Abstract
Thirty-five Staphylococcus aureus strains, including 10 reference strains and 25 strains recovered from clinical specimens and food samples, were analyzed by PCR REA (restriction endonucleases analysis) of the egc operon and spa typing. Nineteen spa types and seven different egc operons, including four putative new egc variants, were revealed. In 13 strains, allelic variants of sei and/or seg were found. By an analysis of their nucleotide sequence identities, a new homogeneous cluster of a sei variant, called the sei variant, was detected in six strains. In addition, the prototype sei was shown to be more polymorphic than assumed so far. Seven strains possessed the recently described seg variant, also exhibiting several nucleotide exchanges. spa typing was more effective than REA egc grouping as a typing technique. Since, in some cases, the REA typing method was able to discriminate strains showing the same spa type, it must be considered for PCR approaches involved in diagnostic procedures and may be useful for epidemiological studies. Hence, the polyphasic approach used in this study can be reliably and advantageously applied for typing egc-positive S. aureus strains.
Staphylococcus aureus is an extraordinarily versatile pathogen causing a wide spectrum of infections, ranging from mild to severe and life-threatening, in humans as well as economically important infections in animals. In addition to superficial lesions and systemic infections, S. aureus is responsible for toxin-mediated diseases, such as toxic shock syndrome (TSS) and staphylococcal food poisoning. The virulence factors causing these toxicoses are members of the family of bacterial pyrogenic toxin superantigens (PTSAgs) comprising the TSS-causing toxins and the staphylococcal enterotoxins (SEs) producing food-borne illness (11, 24). Superantigens bypassing normal antigen presentation stimulate large populations of T cells by binding to a specific variable region of the T-cell antigen receptor beta chain (22). In addition to their nature as superantigens, SEs operate as potent gastrointestinal toxins, causing staphylococcal food poisoning, which has a major public health impact (18, 28). Approximately 1.5 billion dollars are spent annually in the United States because of staphylococcal intoxications (38).
Primarily, five major serological types, SEA through SEE, have been characterized (6). In the past years, many new types of SEs and their coding genes (seg through seu) have been reported (12, 16, 17, 19, 27, 30-33, 37, 37, 39, 40). However, some of the novel SE homologues were shown to be nonemetic, thus actually lacking the defining property of SEs and consequently designated “staphylococcal enterotoxin-like” superantigens (20). For SEC, minor variants have been reported (23). The staphylococcal PTSAgs constitute a large family of structurally related proteins whose genes are associated with mobile genetic elements. SEB, SEC, SEG, SEI, SEM, SEN, SEO, SEK, SEL, SEQ, and TSS toxin 1 are encoded by pathogenicity islands (2, 16, 17, 21). SEA, SEE, and SEP are encoded by prophages (7, 10, 17), whereas SED, SEJ, and SER are encoded by a plasmid known as pIB485 (3, 29, 40). The association with mobile genetic elements implies a horizontal transfer of the PTSAg genes between staphylococcal strains and an important role in the evolution of S. aureus as a pathogen.
Sequencing of the seg-sei intergenic DNA and flanking regions revealed three enterotoxin-like open reading frames related to seg and sei, designated sen, seo, and sem, and two pseudogenes, ϕent1 and ϕent2. Moreover, it was shown that these genes belong to an operon designated the enterotoxin gene cluster (egc), comprising seo, sem, sei, ϕent1-ϕent2, sen, and seg (16). In addition, minor variants for seg and, limited to one strain, for sei were reported recently (1, 8). Due to sequence divergences in the ϕent1-ϕent2 pseudogenes, the seu gene (including a variant) was described as a further part of the egc cluster (19). Consequently, at least three different egc subtypes were suggested: (i) egc1 (harboring seo, sem, sei, ϕent1, ϕent2, sen, and seg), as represented by strain A900322 (GenBank accession number AF285760), (ii) egc2 (containing seu instead of ϕent1 and ϕent2), as represented by strain FRI 137 (GenBank accession number AY205306), and (iii) egc3 (containing sei, seu, sen, and seg variants), as represented by strain 382F (GenBank accession number AY158703).
Recent studies, comprising isolates recovered from different human and veterinary specimens, showed that egc and its carried SE genes are more common in S. aureus strains than assumed so far (4, 5, 15, 26, 36).
Increasingly, S. aureus typing has become an important tool in the study of strain origin, clonal relatedness, and the epidemiology of outbreaks. Although several different phenotypic and, more recently, molecular techniques are available for differentiating S. aureus strains, no method is clearly superior under all conditions.
Previous studies (15-16) have shown that egc SE genes are arranged in tandem orientation in the egc cluster and are coexpressed. egc-encoded superantigens from Staphylococcus aureus are neutralized by human sera much less efficiently than are classical staphylococcal enterotoxins and toxic shock syndrome toxin (14). Moreover SEG and SEI interact differently with major histocompatibility complex class II and stimulate completely different subsets of human and mouse T cells. These characteristics indicate complementary superantigenic activity and suggest an important advantage to staphylococcal strains in producing both SEG and SEI.
Therefore, analyzing the diversity of staphylococcal PTSAgs may enhance our knowledge of the pathogenicity and evolution of S. aureus and improve detection approaches for diagnostic purposes and epidemiological studies.
The purpose of this study was to analyze the genetic variability within the egc cluster by restriction endonucleases analysis (REA) and nucleotide sequencing of a collection of S. aureus strains recovered from different human sources and food samples. The discrimination power of this technique was evaluated by comparing its results with those obtained by spa typing, which is widely used for biotyping S. aureus strains.
MATERIALS AND METHODS
Bacterial strains.
A total of 35 Staphylococcus aureus strains comprising 9 clinical, 16 food-derived, and 10 reference strains (including 4 egc-negative control strains, DSM 20231T, D 4508, ATCC 14458, and ATCC 27664) were investigated. The origins and sources of strains are reported in Table 1.
TABLE 1.
Source, origin and results of egc characterization and spa typing of strains analyzed in this study
| Strain | Sourceb | Originc | PCR result
|
REA egc group | spa type | ||
|---|---|---|---|---|---|---|---|
| seϕ | seu | sem-segd | |||||
| DSM 20231Ta | DSMZ | Human pleural fluid | − | − | − | t011 | |
| D4508a | CNTS | − | − | − | t948 | ||
| ATCC 14458a | ATCC | Feces of child | − | − | − | t008 | |
| ATCC 27664a (FRI326) | CNTS | Chicken tetrazzini | − | − | − | t029 | |
| A900322 | CNTS | Patient with TSS | + | −e | + | 1 | t002 |
| NCTC 9393 | CNTS | + | −e | + | 1 | t002 | |
| RIMD 31092 | CNTS | MRSA strain | + | −e | + | 1 | t002 |
| AS14g | DSAN | NTS (sample A) | + | −e | + | 1 | t209 |
| AS27 | DSAN | NTS (sample A) | + | −e | + | 1 | t209 |
| BS4 | DSAN | NTS (sample B) | + | −e | + | 1 | t209 |
| DS18g | DSAN | NTS (sample D) | + | −e | + | 1 | t209 |
| SI9 | DSAN | MCM (plant A) | + | −e | + | 1 | t164 |
| SI1 | DSAN | MCM (plant A) | + | −e | + | 1 | t164 |
| LA14 | DSAN | WBRM (plant A) | + | −e | + | 1 | t164 |
| R1 | DSAN | WBRM (plant A) | + | −e | + | 1 | t164 |
| ATCC 19095 (FRI137) | CNTS | Leg abscess | + | + | + | 2 | t352 |
| 382F | AFSSA | Unspecified (food) | + | + | + | 3 | t166 |
| ED-3 | DSAN | RPM (sample ED) | + | −e | + | 4 | t078 |
| ED-4 | DSAN | RPM (sample ED) | + | −e | + | 4 | t078 |
| 105 | DSAT | SP (defeathering machine) | + | −e | + | 4 | t002 |
| 106 | DSAT | SP (defeathering machine) | + | −e | + | 4 | t002 |
| ATCC 25923 | ATCC | Clinical isolate | + | + | + | 5 | t021 |
| AB-8802 | DSAN | RPM (sample AB) | + | + | + | 6 | t021 |
| 7645a | IMM | HP (osteomyelitis) | + | + | + | 7 | t021 |
| 109 | DSAT | SP (defeathering machine) | + | + | + | 7 | t012 |
| OM 56/2a | IMM | HP (osteomyelitis) | + | + | + | 7 | t012 |
| K4644/97 | IMM | HP (blood cultures) | + | + | + | 7 | t949 |
| K6278/97 | IMM | HP (blood cultures) | + | + | + | 7 | t298 |
| A652/99 | IMM | HP (nasal swabs) | + | + | + | 7 | t012 |
| A2586/99 | IMM | HP (nasal swabs) | + | + | + | 7 | t947 |
| A1048/98 | IMM | HP (nasal swabs) | + | + | + | 7 | t363 |
| A2812/98 | IMM | HP (nasal swabs) | + | + | + | 7 | t012 |
| A4178/98 | IMM | HP (nasal swabs) | + | + | + | 7 | t019 |
| 10 | DSAT | SP (gutting machine) | + | −c | (+)f | t267 | |
| 107 | DSAT | SP (defeathering machine) | + | −c | (+)f | t1059 | |
egc negative reference strains (used as negative controls).
ATCC, American Type Culture Collection, Rockville, Md.; AFSSA, Agence Francaise de Sécurité Sanitare des Aliments, Maisons-Alfort, France (kindly provided by P. Fach); DSMZ, Deutsche Sammlung für Mikroorganismen und Zellkulturen, Braunschweig, Germany; CNTS, Centre Nationale des Toxemies a Staphylococques, Faculté de Medecine Laennee, Lyon, France (kindly provided by G. Lina); DSAN, Dipartimento di Scienza degli Alimenti, Università degli Studi di Napoli Federico II, Portici, Italy; DSAT, Dipartimento di Scienze degli Alimenti, Università degli Studi di Teramo, Italy (kindly provided by A. Ianieri); IMM, Institut für Medizinische Mikrobiologie, Universitätsklinikum Münster, Münster, Germany.
MRSA, methicillin-resistant S. aureus; NTS, “Napoli-Type” salami; WBRM, water buffalo raw milk; MCM, water buffalo Mozzarella cheese manufacturing (natural whey cultures); RPM, raw poultry meat; SP, slaughterhouse for pigeon; HP, strains were isolated from different patients admitted to the University Hospital of Muenster (1997-1999).
PCR using the primer pair SEI-1 (targeting sem) and SEG-2 (targeting seg) covering a 3,375-bp fragment of the egc cluster.
The size of the amplification band was other than expected.
(+), weak PCR amplification result.
Identification was based on growth characteristics on Columbia agar with 5% (vol/vol) sheep blood (at 37°C) and positive catalase production. The clinical isolates were identified biochemically by the automated ID 32 Staph system (bioMérieux, France). The presence or absence of bound or free coagulase, respectively, was confirmed by latex slide agglutination for clumping factor, protein A, and capsular polysaccharides (Pastorex Staph-Plus; Sanofi Diagnostics Pasteur, France). For all strains, the S. aureus-specific nuc gene was detected as previously described (9).
DNA isolation.
Following overnight incubation in brain heart infusion broth (Oxoid, Basingstoke, Hampshire, United Kingdom), staphylococcal cells were streaked on brain heart infusion agar plates and incubated overnight at 37°C. DNA extraction was carried out from a single colony by using the InstaGene matrix (Bio-Rad Laboratories, Hercules, CA) according to the manufacturer's recommendations. About 25 ng of DNA was used for PCR amplification.
PCR amplification of the S. aureus egc.
The egc operon was detected by using PCR primers SEI-1 and SEG-2 as previously described by McLauchlin et al. (25) (Table 2), amplifying a 3,375-bp fragment comprising 47 nucleotides of sem, the complete nucleotide sequences of sei, ϕent1-ϕent2 (or seu), and sen followed by 610 nucleotides of seg.
TABLE 2.
PCR primers used in this study
| Primer | Sequence (5′-3′) | egc positiona | Reference |
|---|---|---|---|
| SEO-1 | AGT CAA GTG TAG ACC CTA TT | 494-513 | 8 |
| SEO-2 | TAT GCT CCG AAT GAG AAT GA | 1027-1008 | 8 |
| SEM-1 | CCA ATT GAA GAC CAC CAA AG | 1544-1563 | 8 |
| SEM-2 | CTT GTC CTG TTC CAG TAT CA | 2060-2042 | 8 |
| SEI-1 | GAC AAC AAA ACT GTC GAA ACT G | 2087-2108 | 25 |
| SEI-2 | CCA TAT TCT TTG CCT TTA CCA G | 2716-2695 | 25 |
| SEI-4 | GCC CTA GAG ACT TTA AAA TT | 3002-2983 | This study |
| PSE-1 | TGA TAA TTA GTT TTA ACA CTA AAA TGC G | 2919-2946 | 19 |
| PSE-2 | TAA AAT AAA TGG CTC TAA AAT TGA TGGb | 3278-3289 | 19 |
| PSE-4 | CGT CTA ATT GCC ACG TTA TAT CAG T | 3984-3960 | 19 |
| SEN-1 | ATT GTT CTA CAT AGC TGC AA | 3818-3837 | 8 |
| SEN-2 | TTG AAA AAA CTC TGC TCC CA | 4499-4480 | 8 |
| SEG-1 | TGC TAT CGA CAC ACT ACA ACC | 4758-4778 | 25 |
| SEG-2 | CCA GAT TCA AAT GCA GAA CC | 5461-5442 | 25 |
| SEG-4 | AGT TCG AAA CGC ACT TTA TG | 5674-5655 | This study |
According to the sequence of the 6,418-bp DNA fragment of S. aureus A900322 harboring the egc cluster (GenBank accession number AF285760) (16).
The underlined sequence corresponds to the 15-bp insertion allowing the seu gene.
PCR amplifications were performed in a 50-μl total volume, including 5 μl of the target DNA, 5.0 μl of Taq DNA polymerase 10× buffer (Invitrogen, San Giuliano Milanese, Italy), 2.5 μl of 50 mM MgCl2, 0.5 μl of dNTP mix (25 mM each), 0.1 μl of each primer (0.1 mM), and 0.4 μl of Taq DNA polymerase (5 U/μl) (Invitrogen). PCR consisted of 30 cycles at 95°C for 10 s and 55°C for 3.5 min and one additional final cycle at 72°C for 10 min. The PCR amplification fragments were resolved by agarose (1% wt/vol) gel electrophoresis at 100 V for 2 h. The gel was stained with ethidium bromide, and the bands were visualized under UV illumination at 254 nm.
REA of S. aureus egc cluster.
REA of the amplification fragments of the primer pair SEI-1 and SEG-2 was performed by digestion of about 1 μg (30 to 35 μl) of the PCR product with 20 U of the following restriction enzymes (in a total volume of 50 μl): EcoRI, AluI, TaqI, and CfoI (Promega Italia, Milan, Italy). Restriction digests were resolved by agarose (2% wt/vol) gel electrophoresis at 80 V for 3 h.
PCR detection of the individual SE genes of the egc cluster.
seo, sem, sei, sen, and seg genes were detected by using the following primer pairs: SEO-1 and SEO-2, SEM-1 and SEM-2, SEI-1 and SEI-2, SEN-1 and SEN-2, and SEG-1 and SEG-2 (Table 1). The ϕ1-ϕ2 pseudogenes and seu were detected by the use of the primer pairs PSE-1 plus PSE-4 and PSE-2 plus PSE-4, respectively, as previously published by Letertre et al. (19) (Table 2).
Sequencing procedures of sei, and seg genes.
The 3,375-bp SEI-1/SEG-2 amplification fragments were used for sequencing the sei and seg genes. In the case of weak amplification products (Table 3), the primer pairs SEI-1/SEI-4 and SEG-1/SEG-4 (Table 2) were used alternatively. sei/seg PCR was performed as described above, while PCR cycling conditions were 94°C for 3 min, followed by 30 cycles of 94°C for 5 s and 53°C for 1 min and finally 72°C for 5 min.
TABLE 3.
PCR egc characterization of strains showing SEI-1/SEG-2 weak amplification bands
| Strain | Sourcea | PCR resultb
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| sem-segc | seψ | seu | seo | sem |
sei
|
sen |
seg
|
||||
| SEI-1 SEI-2 | SEI-1 SEI-4 | SEG-1 SEG-2 | SEG-1 SEG-4 | ||||||||
| 10 | DSAT | (+) | + | −e | + | + | − | + | + | − | + |
| 107 | DSAT | (+)d | + | −e | + | + | + | + | + | − | + |
DSAT, Dipartimento di Scienze degli Alimenti Università degli Studi di Teramo, Italy (kindly provided by A. Ianieri).
If not specified, PCR results are based on the use of previously published primer pairs (8, 19, 25).
PCR using the primer pair SEI-1 (targeting sem) and SEG-2 (targeting seg) covering a 3,375-bp fragment of the egc cluster.
(+), weak PCR amplification result.
The size of the amplification band was other than expected.
Following purification of the amplification products by using the QIAquick gel extraction kit (QIAGEN, Milan, Italy), the fragments were sequenced, with the same primers used for PCR amplification being applied. DNA sequences were determined by the dideoxy chain termination method using the DNA sequencing kit (Perkin-Elmer Cetus, Emeryville, CA) according to the manufacturer's instructions. The sequences were analyzed, and the GenBank and EMBL databases were queried by using MacDNASIS Pro, version 3.0.7 (Hitachi Software Engineering Europe, Olivet, France).
spa typing.
The x region of the spa gene was amplified by PCR with primers 1095F and 1517R as previously described by Shopsin et al. (35). DNA sequences were obtained by an ABI 377 sequencer (Applied Biosystems, Foster City, Calif.). spa types were determined with the software Ridom StaphType (13) by using the Ridom SpaServer (http://spa.ridom.de/).
Nucleotide sequence accession numbers.
All sequences determined in this study were deposited in GenBank under accession numbers AY920260, AY920261, AY920259, AY920257, AY920262, AY920256, AY920258, DQ778337, DQ778338, AY920264, AY920265, AY920263, AY920269, AY920266, AY920267, and AY920268.
RESULTS
PCR amplification of the S. aureus egc cluster.
Primers SEI-1 and SEG-2 (Table 2) were used to amplify a 3,375-bp fragment representing approximately 65% of the nucleotide sequence of the egc operon. PCR amplification showed the expected fragment in 29 strains, whereas 2 strains (10 and 107) showed reproducible but weak amplification bands and no amplification signal was obtained by the 4 strains used as negative controls (Table 3).
REA of S. aureus egc cluster.
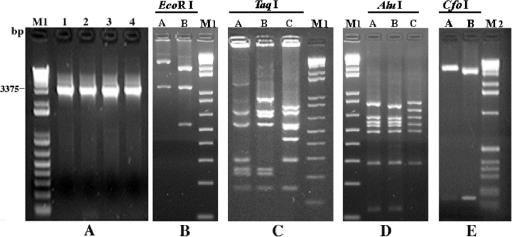
Fig. 1 (panels B, C, D, and E) shows the different REA patterns of the 29 egc-positive strains obtained from digestions of the 3,375-bp PCR fragment using EcoRI, TaqI, AluI, and CfoI. According to the results obtained by REA, the strains were classified into seven groups (Table 4). Three groups were associated with strains possessing the previously described three types of the egc operon (Table 1): (i) REA-1, comprising A900322 known to harbor egc1, other reference strains, and food-borne isolates, (ii) REA-2, restricted to FRI 137 and known to harbor egc2, and (iii) REA-3, restricted to 382F and known to possess egc3. Further REA groups (Table 1), such as REA-4, found in veterinary and food origin isolates, REA-5, shown for ATCC 25923, REA-6, observed for AB8802, as well as REA-7, comprising several isolates of human and veterinary origin, represent S. aureus strains which may harbor putative novel types of the egc operon (designated egc4, egc5, egc6, and egc7, respectively).
FIG. 1.
Specific PCR detection and REA characterization of S. aureus egc. Panel A shows the 3,375-bp PCR fragment obtained by amplifying the S. aureus egc of strains FRI137 (lane 1), NCTC 9393 (lane 2), RIMD 31092 (lane 3), and AB-8802 (lane 4). Panels B, C, D, and E show different restriction endonuclease patterns obtained from digestions of the 3,375-bp PCR fragment from egc-positive strains with EcoRI, TaqI, AluI, and CfoI, respectively. M1, 1-Kb DNA Ladder Plus (Invitrogen SRL); M2, 1-Kb DNA Ladder (Invitrogen SRL).
TABLE 4.
REA egc patterns shown by strains of the same egc group
| egc group | Pattern obtained by
|
|||
|---|---|---|---|---|
| EcoRI | AluI I | TaqI | CfoI | |
| 1 | A | B | B | A |
| 2 | A | A | A | A |
| 3 | B | B | C | A |
| 4 | A | B | C | A |
| 5 | B | B | C | B |
| 6 | B | C | C | B |
| 7 | B | C | B | B |
PCR detection of the individual SE genes of the egc cluster.
Applying the primers PSE-2 and PSE-4, which were previously reported by Letertre et al. (19) to be specific for the detection of seu, a PCR fragment of the expected length was obtained for strains belonging to only REA-2, REA-3, and REA-5 to REA-7, not REA-1 and REA-4 (Table 1). In contrast, a specific PCR fragment was amplified for all egc-positive strains using the PSE-1 and PSE-4 primer set (19), flanking a region covering both the pseudogenes ϕent1-ϕent2 and seu, respectively.
Analysis of strains with weak SEI-1/SEG-2 amplicons.
The two strains providing weak SEI-1/SEG-2 amplification results using primers as previously described (8, 25) tested PCR positive for seo, sem, and sen, negative for seg and seu, and variable for sei. Thus, these strains were grouped into two different toxin genotypes differing in the possession of sei (Table 3). However, a positive sei PCR result was detected in these strains by applying SEI-1 (25) in combination with the newly designed primer SEI-4, which targets an alternative egc position. Also, seg tested PCR positive for these strains by using a further newly designed primer (SEG-4) along with SEG-1 (25).
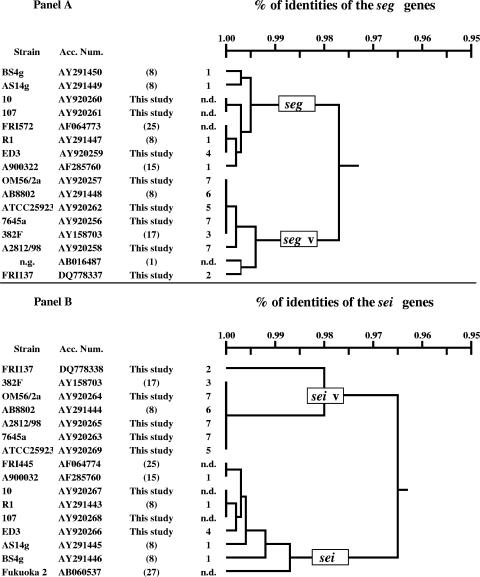
Sequencing of seg and sei fragments.
The sei and seg regions of strains giving weak amplification results were sequenced and compared to the sequencing results of the respective sequences of other strains known to harbor seg and sei (Fig. 2, panel A and B, respectively). The accession numbers of the sei and seg gene sequences analyzed in this study are given in Fig. 2. Dendrograms, as depicted in Fig. 2, demonstrate the relationships of the sequences. In both panels of Fig. 2, strains were clearly gathered in two separate clusters based on the percentage of identities. In addition to the prototype seg and sei, variant types, called the seg and sei variants, were found for both genes. Moreover, a low-grade polymorphism was revealed for seg, the seg variant, and sei, but not for the sei variant. In particular, two nucleotide changes were found for seg (a G→T substitution at position 5442 and a A→G substitution at position 5450; the nucleotide numbering was previously described by Jarraud et al. [16]) in strains 10 and 107 in the binding site of primer SEG-2, explaining the negative amplification results obtained by the SEI-1/SEG-2 and SEG-1/SEG2 primer sets.
FIG. 2.
Nucleotide sequence identities of seg (panel A) and sei (panel B) genes as analyzed in this study. n.d., not determined; n.g., not given.
Finally, a comparison of amino acid sequences, as obtained by virtual translation of the corresponding genes, showed that the SEI variant presents 4 to 5% (10 to 11 amino acids) of amino acid changes in respect to the SEI of strain FRI 445, while the SEG variant presents about 4% (7 to 8 amino acids) of amino acid changes in respect to the SEG of strain FRI 572.
spa typing.
A total of 19 spa types were obtained by analyzing the 35 strains included in this study (Table 1). In both directions, there was no general concordance between the affiliation to a REA egc group and the spa type of a given isolate. On one hand, the strains of the REA egc groups 1 (n = 11), 4 (n = 4), and 7 (n = 10) displayed three, two, and seven different spa types, respectively. On the other hand, some strains exhibiting the same spa type were shown to differ by their REA egc polymorphism. The three reference strains A900322, NCTC 9393, and RIMD 31092 as well as the two veterinary strains, 105 and 106, exhibiting the t002 spa type, fitted into the two different REA egc groups 1 and 4, respectively. Also, the reference strain ATCC 25923, the food-derived strain AB-8802, and the human-derived strain 7645a, harboring the t021 spa type, were shown to belong to three different REA egc groups (REA-5, -6, and -7, respectively). Moreover, in other cases, as expected, strains isolated from the same source showed the same spa type and REA egc group: (i) strains AS14 and AS27 were isolated from the same sample A; (ii) strains SI9, SI1, LA 14, and R1 from the same plant A; (iii) strains ED3 and ED4 from the same sample ED; and (iv) strains 105 and 106 from the same machine.
DISCUSSION
Originally, seg and sei genes were identified in two separate strains (FRI 572 and FRI 445) by Munson et al. (27). Subsequently it was shown that, when present, seg and sei genes coexist in all clinical isolates of S. aureus examined (4, 15), and they are arranged in tandem orientation on the same 3.2-kb DNA fragment (15). Sequence analysis of the seg-sei intergenic DNA and flanking regions revealed three enterotoxin-like open reading frames related to seg and sei, formerly designated sek, sel, and sem, and two pseudogenes, ϕent1 and ϕent2, (16). Furthermore, reverse transcription-PCR experiments revealed that all of these genes belong to one operon, designated the enterotoxin gene cluster (egc) (16). To rule out possible confusion with SEs described at the same time (12, 32), the sek and sel genes were renamed sen and seo, respectively, as published in a correction note (16).
Mempel et al. showed that egc occurs in about 48% of the SE-positive S. aureus strains isolated from atopic eczema (26). Recently, we detected this cluster in S. aureus strains isolated from food samples (8). Moreover, genes of the egc cluster were found in isolates recovered from nasal and blood specimens in high percentages (4, 5). Recently, by applying the microarray technology for simultaneous detection of the SE genes, Sergeev et al. (34) estimated that about 92% of S. aureus strains contain multiple SE genes (especially the egc cluster genes). Surveys of the egc distribution in S. aureus strains of animal origin also demonstrated a high frequency of egc-like genes (36).
However, based on several studies, it has been assumed that differences in the possession of individual genes or nucleotide variations may occur for the SE genes of the egc cluster (4, 5, 8, 14, 16, 19). In particular, Letertre et al. (19) found a 15-bp insertion in ϕent1 and some point mutations in both ϕent1 and ϕent2 of the strains FRI 137 and 382F with respect to these two pseudogenes of strain A900322 originally described by Jarraud et al. (16). Instead of allowing translation of the pseudogenes (ϕent1 and ϕent2), this insertion allowed a putative 261-nucleotide open reading frame (named seu) to be translated (19). In addition, it was demonstrated that egc of strain 382F is characterized by variants of the sei, seu, sen, and seg genes (19). This seg allele was shown to be identical to the previously characterized seg variant (1). A further SEGL29P variant was reported by Jarraud et al. (16).
We recently showed that some egc-positive strains, derived from food samples, were negative using PCR assays designed for sei and/or seg (8). Those strains displayed a egc restriction pattern compared to those of other egc-positive strains. Partial nucleotide sequencing of the sei and seg genes in strain AB-8802 (egc+, sei negative, and seg negative) confirmed the mispriming of both reverse sei primer (SEI-2) and forward seg primer (SEG-1) used in the PCR amplifications. We also found that approximately 30% of the S. aureus strains tested showed a positive sei amplification result by PCR, but they failed in subsequent hybridization reactions with sei-specific probes using a generic DNA enzyme immunoassay system (4). In this study, two strains (10 and 107) of veterinary origin showed weak amplification using primers SEI-1 and SEG-2 but tested PCR positive using newly designed egc primers, suggesting that the binding sites of the original primer may be polymorphic. The last hypothesis was corroborated by the sequence analysis of sei and seg genes of 10 and 107 S. aureus strains. The full sei gene sequences of AB-8802 and 7645a S. aureus strains showed 100% identity with the sei variant gene of strain 382F (GenBank accession number AF064774 [19]), while their full-nucleotide seg gene sequences displayed 99% identity (AB-8802) with the seg variant gene as previously described by Abe et al. (1).
In addition, we analyzed a part of the egc operon (sem-seg segment) of food-borne, human, veterinary, and reference S. aureus strains by REA using EcoRI, CfoI, AluI, and TaqI, allowing the differentiation of at least seven different egc operons. Thus, REA of the egc operon was shown to be successful in typing and analyzing egc-positive S. aureus strains. Sequence analysis of the putative new egc types may provide further insight into the evolution of egc, which was recently identified as a highly prevalent operon of enterotoxin genes, forming a putative nursery of superantigens in S. aureus.
The high level of polymorphism of the S. aureus egc, as detected by REA analysis of the SEI-1/SEG-2 PCR fragment and confirmed by DNA sequencing, was shown. The egc polymorphism seems to be unrelated to the evolution of S. aureus, as demonstrated by spa typing. In addition to the recently described three egc operons, four further variants were characterized. In particular, a variant of sei, called the sei variant, detectable in several S. aureus strains was delineated. Furthermore, the previously described seg variant was shown to occur frequently in a defined S. aureus strain collection. The observed polymorphism was due to point mutations causing loss or generation of restriction endonuclease sites or DNA insertions as described elsewhere. Moreover, this distinct polymorphism may explain the existence of strains possessing only some of the egc-carried genes or pseudogenes. Furthermore, failures in the detection of the egc cluster may occur, especially if the primer pair SEI-1/SEG2 is used. This should be considered, particularly for PCR approaches for diagnostic purposes and epidemiological studies, although it may be avoided by the application of the PSE-1/PSE-4 primers (19), which target regions representing highly conserved parts of the cluster.
During this study, the possibility of exploiting the egc polymorphism for epidemiological studies was evaluated by comparing results obtained by REA of the egc operon with those by spa typing. Although more limited than REA by pulsed-field gel electrophoresis analysis in discriminatory power, spa typing is often used as a screening method for typing S. aureus strains due to its shorter turnaround time, ease of use, and inherent advantages of sequence analysis, storage, and information sharing (35).
Our results reveal higher discriminatory power of spa typing in biotyping S. aureus egc-positive strains than that of REA egc. On the other hand, the REA egc analysis' capability to discriminate, in some cases, among strains showing the same spa type, even though they were geographically related, makes it a reliable typing tool. Therefore, by using a polyphasic approach, including both REA of egc and spa typing analyses, the biotyping of egc-positive S. aureus strains can be reliably achieved, furnishing, at the same time, knowledge of great consequence about the potential toxicity of S. aureus strains.
Acknowledgments
We thank G. Lina of the CNTS (Lyon, France), P. Fach of the AFSSA (Maisons-Alfort, France), and A. Ianieri of the DSA (Teramo, Italy) for kindly providing strains used in this study.
Financial support was provided by the MIPAF of Italy (project SIQUALTECA) and the German Network of Competence “Pathogenomics,” Alliance “Gram-Positive Cocci.”
REFERENCES
- 1.Abe, J., Y. Ito, M. Onimaru, T. Kohsaka, and T. Takeda. 2000. Characterization and distribution of a new enterotoxin-related superantigen produced by Staphylococcus aureus. Microbiol. Immunol. 44:79-88. [DOI] [PubMed] [Google Scholar]
- 2.Baba, T., F. Takeuchi, M. Kuroda, H. Yuzawa, K. Aoki, A. Oguchi, Y. Nagai, N. Iwama, K. Asano, T. Naimi, H. Kuroda, L. Cui, K. Yamamoto, and K. Hiramatsu. 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359:1819-1827. [DOI] [PubMed] [Google Scholar]
- 3.Bayles, K. W., and J. J. Iandolo. 1989. Genetic and molecular analyses of the gene encoding staphylococcal enterotoxin D. J. Bacteriol. 171:4799-4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Becker, K., A. W. Friedrich, G. Lubritz, M. Weilert, G. Peters, and C. von Eiff. 2003. Prevalence of genes encoding pyrogenic toxin superantigens and exfoliative toxins among strains of Staphylococcus aureus isolated from blood and nasal specimens. J. Clin. Microbiol. 41:1434-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Becker, K., A. W. Friedrich, G. Peters, and C. von Eiff. 2004. Systematic survey on the prevalence of genes coding for staphylococcal enterotoxins SElM, SElO, and SElN. Mol. Nutr. Food Res. 48:488-495. [DOI] [PubMed] [Google Scholar]
- 6.Bergdoll, M. S. 1989. Staphylococcus aureus, p. 463-523. In M. P. Doyle (ed.), Foodborne bacterial pathogens. Marcel Dekker, Inc., New York, N.Y.
- 7.Betley, M. J., and J. J. Mekalanos. 1985. Staphylococcal enterotoxin A is encoded by phage. Science 229:185-187. [DOI] [PubMed] [Google Scholar]
- 8.Blaiotta, G., D. Ercolini, C. Pennacchia, V. Fusco, A. Casaburi, O. Pepe, and F. Villani. 2004. PCR detection of staphylococcal enterotoxin genes in Staphylococcus spp. strains isolated from meat and dairy products. Evidence for new variants of seG and seI in S. aureus AB-8802. J. Appl. Microbiol. 97:719-730. [DOI] [PubMed] [Google Scholar]
- 9.Brakstad, O. G., K. Aasbakk, and J. A. Maeland. 1992. Detection of Staphylococcus aureus by polymerase chain reaction amplification of the nuc gene. J. Clin. Microbiol. 30:1654-1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Couch, J. L., M. T. Soltis, and M. J. Betley. 1988. Cloning and nucleotide sequence of the type E staphylococcal enterotoxin gene. J. Bacteriol. 170:2954-2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dinges, M. M., P. M. Orwin, and P. M. Schlievert. 2000. Exotoxins of Staphylococcus aureus. Clin. Microbiol. Rev. 13:16-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fitzgerald, J. R., S. R. Monday, T. J. Foster, G. A. Bohach, P. J. Hartigan, W. J. Meaney, and C. J. Smyth. 2001. Characterization of a putative pathogenicity island from bovine Staphylococcus aureus encoding multiple superantigens. J. Bacteriol. 183:63-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harmsen, D., H. Claus, W. Witte, J. Rothganger, H. Claus, D. Turnwald, and U. Vogel. 2003. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J. Clin. Microbiol. 41:5442-5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holtfreter, S., K. Bauer, D. Thomas, C. Feig, V. Lorenz, K. Roschack, E. Friebe, K. Selleng, S. Lovenich, T. Greve, A. Greinacher, B. Panzig, S. Engelmann, G. Lina, and B. M. Broker. 2004. egc-encoded superantigens from Staphylococcus aureus are neutralized by human sera much less efficiently than are classical staphylococcal enterotoxins or toxic shock syndrome toxin. Infect. Immun. 72:4061-4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jarraud, S., G. Cozon, F. Vandenesch, M. Bes, J. Etienne, and G. Lina. 1999. Involvement of enterotoxins G and I in staphylococcal toxic shock syndrome and staphylococcal scarlet fever. J. Clin. Microbiol. 37:2446-2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jarraud, S., M. A. Peyrat, A. Lim, A. Tristan, M. Bes, C. Mougel, J. Etienne, F. Vandenesch, M. Bonneville, and G. Lina. 2001. egc, a highly prevalent operon of enterotoxin gene, forms a putative nursery of superantigens in Staphylococcus aureus. J. Immunol. 166:669-677. (Correction, 166:4260.) [DOI] [PubMed] [Google Scholar]
- 17.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K. Aoki, Y. Nagai, J. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Ui, N. K. Takahashi, T. Sawano, R. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet 357:1225-1240. [DOI] [PubMed] [Google Scholar]
- 18.Le Loir, Y., F. Baron, and M. Gautier. 2003. Staphylococcus aureus and food poisoning. Genet. Mol. Res. 2:63-76. [PubMed] [Google Scholar]
- 19.Letertre, C., S. Perelle, F. Dilasser, and P. Fach. 2003. Identification of a new putative enterotoxin SEU encoded by the egc cluster of Staphylococcus aureus. J. Appl. Microbiol. 95:38-43. [DOI] [PubMed] [Google Scholar]
- 20.Lina, G., G. A. Bohach, S. P. Nair, K. Hiramatsu, E. Jouvin-Marche, R. Mariuzza, and the International Nomenclature Committee for Staphylococcal Superantigens. 2004. Standard nomenclature for the superantigens expressed by Staphylococcus. J. Infect. Dis. 189:2334-2336. [DOI] [PubMed] [Google Scholar]
- 21.Lindsay, J. A., A. Ruzin, H. F. Ross, N. Kurepina, and R. P. Novick. 1998. The gene for toxic shock toxin is carried by a family of mobile pathogenicity islands in Staphylococcus aureus. Mol. Microbiol. 29:527-543. [DOI] [PubMed] [Google Scholar]
- 22.Llewelyn, M., and J. Cohen. 2002. Superantigens: microbial agents that corrupt immunity. Lancet Infect. Dis. 2:156-162. [DOI] [PubMed] [Google Scholar]
- 23.Marr, J. C., J. D. Lyon, J. R. Roberson, M. Lupher, W. C. Davis, and G. A. Bohach. 1993. Characterization of novel type C staphylococcal enterotoxins: biological and evolutionary implications. Infect. Immun. 61:4254-4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCormick, J. K., J. M. Yarwood, and P. M. Schlievert. 2001. Toxic shock syndrome and bacterial superantigens: an update. Annu. Rev. Microbiol. 55:77-104. [DOI] [PubMed] [Google Scholar]
- 25.McLauchlin, J., G. L. Narayanan, V. Mithani, and G. O'Neill. 2000. The detection of enterotoxins and toxic shock syndrome toxin genes in Staphylococcus aureus by polymerase chain reaction. J. Food Prot. 63:479-488. [DOI] [PubMed] [Google Scholar]
- 26.Mempel, M., G. Lina, M. Hojka, C. Schnopp, H. P. Seidl, T. Schafer, J. Ring, F. Vandenesch, and D. Abeck. 2003. High prevalence of superantigens associated with the egc locus in Staphylococcus aureus isolates from patients with atopic eczema. Eur. J. Clin. Microbiol. Infect. Dis. 22:306-309. [DOI] [PubMed] [Google Scholar]
- 27.Munson, S. H., M. T. Tremaine, M. J. Betley, and R. A. Welch. 1998. Identification and characterization of staphylococcal enterotoxin types G and I from Staphylococcus aureus. Infect. Immun. 66:3337-3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Olsen, S. J., L. C. MacKinnon, J. S. Goulding, N. H. Bean, and L. Slutsker. 2000. Surveillance for foodborne-disease outbreaks—United States, 1993-1997. Morb. Mortal. Wkly. Rep. CDC Surveill. Summ. 49:1-62. [PubMed] [Google Scholar]
- 29.Omoe, K., D. L. Hu, H. Takahashi-Omoe, A. Nakane, and K. Shinagawa. 2003. Identification and characterization of a new staphylococcal enterotoxin-related putative toxin encoded by two kinds of plasmids. Infect. Immun. 71:6088-6094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Omoe, K., M. Ishikawa, Y. Shimoda, D. L. Hu, S. Ueda, and K. Shinagawa. 2002. Detection of seg, seh, and sei genes in Staphylococcus aureus isolates and determination of the enterotoxin productivities of S. aureus isolates harboring seg, seh, or sei genes. J. Clin. Microbiol. 40:857-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Orwin, P. M., J. R. Fitzgerald, D. Y. Leung, J. A. Gutierrez, G. A. Bohach, and P. M. Schlievert. 2003. Characterization of Staphylococcus aureus enterotoxin L. Infect. Immun. 71:2916-2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Orwin, P. M., D. Y. Leung, H. L. Donahue, R. P. Novick, and P. M. Schlievert. 2001. Biochemical and biological properties of staphylococcal enterotoxin K. Infect. Immun. 69:360-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ren, K., J. D. Bannan, V. Pancholi, A. L. Cheung, J. C. Robbins, V. A. Fischetti, and J. B. Zabriskie. 1994. Characterization and biological properties of a new staphylococcal exotoxin. J. Exp. Med. 180:1675-1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sergeev, N., D. Volokhov, V. Chizhikov, and A. Rasooly. 2004. Simultaneous analysis of multiple staphylococcal enterotoxin genes by an oligonucleotide microarray assay. J. Clin. Microbiol. 42:2134-2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shopsin, B., M. Gomez, S. O. Montgomery, D. H. Smith, M. Waddington, D. E. Dodge, D. A. Bost, M. Riehman, S. Naidich, and B. N. Kreiswirth. 1999. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J. Clin. Microbiol. 37:3556-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smyth, D. S., P. J. Hartigan, W. J. Meaney, J. R. Fitzgerald, C. F. Deobald, G. A. Bohach, and C. J. Smyth. 2005. Superantigen genes encoded by the egc cluster and SaPIbov are predominant among Staphylococcus aureus isolates from cows, goats, sheep, rabbits and poultry. J. Med. Microbiol. 54:401-411. [DOI] [PubMed] [Google Scholar]
- 37.Su, Y.-C., and A. C. L. Wong. 1995. Identification and purification of a new staphylococcal enterotoxin, H. Appl. Environ. Microbiol. 61:1438-1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Su, Y.-C., and A. C. L. Wong. 1997. Current perspectives on detection of staphylococcal enterotoxins. J. Food Prot. 60:195-202. [DOI] [PubMed] [Google Scholar]
- 39.Yarwood, J. M., J. K. McCormick, M. L. Paustian, P. M. Orwin, V. Kapur, and P. M. Schlievert. 2002. Characterization and expression analysis of Staphylococcus aureus pathogenicity island 3: implications for the evolution of staphylococcal pathogenicity islands. J. Biol. Chem. 277:13138-13147. [DOI] [PubMed] [Google Scholar]
- 40.Zhang, S., J. J. Iandolo, and G. C. Stewart. 1998. The enterotoxin D plasmid of Staphylococcus aureus encodes a second enterotoxin determinant (sej). FEMS Microbiol. Lett. 168:227-233. [DOI] [PubMed] [Google Scholar]