Abstract
Bacteriophage CEV1 was isolated from sheep resistant to Escherichia coli O157:H7 colonization. In vitro, CEV1 efficiently infected E. coli O157:H7 grown both aerobically and anaerobically. In vivo, sheep receiving a single oral dose of CEV1 showed a 2-log-unit reduction in intestinal E. coli O157:H7 levels within 2 days compared to levels in the controls.
Escherichia coli O157:H7 is a highly virulent food-borne pathogen with an infective dose of ∼100 cells in humans (5, 25). Infections can be severe; symptoms include hemorrhagic colitis, hemolytic-uremic syndrome, thrombotic thrombocytopenic purpura, and even death (23-25, 31). Although E. coli O157:H7 infections can be linked to a variety of sources, primary contamination is usually ruminant related; cattle and other ruminants act as asymptomatic reservoirs (2, 6, 7, 11, 12, 15, 17, 30). Despite extensive intervention strategies, human E. coli O157:H7 infections still occur (7, 9, 14). Thus, recent research has focused on the development of preharvest interventions to reduce the entry of the pathogen into the food chain. Bacteriophages have been used successfully as antibacterial agents in both human and veterinary medicine and are one potential preharvest E. coli O157:H7 control strategy (4, 8, 18, 20, 22, 28, 32-34, 39). In this study we describe a new T4-like bacteriophage, CEV1, isolated from sheep resistant to colonization by E. coli O157:H7, and we discuss its potential to control this pathogen.
Isolation and characterization of bacteriophage CEV1.
Crossbred ewes (n = 22) were orally inoculated with ∼109 CFU/sheep of novobiocin-resistant E. coli O157:H7 (NCTC 12900). In the first 2 days following inoculation, these sheep fecally shed ∼102 CFU/g 12900. By day 3, no viable 12900 was detected. Each sheep was rechallenged with ∼1011 CFU 12900 and monitored, with similar results. The sheep were then challenged with E. coli O157:H7 EDL 933 (109 CFU/sheep). Again, fecal shedding decreased rapidly, and by day 4, EDL933 levels were below the detection limit. These results were unprecedented at our facility, where both strains are routinely used in vivo (typically ∼104 CFU/g is shed fecally for ∼10 days). Feces collected from these ewes tested positive for E. coli O157:H7-infecting phage. By using our standard enrichment protocol, a new bacteriophage, CEV1, was isolated (10).
CEV1 was active against 17/19 E. coli O157:H7 strains tested, 9 strains from 4/5 phylogenetic groups of the E. coli reference (ECOR) collection, lab strains B and K-12, and strains from serogroups O43, O126, O153, O158, and Omulti (Table 1) (27). No representatives from serogroups O15 and O50 were lysed by CEV1. Using a set of isogenic E. coli K-12 mutant pairs, with one member of each pair mutant for a specific outer membrane protein, it was found that CEV1 uses ompA as its receptor (data not shown) (16).
TABLE 1.
Host range of bacteriophage CEV1, as determined by spot testsa
| Type of bacteria | CEV1-sensitive strains | CEV1-resistant strains |
|---|---|---|
| Pathogenic | E. coli O157:H7: EDL 933 (ATCC 43895)b; FDIU | E. coli O157:H7: FDIU strains 207 and 2336 |
| strains 86-24, 2027, 2028, 2029, 2030, 2031, 2255, | E. coli O15:H7 | |
| 2257, 2266, 2309, 2317, 2321, 2324, 6058 | E. coli O15:H25 | |
| E. coli O43:H7 | E. coli O50:H7 | |
| E. coli O126:H7 | Salmonella enterica serovar Newport | |
| E. coli O153:H7 | Salmonella enterica serovar Dublin | |
| E. coli O158:H7 | Salmonella enterica serovar Derby | |
| E. coli Omulti:H7 | Salmonella enterica serovar Enteritidis | |
| Salmonella enterica serovar Typhimurium | ||
| Nonpathogenic | E. coli O157:H7: NCTC 12900 (ATCC 700728)c; 87-23c | ECOR: 1 to 3; 5 to 7, 9, 10, 12, 15, 17 to 32, 34 to 47, |
| ECOR: 4, 8, 11, 13, 14, 16, 33, 48, 56 | 49 to 55, 57 to 72 | |
| E. coli K-12, B |
The efficiencies of plating for all phage-sensitive strains were generally similar to those on B and K12; none differed by more than 1 order of magnitude.
Toxigenic E. coli O157:H7 strain used throughout this study both in vitro and in vivo.
Nontoxigenic biosafety level-1 E. coli O157:H7 strains used throughout this study both in vitro and in vivo. These strains are serologically and physiologically wild-type O157:H7 strains but have the pathogenicity island prophage deleted (Stx−).
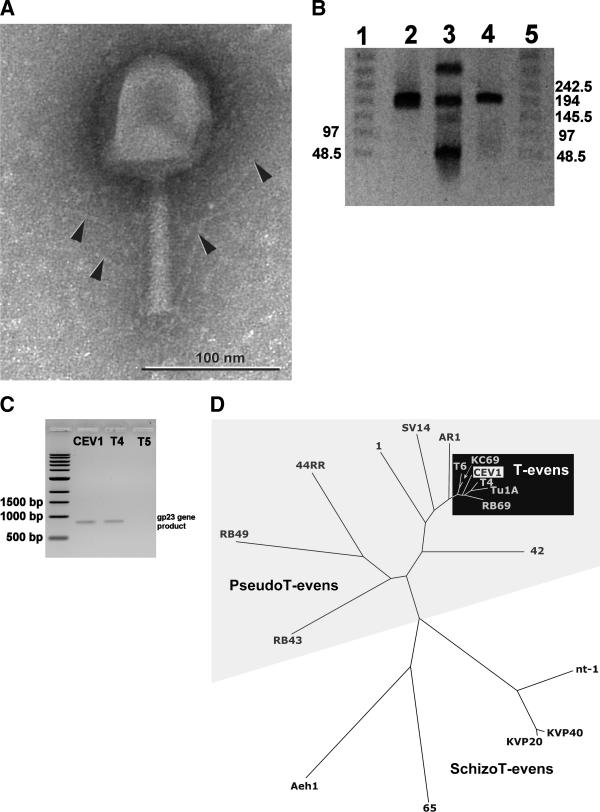
Electron micrographs (Fig. 1A) showed that CEV1 has an icosahedral head, linked by a connector to a contractile tail with fibers at the distal end. The CEV1 genome of ∼180 kb (Fig. 1B) is digested by EcoRV and NdeI but not by EcoRI, BamHI, PstI, SalI, or HindIII, suggesting that CEV1 DNA contains a modified base, probably 5-hydroxymethyl cytosine (data not shown). Based on its morphology and the sequence of a 788-nucleotide DNA fragment (GenBank accession no. AY331985) internal to the major capsid protein gp23 (Fig. 1C), CEV1 is closely related to T4 (95% sequence similarity) (Fig. 1D) (40).
FIG. 1.
Characterization of phage CEV1. (A) Electron micrograph of bacteriophage CEV1. Arrows indicate tail fibers. Dimensions: head, 100 nm by 85 nm; contractile tail, 100 nm by 15 nm. (B) Pulsed-field gel electrophoresis of phage DNA, as observed on an ethidium bromide-stained low-melting-point agarose gel (1%, wt/vol)(0.5× TBE, consisting of 45 mM Tris-borate and 1 mM EDTA [pH 8]) run for 20 h at 6 V/cm and 14°C with a pulse period of 45 to 90 s. Lanes 1 and 5, λ DNA ladder (Bio-Rad); lane 2, T4 DNA (175 kb); lane 3, DNA ladder composed of phage (Pseudomonas φ2a [290 kb], T4 [175 kb], CEV2 [120 kb], and Pseudomonas φ2b [45 kb]) DNA; lane 4, CEV1 DNA. (C) Ethidium bromide-stained agarose (1%, wt/vol) gel of DNA-PCR amplification reactions using primers Cap8 (TGAAGTTACCTTCACCACGACCGG) and Mzia1 (TGTTATIGGTATGGTICGICGTGCTAT) for gp23 (40). First lane, 1-kb DNA ladder (Invitrogen, CA); second lane, phage CEV1; third lane, phage T4 (positive control); fourth lane, phage T5 (negative control). (D) Unrooted phylogenetic tree constructed using the more variable internal region (amino acids 115 to 302) of the gp23 head protein of a number of T4-like phages, including T4 (Protein Data Bank accession no. NP049787), T6 (Q38055), KC69 (AAF61696), Tu1a (AAF61691), SV14 (CAB01542), RB69 (AAF61699), RB49 (CAB01539), RB43 (AAF61698), and AR1 (Q9ZXI0); Enterobacter cloacae phage 1 (AAF61700); Aeromonas salmonicida phages 44RR2.8t (44RR; AAF61693) and 65 (AAF61694); Aeromonas hydrophila phage Aeh1 (AAF61695); Burkholderia cepacia phage 42 (AAF61692); Vibrio natriegens phage nt-1 (AAF61697); and Vibrio parahaemolyticus phages KVP20 (BAA25880) and KVP40 (BAA25567). Subgroups of phage types are indicated by shading: white, schizo T-evens; light gray, pseudo T-evens; black, T-evens.
Infection experiments.
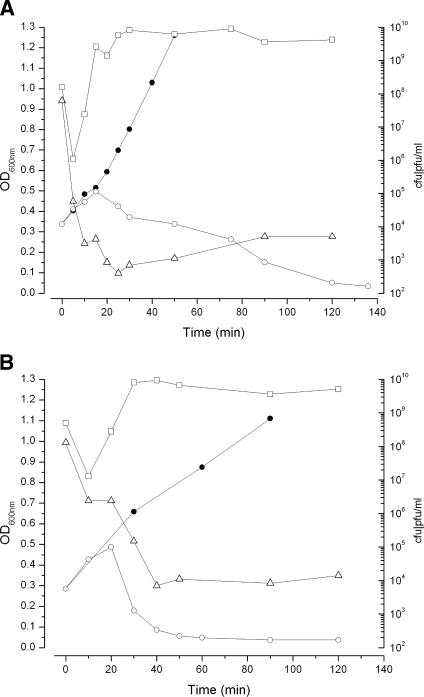
Both aerobic and anaerobic CEV1 infections of 12900 were productive. Aerobic infections of 12900 (optical density at 600 nm [OD600], ∼0.3) were carried out in shake flasks containing tryptic soy broth (TSB) at 37°C with agitation. CEV1 displayed an eclipse period of 18 min, a latent period of 26 min, and a burst size of ∼150 PFU/cell in experiments at a low multiplicity of infection (MOI; ∼0.1) (data not shown). At an MOI of ∼5, >99% of the phages adsorbed, the bacterial population was reduced 5 log units, and progeny phages were observed within 20 min (upon treatment with CHCl3, the burst size was ∼100 PFU/cell) (Fig. 2A). Limited and variable lysis inhibition was observed; lysis began between 30 to 120 min, depending on conditions and the MOI, and took 30 to 60 min to reach completion. Anaerobic infections of 12900 were conducted using adaptations of the Hungate technique (N2 headspace) in 126-ml butyl rubber-sealed serum bottles containing TSB. Within 5 min, >97% of the phages adsorbed, and by 20 min, levels of bacterial survivors fell 4 log units (Fig. 2B). Similarly efficient anaerobic infections were observed in bovine rumen fluid and colon-simulating media (data not shown) (26).
FIG. 2.
CEV1 infections of E. coli O157:H7 NCTC 12900 growing in TSB under conditions of either aerobic respiration (A) or fermentation (B). Infection graphs shown are representative of at least three replicates. Symbols: •, OD600 of controlcultures; ○, OD600 of infected cultures; ▵, CFU of bacterial survivors/ml; □, PFU of phage/ml after the addition of chloroform.
Animal studies.
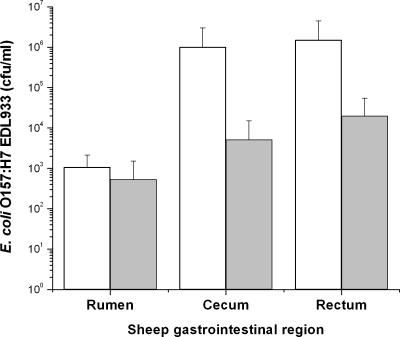
Animal studies were carried out humanely (Southern Plains Agricultural Research Center Institutional Animal Care and Use Committee protocol 01-002). Prior to experimental treatment, 39 crossbred ewes from one flock were screened for O157:H7-infecting phages by using standard enrichment techniques; 19 tested negative (10). The others all contained the same new O157:H7-infecting phage (termed CEV2 and currently undergoing characterization). Sheep (n = 8) that tested negative were inoculated (by oral gavage) with ∼1010 CFU of novobiocin/nalidixic acid-resistant 933; after 3 days, 4/8 sheep were treated with a single oral dose of CEV1 (∼1011 PFU). The animals were humanely euthanized 2 days later; bacterial counts (by plating on MacConkey agar containing 20 μg/ml novobiocin and 25 μg/ml nalidixic acid) of ruminal, cecal, and rectal contents showed that 933 levels were reduced 2 to 3 log units (P > 0.05) in the ceca and rectums of CEV1-treated sheep compared to levels in controls (Fig. 3).
FIG. 3.
Use of CEV1 as a preslaughter treatment to remove resident E. coli O157:H7 from the intestines of ruminants (sheep). Control sheep (white bars) received only E. coli O157:H7 orally on day zero, while experimental sheep (gray bars) orally received E. coli O157:H7 on day zero and then CEV1 on day 3. Data were collected postmortem from various parts of the animals' digestive tracts 5 days after O157:H7 inoculation and 2 days after phage treatment. Error bars, standard deviations.
Over the years, bacteriophages have frequently been isolated from the intestinal tracts of ruminants (1, 19, 29). Swain et al. (1996) studied bovine ruminal phage and concluded that phage help maintain microbial diversity and balance (37). Here we suggest that phage also play a role in controlling the microbial ecology in the colons of livestock—a role that could be adapted as a preharvest treatment to remove E. coli O157:H7 from sheep and other ruminants. In this study, we found two sheep flocks from different regions that harbored O157:H7-infecting phage (group 1 was the CEV1 isolation flock, while group 2 was the CEV1 treatment test flock [20/39 sheep harbored CEV2]). CEV1 was isolated from sheep that cleared E. coli O157:H7 rapidly and were therefore “resistant” to colonization. In view of the origin of CEV1 and its demonstrated in vitro and in vivo activity against these strains, its presence in these sheep is a logical explanation for these atypical results. These data emphasize the importance of screening for phage by enrichment prior to in vivo control/eradication studies.
Extensive characterization of candidate phages is needed before any widespread application is implemented. Host selectivity is key; CEV1 lyses most E. coli O157:H7 strains and a limited number of commensal E. coli strains. This will create minimal disturbance to the gut biota, allowing the simultaneous use of probiotic E. coli to replace O157:H7 within the gastrointestinal niche and/or to maintain sufficient CEV1 levels to clear very low levels of E. coli O157:H7.
Our finding that CEV1 is a close relative of T4 is advantageous; T4-like phages have a long history of safe application and have been intensively studied for >50 years (13, 21, 35, 36). They are exclusively lytic, and none encode pathogenicity islands, virulence/antibiotic resistance genes, or integrases. The ability of CEV1 to efficiently infect E. coli O157:H7 is, however, unusual among T4-like phages; of the ∼100 such phages from our collection, only one infects E. coli O157:H7 at an efficiency of >0.001 (data not shown). Two other reported T4-like O157:H7-infecting phages (AR1 and PPO1) do not infect lab strains B and K-12, unlike CEV1 (AR1 infects 38/72 ECOR strains, as well as some Proteus, Shigella, and Salmonella strains; PPO1 infects 8/72 ECOR strains, but with a specificity pattern different from that of CEV1 (data not shown) (16).
Previous researchers have discussed the applications of O157:H7-infecting phages, but each had some physiological constraint that limited its application. O'Flynn et al. (2004) used a phage cocktail (PPO1, e4/1c, and e11/2) to eliminate E. coli O157:H7 from artificially contaminated meat (28). This postslaughter approach would initially appear promising in reducing the postplant E. coli O157:H7 load, but this application appears impractical (an MOI of ∼106 and a temperature of 37°C were used; no phage infection occurred at lower temperatures). In contrast, our suggested approach with CEV1 is to treat the live animal and thus reduce E. coli O157:H7 populations before entry into the food chain, allowing the use of phage in conjunction with other methods to create a multilevel barrier.
Other preharvest E. coli O157:H7 phage studies have failed to demonstrate efficacy under real-world conditions. For example, Kudva et al. reported in 1999 that their O157:H7-infecting phages effectively killed EDL 932, but only aerobically (MOI, ∼103); thus, these phage were inappropriate for use in the anaerobic gut (20). Bach et al. (2003) isolated phage DC22, which infected all 23 O157:H7 strains tested in the lab but showed little or no efficacy in ruminant gut-simulating chemostats or in lambs (3). This reinforces the importance of understanding infection parameters in vitro under conditions relevant to the ecosystem where the phage will be used. Tanji et al. (2005) found that a three-phage cocktail worked effectively in vitro (aerobically and anaerobically) but was unable to clear mice of E. coli during in vivo studies, again indicating the complexity of this problem (38). In our study, infection of 12900 by CEV1 was productive in TSB aerobically and anaerobically, as well as in both defined and gut-simulating media (data not shown). Further, our in vivo trials show a substantial reduction in intestinal E. coli O157:H7 levels in sheep treated with a single oral dose of CEV1, suggesting that CEV1 shows promise as a component in a treatment for the selective reduction of E. coli O157:H7 levels in food animals.
In summary, although the United States is considered to have one of the world's safest food supply systems, people still become ill from bacterially contaminated food. Our results indicate that phages can reduce intestinal populations of E. coli O157:H7 and could play an important role in future preharvest pathogen reduction strategies. Furthermore, in studies attempting to use intestinal ecological modification, the potential impact of naturally resident phage must not be overlooked.
Acknowledgments
We thank Robert Droleskey for the electron micrographs; Charles Hernandez, Anna Castano, and Luki Goldschmidt for assistance with the PFGE experiments; Travis Steiner, Sarah Perigo, Joe Jardine, Gautam Dutta, Naomi Hoyle, and Matthew Robison, the undergraduates who helped in many experiments throughout this project; Burt Guttman for ongoing helpful discussions; Dan Rice (FDIU strains), Francisco Diez-Gonzalez (strains 86-24 and K-12), and Alison O'Brien (strain 87-23) for kindly providing E. coli O157:H7 strains; and Yasunori Tanji and Larry Goodridge for sharing their E. coli O157:H7-targeting T4-like phages and unpublished data. R.R.R. and R.A.O. thank the personnel and researchers at the USDA Food and Feed Safety Research Unit at College Station, Tex., for their hospitality.
This work was kindly supported by NIH grants 1-R-15 GM63507 and 2-R15 GM063637-02 and by the USDA/ARS.
REFERENCES
- 1.Adams, J. C., J. A. Gazaway, M. D. Brailsford, P. A. Hartman, and N. L. Jacobson. 1966. Isolation of bacteriophages from the bovine rumen. Experientia 22:717-718. [Google Scholar]
- 2.Armstrong, G. L., J. Hollingsworth, and J. G. Morris, Jr. 1996. Emerging foodborne pathogens: Escherichia coli O157:H7 as a model of entry of a new pathogen into the food supply of the developed world. Epidemiol. Rev. 18:29-51. [DOI] [PubMed] [Google Scholar]
- 3.Bach, S. A., T. A. McAllister, D. M. Veira, V. P. J. Gannon, and R. A. Holley. 2003. Effect of bacteriophage DC22 on Escherichia coli O157:H7 in an artificial rumen system (Rusitec) and inoculated sheep. Anim. Res. 52:89-101. [Google Scholar]
- 4.Barrow, P., M. Lovell, and A. Berchieri. 1998. Use of lytic bacteriophage for control of experimental Escherichia coli septicemia and meningitis in chickens and calves. Clin. Diagn. Lab. Immunol. 5:294-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Besser, R. E., P. M. Griffin, and L. Slutsker. 1999. Escherichia coli O157:H7 gastroenteritis and the hemolytic uremic syndrome: an emerging infectious disease. Annu. Rev. Med. 50:355-367. [DOI] [PubMed] [Google Scholar]
- 6.Bielaszewska, M., H. Schmidt, A. Liesegang, R. Prager, W. Rabsch, H. Tschape, A. Cizek, J. Janda, K. Blahova, and H. Karch. 2000. Cattle can be a reservoir of sorbitol-fermenting Shiga toxin-producing Escherichia coli O157:H− strains and a source of human diseases. J. Clin. Microbiol. 38:3470-3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brabban, A. D., D. A. Nelson, E. Kutter, T. S. Edrington, and T. R. Callaway. 2004. Approaches to controlling Escherichia coli O157:H7, a food-borne pathogen and an emerging environmental hazard. Environ. Pract. 6:208-229. [Google Scholar]
- 8.Callaway, T. R., R. C. Anderson, T. S. Edrington, K. J. Genovese, K. M. Bischoff, T. L. Poole, Y. S. Jung, R. B. Harvey, and D. J. Nisbet. 2004. What are we doing about Escherichia coli O157:H7 in cattle? J. Anim. Sci. 82:E93-E99. [DOI] [PubMed] [Google Scholar]
- 9.Callaway, T. R., R. O. Elder, J. E. Keen, R. C. Anderson, and D. J. Nisbet. 2003. Forage feeding to reduce preharvest Escherichia coli populations in cattle: a review. J. Dairy Sci. 86:852-860. [DOI] [PubMed] [Google Scholar]
- 10.Carlson, K. 2005. Appendix. Working with bacteriophages: common techniques and methodological approaches, p. 437-494. In E. Kutter and A. Sulakvelidze (ed.), Bacteriophages: biology and applications. CRC Press, Boca Raton, Fla.
- 11.CDC. 2000. Summary of outbreaks of Escherichia coli O157 and other Shiga toxin-producing E. coli reported to the CDC in 1999. [Online.] http://www.cdc.gov/NCIDOD/DBMD/diseaseinfo/files/ecoli_99summary.pdf.
- 12.Chapman, P. A., C. A. Siddons, D. J. Wright, P. Norman, J. Fox, and E. Crick. 1993. Cattle as a possible source of verocytotoxin-producing Escherichia coli O157 infections in man. Epidemiol. Infect. 111:439-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chibani-Chennoufi, S., A. Bruttin, M. L. Dillmann, and H. Brussow. 2004. Phage-host interaction: an ecological perspective. J. Bacteriol. 186:3677-3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elder, R. O., J. E. Keen, G. R. Siragusa, G. A. Barkocy-Gallagher, M. Koohmaraie, and W. W. Laegreid. 2000. Correlation of enterohemorrhagic Escherichia coli O157 prevalence in feces, hides, and carcasses of beef cattle during processing. Proc. Natl. Acad. Sci. USA 97:2999-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fischer, J. R., T. Zhao, M. P. Doyle, M. R. Goldberg, C. A. Brown, C. T. Sewell, D. M. Kavanaugh, and C. D. Bauman. 2001. Experimental and field studies of Escherichia coli O157:H7 in white-tailed deer. Appl. Environ. Microbiol. 67:1218-1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goodridge, L., A. Gallaccio, and M. W. Griffiths. 2003. Morphological, host range, and genetic characterization of two coliphages. Appl. Environ. Microbiol. 69:5364-5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grauke, L. J., I. T. Kudva, J. W. Yoon, C. W. Hunt, C. J. Williams, and C. J. Hovde. 2002. Gastrointestinal tract location of Escherichia coli O157:H7 in ruminants. Appl. Environ. Microbiol. 68:2269-2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greer, G. G. 2005. Bacteriophage control of foodborne bacteria. J. Food Prot. 68:1102-1111. [DOI] [PubMed] [Google Scholar]
- 19.Klieve, A. V., and T. Bauchop. 1988. Morphological diversity of ruminal bacteriophages from sheep and cattle. Appl. Environ. Microbiol. 54:1637-1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kudva, I. T., S. Jelacic, P. I. Tarr, P. Youderian, and C. J. Hovde. 1999. Biocontrol of Escherichia coli O157 with O157-specific bacteriophages. Appl. Environ. Microbiol. 65:3767-3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kutter, E., K. Gachechiladze, A. Poglazov, E. Marusich, M. Shneider, P. Aronsson, A. Napuli, D. Porter, and V. Mesyanzhinov. 1995. Evolution of T4-related phages. Virus Genes 11:285-297. [DOI] [PubMed] [Google Scholar]
- 22.Loc Carrillo, C., R. J. Atterbury, A. El-Shibiny, P. L. Connerton, E. Dillon, A. Scott, and I. F. Connerton. 2005. Bacteriophage therapy to reduce Campylobacter jejuni colonization of broiler chickens. Appl. Environ. Microbiol. 71:6554-6563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mead, P. S., and P. M. Griffin. 1998. Escherichia coli O157: H7. Lancet 352:1207-1212. [DOI] [PubMed] [Google Scholar]
- 24.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nisbet, D. J., D. E. Corrier, C. M. Scanlan, A. G. Hollister, R. C. Beier, and J. R. DeLoach. 1993. Effect of a defined continuous-flow derived bacterial culture and dietary lactose on Salmonella typhimurium colonization in broiler chickens. Avian Dis. 37:1017-1025. [PubMed] [Google Scholar]
- 27.Ochman, H., and R. K. Selander. 1984. Standard reference strains of Escherichia coli from natural populations. J. Bacteriol. 157:690-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'Flynn, G., R. P. Ross, G. F. Fitzgerald, and A. Coffey. 2004. Evaluation of a cocktail of three bacteriophages for biocontrol of Escherichia coli O157:H7. Appl. Environ. Microbiol. 70:3417-3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Orpin, C. G., and E. A. Munn. 1973. The occurrence of bacteriophages in the rumen and their influence on rumen bacterial populations. Experientia 30:1018-1020. [DOI] [PubMed] [Google Scholar]
- 30.Rasmussen, M. A., W. C. Cray, T. A. Casey, and S. C. Whipp. 1993. Rumen contents as a reservoir of enterohemorrhagic Escherichia coli. FEMS Microbiol. Lett. 114:79-84. [DOI] [PubMed] [Google Scholar]
- 31.Riley, L. W., R. S. Remis, S. D. Helgerson, H. B. McGee, J. G. Wells, B. R. Davis, R. J. Hebert, E. S. Olcott, L. M. Johnson, N. T. Hargrett, P. A. Blake, and M. L. Cohen. 1983. Hemorrhagic colitis associated with a rare Escherichia coli serotype. N. Engl. J. Med. 308:681-685. [DOI] [PubMed] [Google Scholar]
- 32.Smith, H. W., and M. B. Huggins. 1983. Effectiveness of phages in treating experimental Escherichia coli diarrhoea in calves, piglets and lambs. J. Gen. Microbiol. 129:2659-2675. [DOI] [PubMed] [Google Scholar]
- 33.Smith, H. W., M. B. Huggins, and K. M. Shaw. 1987. The control of experimental Escherichia coli diarrhoea in calves by means of bacteriophages. J. Gen. Microbiol. 133:1111-1126. [DOI] [PubMed] [Google Scholar]
- 34.Smith, H. W., M. B. Huggins, and K. M. Shaw. 1987. Factors influencing the survival and multiplication of bacteriophages in calves and in their environment. J. Gen. Microbiol. 133:1127-1135. [DOI] [PubMed] [Google Scholar]
- 35.Sulakvelidze, A., and P. Barrow. 2005. Phage therapy in animals and agribusiness, p. 335-380. In E. Kutter and A. Sulakvelidze (ed.), Bacteriophages: biology and applications. CRC Press, Boca Raton, Fla.
- 36.Sulakvelidze, A., and E. Kutter. 2005. Bacteriophage therapy in humans, p. 381-436. In E. Kutter and A. Sulakvelidze (ed.), Bacteriophages: biology and applications. CRC Press, Boca Raton, Fla.
- 37.Swain, R. A., J. V. Nolan, and A. V. Klieve. 1996. Natural variability and diurnal fluctuations within the bacteriophage population of the rumen. Appl. Environ. Microbiol. 62:994-997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tanji, Y., T. Shimada, H. Fukudomi, K. Miyanaga, Y. Nakai, and H. Unno. 2005. Therapeutic use of phage cocktail for controlling Escherichia coli O157:H7 in gastrointestinal tract of mice. J. Biosci. Bioeng. 100:280-287. [DOI] [PubMed] [Google Scholar]
- 39.Tanji, Y., T. Shimada, M. Yoichi, K. Miyanaga, K. Hori, and H. Unno. 2004. Toward rational control of Escherichia coli O157:H7 by a phage cocktail. Appl. Microbiol. Biotechnol. 64:270-274. [DOI] [PubMed] [Google Scholar]
- 40.Tetart, F., C. Desplats, M. Kutateladze, C. Monod, H.-W. Ackermann, and H. M. Krisch. 2001. Phylogeny of the major head and tail genes of the wide-ranging T4-type bacteriophages. J. Bacteriol. 183:358-366. [DOI] [PMC free article] [PubMed] [Google Scholar]