Abstract
Streptomyces sp. linear plasmids and linear chromosomes usually contain conserved terminal palindromic sequences bound by the conserved telomeric proteins Tap and Tp, encoded by the tap and tpg genes, respectively, as well as plasmid loci required for DNA replication in circular mode when the telomeres are deleted. These consist of iterons and an adjacent rep gene. By using PCR, we found that 8 of 17 newly detected linear plasmids in Streptomyces strains lack typical telomeric tap and tpg sequences. Instead, two novel telomeres in plasmids pRL1 and pRL2 from the eight strains and one conserved telomere in pFRL1 from the other strains were identified, while multiple short palindromes were also found in the plasmids. The complete nucleotide sequence of pRL2 revealed a gene encoding a protein containing two domains, resembling Tap of Streptomyces and a helicase of Thiobacillus, and an adjacent gene encoding a protein similar to Tpg of Streptomyces and a portion of the telomere terminal protein pTP of adenoviruses. No typical iterons-rep loci were found in the three plasmids. These results indicate an unexpected diversity of telomere palindromic sequences and replication genes among Streptomyces linear plasmids.
Streptomyces speciesare gram-positive, high-G+C, mycelium-producing eubacteria. Unlike the case for most eubacteria, linear plasmids and linear chromosomes are common in Streptomyces species (3, 8, 11, 19, 20, 28). The linear plasmids vary in size between 12 kb (16) and 1,700 kb (19). Their telomeres contain long inverted repeat sequences of 44 bp (7) to 180 kb (21), and the 5′ telomeric ends are linked covalently to terminal proteins (Tp) (1, 31). The telomeres of the ∼8-Mb linear Streptomyces chromosomes are 46 bp to 1 Mb long (14, 29). With the exceptions of the telomeres of the large linear plasmid SCP1 and the Streptomyces griseus linear chromosome (9, 18), Streptomyces linear plasmids and linear chromosomes usually contain conserved palindromic DNA sequences at their telomeres (13).
Unlike the terminal protein-capped linear replicons of adenoviruses and bacteriophage Φ29 (25), replication of Streptomyces linear plasmids starts at centrally located loci (27) and proceeds bidirectionally toward the telomeres (5). This leaves an ∼280-nucleotide (nt) single-strand overhang at the 3′ telomeric end of pSLA2 as a replication intermediate (5). To convert the 3′ overhang to a double strand, the terminal 144 nt of the telomere contain short palindromes 1 to 5 (22), with palindromes 2/3 being bound by the conserved telomere-associated protein (Tap) to recruit the conserved telomere terminal protein (Tp) (1, 2).
Streptomyces linear plasmids can also propagate in circular mode when the telomeres are deleted (5, 10, 24, 27). The centrally located locus for replication of pSLA2 consists of a rep-2 gene (encoding a DNA helicase) and its adjacent iterons within a transcribed rep-1 gene (6). The replication loci of plasmids SCP1 and pSLA2-L also consist of rep genes and different iteron sequences (10, 24). Such iterons-rep loci were revealed by complete nucleotide sequencing of other Streptomyces linear plasmids (such as pSCL1 and SLP2 [12, 30]).
Do the conserved telomere palindromic sequences, tap/tpg genes, and iterons-rep loci exist widely among larger populations of Streptomyces linear replicons? By investigating ∼100 Streptomyces strains, we show here that nearly half of the linear plasmid-harboring Streptomyces strains lack highly conserved tap/tpg sequences. Consistent with this, two novel and one conserved telomere palindromic sequence were identified on plasmids in the corresponding Streptomyces strains. The complete nucleotide sequences of the three plasmids showed no typical iterons-rep loci, and one plasmid carried two genes encoding proteins resembling Tap of Streptomyces and a portion of the telomere terminal protein pTP of adenoviruses.
MATERIALS AND METHODS
Bacterial strains, plasmids, and general methods.
Escherichia coli strain DH5α (Life Technologies Inc.) and plasmid pBluescript II SK (Stratagene) were used as the cloning host and vector, respectively. Plasmid isolation, transformation, and PCR amplification were done following the methods of Sambrook et al. (26). Streptomyces lividans ZX7 was the host for propagating plasmids. Streptomyces culture, plasmid isolation, pulsed-field gel electrophoresis, preparation of protoplasts, and transformation were done following the methods of Kieser et al. (17). Isolation of nondenatured plasmid DNA followed the method of Qin and Cohen (22). The ∼80 Streptomyces strains, isolated from Yunnan Province, China, and identified by the standard procedures of actinomycete classification, were provided by Jiang Chenglin and Xu Lihua. The ∼20 Streptomyces strains, isolated from Hunan Province, China, and identified by PCR sequencing of the 16S rRNA gene, were provided by Fang Ping. The sequences of the telomere terminal proteins (Tp) of Streptomyces coelicolor, S. lividans, and Streptomyces rochei were aligned, and the highly conserved amino acid regions were chosen to design a pair of oligonucleotides (for tpgC nt 77 to 96, 5′ CGCAGATGCGGTACCTGGTC 3′; for tpgC nt 281 to 262, 5′ GTGGTTGCCGCCTTCTGCCG 3′). Similarly, oligonucleotides from the conserved sequences of the telomere-associated protein (Tap) were for tapC nt 566 to 585 (5′ GCGGCCTGGTCCTGGACGTG 3′) and tapC nt 1676 to 1657 (5′ AGGTCGGACATCGTGGCGAG 3′).
DNA sequencing and analysis.
About 10 μg DNA of pRL1 and pRL2 was purified from pulsed-field gels. After shotgun cloning, PCR sequencing was done on an Applied Biosystems model 377 genetic analyzer at the Chinese Human Genome Center in Shanghai. Analysis of protein coding regions was performed with FramePlot 3.0 beta (15; http://watson.nih.go.jp/∼jun/cgi-bin/frameplot-3.0b.pl). Sequence comparisons and protein domain searching were done with software from the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/BLAST/). DNA secondary structures (e.g., direct repeats and inverted repeats) were predicted with DNA Folder (www.bioinfo.rpi.edu/applications/mfold/old/dna/) and Clone Manager, version 5.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the complete nucleotide sequences of pRL1, pRL2, and pFRL1 are DQ322649, DQ322650, and DQ322651, respectively.
RESULTS
Identification of linear plasmids among a population of Streptomyces strains.
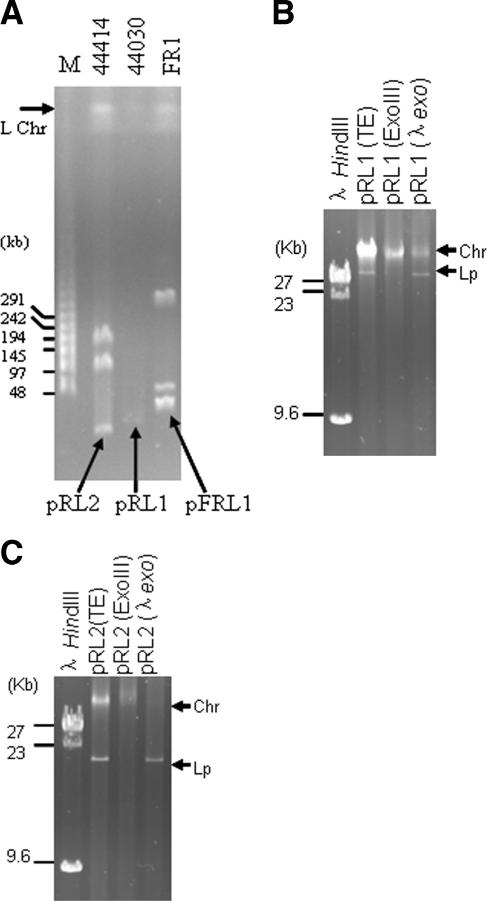
To detect indigenous linear plasmids among a population of Streptomyces strains, mycelia of ∼100 randomly selected strains were embedded in plugs of low-melting-point agarose gel and subjected to pulsed-field gel electrophoresis (17). Both fast-migrating and very large bands were detected (Fig. 1A), suggesting the presence of linear plasmids and linear chromosomes. As summarized in Table 1, various sizes (20 to 500 kb) of linear plasmids were detected; among the 17 strains, most harbored one plasmid, while 3 strains harbored three or four plasmids.
FIG. 1.
Identification of linear plasmids among Streptomyces strains. (A) Linear plasmids were detected by pulsed-field gel electrophoresis (17). The plug-embedded mycelium was electrophoresed in a 1.0% agarose gel at 120 V with a 50- to 90-s switch time at 14°C for 20 h. The DNA marker is in lane M, and linear plasmids pRL2, pRL1, and pFRL1 are indicated by arrows. DNA bands of possible linear chromosomes are indicated (L Chr). (B and C) Confirmation of linearity of pRL1 and pRL2. Nondenatured plasmid DNA was isolated from strains 44030 and 44414(22). Aliquots of the DNA (in Tris-EDTA buffer) were treated with 100 U of E. coli exonuclease III or 10 U of λ exonuclease at 37°C for 1 h and electrophoresed in a 0.5% agarose gel at 25 V for 12 h. λ HindIII DNA was used as a size marker. The position of residual chromosomal DNA (Chr) detected in the lanes after treatment with either λ exonuclease or exonuclease III is indicated. Lp, linear plasmid.
TABLE 1.
Summary of linear plasmids detection among ∼100 Streptomyces strains and PCR results for the conserved regions of the tpg and tap genes
| Strains | Size of linear plasmid(s) detected (kb) | PCR result for conserved regions of the tpg and tap genes |
|---|---|---|
| 44030 | 32 | − |
| 44414 | 20, 160, 260 | − |
| 44463 | 30 | − |
| 44476 | 90 | − |
| 44549 | 50 | − |
| 44380 | 35 | + |
| 44432 | 20, 30, 50, 300 | + |
| 44524 | 280 | + |
| 44554 | 130 | + |
| 44577 | 170 | + |
| 44598 | 500 | + |
| 44628 | 350 | + |
| FR1 | 54, 100, 450 | + |
| F2 | 360 | + |
| F8 | 370 | − |
| F9 | 150 | − |
| F11 | 500 | − |
To verify the conformations of the plasmids, genomic DNA of strain 44030 (harboring the 32-kb plasmid pRL1) was treated with E. coli exonuclease III and λ exonuclease (22) and then electrophoresed in an agarose gel. As shown in Fig. 1B, the 32-kb DNA band was sensitive to E. coli exonuclease III but resistant to λ exonuclease, suggesting that pRL1 DNA was linear, with a free 3′ end but a blocked 5′ end. The same result was obtained with the 20-kb pRL2 plasmid of strain 44414 (Fig. 1C).
PCR testing for conserved telomere tap and tpg sequences in Streptomyces strains.
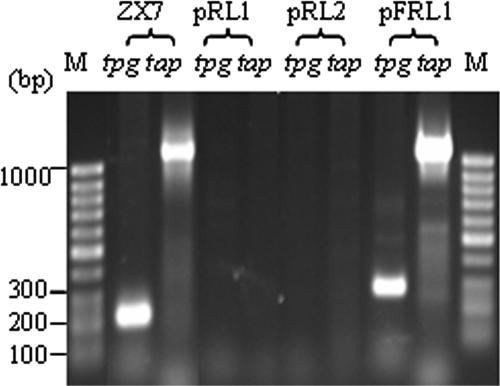
To investigate if the conserved telomere tpg and tap genes exist among the 17 Streptomyces strains harboring linear plasmids, oligonucleotides designed from the highly conserved amino acid sequences of the Tap and Tp proteins were used for PCR (see Materials and Methods). As shown in Fig. 2, PCR-amplified DNA bands of expected sizes were detected for strains FR1 (harboring linear plasmids pFRL1, pFRL2, and pFRL3) and S. lividans ZX7 (as a positive control containing chromosomal tap and tpg sequences) but not for strains 44414 (containing pRL2, pRL3, and pRL4) and 44030 (containing pRL1). As summarized in Table 1, such PCR-amplified products were detected in 9 of 17 strains, indicating that they might contain conserved tpg/tap genes, but not in the other 8 strains, suggesting that they might contain novel tpg/tap genes.
FIG. 2.
Detection of highly conserved tpg and tap sequences among Streptomyces strains by PCR. The genomic DNAs of Streptomyces strains ZX7 (positive control), 44030, 44414, and FR1 were prepared, and PCR amplifications were performed using pairs of oligonucleotides from the tpg/tap sequences (see Materials and Methods). The PCR products were electrophoresed in a 1% agarose gel at 100 V for 2 h. A 100-bp DNA ladder was used as a size marker.
Cloning and sequencing of the novel telomeres of linear plasmids pRL1 and pRL2 and the conserved telomere of pFRL1.
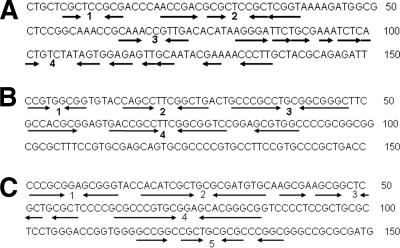
Previous work indicated a correlation between conserved telomere palindromic sequences and conserved telomere binding proteins in Streptomyces (1, 2, 31). To clone putatively novel telomeres of the linear plasmids containing the proposed novel tpg/tap genes, telomere treatment and forced cloning were employed (20). The gel-purified, 32-kb DNA of pRL1 was digested with EcoRI, ligated with E. coli pBluescript II SK treated with EcoRI and SmaI (blunt end cut), and then introduced by transformation into E. coli DH5α. A plasmid, pZR82, containing a 7-kb insert was obtained. The nucleotide sequence extending from the SmaI site of pZR82 (Fig. 3A), presumed to be the telomere of pRL1, contained multiple short palindromes different from those of conserved Streptomyces telomeres (13).
FIG. 3.
Telomere sequences (150 nt) of linear plasmids pRL1 (A), pRL2 (B), and pFRL1 (C). The short palindromes (numbered) are shown by pairs of arrows.
Similarly, a 1.3-kb XhoI fragment of pRL2 was cloned into the XhoI and EcoRV (blunt end cut) sites of pBluescript II SK to obtain pZR11. The corresponding nucleotide sequence of pZR11 (Fig. 3B) also contained multiple short palindromes which were different from those of both pRL1 and conserved Streptomyces telomeres.
A 1.2-kb EcoRI fragment of pFRL1 similarly yielded pZR6. The corresponding sequence closely resembled conserved Streptomyces telomeres [e.g., those of Streptomyces coelicolor A3(2), with an identity of 42/43 bp], especially in the first 43 bp (Fig. 3C).
The complete nucleotide sequence of linear plasmid pRL2 shows that two genes encode proteins, with one resembling Tap of Streptomyces and the helicase of Thiobacillus and one resembling Tpg of Streptomyces and a portion of the telomere terminal protein pTP of adenoviruses.
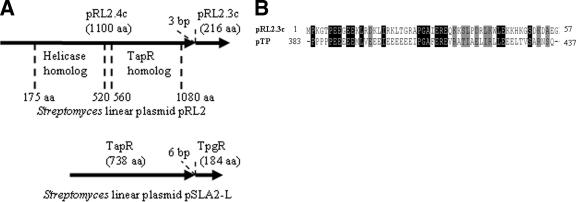
pRL2 was the smallest linear plasmid containing a novel telomere sequence found among Streptomyces strains. PCR sequencing following the “shotgun cloning” of linear plasmid DNA (see Materials and Methods) would miss the end sequences. After combining the internal and telomeric sequences, the complete nucleotide sequence of pRL2 (GenBank accession number DQ322650) consisted of 20,252 bp, with a 70.4% G+C content and a telomere of 427 bp. The 26 open reading frames (ORFs) on pRL2 were predicted by FramePlot 3.0 beta. Interestingly, one ORF, pRL2.4c, encoded a protein of 1,100 amino acids containing two domains which resembled the telomere-associated protein TapR of the Streptomyces rochei linear plasmid pSLA2-L (from aa 560 to 1080; expectation value, 2 × 10−8; identity, 134/640 bp [20%]) and the superfamily II helicase of Thiobacillus denitrificans (from aa 175 to 520; expectation value, 2 × 10−4; identity, 77/325 bp [23%]). As for the cotranscribed Streptomyces tpg and tap genes (2), the spacer between pRL2.4c and the adjacent pRL2.3c ORF was only 3 bp (Fig. 4A). The pRL2.3c-encoded protein of 216 amino acids resembled the telomere terminal protein TpgR of the Streptomyces rochei linear plasmid pSLA2-L (expectation value, 1 × 10−4; identity, 56/184 bp [30%]) and part of the telomere terminal protein pTP of simian adenovirus (Fig. 4B) (expectation value, 3.7; identity, 17/55 bp [30%]). These results indicated that the two genes, pRL2.3c and pRL2.4c, of pRL2 might be cotranscribed and involved in telomere replication.
FIG. 4.
Characteristics of the two genes of pRL2. (A) Comparison between proteins pRL2.4c and pRL2.3c and the TapR/TpgR proteins encoded by Streptomyces linear plasmid pSLA2-L (2). The two functional protein domains (TapR and helicase) of pRL2.4c are indicated. (B) Alignment of the pRL2.3c protein sequence and the telomere terminal protein (pTP) sequence of simian adenovirus.
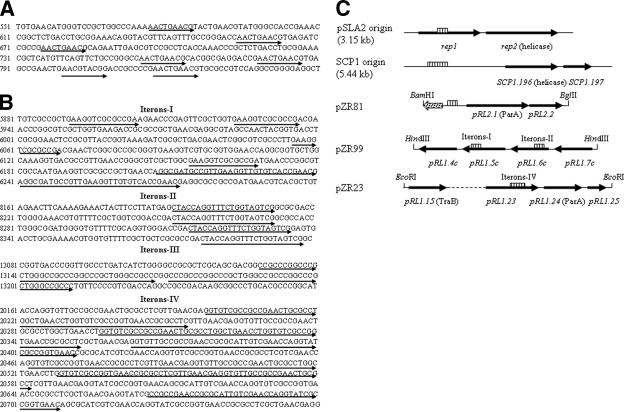
There was one possible predicted iteron near one telomere (Fig. 5A). However, this iteron and its adjacent ORFs (pRL2.1 and pRL2.2) did not propagate when cloned into an E. coli plasmid, pQC156, containing Streptomyces melC and tsr selection markers (23), yielding pZR81, and introduced into S. lividans ZX7 by transformation, suggesting that there was no typical iteron-rep locus in pRL2 for DNA replication (Fig. 5C).
FIG. 5.
Iterons of pRL1 and pRL2 and their comparison with typical iterons-rep loci of Streptomyces linear plasmids pSLA2 and SCP1. (A and B) Iterons (indicated by arrows) at the positions (numbered sequences) of pRL2 and pRL1 predicted by DNA Folder (see Materials and Methods). (C) Comparison of typical iterons-rep loci of pSLA2 (6) and SCP1 (24) with the iterons-ORFs of pRL2 and pRL1. The telomere, iterons, and adjacent genes of pRL2 were cloned into pBluescript II SK to obtain pZR30, treated with BamHI and BglII to release the pRL2 fragment, and cloned into the BamHI site of pQC156 to yield plasmid pZR81. The HindIII fragment (7.0 kb) and EcoRI fragment (9.1 kb) of pRL1 were cloned into pQC156 to yield pZR99 and pZR23, respectively. Iterons are indicated by striped boxes, the telomere is indicated by a dotted arrow, and genes are indicated by thick arrows. The genes encoding known proteins (in parentheses) are shown.
Complete nucleotide sequences of linear plasmids pRL1 and pFRL1 revealed no common iterons-rep loci for replication.
The pRL1 sequence (DQ322649) consisted of 32,759 bp, and its telomere was 3,269 bp long, containing two hypothetical genes. The G+C content was 66.9%, which was lower than that of typical Streptomyces genomes (e.g., 72.1% for S. coelicolor) (4). Thirty-three ORFs were predicted on pRL1, with no homology to conserved tpg/tap or rep genes of Streptomyces linear plasmids. Four different iteron sequences at different positions on pRL1 were predicted (Fig. 5B). As for pRL2, the pRL1 iterons and the adjacent ORFs did not allow propagation when cloned into E. coli plasmid pQC156 to yield pZR23 and pZR99, respectively, and introduced into S. lividans ZX7 by transformation, suggesting a lack of a typical iteron-rep locus on pRL1 for replication (Fig. 5C).
The pFRL1 sequence (DQ322651) consisted of 54,288 bp, with a 72.1% G+C content and a telomere of 421 bp. Fifty-nine ORFs were predicted on pFRL1, including two conserved tpg genes and one tap gene. However, as for pRL1 and pRL2, there was no typical iteron-rep locus on pFRL1.
DISCUSSION
With the exceptions of the large linear plasmid SCP1 and the Streptomyces griseus linear chromosome (9, 18), the telomeres of Streptomyces linear plasmids (e.g., pSLA2, pSLA2-L, pSCL1, SLP2, pSPA1, and SAP1) and linear chromosomes (e.g., those of S. coelicolor, S. lividans, Streptomyces parvulus, Streptomyces lipmanii, and Streptomyces avermitilis) contain conserved palindromic sequences (10, 13, 14, 22, 30). Correspondingly, conserved telomere replication genes (e.g., tap and tpg) are found on several Streptomyces linear plasmids and linear chromosomes (1, 2, 31). However, by investigating a population of Streptomyces strains, we found that nearly one-half (8/17) of the linear plasmid-harboring strains lacked highly conserved tap/tpg sequences. We further identified two novel and one conserved telomere palindromic sequence from the corresponding Streptomyces species, supporting the hypothesis of specificity between telomere palindromic sequences and telomere-binding proteins.
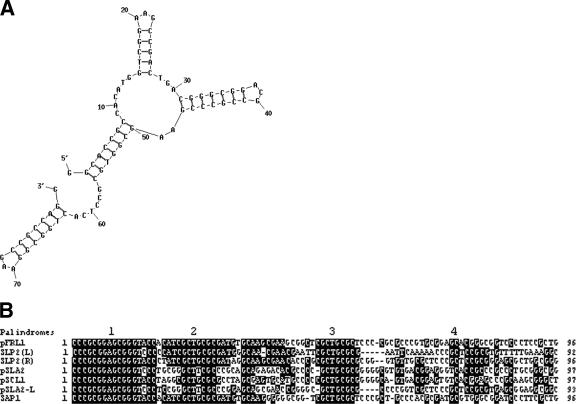
Although the two novel telomere sequences identified here differ from the conserved telomere sequences, they also contain multiple short palindromes. Like the case for linear plasmids pSLA2 and SLP2 (4, 13, 22), the 3′-telomeric palindrome 1 of pRL2 may “fold back” internally (folding energy, −29.1 kcal/mol) to one arm of palindrome 4 (Fig. 6A), suggesting that the “fold-back” is an important step for telomere replication. Interestingly, like the conserved Tap/Tpg proteins, pRL2 encodes two proteins, one of which resembles Tap of Streptomyces and the helicase of Thiobacillus and the other of which resembles Tpg of Streptomyces and the telomere terminal protein pTP of adenoviruses. Recent experiments indicated that the two pRL2 genes are involved in linear plasmid replication (our unpublished data). These results suggest that Streptomyces linear replicons share similar mechanisms of telomere replication, even though the telomere sequences differ, and that the process of “filling in” the 3′ overhang by the proposed telomere terminal protein-priming replication in Streptomyces might be similar to telomere replication in adenoviruses. Replication of linear replicons of adenoviruses starts at the telomeres and undergoes a strand displacement mechanism (25). Although Streptomyces linear plasmids usually contain centrally located origins for starting replication (5, 10, 24, 27), the possibility that replication starts at the telomeres in some linear replicons may not be ruled out. We found that besides the double-stranded DNA band, one high-mobility band could also be detected during the replication of a pRL2-derived plasmid, pZR108, in S. lividans ZX7 (our unpublished data), supporting this suggestion.
FIG. 6.
Characteristics of telomere sequences of linear plasmids pFRL1 and pRL2. (A) Secondary structure of the 80 nt of 3′ pRL2 telomere terminal sequence predicted by DNA Folder. Folding back and base pairing of short palindrome 1 to one arm of palindrome 4 are shown. (B) Multiple sequence alignment of telomere sequences from Streptomyces linear plasmids. The positions of telomere palindromes 1 to 4 are shown.
Previous studies showed that the telomeres of several Streptomyces linear chromosomes and linear plasmids contain conserved palindromes (palindromes 1 to 7) within the first 180 nt (13). Our data on the telomere sequence of linear plasmid pFRL1 and the recently published telomere sequences of linear plasmids pSLA2-L, SLP2 (left telomere), and SAP1 (9, 12, 14) show that the conserved palindromes lie within palindromes 1 to 3 (Fig. 6B). Consistent with this result, Bao and Cohen (2) showed that palindromes 2/3 of the telomere sequence are bound specifically by the Streptomyces telomere-associated protein (Tap).
Even though the telomere of linear plasmid SCP1 differs from those of pSLA2 and pSLA2-L (10, 18), these plasmids all contain typical iterons-rep loci for DNA replication in circular mode when the telomeres are deleted (6, 10, 24). We found that linear plasmid pFRL1, containing the conserved telomere sequence, lacks typical iterons-rep loci, indicating no correlation between the telomere sequences and the iterons-rep loci. Additionally, except for the plasmid transfer gene tra and the partition gene parA, no common genes are found among the three linear plasmids sequenced here, suggesting that the centrally located loci for DNA replication in circular mode are diverse in Streptomyces linear plasmids pRL1, pRL2, and pFRL1.
Acknowledgments
We thank Stanley Cohen and David Hopwood for strains and plasmids and for critically reading the manuscript, and we thank other members of Z. Qin's laboratory for technical help and discussions.
These investigations were supported by grants from the Chinese National and Shanghai Nature Science Foundation (30170019, 30270030, 30325003, and 0202ZA14096) and the national “863” projects (2005AA227020) to Z.Q.
REFERENCES
- 1.Bao, K., and S. N. Cohen. 2001. Terminal proteins essential for the replication of linear plasmids and chromosomes in Streptomyces. Genes Dev. 15:1518-1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bao, K., and S. N. Cohen. 2003. Recruitment of terminal protein to the ends of Streptomyces linear plasmids and chromosomes by a novel telomere-binding protein essential for linear DNA replication. Genes Dev. 17:774-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bentley, S. D., S. Brown, L. D. Murphy, D. E. Harris, M. A. Quail, J. Parkhill, B. G. Barrell, J. R. McCormick, R. I. Santamaria, R. Losick, M. Yamasaki, H. Kinashi, C. W. Chen, G. Chandra, D. Jakimowicz, H. M. Kieser, T. Kieser, and K. F. Chater. 2004. SCP1, a 356,023 bp linear plasmid adapted to the ecology and developmental biology of its host, Streptomyces coelicolor A3(2). Mol. Microbiol. 51:1615-1628. [DOI] [PubMed] [Google Scholar]
- 4.Bentley, S. D., K. F. Chater, A. M. Cerdeno-Tarraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C. H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M. A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141-147. [DOI] [PubMed] [Google Scholar]
- 5.Chang, P. C., and S. N. Cohen. 1994. Bidirectional replication from an internal origin in a linear Streptomyces plasmid. Science 265:952-954. [DOI] [PubMed] [Google Scholar]
- 6.Chang, P. C., E. S. Kim, and S. N. Cohen. 1996. Streptomyces linear plasmids that contain a phage-like, centrally located, replication origin. Mol. Microbiol. 22:789-800. [DOI] [PubMed] [Google Scholar]
- 7.Chen, C. W., T. W. Yu, Y. S. Lin, H. M. Kieser, and D. A. Hopwood. 1993. The conjugative plasmid SLP2 of Streptomyces lividans is a 50 kb linear molecule. Mol. Microbiol. 7:925-932. [DOI] [PubMed] [Google Scholar]
- 8.Chen, C. W. 1996. Complications and implications of linear bacterial chromosomes. Trends Genet. 12:192-196. [DOI] [PubMed] [Google Scholar]
- 9.Goshi, K., T. Uchida, A. Lezhava, M. Yamasaki, K. Hiratsu, H. Shinkawa, and H. Kinashi. 2002. Cloning and analysis of the telomere and terminal inverted repeat of the linear chromosome of Streptomyces griseus. J. Bacteriol. 184:3411-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hiratsu, K., S. Mochizuki, and H. Kinashi. 2000. Cloning and analysis of the replication origin and the telomeres of the large linear plasmid pSLA2-L in Streptomyces rochei. Mol. Gen. Genet. 263:1015-1021. [DOI] [PubMed] [Google Scholar]
- 11.Hirochika, H., and K. Sakaguchi. 1982. Analysis of linear plasmids isolated from Streptomyces: association of protein with the ends of the plasmid DNA. Plasmid 7:59-65. [DOI] [PubMed] [Google Scholar]
- 12.Huang, C. H., C. Y. Chen, H. H. Tsai, C. Chen, Y. S. Lin, and C. W. Chen. 2003. Linear plasmid SLP2 of Streptomyces lividans is a composite replicon. Mol. Microbiol. 47:1563-1576. [DOI] [PubMed] [Google Scholar]
- 13.Huang, C. H., Y. S. Lin, Y. L. Yang, S. W. Huang, and C. W. Chen. 1998. The telomeres of Streptomyces chromosomes contain conserved palindromic sequences with potential to form complex secondary structures. Mol. Microbiol. 28:905-916. [DOI] [PubMed] [Google Scholar]
- 14.Ikeda, H., J. Ishikawa, A. Hanamoto, M. Shinose, H. Kikuchi, T. Shiba, Y. Sakaki, M. Hattori, and S. Omura. 2003. Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat. Biotechnol. 21:526-531. [DOI] [PubMed] [Google Scholar]
- 15.Ishikawa, J., and K. Hotta. 1999. FramePlot: a new implementation of the frame analysis for predicting protein-coding regions in bacterial DNA with a high G + C content. FEMS Microbiol. Lett. 174:251-253. [DOI] [PubMed] [Google Scholar]
- 16.Keen, C. L., S. Mendelovitz, G. Cohen, Y. Aharonowitz, and K. L. Roy. 1988. Isolation and characterization of a linear DNA plasmid from Streptomyces clavuligerus. Mol. Gen. Genet. 212:172-176. [DOI] [PubMed] [Google Scholar]
- 17.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, United Kingdom.
- 18.Kinashi, H., M. Shimaji-Murayama, and T. Hanafusa. 1991. Nucleotide sequence analysis of the unusually long terminal inverted repeats of a giant linear plasmid, SCP1. Plasmid 26:123-130. [DOI] [PubMed] [Google Scholar]
- 19.Kinashi, H., E. Mori, A. Hatani, and O. Nimi. 1994. Isolation and characterization of linear plasmids from lankacidin-producing Streptomyces species. J. Antibiot. (Tokyo) 47:1447-1455. [DOI] [PubMed] [Google Scholar]
- 20.Lin, Y. S., H. M. Kieser, D. A. Hopwood, and C. W. Chen. 1993. The chromosomal DNA of Streptomyces lividans 66 is linear. Mol. Microbiol. 10:923-933. [DOI] [PubMed] [Google Scholar]
- 21.Pandza, S., G. Biukovic, A. Paravic, A. Dadbin, J. Cullum, and D. Hranueli. 1998. Recombination between the linear plasmid pPZG101 and the linear chromosome of Streptomyces rimosus can lead to exchange of ends. Mol. Microbiol. 28:1165-1176. [DOI] [PubMed] [Google Scholar]
- 22.Qin, Z., and S. N. Cohen. 1998. Replication at the telomeres of the Streptomyces linear plasmid pSLA2. Mol. Microbiol. 28:893-903. [DOI] [PubMed] [Google Scholar]
- 23.Qin, Z., M. Shen, and S. N. Cohen. 2003. Identification and characterization of a pSLA2 plasmid locus required for linear DNA replication and circular plasmid stable inheritance in Streptomyces lividans. J. Bacteriol. 185:6575-6582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Redenbach, M., M. Bibb, B. Gust, B. Seitz, and A. Spychaj. 1999. The linear plasmid SCP1 of Streptomyces coelicolor A3(2) possesses a centrally located replication origin and shows significant homology to the transposon Tn4811. Plasmid 42:174-185. [DOI] [PubMed] [Google Scholar]
- 25.Salas, M. 1991. Protein-priming of DNA replication. Annu. Rev. Biochem. 60:39-71. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 27.Shiffman, D., and S. N. Cohen. 1992. Reconstruction of a Streptomyces linear replicon from separately cloned DNA fragments: existence of a cryptic origin of circular replication within the linear plasmid. Proc. Natl. Acad. Sci. USA 89:6129-6133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Volff, J. N., and J. Altenbuchner. 1998. Genetic instability of the Streptomyces chromosome. Mol. Microbiol. 27:239-246. [DOI] [PubMed] [Google Scholar]
- 29.Weaver, D., N. Karoonuthaisiri, H. H. Tsai, C. H. Huang, M. L. Ho, S. Gai, K. G. Patel, J. Huang, S. N. Cohen, D. A. Hopwood, C. W. Chen, and C. M. Kao. 2004. Genome plasticity in Streptomyces: identification of 1 Mb TIRs in the S. coelicolor A3(2) chromosome. Mol. Microbiol. 51:1535-1550. [DOI] [PubMed] [Google Scholar]
- 30.Wu, X., and K. L. Roy. 1993. Complete nucleotide sequence of a linear plasmid from Streptomyces clavuligerus and characterization of its RNA transcripts. J. Bacteriol. 175:37-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang, C. C., C. H. Huang, C. Y. Li, Y. G. Tsay, S. C. Lee, and C. W. Chen. 2002. The terminal proteins of linear Streptomyces chromosomes and plasmids: a novel class of replication priming proteins. Mol. Microbiol. 43:297-305. [PubMed] [Google Scholar]