between these markers. The 20–25 individual F3 plants from each progeny test were characterized with molecular markers flanking the crossover to confirm that the observed segregation in growth habit was determined by variation at the VRN1 locus. G2528 and G1777 were included as controls in each progeny test.
Procedures for genomic DNA extraction, Southern blots, and hybridizations have been described (17). The first 500 F2 plants were screened with flanking restriction fragment length polymorphism markers CDO708 and WG644 (1), which were later replaced by closer PCR markers to screen the complete mapping population. Additional markers were developed for the eight genes present between the PCR markers (Appendix 1, which is published as supporting information on the PNAS web site, www.pnas.org).
Contig Construction and BAC Sequencing.
High-density filters for the BAC libraries from T. monococcum accession DV92 (18), Oryza sativa var. Nipponbare (19), and Sorghum bicolor (20) were screened with segments from the different genes indicated in Fig. 1. Contigs were assembled by using HindIII fingerprinting and confirmed by hybridization of BAC ends obtained by plasmid rescue, inverse PCR (20), or BAC sequencing. Restriction maps using single and double digestions with eight-cutter restriction enzymes, pulsed-field electrophoresis, and hybridization of the Southern blots with different genes were used to order genes within the BACs, select the fragment sequenced from the sorghum BAC, and confirm the assembly results from the BAC sequencing. Shotgun libraries for BAC sequencing were constructed as described (15). Complete T. monococcum BACs 609E6, 719C13, and 231A16 and a 24-kb fragment from sorghum BAC 17E12 were sequenced. Genes were identified by a combination of comparative genomic analysis, blast searches, and gene-finding programs (15).
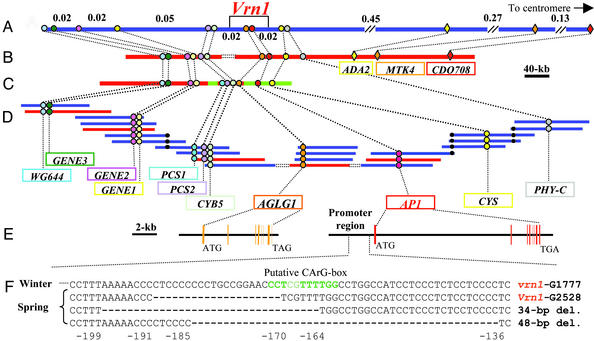
Figure 1.
(A) Genetic map of the VRN1 region on chromosome 5Am of T. monococcum. Genetic distances are in cM (6,190 gametes). (B–D) Physical maps of the colinear VRN1 regions in rice, sorghum, and wheat. Regions indicated in red have been sequenced. Double dot lines indicate gaps in the current physical maps. (B) Sequence of the colinear region in rice chromosome 3. (C) S. bicolor BACs 170F8 (AF503433) and 17E12 (AY188330). (D) T. monococcum physical map. BAC clones order from left to right is: 49I16, 115G1, 136F13, 133P9, 116F2, 89E14, 160C18, 491M20, 328O3, 609E6, 393O11, 719C13, 454P4, 54K21, 579P2, 601A24, 231A16, 638J12, 52F19, 242A12, 668L22, 539M19, and 309P20 (bold indicates sequenced BACs). Black dots indicate validation of BAC connections by hybridization. (E) Gene structure of two MADS-box genes completely linked to the VRN1 gene (AY188331, AY188333). Bars represent exons. (F) Sequence comparison of the AP1 promoter regions from genotypes carrying the Vrn1 and vrn1 alleles and from two T. monococcum accessions with additional deletions. The last two accessions are spring but their genotype has not been determined yet. Numbers indicate distances from the start codon. A putative MADS-box protein-binding site (CArG-box) is indicated in green.
Phylogenetic Analysis.
A phylogenetic study was performed by using the two wheat MADS-box genes found in this study and 24 additional MADS-box genes (Appendix 2, which is published as supporting information on the PNAS web site). Phylogenetic trees were generated from the clustalw sequence alignments of the complete proteins by using multiple distance- and parsimony-based methods available in the MEGA 2.1 software package (21). Distances between each pair of proteins were calculated and a tree was constructed by using the neighbor-joining algorithm. The consensus tree and the confidence values for the nodes were calculated by using 1,000 bootstraps (MEGA 2.1).
RT-PCR and Quantitative PCR.
RNA from leaves, undifferentiated apices, and young spikes was
extracted by using the Trizol method (Invitrogen). RT-PCR procedures
were performed as described (22). Quantitative PCR experiments were
performed in an Applied Biosystems ABI 7700 by using three TaqMan
systems for T. monococcum AP1 and for actin and ubiquitin
as endogenous controls (Appendix 3,
which is published as supporting information on the PNAS web
site). The
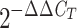 method
(23) was used to normalize and calibrate the
CT values of wheat AP1
relative to the endogenous controls. For the vernalization
time course, RNA was extracted from the youngest fully expanded leaf of
five winter T. monococcum plants (1 month old) immediately
before moving the plants into the cold room, and then after 2, 4, and 6
weeks of vernalization (4°C). The last sample was collected 2 weeks
after moving plants to the greenhouse (20°C). Plants kept in the
greenhouse were sampled as controls at each time point simultaneously
with the plants from the cold room (five plants per time point).
method
(23) was used to normalize and calibrate the
CT values of wheat AP1
relative to the endogenous controls. For the vernalization
time course, RNA was extracted from the youngest fully expanded leaf of
five winter T. monococcum plants (1 month old) immediately
before moving the plants into the cold room, and then after 2, 4, and 6
weeks of vernalization (4°C). The last sample was collected 2 weeks
after moving plants to the greenhouse (20°C). Plants kept in the
greenhouse were sampled as controls at each time point simultaneously
with the plants from the cold room (five plants per time point).
Results
Marker Development.
In the initial genetic map (1) the VRN1 gene was flanked on the distal side by WG644 and on the proximal side by CDO708. These markers were used as anchor points to the rice genome sequence to find additional markers.
Distal region.
WG644 was previously used to identify rice BAC 36I5 that included GENE1 at its proximal end (15). blastn searches of the different rice genome projects using GENE1 and the end sequence of BAC 36I5 (AY013245) identified the connected contig CL013482.168 (12). Two additional genes, Phytochelatin synthetase (PCS, Zea mays AAF24189.1) and Cytochrome B5 (CYB5, NP_173958.1), were discovered and annotated in this contig. These genes were mapped in wheat by restriction fragment length polymorphism (Appendix 1).
Proximal region.
Restriction fragment length polymorphism marker CDO708 was mapped 0.9 cM proximal to VRN1 in the T. monococcum map. The sequence of this clone (AY245605) showed a high homology to a putative RNA-binding protein (AAL58954.1) located in rice BAC AC091811. The end of this rice BAC also included gene MTK4 (putative protein kinase tousled, AAL58952.1), which was converted into a PCR marker and was mapped in wheat (Fig. 1). Rice BAC sequence AC091811 was then connected through contigs CL039395.93, CL039395.83, and CL018222.111.1 (12) to rice BAC sequences AC092556 and AF377947. BAC sequence AC092556 included a transcriptional adaptor gene (ADA2, AJ430205) that was mapped in T. monococcum 0.5 cM from the VRN1 gene (Fig. 1). The last rice BAC sequence, AF377947, included genes Phytochrome C (PHY-C, AAM34402.1), cysteine proteinase (CYS, AAM34401.1) and MADS-box genes AAM34398.1 and AAM34397.1, designated hereafter AP1 and AGLG1 (agamous-like gene from grasses). The rice proximal region included 318 kb of contiguous sequence.
High-Density Genetic Maps of the VRN1 Region.
The PCR markers developed for GENE1 and MTK4 were used to screen 6,190 chromosomes for recombination. Fifty-one recombinant events were detected, and those plants were further characterized by using molecular markers for all of the genes present between these two markers in rice (Fig. 1; Appendix 1). Progeny tests were performed for 32 of the 51 F2 plants to determine the VRN1 genotype of the parental F2 plants. Based on the mapping information, the VRN1 locus was completely linked to AP1 and AGLG1.
On the proximal side, genes PHY-C and CYS flanked the VRN1 locus. The last two genes were completely linked to each other and separated from VRN1 by a single crossover (Fig. 1; Appendix 1). On the distal side, the CYB5 gene was also separated from VRN1 by a single crossover. Comparison of genotypic and phenotypic data from all of the F3 plants used in the 30 progeny tests confirmed that the observed segregation in growth habit was determined by variation at the VRN1 locus. Unvernalized plants homozygous for the G1777 AP1 allele flowered 1–2 months later than G2528, whereas the other plants flowered only 1 week before or after the G2528 control. These results confirmed the simple Mendelian segregation for vernalization requirement in this cross (1).
Physical Maps.
Distal contig.
Genes CYB5 and GENE1 were used to screen the BAC libraries from T. monococcum, rice, and sorghum. T. monococcum BAC clone 609E6 selected with the CYB5 gene was connected to previously sequenced 116F2 (AF459639) by four BACs (Fig. 1). The PCS gene hybridized with two fragments from BAC 609E06 (PCS1 and PCS2, Fig. 1) whereas only one PCS copy was found in the colinear region in rice. No single-copy probes were found in BACs 609E6 or in the unique HindIII fragments from the most proximal BAC 393O11 to continue the chromosome walking toward the proximal region.
Proximal contig.
Screening of the T. monococcum BAC library with PHY-C, CYS, AP1, and AGLG1 yielded 12 BACs organized in two contigs. The largest contig included eight BACs that hybridized with genes PHY-C, CYS, and AP1. The four additional BACs hybridized only with the AGLG1 gene (Fig. 1). The location of the AGLG1 contig within the physical map was determined by the complete linkage between the single-copy genes AGLG1 and AP1 and the proximal location of AGLG1 relative to the single-copy gene CYB5. No additional single-copy probes were found to close the gaps flanking the AGLG1 contig.
The proximal gap between AGLG1 and AP1 was covered by the current rice sequence. However, the distal gap between CYB5 and AGLG1 was also present in the different rice genome sequencing projects. The screening of the Nipponbare BAC libraries with probe CYB5 failed to extend the rice region because of the presence of a gap in the current rice physical maps. Fortunately, sorghum BAC 17E12 included GENE1, PCS1, PCS2, and CYB5 genes from the distal contig, and AGLG1, AP1, and CYS genes from the proximal contig, bridging the gap present in the rice and wheat contigs (Fig. 1). A restriction map of sorghum BAC 17E12 (Appendix 4, which is published as supporting information on the PNAS web site) indicated that the sorghum genes were in the same order as that previously found in rice and wheat and that a 24-kb SwaI–SwaI restriction fragment spanned the region of the rice and wheat gap between CYB5 and AGLG1.
Sequence Analysis.
Annotated sequences from the three T. monococcum BACs (AY188331, AY188332, and AY188333) and the partial sequence of the sorghum BAC 17E12 (AY188330) were deposited in GenBank. Including BACs 115G01 and 116F02 (AF459639), a total of 550 kb was sequenced. Multiple retrotransposons organized in up to four layers of nested elements were the most abundant features, similar to wheat regions analyzed before (13, 14). Annotated retrotransposons and other repetitive elements accounted for 78.4% of the sequence, whereas genes represented only 8.5% of the total. The genes detected in this sequence were in the same order as the ones present in the corresponding regions in rice and sorghum, indicating an almost perfect microcolinearity. The only exception was the duplication of the PCS gene in sorghum and wheat relative to the presence of a single PCS gene in the colinear rice region (Fig. 1).
No additional genes were found in the rice sequence between the two MADS-box genes corresponding to one of the two gaps in the wheat physical map. These two genes were also adjacent in sorghum (Fig. 1; Appendix 4). Similarly, no new genes were found between CYB5 and AGLG1 in the sequence of the 24-kb SwaI–SwaI restriction fragment from sorghum BAC 17E12 (AY188330) that covered the other gap in the wheat physical map. The four genes present in the sorghum sequence were in the same order and orientation as that previously found in rice and wheat (Appendix 4).
The absence of new genes in the colinear regions of rice and sorghum, together with the excellent microcolinearity detected in this region, suggested that it would be unlikely to find additional genes in the current gaps of the wheat physical map. This assumption was also supported by the absence of any new gene in the 324 kb of wheat sequence flanking these gaps. The presence of almost uninterrupted series of nested retrotransposons flanking the gaps also explained the failure to find single-copy probes to close the two gaps.
Classification of the Two MADS-Box Genes.
The AP1 and AGLG1 proteins have MADS-box and K domains characteristic of homeotic genes involved in the flowering process and similar exon structure (Fig. 1; Appendix 5, which is published as supporting information on the PNAS web site) (24). The consensus tree for 26 plant MADS-box proteins (Appendix 2) showed that the closest proteins to wheat AP1 and AGLG1 belonged to the SQUAMOSA (bootstrap 97) and AGL2 groups (bootstrap 95), respectively.
The closest Arabidopsis MADS-box proteins to wheat AP1 were the proteins coded by the three related meristem identity genes AP1, CAL, and FUL (Appendix 2). Two separate clusters were observed in the SQUAMOSA group dividing the Arabidopsis and grass proteins. A similar separation between the monocot and dicot proteins was found in more detailed studies of this group (25). The AP1 protein from T. monococcum was 98.4% similar to previously described Triticum aestivum WAP1, formerly TaMADS11 (26, 27) and 96.0% similar to barley BM5 (28). These two putative orthologous genes were described in papers characterizing the MADS-box family in wheat and barley but were not mapped or associated with the VRN1 gene.
The wheat AGLG1 protein was clustered with members of the AGL2 subgroup and was closely related with the rice AGLG1 orthologue and with rice OsMADS5, OsMADS1, and barley BM7 proteins (bootstrap 87, Appendix 2).
Expression Profiles.
No AP1 transcripts were detected in apices from unvernalized plants of T. monococcum with strong winter growth habit (G3116) even after 10 months in the greenhouse under long-day conditions. However, AP1 transcription was detected in the apices of plants from the same genotype after 6 weeks of vernalization (Fig. 2A, lanes 6 and 7). The same result was obtained in three independent experiments. These apices were morphologically at vegetative stage zero according to the developmental scale of Gardner et al. (29). In T. monococcum accessions with spring growth habit, AP1 transcripts were observed in the apices without previous vernalization.
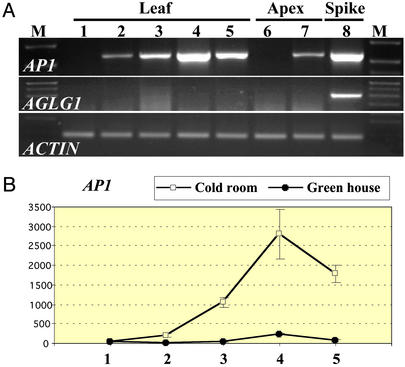
Figure 2.
(A) RT-PCR experiment using T. monococcum
G3116 (winter growth habit) and AP1-,
AGLG1-, and actin-specific primers. The
PCRs for the three genes were performed by using the same cDNA samples.
Lanes 1–5 indicate leaves. Lane 1, before vernalization; lanes 2–4,
2, 4, and 6 weeks of vernalization, respectively; lane 5, 2 weeks after
vernalized plants were returned to the greenhouse; lane 6, unvernalized
apices; lane 7, 6-week vernalized apices; and lane 8, young spikes.
(B) AP1 transcription levels in leaves
relative to actin measured by quantitative PCR. Lanes 1–5, leaves from
plants at the same vernalization stage as samples 1–5 in
A. Units are values linearized by using the
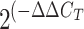 ) method, where
CT is the threshold cycle.
) method, where
CT is the threshold cycle.
Transcripts of AP1 were also detected in the leaves, as reported for WAP1 (26) and BM5 (28). A quantitative PCR experiment using the endogenous controls actin (Fig. 2B) and ubiquitin (Appendix 3) demonstrated that transcription of AP1 in the leaves of the winter genotypes was also regulated by vernalization. The abundance of AP1 transcripts started to increase after the first 2 weeks of vernalization and continued to increase during the 4 additional weeks of the vernalization process (Fig. 2B). AP1 transcripts were also present in the leaves from vernalized plants 2 weeks after their transfer to the greenhouse. At this stage, AP1 transcripts were detected in young and old green leaves (Appendix 3). Control plants kept in the greenhouse showed very low levels of AP1 transcription during the 8 weeks of the experiment (Fig. 2B). In the genotypes with a spring growth habit, AP1 transcripts were observed in the leaves of unvernalized plants that were initiating the transition to flowering.
AGLG1 transcripts were detected only in young spikes (Fig. 2A, lane 8) but were not observed in the same cDNA samples from apices after 6 weeks of vernalization where the AP1 transcripts were already present (Fig. 2A). This finding indicates that AGLG1 transcription is initiated later than AP1. Transcripts from AGLG1 were not detected in the leaves (Fig. 2A).
The expression results, together with the known role of the AP1 homologues in Arabidopsis as meristem identity genes, suggested that AP1 was a better candidate gene for VRN1 than AGLG1.
Allelic Variation.
Four AP1 genes were sequenced from T. monococcum accessions G1777, G3116, and DV92 carrying the vrn1 allele and G2528 carrying the Vrn1 allele (1, 2). The predicted proteins from DV92 and G2528 were identical and differed from the predicted proteins from G3116 and G1777 by a single amino acid (Appendix 5).
Analysis of the 1,024-bp region upstream from the AP1 start codon and up to the insertion point of a large repetitive element (AY188331) showed the presence of five polymorphic sites. Two of them differentiated G2528 from the three accessions carrying the vrn1 allele for winter growth habit. One was a 1-bp insertion located 728 bp upstream from the start codon, and the other one was a 20-bp deletion located 176 bp upstream from the start codon (Fig. 1; Appendix 5). No difference were detected in the first 600 bp of the AP1 3′ region between the vrn1 and Vrn1 alleles.
A PCR screening of a collection of cultivated T. monococcum accessions with primers flanking the 20-bp deletion region revealed the presence of deletions of different sizes in agarose gels (Appendix 5). Sequencing of these lines showed the presence of two new deletions that overlapped with the 20-bp deletion from G2528. These new deletions included a putative MADS-box protein-binding site adjacent to the 20-bp deletion (Fig. 1; Appendix 5).
No DNA differences were detected between accessions DV92 (vrn1) and G2528 (Vrn1) in the coding region, or the 5′ (365 bp) and 3′ (583 bp) UTRs of the AGLG1 gene.
Discussion
Genetic and Physical Maps of the VRN1 Region.
Only eight genes were found in the 556-kb sequence from the T. monococcum VRN1 region, resulting in an estimated gene density of one gene per 70 kb. The low gene density observed in this region was paralleled by a high ratio between physical and genetic distances. Excluding the two gaps in the physical map, a minimum ratio of 6,250 kb⋅cM−1 was estimated for the region between WG644 and PHY-C. This value is two times larger than the average genomewide estimate of 3,000 kb⋅cM−1 (30) and four times larger than the 1,400 kb⋅cM−1 reported for the telomeric region of chromosome 1A (31). Previous cytogenetic studies demonstrated that recombination in the wheat chromosomes decreases exponentially with distance from the telomere (32, 33), predicting an increase of the ratio between physical and genetic distance in the same direction. The region studied here is located between the breakpoints in deletion lines 5AL-6 (FL 0.68) and 5AL-17 (FL 0.78), in a more proximal location than regions used before to estimate ratios between physical and genetic distances in wheat. This result suggests that positional cloning projects in the proximal regions of wheat will be difficult and would greatly benefit from the use of the rice genomic sequence to jump over large blocks of repetitive elements.
Despite the low recombination rate found in this region, the large number of evaluated gametes was sufficient to find crossovers between most of the genes or at least between pairs of adjacent genes. This detailed genetic study showed that the variation in growth habit determined by the VRN1 gene was completely linked to only two genes. Although the possibility that additional genes would be found in the two current gaps and unsequenced regions of our T. monococcum physical maps cannot be ruled out, this seems unlikely based on the comparative studies with rice and sorghum and the absence of any additional genes in the 324-kb wheat sequence between CYB5 and CYS.
The genetic data reduced the problem of the identification of VRN1 to the question of which of the two MADS-box genes was the correct candidate. However, because no recombination was found between AGLG1 and AP1 it was not possible to answer this question based on the available genetic results. Therefore, the relationship between AGLG1 and AP1 with MADS-box genes from other species was established as a first step to predict their function from the known function of the related genes.
Phylogenetic Relationships of the VRN1 Candidate Genes.
The similarity between the wheat AP1 gene and the Arabidopsis meristem identity genes AP1, CAL, and FUL provided an indication that the wheat AP1 gene was a good candidate for VRN1. These Arabidopsis genes are expressed in the apices and are required for the transition between the vegetative and reproductive phases (34). The triple Arabidopsis mutant ap1-cal-ful never flowers under standard growing conditions (34). In wheat, the VRN1 gene is also responsible, directly or indirectly, for the transition between vegetative and reproductive apices. This transition is greatly accelerated by vernalization in the wheat plants carrying the vrn1 allele for winter growth habit. Therefore, it is reasonable to speculate that the sequence similarity between the wheat AP1 gene and the Arabidopsis meristem identity genes may indicate similar functions. An evolutionary change in the promoter region of AP1 may be sufficient to explain the regulation of AP1 by vernalization in wheat (see model below).
The close relationship of wheat AGLG1 to members of the AGL2 subgroup suggested that AGLG1 was a less likely candidate for VRN1 than AP1 because transcripts from genes included in this group are usually not observed in the apices in the vegetative phase (25). Expression of Arabidopsis AGL2, AGL4, and AGL9 begins after the onset of expression of floral meristem identity genes but before the activation of floral organ identity genes, suggesting that members of the AGL2 clade may act as intermediaries between the meristem identity genes and the organ identity genes (35–37). This seems to be valid also for OsMADS1, which is more closely related to AGLG1 than the Arabidopsis members of the AGL2 clade. In situ hybridization experiments of young rice inflorescences with OsMADS1 showed strong hybridization signals in flower primordia but not in other tissues (38).
If the functions of wheat AP1 and AGLG1 were similar to the function of the related genes from Arabidopsis, the initiation of transcription of AP1 should precede the initiation of transcription of AGLG1 in wheat.
Transcription Profiles of the VRN1 Candidate Genes.
RT-PCR experiments using RNA samples from vernalized apices showed transcription of AP1 but not of AGLG1 (Fig. 2A), indicating that transcription of AGLG1 occurs after the initiation of transcription of AP1. The similar timing and order of transcription suggests that the wheat genes might perform similar functions to the related Arabidopsis genes.
It could be argued that any gene in the flowering regulatory pathway would be up-regulated by the initiation of flowering caused by the vernalization process. However, the up-regulation of AP1 transcription in the leaves by vernalization (Fig. 2B) indicated a more direct role of the vernalization pathway in the regulation of wheat AP1 gene. Four additional characteristics of the transcription profile of AP1 paralleled the predicted expression of a vernalization gene. First, vernalization was required to initiate AP1 transcription in the plants with winter growth habit but not in the plants with spring growth habit. Second, AP1 transcription was initiated after only 2 weeks in the cold room, and a minimum of 2 weeks of vernalization is required by many winter wheat varieties to produce any significant acceleration of flowering (39). Third, the progressive increase of AP1 transcripts after the second week of vernalization (Fig. 2B) is consistent with the progressive effect of the length of the vernalization period in the acceleration of flowering time (39). Finally, a high level of AP1 transcripts was observed after the plants were moved from the cold to room temperature, indicating that AP1 is not just a cold stress-induced gene.
Allelic Variation.
No differences were found in the AGLG1 coding region or in its 5′ and 3′ regions between T. monococcum accessions G2528 (Vrn1) and DV92 (vrn1), confirming that AGLG1 was not a good candidate to explain the observed differences in growth habit.
Although no differences were detected in the AP1 coding sequences and 3′ region, the spring and winter accessions differed in their promoter sequence. The first 600 bp upstream from the start codon were identical among the four genotypes analyzed in this study except for a 20-bp deletion located close to the start of transcription and adjacent to a putative MADS-box protein binding site (CArG-box) in G2528 (ref. 40; Appendix 5). Two additional overlapping deletions were discovered in the same region of the promoter in spring accessions of cultivated T. monococcum (Fig. 1). The presence of a putative CArG-box in this region suggests the possibility that a transacting factor may bind to this site and repress AP1 transcription until vernalization occurs. This is similar to the case for FLC in Arabidopsis, which was recently shown to bind to MADS-box gene SOC1 and repress its transcription before vernalization (41).
A Model for the Regulation of Flowering by Vernalization in Wheat.
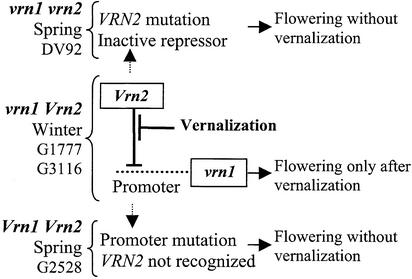
The results presented in this study can be included in an integrated model (Fig. 3) based on the known epistatic interactions between VRN1 and VRN2 (2) and the available information about the evolution of the vernalization requirement in the Triticeae. The significant epistatic interactions observed between VRN1 and VRN2 indicate that these two genes act in the same pathway (2). According to the model presented here (Fig. 3), VRN2 codes for a dominant repressor of flowering that acts directly or indirectly to repress VRN1. As the vernalization process reduces the abundance of the VRN2 gene product, VRN1 transcription gradually increases, leading to the competence to flower (Fig. 3 Middle).
Figure 3.
A model of the regulation of flowering initiation by vernalization in wheat.
The growth habit of plants homozygous for the recessive vrn2 allele for spring growth habit (Fig. 3 Top) is independent of variation at the VRN1 locus (2). According to this model, the vrn2 allele represents a null or defective repressor that cannot interact with the VRN1 promoter. Therefore, variation in the promoter of the VRN1 gene would have no effect on flowering time in homozygous vrn2 plants. This can be illustrated by the expression pattern of AP1 in T. monococcum DV92 (vrn1 vrn2). In this genotype, the initiation of AP1 transcription in leaves and apices did not require vernalization despite the presence of a recessive vrn1 allele. This result indicated that the VRN1 gene acts downstream of VRN2 (Fig. 3).
Conversely, plants homozygous for the Vrn1 allele for spring growth habit showed no significant effects of the VRN2 gene on flowering time (2). According to the model in Fig. 3 Bottom, the VRN2 repressor will have no effect on flowering in genotypes carrying the Vrn1 allele because of the lack of the recognition site in the VRN1 promoter region. This part of the model can be used to explain the AP1 expression profile of G2528 (Vrn1 Vrn2). In this genotype, transcription of AP1 in leaves and apices is initiated without a requirement for vernalization despite the presence of an active VRN2 repressor. This finding suggested that the active repressor could not interact with the G2528 AP1 promoter region, possibly because of the presence of the 20-bp deletion.
This model also provides an explanation for the parallel evolution of VRN1 spring alleles in three different Triticeae lineages. A vernalization gene with a dominant spring growth habit has been mapped in the same map location in diploid wheat (1), barley (42), and rye (43). Most of the wild Triticeae have a winter growth habit, suggesting that the recessive vrn1 allele is the ancestral character (44–46). This notion is also supported by the fact that it is unlikely that a vernalization requirement would be developed independently at the same locus in the three different lineages from an ancestral spring genotype. According to the model presented here, independent mutations in the promoter regions of winter wheat, barley, and rye genotypes have resulted in the loss of the recognition site of the VRN2 repressor (or an intermediate gene) and therefore, in a dominant spring growth habit (Vrn1 allele). Because this is a loss rather than a gain of a new function it is easier to explain its recurrent occurrence in the different Triticeae lineages.
In summary, this article presents the delimitation of the candidate genes for Vrn1 to AP1 and AGLG1 by a high-density genetic map, and the identification of AP1 as the most likely candidate based on its similar sequence to meristem identity genes, its transcription profile, and its natural allelic variation. A model is presented to integrate the results from this study with the previous knowledge about the epistatic interactions between vernalization genes and the evolution of vernalization in the Triticeae. Confirmation of the hypotheses generated by the model presented here will contribute to unraveling the complex net of relationships responsible for the regulation of heading date in the temperate cereals.
Supplementary Material
Acknowledgments
We thank Drs. P. Gepts and M. Soria for their valuable suggestions; R. Shao, A. Sanchez, S. Olmos, and C. Busso for excellent technical assistance; and Dr. W. Ramakrishna and J. Bennetzen for screening the sorghum BAC library. This work was supported by U.S. Department of Agriculture–National Research Initiative Grant 2000-1678 and National Science Foundation Grant PGRP-99-75793.
Abbreviations
- BAC
bacterial artificial chromosome
- cM
centimorgan
Footnotes
References
- 1.Dubcovsky J, Lijavetzky D, Appendino L, Tranquilli G. Theor Appl Genet. 1998;97:968–975. [Google Scholar]
- 2.Tranquilli G E, Dubcovsky J. J Hered. 1999;91:304–306. doi: 10.1093/jhered/91.4.304. [DOI] [PubMed] [Google Scholar]
- 3.Law C N, Worland A J, Giorgi B. Heredity. 1975;36:49–58. [Google Scholar]
- 4.Galiba G, Quarrie S A, Sutka J, Morgounov A. Theor Appl Genet. 1995;90:1174–1179. doi: 10.1007/BF00222940. [DOI] [PubMed] [Google Scholar]
- 5.Iwaki K, Nishida J, Yanagisawa T, Yoshida H, Kato K. Theor Appl Genet. 2002;104:571–576. doi: 10.1007/s00122-001-0769-0. [DOI] [PubMed] [Google Scholar]
- 6.Barrett B, Bayram M, Kidwell K. Plant Breed. 2002;121:400–406. [Google Scholar]
- 7.Kato K, Miura H, Sawada S. Genome. 1999;42:204–209. [Google Scholar]
- 8.Kato K, Kidou S, Miura H, Sawada S. Theor Appl Genet. 2002;104:1071–1077. doi: 10.1007/s00122-001-0805-0. [DOI] [PubMed] [Google Scholar]
- 9.Michaels S D, Amasino R M. Plant Cell. 1999;11:949–956. doi: 10.1105/tpc.11.5.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sheldon C C, Rouse D T, Finnegan E J, Peacock W J, Dennis E S. Proc Natl Acad Sci USA. 2000;97:3753–3758. doi: 10.1073/pnas.060023597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johanson U, West J, Lister C, Michaels S, Amasino R, Dean C. Science. 2000;290:344–347. doi: 10.1126/science.290.5490.344. [DOI] [PubMed] [Google Scholar]
- 12.Goff S A, Ricke D, Lan T H, Presting G, Wang R L, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H, et al. Science. 2002;296:92–100. doi: 10.1126/science.1068275. [DOI] [PubMed] [Google Scholar]
- 13.Wicker T, Stein N, Albar L, Feuillet C, Schlagenhauf E, Keller B. Plant J. 2001;26:307–316. doi: 10.1046/j.1365-313x.2001.01028.x. [DOI] [PubMed] [Google Scholar]
- 14.SanMiguel P, Ramakrishna W, Bennetzen J L, Busso C S, Dubcovsky J. Funct Integr Genomics. 2002;2:70–80. doi: 10.1007/s10142-002-0056-4. [DOI] [PubMed] [Google Scholar]
- 15.Dubcovsky J, Ramakrishna W, SanMiguel P, Busso C, Yan L, Shiloff B, Bennetzen J. Plant Physiol. 2001;125:1342–1353. doi: 10.1104/pp.125.3.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramakrishna W, Dubcovsky J, Park Y J, Busso C S, Emberton J, SanMiguel P, Bennetzen J L. Genetics. 2002;162:1389–1400. doi: 10.1093/genetics/162.3.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dubcovsky J, Galvez A F, Dvorak J. Theor Appl Genet. 1994;87:957–964. doi: 10.1007/BF00225790. [DOI] [PubMed] [Google Scholar]
- 18.Lijavetzky D, Muzzi G, Wicker T, Keller B, Wing R, Dubcovsky J. Genome. 1999;42:1176–1182. [PubMed] [Google Scholar]
- 19.Zhang H-B, Choi S, Woo S S, Li Z, Wing R A. Mol Breeding. 1996;2:11–24. [Google Scholar]
- 20.Woo S S, Jiang J, Gill B S, Paterson A H, Wing R A. Nucleic Acids Res. 1994;22:4922–4931. doi: 10.1093/nar/22.23.4922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar S, Tamura K, Nei M. Comput Appl Biosci. 1994;10:189–191. doi: 10.1093/bioinformatics/10.2.189. [DOI] [PubMed] [Google Scholar]
- 22.Yan L, Echenique V, Busso C, SanMiguel P, Ramakrishna W, Bennetzen J L, Harrington S, Dubcovsky J. Mol Genet Genomics. 2002;268:488–499. doi: 10.1007/s00438-002-0765-3. [DOI] [PubMed] [Google Scholar]
- 23.Livak K J, Schmittgen T D. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Ng M, Yanofsky M F. Nat Rev Genet. 2001;2:186–195. doi: 10.1038/35056041. [DOI] [PubMed] [Google Scholar]
- 25.Johansen B, Pedersen L B, Skipper M, Frederiksen S. Mol Phylogenet Evol. 2002;23:458–480. doi: 10.1016/s1055-7903(02)00032-5. [DOI] [PubMed] [Google Scholar]
- 26.Murai K, Murai R, Ogihara Y. Genes Genet Syst. 1997;72:317–321. doi: 10.1266/ggs.72.317. [DOI] [PubMed] [Google Scholar]
- 27.Murai K, Takumi S, Koga H, Ogihara Y. Plant J. 2002;29:169–181. doi: 10.1046/j.0960-7412.2001.01203.x. [DOI] [PubMed] [Google Scholar]
- 28.Schmitz J, Franzen R, Ngyuen T H, Garcia-Maroto F, Pozzi C, Salamini F, Rohde W. Plant Mol Biol. 2000;42:899–913. doi: 10.1023/a:1006425619953. [DOI] [PubMed] [Google Scholar]
- 29.Gardner J S, Hess W M, Trione E J. Am J Bot. 1985;72:548–559. [Google Scholar]
- 30.Bennett M D, Smith J B. Philos Trans R Soc London B. 1991;334:309–345. [Google Scholar]
- 31.Stein N, Feuillet C, Wicker T, Schlagenhauf E, Keller B. Proc Natl Acad Sci USA. 2000;97:13436–13441. doi: 10.1073/pnas.230361597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dvorak J, Chen K-C. Genetics. 1984;106:325–333. doi: 10.1093/genetics/106.2.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lukaszewski A J, Curtis C A. Theor Appl Genet. 1993;84:121–127. doi: 10.1007/BF00223816. [DOI] [PubMed] [Google Scholar]
- 34.Ferrandiz C, Gu Q, Martienssen R, Yanofsky M F. Development (Cambridge, UK) 2000;127:725–734. doi: 10.1242/dev.127.4.725. [DOI] [PubMed] [Google Scholar]
- 35.Flanagan C A, Ma H. Plant Mol Biol. 1994;26:581–595. doi: 10.1007/BF00013745. [DOI] [PubMed] [Google Scholar]
- 36.Savidge B, Rounsley S D, Yanofsky M F. Plant Cell. 1995;7:721–733. doi: 10.1105/tpc.7.6.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mandel M A, Yanofsky M F. Sex Plant Reprod. 1998;11:22–28. [Google Scholar]
- 38.Chung Y Y, Kim S R, Finkel D, Yanofsky M F, An G H. Plant Mol Biol. 1994;26:657–665. doi: 10.1007/BF00013751. [DOI] [PubMed] [Google Scholar]
- 39.Limin A E, Fowler D B. Ann Bot (London) 2002;89:579–585. doi: 10.1093/aob/mcf102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tilly J J, Allen D W, Jack T. Development (Cambridge, UK) 1998;125:1647–1657. doi: 10.1242/dev.125.9.1647. [DOI] [PubMed] [Google Scholar]
- 41.Hepworth S R, Valverde F, Ravenscroft D, Mouradov A, Coupland G. EMBO J. 2002;21:4327–4337. doi: 10.1093/emboj/cdf432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Laurie D A, Pratchett N, Bezant J H, Snape J W. Genome. 1995;38:575–585. doi: 10.1139/g95-074. [DOI] [PubMed] [Google Scholar]
- 43.Plaschke J, Börner A, Xie D X, Koebner R M D, Schlegel R, Gale M D. Theor Appl Genet. 1993;85:1049–1054. doi: 10.1007/BF00215046. [DOI] [PubMed] [Google Scholar]
- 44.Kihara H, Tanaka M. Preslia. 1958;30:241–251. [Google Scholar]
- 45.Halloran G M. Genetics. 1967;57:401–407. doi: 10.1093/genetics/57.2.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goncharov N P. Euphytica. 1998;100:371–376. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.