Abstract
In order to investigate the prevalence of tick-borne infectious agents among ticks, ticks comprising five species from two genera (Hemaphysalis spp. and Ixodes spp.) were screened using molecular techniques. Ticks (3,135) were collected from small wild-caught mammals or by dragging/flagging in the Republic of Korea (ROK) and were pooled into a total of 1,638 samples (1 to 27 ticks per pool). From the 1,638 tick samples, species-specific fragments of Anaplasma phagocytophilum (1 sample), Anaplasma platys (52 samples), Ehrlichia chaffeensis (29 samples), Ehrlichia ewingii (2 samples), Ehrlichia canis (18 samples), and Rickettsia rickettsii (28 samples) were amplified by PCR assay. Twenty-one pooled and individual tick samples had mixed infections of two (15 samples) or three (6 samples) pathogens. In addition, 424 spleen samples from small captured mammals (389 rodents, 33 insectivores, and 2 weasels) were screened for selected zoonotic pathogens. Species-specific DNA fragments of A. phagocytophilum (110 samples), A. platys (68 samples), E. chaffeensis (8 samples), E. ewingii (26 samples), E. canis (51 samples), and Rickettsia sp. (22 samples) were amplified by PCR assay. One hundred thirty small mammals had single infections, while 4, 14, and 21 striped field mice (Apodemus agrarius) had mixed infections of four, three, and two pathogens, respectively. Phylogenetic analysis based on nucleotide sequence comparison also revealed that Korean strains of E. chaffeensis clustered closely with those from China and the United States, while the Rickettsia (rOmpA) sequences clustered within a clade together with a Chinese strain. These results suggest that these agents should be considered in differential diagnosis while examining cases of acute febrile illnesses in humans as well as animals in the ROK.
Ticks are notorious vectors of various pathogenic protozoa, rickettsiae, bacteria, and viruses that cause serious and life-threatening illnesses in humans and animals worldwide (2, 11, 15, 37, 40, 47). Screening of ticks for such pathogens by using molecular epidemiological tools may disclose the prevalence of tick-borne pathogens in particular geographic environments. Some of these agents, such as Rickettsia prowazekii (typhus fever), Yersinia pestis (plague), Francisella tularensis (tularemia), Coxiella burnetii (Q fever), West Nile virus, Rift Valley fever virus, and hantavirus, are now recognized as important emerging vector-borne infections as well as agents of bioterrorism worldwide. Recently, ehrlichial and rickettsial infections have been reported to exist in a broad band across Europe (15), Asia (9, 27, 29), Africa (3), and the Americas (13, 17). Other tick-borne organisms, including some Borrelia and Bartonella spp., have also been shown to cause disease in animals and humans (23, 44, 45).
In the United States and Korea, rodents (e.g., the white-footed mouse [Peromyscus leucopus]) and white-tailed deer (Odocoileus virginianus) are reservoirs of Ehrlichia and Anaplasma spp. (9, 12, 35, 51, 53). In Europe, several rodent species are implicated as natural reservoirs for Ehrlichia and Anaplasma spp. (38). Additionally, Ehrlichia spp. have been isolated from wild mice in Japan (25).
The examination of ticks for tick-borne pathogens by molecular tools such as PCR is commonly used for collecting and assessing data regarding the prevalence of these agents. Although no cases of human anaplasmosis (formerly called human granulocytic ehrlichiosis [HGE]) or human monocytic ehrlichiosis have been reported, seroepidemiological findings suggest the presence of human monocytic ehrlichiosis and HGE agents in the Republic of Korea (ROK) (18, 40). In 2000, the first suspected case of Ehrlichia chaffeensis was reported for an active-duty American soldier stationed in the ROK (43). Subsequently, Heo et al. (18) identified antibodies against E. chaffeensis and Anaplasma phagocytophilum among serum samples from patients with febrile illnesses of otherwise unknown etiology in the ROK by an indirect fluorescent antibody test and Western blotting. Recently, Rickettsia japonica was identified in Haemaphysalis longicornis ticks by PCR (29). Antibodies against R. japonica were also detected by an indirect fluorescent antibody test in human patients with acute febrile illness in the ROK (24). We previously reported molecular evidence of the presence of E. chaffeensis and A. phagocytophilum by using genus-specific TaqMan PCR and a species-specific PCR with ticks collected from animals and grass vegetation in the ROK (27) and gave preliminary reports of other tick-borne pathogens, including Anaplasma platys, Ehrlichia ewingii, Ehrlichia canis, and Ehrlichia muris. In 2005, Lee et al. (30) identified E. chaffeensis in H. longicornis ticks from the ROK by PCR. Also, Kim et al. (26) reported the detection of Bartonella species in ticks, mites, and small mammals in Korea. However, the prevalence of tick-borne pathogens, including Rickettsia rickettsii and R. japonica, has yet to be determined by molecular methods.
Recently, advanced molecular techniques such as TaqMan PCR became widely used as rapid and effective tools for the detection and identification of tick-borne pathogens in arthropods, including ticks (19, 27, 31, 41). This study was therefore undertaken to investigate the prevalence of tick-borne pathogens in small mammals and various tick species from the ROK. Conventional and TaqMan PCR assays were applied for rapid screening of ticks for detection of selected pathogens, followed by specific identification of the pathogens using species-specific and genus-specific primers. Using these approaches, we report herein the infection rates for A. phagocytophilum, A. platys, E. chaffeensis, E. ewingii, and Rickettsia spp. in ticks as well as wild-collected small mammals.
The purpose of the present study is to provide a disease risk assessment for various tick-borne pathogens, including Ehrlichia, Anaplasma, Borrelia, and Rickettsia spp., for humans and animals based on their potential exposure to ticks and tick-borne disease pathogens in field environments.
MATERIALS AND METHODS
Sample collection.
Ticks were collected by dragging and flagging grass vegetation with a 1 m-by-1 m cotton cloth and removing attached ticks from various species of live wild rodents and insectivores that were trapped near the demilitarized zone (DMZ) and other U.S. military installations in the ROK. During 2001 through 2003, a total of 3,135 ticks were collected from wild rodents (297 ticks) and grass vegetation (2,838 ticks) at 19 sites near or at U.S. military installations and training sites in the ROK. Based on microscopic examination, ticks were identified to the species level and classified morphologically into various developmental stages (54). Subsequently, different species of ticks were pooled by species and stage of development (larvae and nymphs) into 1,638 sample pools (1 to 27 ticks per sample pool; pools included ticks from wild rodents [40 sample pools] and those from grass vegetation [1,638 sample pools]) and stored at −70°C in 1.5-ml microcentrifuge tubes until assayed. Ninety-four mites were collected from wild-caught rodents and insectivores and pooled into 21 samples (four or five mites/sample pool).
During 2001 through 2004, a total of 3,564 small mammals belonging to 11 species and 9 genera were collected throughout the ROK at U.S. military installations and training sites as part of the 18th Medical Command Hantavirus Surveillance Program. Of those mammals, 389 rodents, 33 insectivores, and 2 weasels were selected for assessment of tick-borne pathogens. Live-caught rodents, insectivores, and weasels were transported to the central laboratory (Korea University), where they were killed in accordance with approved animal use protocols. After blood samples were taken, the abdominal cavity was opened aseptically, and spleen and other tissue samples were collected. A subsample of spleen samples were sent to Chonbuk National University packed in dry ice, where they were stored individually at −70°C until assayed. Samples were chosen to balance analysis to include all collection areas and mammalian species.
DNA preparation.
For extraction of PCR-amplifiable DNA, the ticks were pooled into a total of 1,638 sample pools (1 to 27 ticks per pool), including 313 pools of larval stages (1 to 27 ticks per pools), 1,176 pools of nymphal stages (1 to 3 ticks/pool), and 151 adult ticks (assayed individually). Purified DNAs were used for the detection of tick-borne pathogens by standardized TaqMan PCR techniques. Individuals (adults) or pools (larvae/nymphs) of ticks and mites (four or five mites/pool) were mechanically homogenized in 1-ml cryovials by using sterile scissors. DNA extraction was performed with a DNeasy tissue kit (QIAGEN, Germany) according to the instructions provided by the manufacturer.
PCR for Ehrlichia and Anaplasma spp.
The extracted DNAs were subjected to an initial screening by TaqMan PCR as previously described, using a set of primers with a probe that amplified the 116-bp fragment of the 16S rRNA genes of bacteria within the family Anaplasmataceae, including the genera Ehrlichia and Anaplasma (27) (Table 1). The fluorescence data were analyzed using PE 7700 sequence detection system software (version 1.7; ABI).
TABLE 1.
Oligonucleotide primers used for detection of tick-borne pathogens
| Target genus or species | Oligonucleotide primer | Primer sequence (5′-3′) | Annealing temp (°C) | Target gene (expected amplicon size [bp]) | Reference |
|---|---|---|---|---|---|
| Ehrlichia and Anaplasma | ESP-F | AGTCCACGCTGTAAACGATGAG | 55 | 16S rRNA (116) | 27 |
| ESP-R | TTCCTTTGAGTTTTAGTCTTGCGAC | 55 | |||
| ESP-P (probe) | FAM-ACGCGTTAAGCACTCCGCCTGG-TAMRA | 55 | |||
| A. phagocytophilum | EE-1 | TCCTGGCTCAGAACGAACGCTGGCGGC | 50 | 16S rRNA (926) | 6 |
| EE-2 | AGTCACTGACCCAACCTTAAATGGCTG | 50 | |||
| EE-3 | GTCGAACGGATTATTCTTTATAGCTTGC | 50 | |||
| EE-4 | CCCTTCCGTTAAGAAGGATCTAATCTCC | 50 | |||
| A. platys, E. chaffeensis, E. ewingii, and E. canis | ECC (outer primer) | AGAACGAACGCTGGCGGCAAGC | 65 | 16S rRNA (450) | 37 |
| ECB (outer primer) | CGTATTACCGCGGCTGCTGGCA | 65 | |||
| A. platys | EPLAT5 (inner primer) | TTTGTCGTAGCTTGCTATGAT | 55 | 16S rRNA (359) | 36 |
| EPLAT3 (inner primer) | CTTCTGTGGGTACCGTC | 55 | |||
| E. chaffeensis | HE1 (inner primer) | CAATTGCTTATAACCTTTTGGTTATAAAT | 55 | 16S rRNA (390) | 37 |
| HE3 (inner primer) | TATAGGTACCGTCATTATCTTCCCTAT | 55 | |||
| E. ewingii | EE52 (inner primer) | CGAACAATTCCTAAATAGTCTCTGAC | 55 | 16S rRNA (350) | 37 |
| HE3 (inner primer) | TATAGGTACCGTCATTATCTTCCCTAT | 55 | |||
| E. canis | ECAN5 (inner primer) | CAATTATTTATAGCCTCTGGCTATAGGA | 55 | 16S rRNA (365) | 37 |
| HE3 (inner primer) | TATAGGTACCGTCATTATCTTCCCTAT | 55 | |||
| E. muris | CAN M61F | TTATCTGTTTATGTTATATAAGC | 50 | gltA (288) | 21 |
| MUR SPR1 | TAAATCTACTATGTTATGTCC | 50 | |||
| B. burgdorferi | BBOSPF | AAAGAATACATTAAGTGCGATATT | 54 | ospC (597) | 52 |
| BBOSPR | GGGCTTGTAAGCTCTTTAACTG | 54 | |||
| R. rickettsii | Rr190k. 71p | TGGCGAATATTTCTCCAAAA | 48 | ompA (532) | 42 |
| Rr190k. 602n | AGTGCAGCATTCGCTCCCCCT | 48 | |||
| R. japonica | Rj10 | ATTCTAAAAACCATATACTG | 57 | 17K antigen (357) | 16 |
| Rj5 | CGCCATTCTACGTTACTACC | 57 |
TaqMan PCR-positive DNA samples were used for specific identification of A. phagocytophilum, A. platys, E. chaffeensis, E. ewingii, E. canis, and E. muris by nested and single-tube conventional PCRs, as previously described, using species-specific primers (Table 1). The 16S rRNA gene fragment of A. phagocytophilum was amplified by a nested PCR assay according to the procedure of Barlough et al. (6). PCR for E. muris was performed using primers CAN M61f and MUR SPR1, targeting the gltA gene of E. muris (21). The primer sets for nested PCRs for A. platys, E. chaffeensis, E. ewingii, and E. canis DNAs were derived from the 16S rRNA gene sequences, with the same pair of outer primers and different sets of inner primers (36, 37). The oligonucleotide sequences for each pair of genus- and species-specific primers are shown in Table 1. PCR amplification of genomic DNAs of R. rickettsii, R. japonica, and Borrelia burgdorferi was performed with species-specific primers (Table 1) as previously described (16, 42, 52).
Nested and single PCRs were performed as previously described in a total volume of 25 μl. Each PCR mixture consisted of 2 pmol of each primer, a 200 μM concentration of each deoxynucleoside triphosphate, PCR buffer I (Super Bio, Korea), 0.75 U of SuperTaq DNA polymerase (Super Bio, Korea), and 50 to 100 ng of sample DNA for the first PCR or 1 μl of the first PCR product for the second PCR. PCR products were electrophoresed in 1% agarose gels, stained with ethidium bromide, and photographed using a still-video documentation system (Gel Doc 2000; Bio-Rad). Unless specified otherwise, PCR products were purified with a GFX PCR DNA purification kit for cloning and sequencing (Amersham Biosciences, United Kingdom). Prevention of cross-contamination and false-negative and -positive results was managed by using plugged tips, performing PCR in a separate room from that used for DNA extraction, and including a negative (water) control in each run. A positive control was run for each master mix batch.
Cloning, nucleotide sequencing, and phylogenetic analysis.
PCR products were purified using the Wizard Plus DNA purification system (Promega), ligated into the pGEM-T Easy vector (Promega), and transformed into TOP10F competent cells. The recombinant clones were verified by colony PCR for the respective clones. Two clones of each isolate were arbitrarily chosen for sequencing of the forward and reverse strands. Plasmid DNA for sequencing was prepared using the SV Minipreps DNA purification system (Promega) according to the manufacturer's instructions.
Amplified and purified DNAs were prepared for direct sequencing using a GFX PCR DNA purification kit (Amersham Biosciences, United Kingdom) and were sequenced by dideoxy termination with an automatic sequencer (ABI Prism 3700 DNA analyzer). Sequence data were collected using ABI Prism data collection software (version 2.1) and analyzed by ABI Prism sequence analysis software (version 2.1.1) and Chromas software (version 1.51; Technelysium Pty., Ltd., Mt. Gravatt Plaza, Queensland, Australia). Sequence homology searches were made via the National Center for Biotechnology Information (National Institutes of Health) BLAST network service. The sequences were aligned initially using ClustalX 1.60 (49). 16S rRNA and rickettsia ompA gene sequences were used for phylogenetic analyses. Aligned sequences were examined with a similarity matrix. Relationships between individuals were assessed by the neighbor-joining method with nucleotide distances (P distance) for 100 replications in the bootstrap test. Phylogenetic analyses based on the obtained sequences were conducted using the maximum likelihood method (PAUP* 4.0b for Macintosh) (48).
Nucleotide sequence accession numbers.
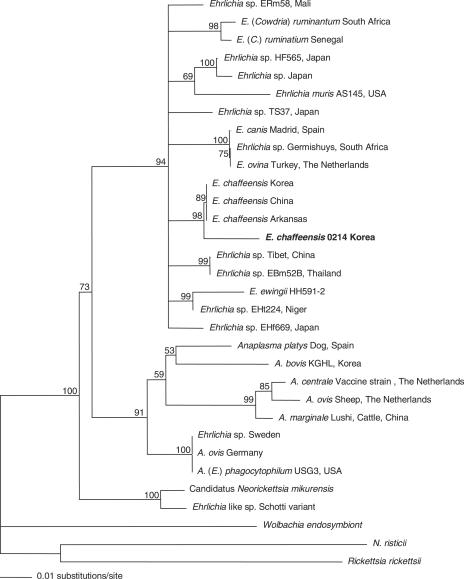
The 16S rRNA gene sequences for the following organisms (with GenBank accession numbers) were used: Ehrlichia sp. strain ERm58 from Rhipicephalus muhsame in Mali (AF311967), Ehrlichia (Cowdria) ruminantium Omatjenne from South Africa (U03776), E. ruminantium from Senegal (X62432), Ehrlichia sp. strain HF565 from Japan (AB024928), Ehrlichia sp. from Ixodes ovatus in Japan (AB028319), E. muris AS145 from Eothenomys kageus in the United States (U15527), Ehrlichia sp. strain TS37 from Japan (AB074459), E. canis from Madrid, Spain (AY394465), Ehrlichia sp. strain Germishuys from South Africa (U54805), Ehrlichia ovina from Turkey and The Netherlands (AF318946), Ehrlichia chaffeensis from H. longicornis in Korea (AY350424), E. chaffeensis from China (AF147752), E. chaffeensis from Arkansas (M73222), E. chaffeensis 0214 from Korea (DQ402484), Ehrlichia sp. strain Tibet from Boophilu micropus in China (AF414399), Ehrlichia sp. strain EBm52 from B. micropus in Thailand (AF497581), E. ewingii HH591-2 from the United States (AY093440), Ehrlichia sp. strain EHt224 from Hyalomma truncatum in Niger (AF311968), Ehrlichia sp. strain EHf669 from Hemaphysalis sp. in Japan (AY309969), A. platys from a dog in Spain (AY530806), Anaplasma bovis AB-KGHL from H. longicornis in Korea (AF470698), Anaplasma centrale vaccine strain from The Netherlands (AF318944), Anaplasma ovis from sheep (AF318945), Anaplasma marginale Lushi from cattle in China (AJ633048), Ehrlichia sp. from Sweden (AJ242785), A. ovis from Germany (AY262124), A. phagocytophilum USG3 from the United States (AY055469), “Candidatus Neorickettsia mikurensis” (AB074460), Ehrlichia-like sp. Schotti variant (AF104680), Wolbachia endosymbiont (AM180551), Neorickettsia risticii (M21290), and Rickettsia rickettsii (DQ150688) (Fig. 1).
FIG. 1.
Phylogenetic tree showing the position of Ehrlichia chaffeensis 0214 from Korea. The tree was made using PAUP* 4.0b software after alignment of 16S rRNA gene fragments obtained (390 bp) from GenBank and sequenced during this study by the ClustalX program. The scale represents 0.01 substitution per base per indicated horizontal distance. The numbers present at nodes of the tree represent the numbers of bootstrap replicates of 100 that display the indicated sequence groupings.
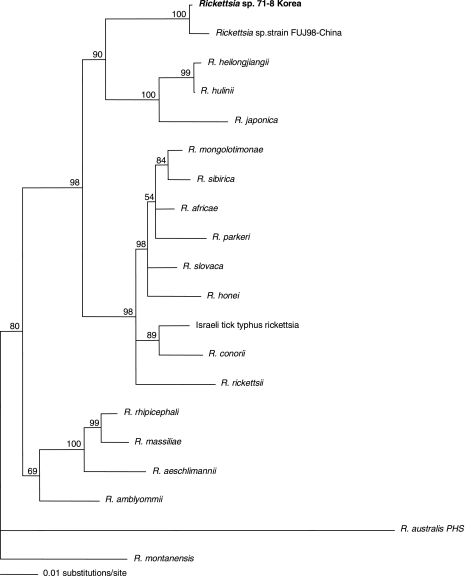
The ompA gene sequences for the following rickettsiae (with GenBank accession numbers) were used: Rickettsia sp. strain 71-8 from Korea (DQ402485), Rickettsia sp. strain FUJ98 from China (AF169629), Rickettsia heilongjiangii HLJ-054 (AF179362), Rickettsia hulinii (AF179364), R. japonica (D28766), Rickettsia mongolotimonae (U43796), Rickettsia sibirica (U43807), Rickettsia africae (U43790), Rickettsia parkeri (U43802), Rickettsia slovaca (U43808), Rickettsia honei (AF018075), Israeli tick typhus rickettsia (AY197564), Rickettsia conorii (U43794), R. rickettsii (U55822), Rickettsia rhipicephali (U43803), Rickettsia massiliae (U43799), Rickettsia aeschlimannii (U43800), Rickettsia amblyommii (AY062007), Rickettsia australis (AF149108), and Rickettsia montanensis (U43801) (Fig. 2).
FIG. 2.
Phylogenetic tree showing the position of Rickettsia sp. strain 71-8 from Korea. The tree was made using PAUP* 4.0b software after alignment of ompA gene fragments obtained (532 bp) from GenBank and sequenced during this study by the ClustalX program. The scale represents 0.01 substitution per base per indicated horizontal distance. The numbers present at nodes of the tree represent the numbers of bootstrap replicates of 100 that display the indicated sequence groupings.
RESULTS
A total of 3,135 ticks, including five species from two genera (2,701 H. longicornis, 115 H. flava, 9 Ixodes turdus, 3 Ixodes persulcatus, 20 Ixodes nipponensis, and 287 Ixodes sp. ticks), were collected from wild rodents and insectivores (297 ticks) and from grass/vegetation (2,838 ticks) from 2001 through 2003 (Table 2). H. longicornis ticks were the most commonly collected species, and irrespective of species, most of the ticks were collected in the nymphal stage of development (Table 2). All ticks collected from wild rodents and insectivores during this period were Ixodes spp. All of the mesostigmatid mite samples included in this study were collected from rodents and insectivores and assayed for the same pathogens examined in ticks.
TABLE 2.
Results of TaqMan and species-specific tick-borne pathogen PCR for selected tick species and stages of development collected at or near U.S. military installations and training sites in the ROK, 2001 to 2003
| Species | Stage | n | No. (%) of PCR-positive samples
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ehrlichia/Anaplasmah | A. phagocytophilum | A. platys | E. chaffeensis | E. ewingii | E. canis | E. muris | B. burgdorferii | Rickettsia sp.i | R. japonicai | |||
| H. longicornis | Larva poolsa | 274 | 229 | 0 | 20 | 15 | 0 | 10 | 0 | 0 | 0 | 0 |
| Nymphb | 1,099 | 510 | 0 | 23 | 10 | 1 | 6 | 0 | 0 | 27 | 0 | |
| Male | 53 | 28 | 0 | 4 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | |
| Female | 77 | 33 | 0 | 4 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | |
| Subtotal | 1,503 | 800 | 0 | 51 | 28 | 2 | 17 | 0 | 0 | 28 | 0 | |
| H. flava | Larva poolsc | 11 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Nymph | 53 | 21 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Male | 7 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Female | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Subtotal | 74 | 24 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| I. turdus | Nymph | 8 | 4 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| Female | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Subtotal | 9 | 4 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | |
| I. nipponensis | Nymph | 13 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Male | 4 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Female | 3 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Subtotal | 20 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| I. persulcatus | Male | 2 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Female | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Subtotal | 3 | 3 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Ixodes spp. | Larva poolsd | 26 | 19 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Nymph | 3 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Subtotal | 29 | 21 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Total | Larva poolse | 313 | 245 | 0 | 20 | 15 | 0 | 10 | 0 | 0 | 0 | 0 |
| Nymphf | 1,176 | 536 | 0 | 23 | 10 | 1 | 7 | 0 | 0 | 27 | 0 | |
| Male | 66 | 66 | 1 | 5 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | |
| Female | 85 | 85 | 0 | 4 | 3 | 0 | 1 | 0 | 0 | 0 | 0 | |
| Totalg | 1,638 | 857 (52.3) | 1 (0.1) | 52 (3.2) | 29 (1.8) | 2 (0.1) | 18 (1.1) | 0 | 0 | 28 (1.7) | 0 | |
Two to seven ticks per pool (1,445 ticks).
One to three ticks per pool (1,127 ticks).
Two to five ticks per pool (52 ticks).
One to 27 ticks per pool (284 ticks).
One to 27 ticks per pool (1,780 ticks).
One to three ticks per pool (1,204 ticks).
One to 27 ticks per pool (3,135 ticks).
TaqMan PCR-positive samples were tested for A. phagocytophilum, A. platys, E. chaffeensis, E. ewingii, E. canis, and E. muris, and the numbers and percentages were calculated based on the numbers of TaqMan PCR-positive samples for the respective species.
The numbers and percentages of positive samples for B. burdgorferi, R. rickettsii, and R. japonica were calculated based on the total numbers of ticks tested.
A total of 424 small mammals (six rodent species belonging to five genera [373 Apodemus agrarius, 3 Apodemus peninsulae, 1 Cricetulus triton, 9 Eothenomys regulus, 1 Mus musculus, and 2 Rattus rattus animals], one insectivore species [33 Crosidura lasiura animals], and one mustelid species [2 Mustela sibirica animals]) were collected from 2001 through 2004 at U.S. military installations and training sites near the DMZ and at other U.S. military installations south of Seoul, ROK. A. agrarius was the most commonly collected mammal and accounted for 88% (373/424) of the total samples from small mammals (Table 3).
TABLE 3.
Tick-borne pathogens identified by DNA analysis in small mammals collected at U.S. military installations and training sites in the ROK, 2001 to 2004
| Species | n | No. (%) of PCR-positive samples
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Ehrlichia/Anaplasma | A. phagocytophilum | A. platys | E. chaffeensis | E. ewingii | E. canis | E. muris | B. burgdorferi | Rickettsia sp. | R. japonica | ||
| Apodemus agrarius | 373 | 270 | 88 | 68 | 8 | 26 | 50 | 0 | 0 | 22 | 0 |
| Crosidura lasiura | 33 | 26 | 21 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| Eothenomys regulus | 9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Apodemus peninsulae | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Rattus rattus | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Mustela sibirica | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Cricetulus triton nestor | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Mus musculus | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Total | 424 | 299 (70.5) | 110 (25.9) | 68 (16) | 8 (1.9) | 26 (6.1) | 51 (12.0) | 0 | 0 | 22 (5.2) | 0 |
TaqMan PCR signals for Ehrlichia and Anaplasma spp. were detected from all tick species sampled and from five of eight species of small wild mammals (A. agrarius, C. lasiura, M. sibirica, C. triton nestor, and M. musculus). The prevalence of Ehrlichia and Anaplasma parasites in ticks and all small mammals was 52.3% (857/1,638 tick pools), with minimum infection rates of 27.3% (857/3,135 total ticks) and 70.5% (299/424 mammals), respectively (Tables 2 and 3).
Species-specific PCR assays were conducted with DNA samples from 857 tick pools and 277 small mammals that were positive by TaqMan PCR. Except for E. muris, five of six tick-borne pathogens evaluated in this study were detected in ticks (A. phagocytophilum [1], A. platys [52], E. chaffeensis [29], E. ewingii [2], and E. canis [18]) and small mammals (A. phagocytophilum [110], A. platys [68], E. chaffeensis [8], E. ewingii [26], and E. canis [51]) by nested PCR (Tables 2 and 3). The most frequently isolated species in ticks was A. platys (52 [3.2%]), followed by E. chaffeensis (29 [1.8%]), E. canis (18 [1.1%]), E. ewingii (2 [0.1%]), and A. phagocytophilum (1 [<0.1%]). In the case of small mammals, the most frequently detected species (n = 424) was A. phagocytophilum (110 [25.9%]), followed by A. platys (68 [16%]), E. canis (51 [12.0%]), E. ewingii (26 [6.1%]), and E. chaffeensis (8 [1.9%]). In an additional study, all of the DNA samples from 1,638 tick pools and 424 small wild mammals were examined for three tick-borne pathogens, B. burgdorferi, R. rickettsii, and R. japonica, directly by single PCRs with specific primers. Twenty-eight H. longicornis (1.7%) ticks and 22 A. agrarius (5.2%) animals were positive for R. rickettsii. However, DNA bands specific for B. burgdorferi and R. japonica were not observed for ticks or small mammals examined during this study. None of the 297 Ixodes spp. (40 pools) collected from small mammals were found to be infected with any of the nine tick-borne pathogens examined in this study. Among 21 mite samples collected from wild rodents, 4 (19%) were PCR positive for Ehrlichia and Anaplasma spp. by TaqMan PCR, while a specific DNA band was not observed for six Ehrlichia and Anaplasma spp., B. burgdorferi, R. rickettsii, or R. japonica.
A total of 21 pools/individual ticks represented multiple infections (Table 4). Pools of larvae and nymphs with multiple infections may represent individual infections from ticks within each of the samples. However, two and one adult ticks, assayed individually, were positive for two and three Ehrlichia/Anaplasma pathogens, respectively. One hundred thirty small mammals had single infections, while 4, 14, and 21 A. agrarius animals had mixed infections of four, three, or two pathogens, respectively (Table 5).
TABLE 4.
Mixed infections of tick-borne pathogens among individual ticks and pools of ticks collected at or near U.S. military installations and training sites near the DMZ, ROK, 2001 to 2003a
| Species | Stage | No. of infected samples
|
|||||
|---|---|---|---|---|---|---|---|
| A. phagocytophilum/A. platys | E. chaffeensis/E. canis | E. chaffeensis/A. platys | E. chaffeensis/ E. canis/A. platys | E. chaffeensis/ E. canis/E. ewingii | Total | ||
| H. longicornis | Larva pools | 0 | 10 | 2 | 0 | 0 | 12 |
| Nymph | 0 | 0 | 1 | 4 | 1 | 6 | |
| Male | 0 | 0 | 1 | 0 | 0 | 1 | |
| Female | 0 | 0 | 0 | 0 | 1 | 1 | |
| Subtotal | 0 | 10 | 4 | 4 | 2 | 20 | |
| I. persulcatus | Female | 1 | 0 | 0 | 0 | 0 | 1 |
| Subtotal | 1 | 0 | 0 | 0 | 0 | 1 | |
| Total | Larva pools | 0 | 10 | 2 | 0 | 0 | 12 |
| Nymph | 0 | 0 | 1 | 4 | 1 | 6 | |
| Male | 0 | 0 | 1 | 0 | 0 | 1 | |
| Female | 1 | 0 | 0 | 0 | 1 | 2 | |
| Total | 1 | 10 | 4 | 4 | 2 | 21 | |
Adult male and female ticks were assayed individually. Pools may represent single infections among ticks from the same sample.
TABLE 5.
Mixed infections of tick-borne pathogens in small mammals (Apodemus agrarius) collected at U.S. military installations and training sites, ROK, 2001 to 2004
| Species | No. of mixed infections | Total no. of infections for groupa |
|---|---|---|
| A. phagocytophilum/A. platys/E. canis/ E. ewingii | 3 | |
| A. platys/E. canis/E. ewingii/E. chaffeensis | 1 | 4 |
| A. platys/E. ewingii/E. canis | 7 | |
| A. platys/E. canis/E. chaffeensis | 2 | |
| A. phagocytophilum/E. ewingii/E. canis | 1 | |
| A. phagocytophilum/E. chaffeensis/ E. ewingii | 4 | 14 |
| A. phagocytophilum/A. platys | 1 | |
| A. platys/E. ewingii | 1 | |
| A. platys/E. canis | 13 | |
| E. ewingii/E. canis | 2 | |
| A. phagocytophilum/E. ewingii | 1 | |
| A. phagocytophilum/R. rickettsii | 1 | |
| E. chaffeensis/R. rickettsii | 1 | |
| E. ewingii/R. rickettsii | 1 | 21 |
| Total | 39 | 39 |
Mixed infections are divided into groups of infections with four, three, or two organisms.
Among 19 tick collection sites, Ehrlichia and Anaplasma DNAs were detected from all sites, while selected pathogens tested by species-specific PCR in this study were recovered from only 13 sites. Of those, A. platys was detected at the most sites (11), followed by E. chaffeensis (5 sites), R. rickettsii (3 sites), E. canis (2 sites), A. phagocytophilum (1 site), and E. ewingii (1 site).
During the 3-year period, monthly infection rates for Ehrlichia and Anaplasma spp. in ticks were higher in October (148/172 [86%]) than during earlier months (25% [2/8] in March, 42.8% [511/1,193] in June, 63.3% [19/30] in August, and 73.6% [170/231] in September), except for April (1/1) and July (3/3). Infection rates for small mammals were higher in February (46/49 [93.9%]) than in the other 5 months of sampling (60.3% [38/63] in March, 74.8% [92/123] in April, 83.9% [26/31] in May, 68.8% [44/64] in June, and 56.4% [53/94] in October). The infection rates among ticks positive for Ehrlichia or Anaplasma spp. were higher in September (n = 231) (28 A. platys infections [12.1%], 23 E. chaffeensis infections [10.0%], 2 E. ewingii infections [0.9%], and 16 E. canis infections [6.9%]), while positive rates for small mammals were higher in April (n = 123) (3 A. phagocytophilum infections [2.4%], 42 A. platys infections [34.1%], 2 E. chaffeensis infections [1.6%], 9 E. ewingii infections [7.3%], and 22 E. canis infections [1.9%]). R. rickettsii was observed in ticks collected during June (28) and in small mammals collected during March (19) and April (3).
E. chaffeensis sequences were compared with those of other isolates of E. chaffeensis available in GenBank. The E. chaffeensis 0214 sequence (present study) was homologous to those of other Korean (99.7%) (AY350424), U.S. (99.7%) (AF416764), and Chinese (99.7%) (AF147752) isolates. The homology level between the two nucleotide sequences determined in this study varied from 96.2% to 100%. Comparative analysis of nucleotide sequences of the Korean strains determined in this study and the 16S rRNA sequences of 14 known Ehrlichia spp. available in the GenBank database is shown in Table 6. Phylogenetic analysis showed that E. chaffeensis 0214 clustered together with Korean, Arkansan, and Chinese strains of E. chaffeensis (Fig. 1).
TABLE 6.
Homology comparison of Korean Ehrlichia chaffeensis 16S rRNA gene fragment (390-bp) sequences with those of other strains
| Sequence | % Identity or no. of nucleotide differencesa
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | |
| 1 | 99.7 | 99.7 | 99.7 | 98.2 | 98.2 | 97.9 | 97.4 | 97.4 | 97.2 | 97.2 | 97.2 | 96.7 | 96.2 | 96.2 | |
| 2 | 1 | 100 | 100 | 98.5 | 98.5 | 98.2 | 97.7 | 97.7 | 97.4 | 97.4 | 97.4 | 96.9 | 96.4 | 96.4 | |
| 3 | 1 | 0 | 100 | 98.5 | 98.5 | 98.2 | 97.7 | 97.7 | 97.4 | 97.4 | 97.4 | 96.9 | 96.4 | 96.4 | |
| 4 | 1 | 0 | 0 | 98.5 | 98.5 | 98.2 | 97.7 | 97.7 | 97.4 | 97.4 | 97.4 | 96.9 | 96.4 | 96.4 | |
| 5 | 7 | 6 | 6 | 6 | 100 | 97.7 | 99.2 | 99.2 | 99.0 | 99.0 | 99.0 | 96.2 | 96.4 | 96.4 | |
| 6 | 7 | 6 | 6 | 6 | 0 | 97.7 | 99.2 | 99.2 | 99.0 | 98.5 | 96.7 | 96.2 | 96.4 | 96.4 | |
| 7 | 8 | 7 | 7 | 7 | 9 | 9 | 97.4 | 97.4 | 96.9 | 98.2 | 97.2 | 96.7 | 96.7 | 97.2 | |
| 8 | 9 | 8 | 8 | 8 | 3 | 3 | 10 | 100 | 98.2 | 97.7 | 96.9 | 96.4 | 96.1 | 96.6 | |
| 9 | 9 | 8 | 8 | 8 | 3 | 3 | 10 | 0 | 98.2 | 97.7 | 96.9 | 96.4 | 96.1 | 96.6 | |
| 10 | 11 | 8 | 8 | 8 | 4 | 3 | 12 | 7 | 7 | 97.4 | 96.2 | 95.9 | 95.6 | 95.6 | |
| 11 | 11 | 9 | 9 | 9 | 4 | 5 | 7 | 9 | 9 | 10 | 95.9 | 95.4 | 95.9 | 96.4 | |
| 12 | 11 | 9 | 9 | 9 | 4 | 12 | 11 | 12 | 12 | 16 | 16 | 99.5 | 95.9 | 95.1 | |
| 13 | 13 | 9 | 9 | 9 | 15 | 15 | 13 | 14 | 14 | 18 | 18 | 2 | 95.4 | 94.6 | |
| 14 | 15 | 12 | 12 | 12 | 13 | 14 | 12 | 15 | 15 | 17 | 16 | 15 | 18 | 95.4 | |
| 15 | 15 | 14 | 14 | 14 | 14 | 14 | 11 | 13 | 13 | 17 | 14 | 19 | 22 | 18 | |
Percentages of identity between 16S rRNA gene fragment sequences are shown in the upper matrix. The lower matrix shows numbers of nucleotide differences. The following sequences were compared: 1, E. chaffeensis 0214 from Haemaphysalis longicornis in Korea; 2, AY350424 (E. chaffeensis from Korea); 3, AF416764 (E. chaffeensis from Arkansas); 4, AF147752 (E. chaffeensis from China); 5, AF414399 (Ehrlichia sp. from Boophilu micropus from Tibet (in China); 6, AF497581 (Ehrlichia sp. strain EBm52 from Boophilus microplus in Thailand); 7, AY309969 (Ehrlichia sp. strain EHf669 from Haemaphysalis sp. in Japan); 8, AF311967 (Ehrlichia sp. strain ERm58 from Africa); 9, AF311967 (E. chaffeensis from France); 10, AF311968 (Ehrlichia sp. strain EHt224 from Hyalomma truncatum in Niger); 11, AB074459 (Ehrlichia sp. strain TS37 from Japan); 12, AB024928 (Ehrlichia sp. strain HF565); 13, AB028319 (Ehrlichia sp. strain Anan); 14, AF318946 (E. ovina [isolate from turkey ruminant in The Netherlands]); 15, U03776 (C. ruminantium from Omatjenne in South Africa).
Sequence comparison and alignment of ompA gene nucleotide sequences from Rickettsia sp. strain 71-8 (present study) revealed >99% homology to previously reported sequences for Korean and Chinese strains of Rickettsia spp. (Table 7). The sequence similarity of Rickettsia sp. strain 71-8 with an R. rickettsii isolate from Japan was <94%. However, phylogenetic analysis showed that Rickettsia sp. strain 71-8 formed a single cluster with a Rickettsia sp. Chinese isolate, while an R. rickettsii U.S. isolate was placed in a different cluster. The close clustering of Chinese and Korean strains of Rickettsia spp. may indicate a close epidemiological link between these strains (Fig. 2).
TABLE 7.
Homology comparison of Rickettsia sp. ompA gene fragment (532-bp) sequences
| Sequence | % Identity or no. of nucleotide differencesa
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | |
| 1 | 99.4 | 94.7 | 94.5 | 94.3 | 93.9 | 93.8 | 93.8 | 93.8 | 93.8 | 93.8 | 93.8 | 93.2 | 92.6 | 92.4 | |
| 2 | 3 | 94.5 | 94.3 | 93.8 | 93.4 | 93.6 | 93.2 | 93.6 | 93.2 | 93.2 | 93.2 | 92.6 | 92.0 | 91.8 | |
| 3 | 26 | 28 | 99.8 | 94.5 | 94.7 | 97.3 | 93.9 | 93.9 | 94.1 | 94.3 | 93.8 | 94.1 | 93.0 | 92.2 | |
| 4 | 27 | 29 | 1 | 94.3 | 94.5 | 97.1 | 93.8 | 93.8 | 93.9 | 93.9 | 93.6 | 93.9 | 92.8 | 92.0 | |
| 5 | 28 | 32 | 28 | 28 | 98.6 | 93.6 | 97.9 | 97.5 | 98.0 | 98.2 | 98.0 | 96.9 | 93.4 | 93.2 | |
| 6 | 28 | 34 | 27 | 27 | 6 | 93.8 | 97.7 | 98.2 | 98.6 | 98.8 | 98.2 | 96.7 | 93.4 | 93.2 | |
| 7 | 28 | 33 | 14 | 15 | 33 | 32 | 93.4 | 93.0 | 93.2 | 93.4 | 92.8 | 93.2 | 92.2 | 91.4 | |
| 8 | 32 | 35 | 31 | 32 | 11 | 12 | 34 | 96.5 | 97.5 | 97.7 | 97.1 | 96.3 | 93.2 | 93.0 | |
| 9 | 32 | 33 | 31 | 31 | 13 | 9 | 35 | 18 | 97.5 | 98.0 | 96.9 | 95.5 | 92.6 | 92.4 | |
| 10 | 32 | 35 | 29 | 31 | 10 | 7 | 35 | 11 | 12 | 99.0 | 97.3 | 96.5 | 93.2 | 93.0 | |
| 11 | 32 | 35 | 28 | 30 | 9 | 6 | 33 | 12 | 9 | 5 | 97.5 | 96.7 | 93.4 | 93.2 | |
| 12 | 32 | 35 | 32 | 33 | 10 | 9 | 37 | 15 | 15 | 13 | 13 | 96.5 | 92.8 | 92.6 | |
| 13 | 32 | 38 | 29 | 31 | 16 | 17 | 35 | 19 | 23 | 18 | 16 | 18 | 92.4 | 92.2 | |
| 14 | 40 | 41 | 35 | 37 | 34 | 34 | 38 | 35 | 37 | 35 | 34 | 37 | 39 | 98.8 | |
| 15 | 40 | 42 | 39 | 41 | 35 | 35 | 43 | 36 | 38 | 36 | 35 | 38 | 41 | 6 | |
Percentages of identity between sequences of ompA gene fragments are shown in the upper matrix. The lower matrix shows the numbers of nucleotide differences. The following sequences were used: 1, Rickettsia sp. strain 71-8 from Korea; 2, AF169629 (Rickettsia sp. from China); 3, AF179364 (Rickettsia hulinii from France); 4, AF179362 (Rickettsia heilongjiangii from France); 5, U43808 (Rickettsia slovaca from France); 6, U43790 (Rickettsia africae from France); 7, D28766 (Rickettsia japonica from Japan); 8, AY197564 (Israeli tick typhus rickettsia from Italy); 9, U43802 (Rickettsia parkeri from France); 10, U43807 (Rickettsia sibirica from France); 11, U43796 (Rickettsia mongolotimonae from France); 12, AF018075 (Rickettsia honei from the United States); 13, U55822 (Rickettsia rickettsii from the United States); 14, U43803 (Rickettsia rhipicephali from France); 15, U43799 (Rickettsia massiliae from France); 16, AY062007 (Rickettsia amblyommii from the United States); 17, U43800 (Rickettsia aeschlimannii from France).
DISCUSSION
An analysis of the prevalence of selected tick-borne pathogens in ticks and mites collected from wild-caught rodents/insectivores and by dragging and flagging vegetation at or near U.S. military installations in the ROK demonstrated a high rate of infection with Ehrlichia and Anaplasma sp. pathogens. Most Ehrlichia and Anaplasma spp. have been recovered from Ixodes sp. ticks in the Americas and Europe (1, 50), while in Asia, Ehrlichia spp. were identified from both H. and Ixodes sp. ticks (22, 27). H. longicornis is the most commonly collected species in tick drag-and-flag collections in the ROK, accounting for >98% of all ticks collected, especially in and around pastures for grazing cattle or where deer congregate. H. flava is more commonly collected in pine forests, accounting for >95% of all ticks collected in this environment. Ixodes spp. are infrequently collected in grassy vegetation and pine forests, generally making up <2% of the collected ticks (data not shown). However, in deciduous forests with an abundance of leaf litter, collection of I. nipponensis may exceed 5% of the collected ticks during tick dragging and flagging (H. C. Kim, personal communication). Another sampling technique that used CO2-baited traps proved to be unsuccessful for capturing ticks. In that investigation, all of the ticks (297) taken from wild rodents and insectivores belonged to the genus Ixodes. Recently, all ticks (2,760) taken from rodents and insectivores captured at the same sites during 2004 and 2005 were identified as I. nipponensis, except for one H. flava tick (Kim, unpublished data). Thus, it is likely that the ixodid ticks tested herein for zoonotic pathogens were I. nipponensis. Human cases of tick bite are more commonly reported for Ixodes spp. (20, 28). Thus, while small numbers of Ixodes spp. were collected through dragging and flagging vegetation, this method may represent a possible bias for attracting members of the genus Hemaphysalis, or the bites from ixodid ticks may be of greater severity and thus reported more frequently.
Simple and easy methods for the identification of tick-borne pathogens in ticks and wild animals are necessary for a rapid analysis of disease-causing agents in developing an accurate risk assessment for soldiers training in the field. TaqMan PCR has proven to be a relatively easy and rapid method for the detection and identification of microorganisms in field samples, such as ticks (32), and in a previous report, its sensitivity was identical to that of nested PCR (6). These techniques can also be applied for rapid diagnosis with other samples, e.g., blood from patients suffering from a febrile disease with an unknown etiology.
16S rRNA gene sequence analysis is a widely used method for characterization of pathogenic bacteria, especially for the genera Ehrlichia and Anaplasma. Primers ECC and ECB (12) were used to amplify a segment of the 16S rRNA gene, which includes the specific primer regions of four Ehrlichia/Anaplasma-related pathogens (A. platys, E. chaffeensis, E. ewingii, and E. canis) examined in this study (Table 1). For A. phagocytophilum and E. muris, we designed separate primers from ECC and ECB because there are no species-specific regions between the ECC and ECB sequences. While these primers effectively identified pathogens to the species level, approximately two-thirds of the Anaplasma/Ehrlichia-positive samples were unidentified. Some of these positive samples may represent other infectious agents previously identified in Korea, i.e., E. bovis, A. marginale, and A. centrale, or may represent unknown or previously unreported pathogens.
Ticks of the genera Hemaphysalis and Ixodes collected from grass were found to be infected with one or more of the six tick-borne pathogens tested (A. phagocytophilum, A. platys, E. chaffeensis, E. ewingii, E. canis, and R. rickettsii). Three pathogens identified in this study have been reported previously in the ROK (9, 27, 29, 30). To the best of our knowledge, this is the first report of A. platys, E. ewingii, and E. canis in the ROK (9). Screening of ticks as well as small mammalian spleens for the presence of Rickettsia sp. infections corroborated well with earlier findings in the ROK (Joon-Seok Chae, unpublished data). In this study, larval ticks were positive for various Ehrlichia/Anaplasma sp. infections. While transovarial transmission in Amblyomma americanum has not been reported, our results for transovarial transmission of E. chaffeensis parasites in female ticks is undetermined, as 15 pools of larval H. longicornis ticks were positive for E. chaffeensis (33). However, some larval ticks may have been collected either partially or fully engorged, and it is possible that the DNA detected might have been in the host's blood that was ingested by the ticks. While H. longicornis larvae were positive, there were no larval Ixodes spp. taken from rodents positive for E. chaffeensis, even though they had been attached and partially blood fed and 3% of the striped field mice were positive for this pathogen. More studies have to be conducted to determine the status of transovarial transmission of E. chaffeensis and other zoonotic tick-borne pathogens among various species of ticks found in Korea.
There were observed differences in the infection rates of ticks versus small-mammal tissues. For example, only 1 of 1,638 pools of ticks was positive for A. phagocytophilum, while 24% (88/373) of the A. agrarius and 64% (21/33) of Crosidura lasiura (shrew) animals were positive. However, this may be a result of the fact that most of the ticks sampled from tick dragging and flagging were from the genus Hemaphysalis, while nearly all ticks (>99.9%) taken from small mammals were I. nipponensis. Soft ticks, which were not sampled during this study, also may be vectors of various pathogens. More investigations are needed to resolve these differences.
While there is no evidence of Rickettsia sp. infections in animals and only a single case reported among Koreans, the results based on the present study, combined with previous serologic and molecular evidence (24, 29), suggest that cases of febrile illness by spotted fever group Rickettsia in the ROK could be missed during the diagnosis of such illnesses. This is supported by retrospective studies by Song et al. (46) and Baek et al. (5), who demonstrated that a large proportion of patients with suspected hemorrhagic fever with renal syndrome were serologically positive for spotted fever group agents while being negative for hemorrhagic fever with renal syndrome. Specific DNAs of B. burgdorferi and R. japonica were not amplified in this study, although there are previous reports of these infections in Korean patients and ticks (24, 39). Therefore, it appears that cases of febrile illness of unknown etiology in humans as well as animals should always be considered possible tick-borne infections in differential diagnosis.
Since human granulocytic and monocytic ehrlichioses were first reported in 1994 and 1987, respectively, they have been found in many countries by molecular and serologic surveys (8, 10, 34). In particular, tick infestations of wild animals have often been investigated because these animals have a high risk of infections. Epidemiological, molecular, and serological studies provided evidence that wild rodents are the reservoir of Ehrlichia and Anaplasma spp. in the United States, Europe, and Asia. Natural reservoirs for A. phagocytophilum were demonstrated to be white-footed mice (Peromyscus leucopus), eastern chipmunks (Tamias striatus), southern red-blacked voles (Clethrionomys gapperi), and insectivorous shrews (Blarina brevicauda and Sorex cinereus) in Minnesota (51). In Japan, while antibodies against E. muris were detected in Apodemus speciosus and Apodemus argenteus mice (25), they were observed neither in rodent tissues nor from ticks collected near the DMZ in Korea. Our results demonstrate that small mammals captured in the ROK were infected with Ehrlichia/Anaplasma and/or Rickettsia parasites. Infections with Ehrlichia and Anaplasma spp. have generally been considered host specific. However, our studies suggest that several Ehrlichia and Anaplasma spp. can be transmitted to a variety of hosts in nature. Therefore, additional efforts to define the spectrum of host susceptibility in domestic and wild animals are appropriate.
The spotted fever group rickettsiae in Korea were identified using a primer for R. rickettsii. However, phylogenetic analysis revealed that Rickettsia sp. strain 71-8 (present study) formed a single cluster with a Chinese Rickettsia sp., while a U.S. R. rickettsii strain was placed in a different cluster and most likely represents a distinct species. Close clustering with the Rickettsia sp. from China indicated that although Korean strains differed significantly from those of R. rickettsii, there is a potential epidemiological link between the Chinese and Korean strains. Similarly, E. chaffeensis 0214 clustered together with E. chaffeensis strains from Korea, Arkansas, and China, which also indicates the possibility of an epidemiological link between Korea and China. Further studies that provide for molecular characterization and identify the epidemiology, human infection rates, and potential human health hazards of these pathogens are needed.
Previous studies have implicated mites as potential vectors of Ehrlichia and Anaplasma pathogens in Spain (14). In Korea, mites have not been well studied to determine their potential impact on the zoonotic maintenance and transmission of Ehrlichia, Anaplasma, or Rickettsia pathogens. Of 21 mite samples assayed during this study, 4 (19%) were positive for Ehrlichia and Anaplasma spp. However, species-specific DNAs examined in this study were not amplified and may represent Anaplasma or Ehrlichia pathogens previously described from Korea but not evaluated here, unidentified emerging pathogens, or previously unreported pathogens. Further investigation is needed to determine the role of mites in maintenance and transmission of zoonotic vector-borne pathogens. However, the association between positive TaqMan PCR and negative PCR for nine tick-borne pathogens has not been determined.
Ixodid ticks play an important role as a reservoir for latent infections with various tick-borne pathogens (4, 7). In 2003, three members of the family Anaplasmataceae, including E. chaffeensis, A. phagocytophilum, and A. bovis, were initially described in the ROK (27). Until now, there have been very few reports of epidemiological studies for tick-borne disease surveillance in the ROK, in contrast with numerous reports throughout the world. These studies have enabled us to provide further information on the epidemiology of tick-associated pathogens in the ROK, where little information on the subject exists. Further studies are required for a detailed understanding of these newly emerging tick-borne diseases in the ROK.
Acknowledgments
Funding for portions of this work was provided by the U.S. Department of Defense Global Emerging Infections Surveillance and Response System, Silver Spring, MD, the Armed Forces Medical Intelligence Center, Ft. Detrick, MD, the international collaborative research fund of Chonbuk National University (2004), and the Brain Korea 21 Project in E007.
REFERENCES
- 1.Adelson, M. E., R. V. S. Rao, R. C. Tilton, K. Cabets, E. Eskow, L. Fein, J. L. Occi, and E. Mordechai. 2004. Prevalence of Borrelia burgdorferi, Bartonella species, Babesia microti, and Anaplasma phagocytophila in Ixodes scapularis ticks collected in northern New Jersey. J. Clin. Microbiol. 42:2799-2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alekseev, A. N., H. V. Dubinina, I. van de Pol, and L. M. Schouls. 2001. Identification of Ehrlichia species and Borrelia burgdorferi in Ixodes ticks in the Baltic regions of Russia. J. Clin. Microbiol. 39:2237-2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allsopp, M. T. E. P., and B. A. Allsopp. 2001. Novel Ehrlichia genotype detected in dogs in South Africa. J. Clin. Microbiol. 39:4204-4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson, B. E., K. G. Sims, and J. G. Olson. 1993. Amblyomma americanum: a potential vector of human ehrlichiosis. Am. J. Trop. Med. Hyg. 49:239-244. [DOI] [PubMed] [Google Scholar]
- 5.Baek, L. J., J. W. Song, and H. W. Lee. 1989. Serologic diagnosis of acute febrile hemorrhagic disease patients in Korea, 1988. J. Korean Soc. Virol. 19:117-125. [Google Scholar]
- 6.Barlough, J. E., J. E. Madigan, E. Derock, and L. Bigornia. 1996. Nested polymerase chain reaction for detection of Ehrlichia equi genomic DNA in horses and ticks (Ixodes pacificus). Vet. Parasitol. 63:319-329. [DOI] [PubMed] [Google Scholar]
- 7.Cao, W. C., Y. M. Gao, and P. H. Zhang. 2000. Identification of Ehrlichia chaffeensis by nested PCR in ticks from southern China. J. Clin. Microbiol. 38:2778-2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Castro, M. B., W. L. Nicholson, L. C. Whitworth, J. C. Fox, and A. A. Kocan. 2001. Persistent infection in Neotoma fuscipes (Muridae: Sigmodontinae) with Ehrlichia phagocytophila sensu lato. Am. J. Trop. Med. Hyg. 65:261-267. [DOI] [PubMed] [Google Scholar]
- 9.Chae, J. S., C. M. Kim, E. H. Kim, E. J. Hur, T. A. Klein, T. K. Kang, H. C. Lee, and J. W. Song. 2003. Molecular epidemiological study for tick-borne disease (Ehrlichia and Anaplasma species) surveillance at selected U.S. military training sites/installations in Korea. Ann. N. Y. Acad. Sci. 990:118-125. [DOI] [PubMed] [Google Scholar]
- 10.Chen, S. M., J. S. Dumler, J. S. Bakker, and D. H. Walker. 1994. Identification of a granulocytotropic Ehrlichia species as the etiologic agent of human disease. J. Clin. Microbiol. 32:589-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiba, N., M. Osada, K. Komoro, T. Mizutani, H. Kariwa, and I. Takashima. 1999. Protection against tick-borne encephalitis virus isolated in Japan by active and passive immunization. Vaccine 17:1532-1539. [DOI] [PubMed] [Google Scholar]
- 12.Dawson, J. E., D. E. Stallknecht, E. W. Howerth, C. Warner, K. Biggie, W. R. Davidson, J. M. Lockhart, V. F. Nettles, J. G. Olson, and J. E. Childs. 1994. Susceptibility of white-tailed deer (Odocoileus virginianus) to infection with Ehrlichia chaffeensis, the etiologic agent of human ehrlichiosis. J. Clin. Microbiol. 32:2725-2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dawson, J. E., K. L. Biggie, C. K. Warner, K. Cookson, S. Jenkins, J. F. Levine, and J. G. Olson. 1996. Polymerase chain reaction evidence of Ehrlichia chaffeensis, an etiologic agent of human ehrlichiosis, in dogs from southeast Virginia. Am. J. Vet. Res. 57:1175-1179. [PubMed] [Google Scholar]
- 14.Fernandez, S. P., S. R. Perez, and G. A. Encinas. 2001. Molecular detection of Ehrlichia phagocytophila genogroup organisms in larvae of Neotrombicula autumnalis (Acari: Trombiculidae) captured in Spain. J. Parasitol. 87:1482-1483. [DOI] [PubMed] [Google Scholar]
- 15.Fournier, P. E., and D. Raoult. 2004. Suicide PCR on skin biopsy specimens for diagnosis of rickettsioses. J. Clin. Microbiol. 42:3428-3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Furuya, Y., T. Katayama, Y. Yoshida, and I. Kaiho. 1995. Specific amplification of Rickettsia japonica DNA from clinical specimens by PCR. J. Clin. Microbiol. 33:487-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galvao, M. A., J. A. Lamounier, E. Bonomo, M. S. Tropia, E. G. Rezende, S. B. Calic, C. B. Chamone, M. C. Machado, M. E. Otoni, R. C. Leite, C. Caram, C. L. Mafra, and D. H. Walker. 2002. Emerging and reemerging rickettsiosis in an endemic area of Minas Gerais State, Brazil. Cad. Saude Publica 18:1593-1597. [DOI] [PubMed] [Google Scholar]
- 18.Heo, E. J., J. H. Park, J. R. Koo, M. S. Park, M. Y. Park, J. S. Dumler, and J. S. Chae. 2002. Serologic and molecular detection of Ehrlichia chaffeensis and Anaplasma phagocytophila (human granulocytic ehrlichiosis agent) in Korean patients. J. Clin. Microbiol. 40:3082-3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hulinska, D., K. Langrova, M. Pejcoch, and I. Pavlasek. 2004. Detection of Anaplasma phagocytophilum in animals by real-time polymerase chain reaction. APMIS 112:239-247. [DOI] [PubMed] [Google Scholar]
- 20.Im, K. I., I. Y. Lee, and W. J. Lee. 1998. A human case of tick bite by Ixodes persulcatus. Korean J. Parasitol. 36:63-65. [DOI] [PubMed] [Google Scholar]
- 21.Inokuma, H., I. Brouqui, M. Drancourt, and D. Raoult. 2001. Citrate synthase gene sequence: a new tool for phylogenetic analysis and identification of Ehrlichia. J. Clin. Microbiol. 39:3031-3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Inokuma, H., T. Beppu, M. Okuda, Y. Shimada, and Y. Sakata. 2001. Detection of ehrlichial DNA in Haemaphysalis ticks recovered from dogs in Japan that is closely related to a novel Ehrlichia sp. found in cattle ticks from Tibet, Thailand, and Africa. J. Clin. Microbiol. 42:1353-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ishiguro, F., N. Takada, T. Masuzawa, and T. Fukui. 2000. Prevalence of Lyme disease Borrelia species in ticks from migratory birds on the Japanese mainland. Appl. Environ. Microbiol. 66:982-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jang, W. J., J. H. Kim, Y. J. Choi, K. D. Jung, Y. G. Kim, S. H. Lee, M. S. Choi, I. S. Kim, D. H. Walker, and K. H. Park. 2004. First serologic evidence of human spotted fever group rickettsiosis in Korea. J. Clin. Microbiol. 42:2310-2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawahara, M., T. Ito, C. Suto, S. Shibata, Y. Rikihisa, K. Hata, and K. Hirai. 1999. Comparison of Ehrlichia muris strains isolated from wild mice and ticks and serologic survey of humans and animals with E. muris as antigen. J. Clin. Microbiol. 37:1123-1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim, C. M., J. Y. Kim, Y. H. Yi, M. J. Lee, M. R. Cho, D. H. Shah, T. A. Klein, H. C. Kim, J. W. Song, S. T. Chong, M. L. O'Guinn, J. S. Lee, I. Y. Lee, J. H. Park, and J. S. Chae. 2005. Detection of Bartonella species from ticks, mites and small mammals in Korea. J. Vet. Sci. 6:327-334. [PubMed] [Google Scholar]
- 27.Kim, C. M., M. S. Kim, M. S. Park, J. H. Park, and J. S. Chae. 2003. Identification of Ehrlichia chaffeensis, Anaplasma phagocytophilum, and A. bovis in Haemaphysalis longicornis and Ixodes persulcatus ticks from Korea. Vector Borne Zoonotic Dis. 3:17-26. [DOI] [PubMed] [Google Scholar]
- 28.Ko, J. H., D. Y. Cho, B. S. Chung, and S. I. Kim. 2002. Two human cases of tick bite caused by Ixodes nipponensis. Korean J. Parasitol. 40:199-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee, J. H., H. S. Park, K. D. Jung, W. J. Jang, S. E. Koh, S. S. Kang, I. Y. Lee, W. J. Lee, B. J. Kim, Y. H. Kook, K. H. Park, and S. H. Lee. 2003. Identification of the spotted fever group rickettsiae detection from Haemaphysalis longicornis in Korea. Microbiol. Immunol. 47:301-304. [DOI] [PubMed] [Google Scholar]
- 30.Lee, S. O., D. K. Na, C. M. Kim, Y. H. Li, Y. H. Cho, J. H. Park, J. H. Lee, S. K. Eo, T. A. Klein, and J. S. Chae. 2005. Identification and prevalence of Ehrlichia chaffeensis infection in Haemaphysalis longicornis ticks from Korea by PCR, sequencing and phylogenetic analysis based on 16S rRNA gene. J. Vet. Sci. 6:151-155. [PubMed] [Google Scholar]
- 31.Leutenegger, C. M., N. Pusteria, R. Wicki, and H. Lutz. 2002. New molecular biology detection methods for tick-borne infectious agents. Schweiz Arch. Tierheilkd. 144:395-404. [DOI] [PubMed] [Google Scholar]
- 32.Loftis, A. D., R. F. Massung, and M. L. Levin. 2003. Quantitative real-time PCR assay for detection of Ehrlichia chaffeensis. J. Clin. Microbiol. 41:3870-3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Long, S. W., X. Zhang, J. Zhang, R. P. Ruble, P. Teel, and X. J. Yu. 2003. Evaluation of transovarial transmission and transmissibility of Ehrlichia chaffeensis (Rickettiales: Anaplasmataceae) in Amblyoma americanus (Acari: Ixodidae). J. Med. Entomol. 40:1000-1004. [DOI] [PubMed] [Google Scholar]
- 34.Maeda, K., N. Markowitz, R. C. Hawley, M. Ristic, D. Cox, and J. E. Mcdade. 1987. Human infection with Ehrlichia canis, a leukocytic rickettsia. N. Engl. J. Med. 316:853-856. [DOI] [PubMed] [Google Scholar]
- 35.Magnarelli, L. A., J. F. Anderson, K. C. Stafford, and J. S. Dumler. 1997. Antibodies to multiple tick-borne pathogens of babesiosis, ehrlichiosis, and Lyme borreliosis in white-footed mice. J. Wildl. Dis. 33:466-473. [DOI] [PubMed] [Google Scholar]
- 36.Mathew, J. S., S. A. Ewing, G. L. Murphy, K. C. Kocan, R. E. Cortvet, and J. C. Fox. 1997. Characterization of a new isolate of Ehrlichia platys (order Rickettsiales) using electron microscopy and polymerase chain reaction. Vet. Parasitol. 68:1-10. [DOI] [PubMed] [Google Scholar]
- 37.Murphy, G. L., S. A. Ewing, L. C. Whitworth, J. C. Fox, and A. A. Kocan. 1998. A molecular and serologic survey of Ehrlichia canis, E. chaffeensis, and E. ewingii in dogs and ticks from Oklahoma. Vet. Parasitol. 79:325-339. [DOI] [PubMed] [Google Scholar]
- 38.Ogden, N. H., K. Bown, and B. K. Horrocks. 1998. Granulocytic Ehrlichia infection in ixodid ticks and mammals in woodlands and uplands of the U.K. Med. Vet. Entomol. 12:423-429. [DOI] [PubMed] [Google Scholar]
- 39.Oh, S. H., Y. H. Song, D. H. Yoo, S. Y. Kim, and H. Lee. 1993. Distribution of Borrelia burgdorferi specific antibody among patients with juvenile rheumatoid arthritis in Korea. J. Korean Med. Sci. 8:405-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park, J. H., E. J. Heo, K. S. Choi, J. S. Dumler, and J. S. Chae. 2003. Detection of antibodies to Anaplasma phagocytophilum and Ehrlichia chaffeensis antigens in sera of Korean patients by Western immunoblotting and indirect immunofluorescence assays. Clin. Diagn. Lab. Immunol. 10:1059-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pusterla, N., J. B. Huder, C. M. Leutenegger, U. Braun, E. John, J. E. Madigan, and H. Lutz. 1999. Quantitative real-time PCR for detection of members of the Ehrlichia phagocytophila genogroup in host animals and Ixodes ricinus ticks. J. Clin. Microbiol. 37:1329-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Regnery, R. L., C. L. Spruill, and B. D. Plikaytis. 1991. Genotypic identification of rickettsiae and estimation of intraspecies sequence divergence for portions of two rickettsial genes. J. Bacteriol. 173:1576-1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sachar, D. S. 2000. Ehrlichia chaffeensis infection in an active duty soldier stationed in Korea. MSMR 6:9-11. [Google Scholar]
- 44.Schouls, L. M., I. Van De Pol, S. G. Rijpkema, and C. S. Schot. 1999. Detection and identification of Ehrlichia, Borrelia burgdorferi sensu lato, and Bartonella species in Dutch Ixodes ricinus ticks. J. Clin. Microbiol. 37:2215-2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scola, B. L., Z. Liang, Z. Zeaiter, P. Houpikian, P. A. D. Grimont, and D. Raoult. 2002. Genotypic characteristics of two serotypes of Bartonella henselae. J. Clin. Microbiol. 40:2002-2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Song, G., C. S. Hang, X. Z. Qui, D. S. Ni, H. X. Liao, G. Z. Gao, Y. L. Du, J. K. Xu, Y. S. Wu, J. N. Zhao, B. X. Kong, Z. S. Wang, Z. Q. Zhang, H. K. Shen, and N. Zhou. 1983. Etiologic studies of epidemic hemorrhagic fever (hemorrhagic fever with renal syndrome). J. Infect. Dis. 147:654-659. [DOI] [PubMed] [Google Scholar]
- 47.Sparagano, O. A. E., M. T. E. P. Allsopp, R. A. Mank, S. G. T. Rijpkema, J. V. Figueroa, and F. Jongejan. 1999. Molecular detection of pathogen DNA in ticks (Acari: Ixodidae): a review. Exp. Appl. Acarol. 23:929-960. [DOI] [PubMed] [Google Scholar]
- 48.Swofford, D. L. 2002. PAUP*. Phylogenetic analysis using parsimony (*and other methods), version 4. Sinauer Associates, Sunderland, Mass.
- 49.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.von Loewenich, F. D., G. Stumpf, B. U. Baumgarten, M. Rollinghoff, J. S. Dumler, and C. Bogdan. 2003. A case of equine granulocytic ehrlichiosis provides molecular evidence for the presence of pathogenic Anaplasma phagocytophilum (HGE agent) in Germany. Eur. J. Clin. Microbiol. Infect. Dis. 22:303-305. [DOI] [PubMed] [Google Scholar]
- 51.Walls, J. J., B. Greig, D. F. Neitzel, and J. S. Dumler. 1997. Natural infection of small mammal species in Minnesota with the agent of human granulocytic ehrlichiosis. J. Clin. Microbiol. 35:853-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang, I. N., D. E. Dykhulzen, W. Qiu, J. J. Dunn, E. M. Bosler, and B. J. Luft. 1999. Genetic diversity of ospC in a local population of Borrelia burgdorferi sensu stricto. Genetics 151:15-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yabsley, M. J., A. S. Varela, and C. M. Tate. 2002. Ehrlichia ewingii infection in white-tailed deer (Odocoileus virginianus). Emerg. Infect. Dis. 8:668-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yamaguti, N., V. J. Tipton, H. L. Keegan, and S. Toshiaoka. 1971. Ticks of Japan, Korea, and the Ryukyu Islands. Brigham Young Univ. Sci. Bull. Biol. Ser. 15:1-225. [Google Scholar]