Abstract
The free-living amoeboflagellate genus Naegleria includes one pathogenic and two potentially pathogenic species (Naegleria fowleri, Naegleria italica, and Naegleria australiensis) plus numerous benign organisms. Monitoring of bathing water, water supplies, and cooling systems for these pathogens requires a timely and reliable method for identification, but current DNA sequence-based methods identify only N. fowleri or require full sequencing to identify other species in the genus. A novel closed-tube method for distinguishing thermophilic Naegleria species is presented, using a single primer set and the DNA intercalating dye SYTO9 for real-time PCR and melting-curve analysis of the 5.8S ribosomal DNA gene and flanking noncoding spacers (ITS1, ITS2). Collection of DNA melting data at close temperature intervals produces highly informative melting curves with one or more recognizable melting peaks, readily distinguished for seven Naegleria species and the related Willaertia magna. Advantages over other methods used to identify these organisms include its comprehensiveness (encompassing all species tested to date), simplicity (no electrophoresis required to verify the product), and sensitivity (unambiguous identification from DNA equivalent to one cell). This approach should be applicable to a wide range of microorganisms of medical importance.
The thermophilic amoeboflagellate Naegleria fowleri, responsible for the severe waterborne disease primary amoebic meningoencephalitis, is both a natural aquatic organism and an opportunistic pathogen (7). The free-living nature of this protozoan has two important practical consequences for public health management. The first is that there are numerous closely related species from which N. fowleri must be distinguished in environmental surveys. Most are benign aquatic species, although some strains of Naegleria italica and Naegleria australiensis can infect laboratory animals (7). A second consequence is that N. fowleri can colonize and proliferate in artificial aquatic environments such as swimming pools, cooling systems, and water supplies when temperatures are suitable, 25°C and above, becoming progressively more selective up to approximately 42°C (7, 9). Analysis of environmental samples may have an epidemiological purpose, identifying sources of recent N. fowleri infection (15), or be prospective, as an element of risk assessment (3). In either case, the timeliness of identifications can be critical.
In our laboratory, the density of thermophilic amoebae is estimated by differential plaque counts, with identification of discrete plaque types by biochemical or molecular criteria. Until recently, identification was based on a well-validated data set of isozyme profiles (1, 9, 25). While isozyme analysis has a number of attractions, it is less sensitive than DNA sequence-based methods, requiring 106 to 107 cells for detection of sufficient enzyme activity. In moving toward sequence-based identification, we have several criteria for an effective method. Foremost, the method must be definitive in identifying the pathogenic and potentially pathogenic species. Second, to facilitate multiple identifications of strains to enable quantitation, the method should be simple and reliable to execute, with as few steps and requiring as few specific molecular tools as possible. Finally, it is desirable that the method provide simultaneous identification of as many Naegleria species as possible for the collection of ecologically useful data. The published PCR methods for Naegleria (16, 19, 23, 26, 27) meet these criteria only to a limited extent, as all require end-point analysis using agarose gel electrophoresis and none identifies species other than N. fowleri without additional sequencing.
DNA melting-curve analysis is a relatively new analytical technique that can be applied post-PCR to provide information on the characteristics of the amplification products. To date, this technique has been used principally as an alternative to gel electrophoresis to verify the identity of a desired PCR product (21, 24, 32), or to differentiate the matching or mismatching of particular types of hybridization probes, for example, in fluorescence resonance energy transfer (22, 31). A few publications have explored the potential of melting analysis to differentiate the products of a PCR in which the primers are conservative but the amplified material spans a region of variable sequence (11, 14, 18). The most commonly used dye for DNA melting-curve analysis is SYBR green I; however, melting curves generated using this dye are sensitive to changes in either dye or DNA concentration (17, 24). Recently, two alternative dyes, LCGreen (30) and SYTO9 (17), have been reported to provide melting-curve-analysis performance superior to that of SYBR green I.
Currently, the soundest systematic scheme for Naegleria species is based on the internal transcribed spacer (ITS) region of the ribosomal DNA (rDNA) complex, comprising ITS1-5.8S rDNA-ITS2 within which the sequence of individual species is almost invariant but divergent from other species (6, 8). The consensus primers for this region were originally designed to amplify DNA for sequencing from a range of Naegleria species. The published data suggested to us that the sequence differences in this region might be manifested in distinguishable, species-specific melting curves. In this paper, we report a discriminating and sensitive method for identifying thermophilic Naegleria species, based on real-time PCR coupled with melting-curve analysis of the ITS amplicon using the intercalating dye SYTO9.
MATERIALS AND METHODS
Species and strains.
The genus Naegleria currently includes more than 30 named species and at least an equal number of partially characterized but unnamed groups (8, 25; B. Robinson, unpublished results). For this study, we concentrated on the confirmed and potential pathogens N. fowleri, N. italica, and N. australiensis, and the most frequently detected nonpathogenic species Naegleria lovaniensis and Naegleria carteri. We also characterized the widely studied mesophilic type species Naegleria gruberi, the recently described Naegleria byersi, and the only species in the sister genus Willaertia, Willaertia magna, which is also thermophilic and can be mistaken for a Naegleria species by an inexperienced microscopist. For initial characterization of the melting curve of each species, a single well-established reference strain was used (Table 1). In testing the reproducibility of the melting analysis, we expanded this set of organisms to 20 isolates of N. fowleri, 20 isolates of N. lovaniensis, 20 isolates of N. australiensis, 16 isolates of N. carteri, 5 isolates of N. italica, 3 isolates of N. byersi, 3 isolates of N. gruberi, and 6 isolates of Willaertia magna (all strains from the collection of the Australian Water Quality Centre; full list available from the authors). There are published sequence data for at least one strain in each species set (usually the strain in Table 1), and all others clustered closely with the reference strain by well-validated isozyme criteria (1, 9, 25). Most Australian strains were isolated originally from water samples of 0.5 to 1.0 liter which were coconcentrated with added live Escherichia coli (ATCC 11775) by centrifugation for 15 min at 3,000 relative centrifugal force. The concentrate was plated as an overlay on 1.5% nonnutrient agar (NNA) and incubated for 48 h at 42°C or 44°C for plaque development (Australian Water Quality Centre, unpublished method).
TABLE 1.
Positions of diagnostic melting curve peaks for reference strains of Naegleria species and Willaertia magna
| Species (n) | Strain | Peak (°C) for melting curvea:
|
||
|---|---|---|---|---|
| Tm1 | Tm2 | Tm3 | ||
| N. fowleri (4) | NG166 | 80.46 ± 0.11 | 82.25 ± 0.0 | 84.75 ± 0.0 |
| N. lovaniensis (4) | NG034 | 81.55 ± 0.05 | 84.75 ± 0.0 | |
| N. italica (4) | NG073 | 82.25 ± 0.0 | 84.75 ± 0.0 | |
| N. australiensis (4) | NG035 | 83.75 ± 0.0 | 85.15 ± 0.0 | |
| N. gruberi (4) | NG008 | 82.4 ± 0.0 | 84.85 ± 0.025 | |
| N. byersi (4) | NG597 | 83.64 ± 0.05 | 85.1 ± 0.05 | |
| N. carteri (4) | NG055 | 82.25 ± 0.0 | 83.25 ± 0.0 | 84.33 ± 0.09 |
| Willaertia magna (4) | WT006 | 79.25 ± 0.0 | 82.0 ± 0.0 | 84.8 ± 0.09 |
Highest peaks are shown in boldface type.
Cultivation and DNA extraction.
For preparation of DNA, all strains were cultivated on 90-mm plates containing NNA prespread with E. coli (NNA/E. coli), at 35°C. For qualitative demonstration of amplification and melting analysis, analogous to characterization of a new environmental isolate, DNA was extracted from an undefined number of cells (usually a few 10s) transferred from the edge of a plaque using a sterile, disposable bacteriological loop into 25 μl Instagene (Bio-Rad, Hercules, CA) and following the manufacturer's instructions. For sensitivity experiments, amoebae of selected strains were harvested from confluent NNA/E. coli cultures (approximately 1 × 107 cells) and suspended to 50 ml in PCR buffer II. A 50-μl aliquot was used for a 10-fold serial dilution to determine the actual number of cells by plaque count. The remaining volume was concentrated by centrifugation and DNA extracted from the pellet by boiling for 10 min. After centrifugation for 5 min at 13,000 relative centrifugal force, the supernatant was removed and stored at −20°C, and the volume recorded. DNA dilutions were made where appropriate using nuclease-free water (Promega, Madison, WI).
PCR and melting analysis.
Real-time PCR and melting-curve analysis were carried out in a RotorGene 3000 (Corbett Research, Sydney, Australia). An advantageous feature of this instrument for our purpose is that it permits the operator to manipulate key conditions of the melting analysis, including the temperature interval and the holding time before fluorescence data are collected at each step. The 20-μl reaction mixture consisted of 200 μM concentrations of deoxynucleoside triphosphates (Promega, Madison, WI), 200 nM forward and reverse primers (NGITSF, 5′-AACCTGCGTAGGGATCATTT, and NGITSR, 5′-TTTCCTCCCCTTATTAATAT (6), PCR buffer II, 2.5 mM MgCl2 (Applied Biosystems, Branchburg, NJ), 2.0 μM SYTO9 (Molecular Probes, Eugene, OR), 1 U of AmpliTaq Gold (Applied Biosystems), and 2.0 μl of sample DNA in Instagene supernatant or nuclease-free water. A range of conditions for real-time PCR were tested. Unless otherwise stated, the conditions for experiments reported here were 10 min at 95°C to activate Taq polymerase, followed by 50 cycles of 20 s at 94°C, 20 s at 50°C, and 20 s at 72°C. The ramping between the extension and melting steps included a 1-s pause at 80°C for acquisition of fluorescence data (6-carboxyfluorescein channel, excitation at 470 nm, detection at 510 nm, gain set to maximum of 10). Upon completion of amplification, the program continued directly to a melting-curve analysis in which temperature was ramped from 75 to 95°C in steps of 0.5°C. Fluorescence data were collected after pauses of 60 s on the first step and 20 s on subsequent steps to allow the melting DNA structure to stabilize, again using the 6-carboxyfluorescein channel, but with two gain settings, 3 and 5, to allow for variations in the amount of product amplified. For fine-scale resolution of the melting analysis, data were collected in some experiments at 0.2°C intervals, with pauses of 60 s on the first and 10 s on subsequent steps. The method of analysis of the DNA melting-curve data is critical for the resolution of multiple melting domains. The RotorGene software uses bicubic interpolation to estimate fluorescence between data points when graphing the negative of the first derivative of the raw DNA melting data and provides the option of curve smoothing. For all analyses reported here, no smoothing was used in the generation of melting curves (i.e., the digital filter was set to none).
Sensitivity.
To determine the sensitivity of the method, serial dilutions of DNA extracted from a known number of log-phase cells were run in triplicate in the real-time PCR for N. fowleri NG166 (several experiments using separate DNA extracts), N. lovaniensis, and N. carteri. This quantitative PCR was calibrated to cell number because the copy number of rRNA genes is known only for N. gruberi (5) and because it provided the sensitivity measure most useful in assessing the utility of the assay for identifying environmental isolates. The proprietary software of the RotorGene 3000 calculated the best fit of threshold cycle (CT) as a function of cell number by linear regression to produce a standard curve.
RESULTS
Specificity and reproducibility.
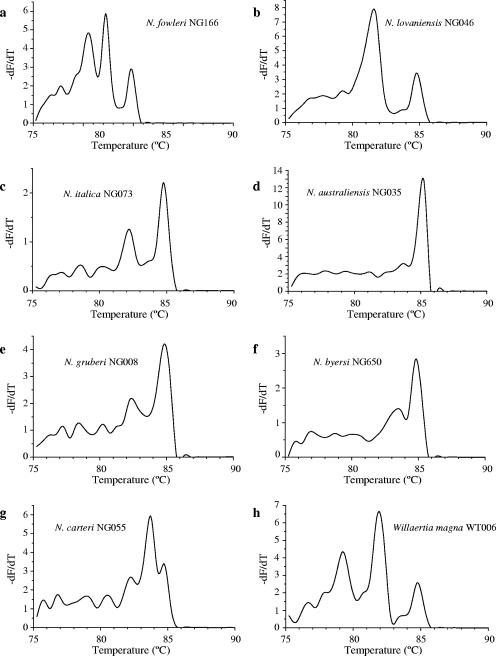
The amplification of the Naegleria ITS region using the published consensus primers (6) was adapted readily to a real-time PCR format. Real-time PCR produced amplicons of the expected size when analyzed by agarose gel electrophoresis (data not shown). DNA from all Naegleria and Willaertia strains amplified satisfactorily, while DNA extracted from the food organism E. coli did not. Melting curves had one to three informative peaks in the interval 79 to 86°C when resolved at 0.5°C intervals (Fig. 1) and were distinguishable among species from the positions and relative heights of the peaks (melting temperatures [Tms] listed in Table 1). For example, N. fowleri gave a melting curve with three peaks (at 80.45°C, 82.25°C, and 84.75°C), with peak 2 consistently highest (Fig. 1a), whereas N. lovaniensis (Fig. 1b) and N. italica (Fig. 1c) gave melting curves with two peaks, but with higher left and right peaks, respectively. The melting curve of N. australiensis (Fig. 1d) was dominated by a large peak, with a small but reproducible peak to the left. N. byersi (Fig. 1f) produced a melting curve similar to that of N. australiensis, but the secondary peak was more prominent. The mesophilic species N. gruberi also produced a melting curve consisting of two peaks (Fig. 1e), similar in position and height to those of N. italica, but with peak 1 reproducibly 0.1 to 0.2°C to the right (mean 82.4°C ± 0.0 versus 82.25°C ± 0.0, n = 4). N. carteri and W. magna gave melting curves with three peaks (Fig. 1g and h), differing in spacing and relative heights from those of N. fowleri. We have annotated the positions of these peaks as Tm1, Tm2, etc. (see justification in Discussion).
FIG. 1.
Melting curves of the 5.8S rDNA/ITS product of seven Naegleria species: N. fowleri (a), N. lovaniensis (b), N. italica (c), N. australiensis (d), N. gruberi (e), N. byersi (f) N. carteri (g), and Willaertia magna (h). Diagnostic features include the number and position of peaks and their relative heights.
Reproducibility was investigated by running four replicate extracts for the principal strains studied. Within-batch standard deviations for Tms were 0.11 or lower (Table 1); in several cases, the machine-interpolated Tms expressed at 0.05°C resolution were indistinguishable. Between PCR mixture batches and between runs, Tms of reference extracts rarely varied outside a 0.2°C range spanning the mean values reported here. During development of the assay, we noted a systematic drift of approximately 0.2°C in Tms that was attributable to differences in temperature calibration between instruments (since corrected) and to a software upgrade that altered the way in which the Rotorgene software calculated the first derivatives from the DNA melting curves. Over this period, Tms for unknown strains were almost invariably within 0.1°C of a reference extract (see “Utility” below). Some exceptions to the position of melting peaks or to the shapes of the curves were observed. Investigation showed that occasional shifts in Tm resulted from failure to remove the Instagene completely following DNA extraction, but other differences were reproducible and appeared to represent novel strains (data not shown). We have not explored the specificity of the PCR itself beyond Naegleria and Willaertia to related genera of amoebae and amoeboflagellates. For the present, therefore, species level identification depends on the initial assignment of the cultured organism to one of these genera.
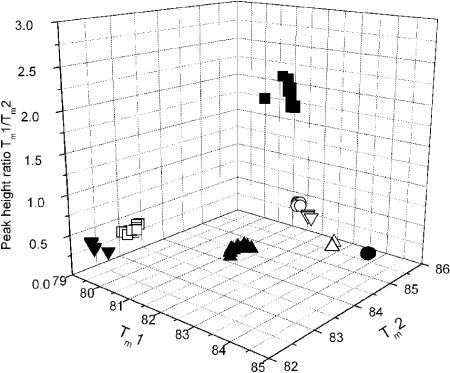
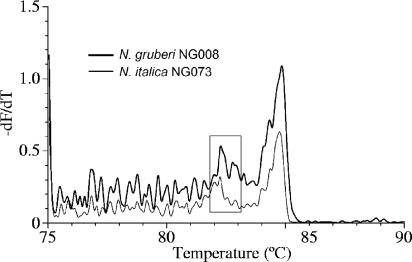
The species specificity of the melting curves is confirmed by their concordance with isozyme- or sequence-based identification for all strains tested (between 3 and 20 per species). The reproducibility among strains is illustrated by plotting Tm1, Tm2, and the ratio of the peak heights in three-dimensional space (Fig. 2): the combination of these key features of the melting curves is clearly discrete for each species. We investigated the small but reproducible difference in the position of peak 1 for N. italica and N. gruberi by running melting analysis at 0.2°C intervals. Each melting curve was resolved into a series of smaller peaks (Fig. 3), which we term cryptic peaks, since they are hidden at the temperature resolution of the usual diagnostic test. In particular, the position of peak 1 at 0.5°C resolution is seen to be determined by the weighted average of several cryptic peaks which have coincident positions but different relative heights in the two species (Fig. 3).
FIG. 2.
Diagnostic features of the melting curves of multiple strains of Naegleria species and Willaertia magna, plotted in three-dimensional space: N. fowleri (□, n = 20), N. lovaniensis (▪, n = 20), N. carteri (▴, n = 16), N. italica (○, n = 5), N. australiensis (•, n = 20), N. gruberi (▿, n = 3), N. byersi (▵, n = 3), and Willaertia magna (▾, n = 6). Note that many of the points are nearly coincident.
FIG. 3.
Melting curves of the 5.8S/ITS product of N. italica NG073 (thin line) and N. gruberi NG008 (thick line) resolved at 0.2°C. The boxed area highlights the cryptic peaks that contribute to the small difference in position of peak 1 at 0.5°C resolution (compare Fig. 1c and e).
Sensitivity.
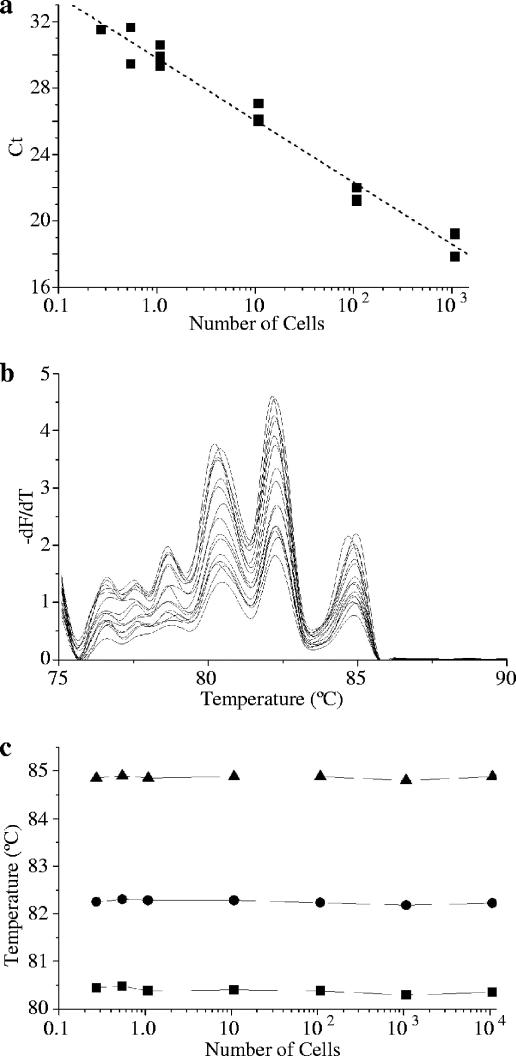
Sensitivity was demonstrated for N. fowleri, N. lovaniensis, and N. carteri by running PCR and melting curve analysis on a serial dilution of DNA extracted from a known number of cells, calibrated by plaque count assay. Since plaque count corresponded closely to a hemocytometer count for log-phase cells, the contribution of nonviable cells to extracted DNA was considered negligible. The cycle at which significant fluorescence appeared (CT) was inversely proportional to the log of the DNA concentration and, hence, to the number of cells (illustrated for N. fowleri in Fig. 4a) (r2 = −0.976). The reaction efficiency calculated from the standard curve was 0.92 for N. fowleri. For each of these species, a product was amplified from extracted DNA equivalent to 0.1 to 0.2 cells, with unambiguously recognizable melting curves from even the lowest concentration of DNA (Fig. 4b). In particular, numerical values of Tm were essentially unaffected by the starting template concentration or by the relative concentration of DNA and SYTO9 (Fig. 4c). The linear relationship of CT to cell number indicates that quantitative PCR is likely to be a valuable tool to measure the growth kinetics of Naegleria species. The method presented here is not suitable for direct quantitation of Naegleria spp. in environmental samples: the use of consensus primers would permit amplification of multiple products from mixed populations, with consequent hybrid melting profiles that would be difficult to interpret.
FIG. 4.
Sensitivity of real-time PCR and integrity of melting curves for N. fowleri NG166. (a) Relationship of CT to DNA concentration, expressed as the log-equivalent number of cells (r = −0.987); (b) melting curves from the corresponding melting analyses; (c) lack of effect of starting template concentration on the position of melting peaks.
Utility.
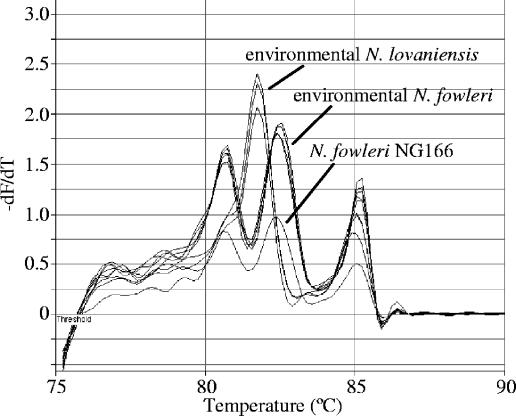
We have tested the method described here with samples from a number of aquatic environments colonized by simple or mixed populations of thermophilic Naegleria species and other amoebae, identifying more than 1,500 Naegleria isolates in total. Figure 5 illustrates the identification of multiple plaques from the quantitative analysis of samples from an industrial cooling tower in which the thermophilic Naegleria community was dominated by N. fowleri (occasionally reaching thousands of organisms per liter) but also included N. lovaniensis. For most sample types, a simple Instagene extraction of a few cells (10s) from the edge of a primary plaque provided suitable DNA for real-time PCR. The major exceptions were samples from cooling systems, among which PCR from primary cultures often failed, probably due to inhibitory materials from the sample (perhaps metal ions) persisting in the overlay in which the plaques grow. In these cases, it was necessary to make a 10-fold dilution of the extracted DNA or to repeat the extraction from a clean subculture for successful amplification and melting analysis.
FIG. 5.
Melting curves of amplified DNA from multiple Naegleria plaques from a survey of an industrial cooling system, showing unambiguous identification of N. fowleri and N. lovaniensis (Rotorgene-generated graphic).
DISCUSSION
Medical interest in the genus Naegleria focuses on the human pathogen N. fowleri and to a lesser extent on the potential pathogens N. australiensis and N. italica. All other well-characterized species, both thermophilic and mesophilic, are benign (7). Species-level radiation in Naegleria has involved adaptation to a series of thermal niches, with the majority of species unable to grow above 40°C (7, 9, 25). Incubation temperature remains the sole basis for partially selective isolation of N. fowleri, and a definitive, genetically sound identification step is still required to distinguish this organism from other thermophilic species.
The current expanded taxonomy of Naegleria, which recognizes more than 30 species, is based on sequence divergence in the 5.8S rRNA gene and the flanking noncoding spacers, particularly the longer ITS2 (7, 8). Comparison of large- and small-subunit (LSU and SSU) sequences returns similar relationships among Naegleria species, but these genes have not been sequenced for some of the more recently named species. Since the rRNA genes in Naegleria and related organisms are encoded on an extrachromosomal circular DNA element with a copy number estimated at 3 × 103 to 5 × 103 in N. gruberi (4, 5), these genes make ideal targets for a sensitive, sequence-based identification method.
While the published primers have been used principally to amplify DNA for sequencing, Pélandakis and Pernin (19) developed a multiplex PCR using these consensus primers to identify Naegleria collectively and species-specific primers to identify N. fowleri. In other published sequence-based identification methods for N. fowleri, the PCR targets have been an ATPase 6-subunit homologue (16, 27) or genes from clone libraries which have unknown functions but have been shown by hybridization to be N. fowleri specific (12, 23).
Recently, there has been a move toward direct detection of N. fowleri by extraction of DNA from environmental samples and amplification by N. fowleri-specific PCR (15, 26). Such a method is attractive because it has the potential to be more rapid, since it eliminates the cultivation step, and because it removes the need for microscopists experienced in the generic identification of Naegleria species. Unfortunately, DNA-based detection also presents several difficulties. First, the potential for coconcentration of environmental materials that are inhibitory to PCR makes it necessary to design inhibition controls to interpret negative results correctly (and, if quantitative PCR is attempted, to ensure that the quantity of the product can be reliably related to a standard curve). Second, the stability of genomic DNA and the high copy number of rDNA targets make the detection of nonviable organisms highly likely; this may not be a problem if the analysis has a forensic purpose but is not helpful in prospective investigations of risk or in the monitoring of water treatment processes. All of the methods reviewed here require gel electrophoresis to verify the PCR product and none identifies Naegleria species other than N. fowleri without additional sequencing.
Our approach to identification is based on an idealization that analysis of environmental samples should detect only viable N. fowleri, that it should provide estimates of organism density, that it should be simple, rapid, and inexpensive, and that, if possible, it should identify the other potential pathogens and benign Naegleria species. The requirements for quantitation and identification of other species have an ecological basis. N. fowleri is far more dynamic in aquatic environments than truly parasitic protozoa such as Cryptosporidium or Giardia, with marked spatial and temporal density variations arising from proliferation, mortality, and conversion between amoeboid and flagellate forms. Observation of these patterns and interpretation of their significance for the risk of infection demands a quantitative approach. The occurrence of other Naegleria species, particularly those in the same or adjoining thermal niches, adds another dimension to ecological questions; understanding the seasonal and spatial distribution of this community of organisms can lead to more intelligent design of investigative and monitoring programs.
Melting-curve analysis is growing in precision as thermal cyclers continue to improve in the precision and reproducibility of temperature ramping. In its simplest form, melting analysis employs an intercalating fluorogenic dye specific for double-stranded DNA, most commonly SYBR green I, to monitor the amount of duplex DNA during both the amplification phase of the real-time PCR and the subsequent controlled melting. Melting of a PCR product is predicted to depend on its size and GC content, and melting temperature (Tm) is increasingly being used to verify that a particular target sequence has been amplified, replacing gel electrophoresis and reducing the repeat handling of samples and PCR products in some assays (24). Under carefully controlled conditions, the separate products of a real-time multiplex PCR can be recognized as separate peaks in a more complex melting curve (17). SYBR green I has a number of limitations, including inhibition of PCR at certain concentrations (17), selective attachment to higher-temperature-melting products, and dye redistribution during melting (10). Exploration of the utility of other dyes, including LC Green (30) and SYTO9 (17), is in its early stages.
A few recent studies have reported the discrimination of closely related microorganisms using SYBR green I and melting analysis coupled to PCR with a single set of consensus primers. Where the PCR product spans a region whose sequence has diverged among the organisms of interest, the differences may be sufficient to be manifested as distinguishable curves. Nicolas et al. (18) amplified part of the Leishmania kinetoplast minicircle DNA (kDNA) that varies in length among species from 650 to 750 bp. The amplicons melted at characteristic and distinguishable Tms for Leishmania major and Leishmania donovani, while Leishmania tropica and Leishmania infantum melted at a third Tm but were not distinguishable from each other. The melting profile was simple, comprising only a single peak for each species, and Tms were reproducible (<1% variation between different runs) and not greatly affected by DNA concentration in the case of L. major. Helps et al. (11) reported the differentiation of feline caliciviruses from the Tms of a 76-bp amplicon spanning a variable region. The melting profile for this amplicon resulted in a single peak, and the maximum variation in Tm between replicates for any given isolate was 0.2°C. The authors did not investigate the effect of different starting numbers of virus on Tm and attributed the variation in Tm to sequence variation between isolates. Most recently, Mangold et al. (14) distinguished four Plasmodium species using the Tms of partial SSU rDNA duplexes amplified using consensus primers and labeled with SYBR green I. As with the other reported assays, the melting profile consisted of a single peak. The variation in Tm among replicates was higher than that reported in the two other studies, and the authors observed differences in Tm between plasmid clones and patient specimens for the same amplicon. The effect of DNA concentration on Tm was not investigated and could have been a contributing factor to the variation that they observed. Melting curve analysis has also been used to differentiate Cryptosporidium species that have different host specificities (13, 28).
In the study reported here, we were able to distinguish the pathogenic Naegleria species N. fowleri, potential pathogens N. italica and N. australiensis, four benign Naegleria species, and the related organism Willaertia magna. This assay represents a considerable advance on other published diagnostic applications of melting analysis described above, which we attribute to the information-rich and reproducible melting profiles observed. There is little recognition in published reports that the step-wise, sequence-dependent melting of double-stranded DNA might be manifested in distinguishable melting maxima within a single amplicon. Indeed, the conventional definition of Tm as “the temperature where one-half of the nucleotides are paired and one-half are unpaired” (29) does not encourage the exploration of this possibility. The complexity of DNA melting has long been recognized in physical chemistry; most of the experimental work in this area has been directed at understanding the subtransitions that occur when specific (usually synthetic) sequences melt over narrow temperature intervals (2). The only significant discussion in the diagnostic literature is that of Wittwer et al. (30), who recognize that distinguishable melting domains of 50 to 300 bp may occur within larger amplicons. Here, we treat the reproducible peaks as local maxima in the melting process, with the annotation Tm1, Tm2, etc.
We believe that the melting curves reported here are more informative than any previously published. This has been made possible by the selection of an appropriate intercalating dye, the size and specific sequences of the PCR products, the temperature resolution (i.e., the interval between fluorescence measurements), and the method of graphing the first derivative of the fluorescence data without smoothing. The latter is particularly important because data smoothing, which is often a default setting of the software used to operate real-time PCR platforms, will result in a loss of peak resolution.
Since a number of experimental variables can influence Tm, including choice of dye, Mg2+ concentration, ramping rate, and holding time at each temperature step, the values reported here are not absolute characteristics of the species studied. Nevertheless, the assay is highly reliable and lends itself readily to making multiple identifications in conjunction with a plaque count method to enable species quantitation. Application to environmental samples has resulted in successful identification of a large number of isolates, with a relatively small number showing divergent melting curves. We suspect that these melting curves represent different species and have archived selected isolates for further study.
At present, the melting domains cannot be assigned to specific sequence domains, but we believe that there are real prospects for doing so. For example, the observation that N. fowleri, which has the most AT-rich ITS1 in the genus (6), also has the most significant leftward (low temperature) peak suggests that this peak may correspond to (or incorporate or overlap with) ITS1. We postulate that the currently recognized N. fowleri genotypes that vary in the number of tandem repeats in ITS1 (6, 20, 33) might be distinguishable in this part of the melting curve. We intend to pursue the theoretical interpretation of the melting curves reported here and to explore the identification of the N. fowleri genotypes and additional Naegleria species by similar criteria.
Acknowledgments
We are grateful to the Australian Water Quality Centre and the South Australian Water Corporation for their support for this project.
REFERENCES
- 1.Adams, M., R. H. Andrews, B. Robinson, P. Christy, P. R. Baverstock, P. J. Dobson, and S. J. Blackler. 1989. A genetic approach to species criteria in the amoeba genus Naegleria using allozyme electrophoresis. Int. J. Parasitol. 19:823-834. [DOI] [PubMed] [Google Scholar]
- 2.Blake, R. D., and S. G. Delcourt. 1998. Thermal stability of DNA. Nucleic Acids Res. 26:3323-3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cabanes, P.-A., F. Wallet, E. Pringuez, and P. Pernin. 2001. Assessing the risk of primary amoebic meningoencephalitis from swimming in the presence of environmental Naegleria fowleri. Appl. Environ. Microbiol. 67:2927-2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clark, C. G., and G. A. M. Cross. 1988. Circular ribosomal RNA genes are a general feature of Schizopyrenid amoebae. J. Protozool. 35:326-329. [DOI] [PubMed] [Google Scholar]
- 5.Clark, C. G., and G. A. M. Cross. 1987. rRNA genes of Naegleria gruberi are carried exclusively on a 14-kilobase-pair plasmid. Mol. Cell. Biol. 7:3027-3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Jonckheere, J. F. 1998. Sequence variation in the ribosomal internal transcribed spacers, including the 5.8S rDNA, of Naegleria spp. Protist 149:221-228. [DOI] [PubMed] [Google Scholar]
- 7.De Jonckheere, J. F. 2002. A century of research on the amoeboflagellate genus Naegleria. Acta Protozool. 41:309-342. [Google Scholar]
- 8.De Jonckheere, J. F. 2004. Molecular definition and the ubiquity of species in the genus Naegleria. Protist 155:89-103. [DOI] [PubMed] [Google Scholar]
- 9.Dobson, P. J., B. S. Robinson, and B. Rowan-Kelly. 1997. New thermophilic Naegleria species (Heterolobosea: Vahlkampfiidae) from Australia and Asia: allozyme, morphometric and physiological characterisation. Acta Protozool. 36:261-271. [Google Scholar]
- 10.Giglio, S., P. T. Monis, and C. P. Saint. 2003. Demonstration of preferential binding of SYBR green I to specific DNA fragments in real-time multiplex PCR. Nucleic Acids Res. 31:e136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Helps, C., P. Lait, S. Tasker, and D. Harbour. 2002. Melting curve analysis of feline calicivirus isolates detected by real-time reverse transcription PCR. J. Virol. Methods 106:241-244. [DOI] [PubMed] [Google Scholar]
- 12.Kilvington, S., and J. Beeching. 1995. Development of a PCR for identification of Naegleria fowleri from the environment. Appl. Environ. Microbiol. 61:3764-3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Limor, J. R., A. A. Lal, and L. Xiao. 2002. Detection and differentiation of Cryptosporidium parasites that are pathogenic for humans by real-time PCR. J. Clin. Microbiol. 40:2335-2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mangold, K. A., R. U. Manson, E. S. C. Koay, L. Stephens, M. Regner, R. B. Thomson, L. R. Peterson, and K. L. Kaul. 2005. Real-time PCR for detection and identification of Plasmodium spp. J. Clin. Microbiol. 43:2435-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marciano-Cabral, F., R. MacLean, A. Mensah, and L. LaPat-Polasko. 2003. Identification of Naegleria fowleri in domestic water sources by nested PCR. Appl. Environ. Microbiol. 69:5864-5869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McLaughlin, G. L., M. H. Vodkin, and H. W. Huizinga. 1991. Amplification of repetitive DNA for the specific detection of Naegleria fowleri. J. Clin. Microbiol. 29:227-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Monis, P. T., S. Giglio, and C. P. Saint. 2005. Comparison of SYTO9 and SYBR green I for real-time polymerase chain reaction and investigation of the effect of dye concentration on amplification and DNA melting curve analysis. Anal. Biochem. 370:24-34. [DOI] [PubMed] [Google Scholar]
- 18.Nicolas, L., G. Milon, and E. Prina. 2002. Rapid differentiation of Old World Leishmania species by LightCycler polymerase chain reaction and melting curve analysis. J. Microbiol. Methods 51:295-299. [DOI] [PubMed] [Google Scholar]
- 19.Pélandakis, M., and P. Pernin. 2002. Use of multiplex PCR and PCR restriction enzyme analysis for detection and exploration of the variability in the free-living amoeba Naegleria in the environment. Appl. Environ. Microbiol. 68:2061-2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pelandakis, M., S. Serre, and P. Pernin. 2000. Analysis of the 5.8S rRNA gene and the internal transcribed spacers in Naegleria spp. and in N. fowleri. J. Eukaryot. Microbiol. 47:116-121. [DOI] [PubMed] [Google Scholar]
- 21.Piatek, A. S., S. Tyagi, A. C. Pol, A. Telenti, L. P. Miller, F. R. Kramer, and D. Alland. 1998. Molecular beacon sequence analysis for detecting drug resistance in Mycobacterium tuberculosis. Nat. Biotechnol. 16:359-363. [DOI] [PubMed] [Google Scholar]
- 22.Rauter, C., R. Oehme, I. Diterich, M. Engele, and T. Hartung. 2002. Distribution of clinically relevant Borrelia genospecies in ticks assessed by a novel, single-run, real-time PCR. J. Clin. Microbiol. 40:36-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Réveiller, F. L., P. A. Cabanes, and F. Marciano-Cabral. 2002. Development of a nested PCR assay to detect the pathogenic free-living amoeba Naegleria fowleri. Parasitol. Res. 88:443-450. [DOI] [PubMed] [Google Scholar]
- 24.Ririe, K. M., R. P. Rasmussen, and C. T. Wittwer. 1997. Product differentiation by analysis of DNA melting curves during the polymerase chain reaction. Anal. Biochem. 245:154-160. [DOI] [PubMed] [Google Scholar]
- 25.Robinson, B. S., P. Christy, S. J. Hayes, and P. J. Dobson. 1992. Discontinuous genetic variation among mesophilic Naegleria isolates: further evidence that N. gruberi is not a single species. J. Protozool. 39:702-712. [DOI] [PubMed] [Google Scholar]
- 26.Sheehan, K. B., J. A. Fagg, M. J. Ferris, and J. M. Henson. 2003. PCR detection and analysis of the free-living amoeba Naegleria in hot springs in Yellowstone and Grand Teton National Parks. Appl. Environ. Microbiol. 69:5914-5918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sparagano, O. 1993. Differentiation of Naegleria fowleri and other Naegleriae by polymerase chain reaction and hybridization methods. FEMS Microbiol. Lett. 110:325-330. [DOI] [PubMed] [Google Scholar]
- 28.Tanriverdi, S., A. Tanyeli, F. Baslamisli, F. Koksal, Y. Kilinc, X. Feng, G. Batzer, S. Tzipori, and G. Widmer. 2002. Detection and genotyping of oocysts of Cryptosporidium parvum by real-time PCR and melting curve analysis. J. Clin. Microbiol. 40:3237-3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.von Ahsen, N., C. T. Wittwer, and E. Schutz. 2001. Oligonucleotide melting temperatures under PCR conditions: nearest-neighbor corrections for Mg2+, deoxynucleotide triphosphate, and dimethyl sulfoxide concentrations with comparison to alternative empirical formulas. Clin. Chem. 47:1956-1961. [PubMed] [Google Scholar]
- 30.Wittwer, C. T., G. H. Reed, C. N. Gundry, J. G. Vandersteen, and R. J. Pryor. 2003. High-resolution genotyping by amplicon melting analysis using LCGreen. Clin. Chem. 49:853-860. [DOI] [PubMed] [Google Scholar]
- 31.Wolk, D. M., S. K. Schneider, N. L. Wengenack, L. M. Sloan, and J. E. Rosenblatt. 2002. Real-time PCR method for detection of Encephalitozoon intestinalis from stool specimens. J. Clin. Microbiol. 40:3922-3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Woo, T. H., B. K. Patel, L. D. Smythe, M. L. Symonds, M. A. Norris, R. S. Weyant, and M. F. Dohnt. 1998. Identification of Leptospira inadai by continuous monitoring of fluorescence during rapid cycle PCR. Syst. Appl. Microbiol. 21:89-96. [DOI] [PubMed] [Google Scholar]
- 33.Zhou, L., R. Sriram, G. S. Visvesvara, and L. Xiao. 2003. Genetic variations in the internal transcribed spacer and mitochondrial small subunit rRNA gene of Naegleria spp. J. Eukaryot. Microbiol. 50:522-526. [DOI] [PubMed] [Google Scholar]