Abstract
The dehalorespiring Desulfitobacterium hafniense strain Y51 efficiently dechlorinates tetrachloroethene (PCE) to cis-1,2-dichloroethene (cis-DCE) via trichloroethene by PceA reductive dehalogenase encoded by the pceA gene. In a previous study, we found that the significant growth inhibition of strain Y51 occurred in the presence of commercial cis-DCE. In this study, it turned out that the growth inhibition was caused by chloroform (CF) contamination of cis-DCE. Interestingly, CF did not affect the growth of PCE-nondechlorinating SD (small deletion) and LD (large deletion) variants, where the former fails to transcribe the pceABC genes caused by a deletion of the promoter and the latter lost the entire pceABCT gene cluster. Therefore, PCE-nondechlorinating variants, mostly LD variant, became predominant, and dechlorination activity was significantly reduced in the presence of CF. Moreover, such a growth inhibitory effect was also observed in the presence of carbon tetrachloride at 1 μM, but not carbon dichloride even at 1 mM.
Chlorinated organic solvents, such as tetrachloroethene (PCE) and trichloroethene (TCE), have been recognized worldwide as some of the most serious environmental pollutants. Because highly chlorinated chemicals are typically not degraded through oxygenation by aerobic bacteria, anaerobic dehalorespiring bacteria have received increased attention in the last decade. Dehalorespiration is an efficient dechlorination mechanism in which halogenated compounds can be used as the final electron acceptor. This reductive dechlorination is coupled with energy-yielding phosphorylation. Dehalorespiring bacteria are believed to play an important role in the degradation of chlorinated environmental pollutants. Since Desulfomonile tiedjei DCB-1, capable of the reductive dechlorination of chlorobenzoates, was first reported (5, 16), a number and variety of dehalorespiring bacteria have been isolated to date (4, 10, 11, 13, 17, 18, 22). Desulfitobacterium hafniense strain Y51 is one of these bacteria that dechlorinates PCE to cis-1,2-dichloroethene (cis-DCE) via TCE (19).
We previously purified and characterized the reductive PCE dehalogenase PceA and cloned the corresponding pceA gene from Desulfitobacterium hafniense strain Y51 (GenBank accession no. AY706985) (20). Subsequently, the rest of the pce genes, such as pceB, pceC, and pceT, were cloned (GenBank accession no. AY706985) (7, 20). The pceABCT gene cluster was situated between the two nearly identical IS elements termed ISDesp1 and ISDesp2, which belong to the IS256 family. Thus, the set of these genes could form a composite transposon (8). We isolated two different PCE-nondechlorinating variants that emerged during repeated subculturing of strain Y51 and named them SD (small deletion) and LD (large deletion) (8). Sequencing analysis revealed that the SD variant lost the ISDesp1 element (ca. 1.6-kb DNA region upstream of pceA), which includes the −35 promoter sequence of the pceA gene, so that the SD variant failed to transcribe the pceA gene. On the other hand, the LD variant lost ca. 6.5 kb including the pceABCT genes. This deletion took place by homologous recombination between ISDesp1 and ISDesp2 (8). Moreover, we found a significant growth inhibition when strain Y51 was cultured in the presence of 5 mM cis-DCE (7). After an extended lag time, the cells grew normally. This growth inhibition was observed to a lesser extent when the cells were grown with 5 mM PCE, but not with 5 mM TCE (7). In this study, we happened to find that the cis-DCE used in the previous study contained significant amounts of chloroform (CF). These findings led us to investigate how CF affects the growth of strain Y51 and the stability of the pce genes. We now report CF inhibits the growth of strain Y51, but not SD and LD variants, so that these deletion variants become predominant in the presence of CF. These findings should be important to the bioremediation and natural attenuation in the cocontaminated site by chloroethenes and chloromethanes.
MATERIALS AND METHODS
Chemicals.
Chloroethenes and chloromethanes were purchased from the following companies: PCE, Ishizu Seiyaku (Osaka, Japan); TCE, Katayama Chemical (Osaka, Japan); cis-DCE, Tokyo Kasei Kogyo (Tokyo, Japan); all chloromethanes, GL Sciences (Tokyo, Japan). Quantitative analyses of contaminants in PCE, TCE, and cis-DCE were done at the Shimadzu Analytical and Measuring Center. CF contaminated the chloroethenes as follows: 0.62% (mol/mol) in cis-DCE, 0.00026% (mol/mol) in TCE, and 0.0017% (mol/mol) in PCE.
Strain and cultivation.
Desulfitobacterium sp. strain Y51 was identified as Desulfitobacterium hafniense strain Y51 in a previous study (15). The SD and LD variants were isolated and characterized in a previous study (8). The wild-type (wt) strain Y51 and its SD and LD variants were anaerobically grown at 30°C in minimum medium containing yeast extract, pyruvate, and fumarate (MMYPF medium) (45.9 mM K2HPO4, 8.8 mM KH2PO4, 1.7 mM sodium citrate, 0.4 mM MgSO4 · 7H2O, yeast extract [2.0 g/liter], 68.2 mM sodium pyruvate, 5.0 mM sodium fumarate, and 4.0 μM resazurin sodium salt at pH 7.2) with or without chloroethenes or chloromethanes (19). Chloroethenes and chloromethanes dissolved in N,N-dimethylformamide were added to the culture medium after inoculation. The growth of strain Y51 and its variants was measured by their optical density at 660 nm. Growth was measured at least three times independently.
Production of biogenic cis-DCE.
cis-DCE was produced biologically from TCE using a culture of strain Y51. Strain Y51 was grown in 1,000 ml MMYPF medium and harvested at the stationary phase by centrifugation at 4,000 × g for 15 min. The cells were suspended in 160 ml MMYP medium, which is a variant of MMYPF, lacking fumarate, and TCE was added at 6 mM. The complete conversion of TCE to biogenic cis-DCE was confirmed by gas chromatography as previously described (19). Samples of the liquid phase were extracted by a sterile syringe equipped with a 0.45-μm filter and added to fresh MMYPF cultures of strain Y51 to achieve appropriate cis-DCE concentrations.
Stability of the pceA and tnpA genes.
The cells cultivated in MMYPF medium with or without CF were harvested at the stationary phase, and the genomic DNA was isolated according to the established procedures (21). The genomic DNA (500 ng) digested with EcoRI was subjected to the 0.8% agarose gel electrophoresis. Hybridization with digoxigenin (DIG)-labeled (Roche, Penzberg, Germany) pceA and tnpA as DNA probes was done to detect the presence or absence of the pceA and tnpA genes. This experiment was done in duplicate.
Detection of the pceA deletion variants.
Strain Y51 was inoculated into MMYPF medium with or without chloromethanes and anaerobically cultivated until the cells reached the stationary phase. The cells were then spread onto the MMYPF agar medium. The agar (Sigma Aldrich Japan, Tokyo, Japan) was added to the MMYPF medium at 1.5%. Two hundred colonies were subjected to colony hybridization using the DIG-labeled pceA gene as a probe to count the pceA deletion variants. The frequency of loss of the pceA gene (as a percentage) was calculated as the ratio of the number of pceA-deleted colonies to the total number of colonies. This experiment was done three times independently.
PCE dehalogenase activity.
The crude cell extracts were prepared from Y51 cells grown to stationary phase as previously described (19). PCE dehalogenase activity was measured in the following mixture (1.0 ml in a 5-ml serum vial): cell extracts (200 μg protein/ml), 2.5 mM sodium pyruvate, 0.2 mM NADH, 0.2 mM NADPH, 10 mM flavin adenine dinucleotide, 0.4 mM FeSO4, 3 mM methyl viologen, 25 mM imidazole, 5 mM dithiothreitol, and 10% glycerol at pH 7.5. The cell extract (200 μg protein/ml) of Clostridium saccharoperbutylacetonicum ATCC 13564 containing hydrogenase was added to reduce the methyl viologen. The vials were sealed with butyl rubber stoppers and crimped. PCE was added to get a final concentration of 600 μM. The reaction mixtures were incubated at 30°C with gentle shaking. The chloroethenes generated by the PCE dechlorination reaction were quantified by gas chromatography (19).
Immunoblotting of PCE dehalogenase.
The cell extracts of strain Y51 (5 μg) were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The PCE reductive dehalogenase (PceA) was detected by immunoblotting using the anti-PceA antiserum as described previously (20).
RT-PCR of the pce transcripts.
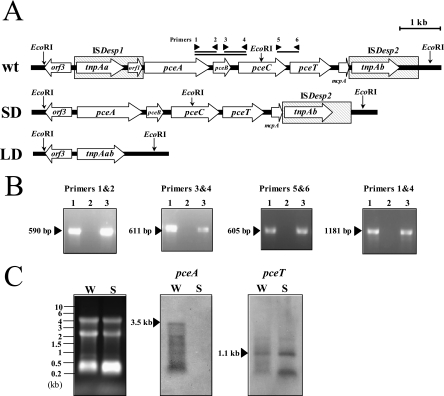
Cells of wt Y51 and SD variant cultivated in MMYPF medium were harvested at the mid-logarithmic phase. Total RNA from these cells was isolated by TRIzol reagent (GIBCO-BRL) according to the manufacturer's protocol. The resultant RNA was treated with DNase I for 1 h at 37°C and used as a template RNA for the reverse transcriptase PCR (RT-PCR) using the SuperScript III One Step RT-PCR synthesis system with Platinum Taq (Invitrogen) according to the manufacturer's protocol. For amplification of the intergenic regions between pceA and pceB, pceB and pceC, pceC and pceT, and pceA and pceC (see Fig. 4A), four sets of forward and reverse primers were chosen as follows: primer 1 (5′-TGGCATCTTGATTCCAACCG-3′) and primer 2 (5′-AATCGCAGGAATAAGTGCCG-3′), primer 3 (5′-CGGCACTTATTCCTGCGATT-3′) and primer 4 (5′-GCTCCTTGATTGCAAGACCT-3′), primer 5 (5′-GGTGACTATGGGACATTGGA-3′) and primer 6 (5′-TAACCGGTACTGTTGCATCC-3′), and primer 1 and primer 4, respectively.
FIG. 4.
(A) Gene organization around the pce gene cluster of wt strain Y51 and SD and LD variants. Primers for RT-PCR used in this study are depicted as arrowheads. The EcoRI sites are shown by vertical arrows. The mRNA transcripts detected by RT-PCR are shown as solid bars. (B) RT-PCR analyses for total RNA from wt strain Y51. RT-PCR was done targeting intergenic regions between pceA and pceB (primers 1 and 2), pceB and pceC (primers 3 and 4), pceC and pceT (primers 5 and 6), and pceA and pceC (primers 1 and 4), respectively. Lane 1, genomic DNA as the template; lane 2, RNA as the template, no RT; lane 3, RNA as the template, with RT. The sizes of RT products are indicated to the left of the blots. (C) Northern blot analyses using pceA and pceT as the probes for wt strain Y51 (W) and SD variant (S). Total RNAs from wt Y51 and SD variant were electrophoresed with RNA size markers (leftmost blot).
Northern blot analysis of the pce transcripts.
Total RNA (20 μg) was electrophoresed through a 1.0% agarose gel with 15% (vol/vol) formaldehyde and transferred to a Hybond N membrane (Amersham Biosciences, Buckinghamshire, United Kingdom). Hybridization with pceA and pceT labeled with DIG as DNA probes and subsequent detection with the nitroblue tetrazolium-bromochloroindolylphosphate (Roche, Penzberg, Germany) were done according to the manufacturer's instructions.
RESULTS
Comparison of inhibitory effect between commercial and biogenic cis-DCE on the growth of D. hafniense strain Y51.
In the previous study, 5 mM commercial cis-DCE inhibited the growth of strain Y51. However, it turned out that the commercial cis-DCE is contaminated by CF at 0.62% (mol/mol). Therefore, to investigate whether the commercial cis-DCE is intrinsically inhibitory, we compared the inhibitory effect between commercial and biogenic cis-DCE on the growth of strain Y51 (data not shown). The addition of commercial cis-DCE at 20 μM did not affect the growth of strain Y51. However, a prolonged lag period (ca. 24 h) was observed at 200 μM commercial cis-DCE, where the CF concentration was 1.2 μM in the culture medium. On the other hand, 200 μM biogenic cis-DCE did not affect growth. This result indicates that cis-DCE is not intrinsically inhibitory for the growth of strain Y51.
Inhibitory effect of CF on the growth of strain Y51.
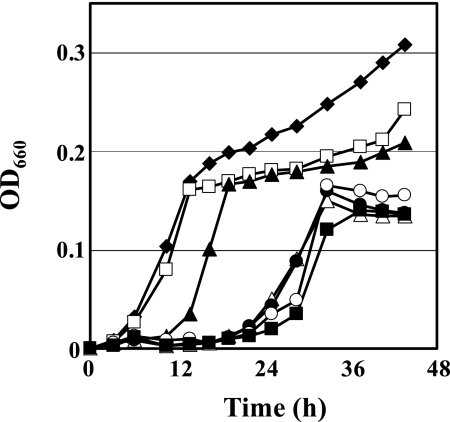
We investigated the effect of CF on the growth of strain Y51 (Fig. 1). Strain Y51 was inoculated into MMYPF medium, and CF was added at various concentrations. No significant growth delay was observed at 0.01 μM. However, a growth lag of ca. 10 h was observed at 0.1 μM CF. The growth lag was extended for ca. 24 h at 1, 10, 100 and 1,000 μM, and the cells grew normally after a prolonged lag phase in these cases.
FIG. 1.
Effect of CF on the growth of wt strain Y51. Strain Y51 was cultivated in MMYPF medium alone (⧫) or supplemented with 0.01 μM CF (□), 0.1 μM CF (▴), 1 μM CF (▵), 10 μM CF (•), 100 μM CF (○), or 1,000 μM CF (▪). OD600, optical density at 660 nm.
PCE dehalogenase activity of the cells grown with CF.
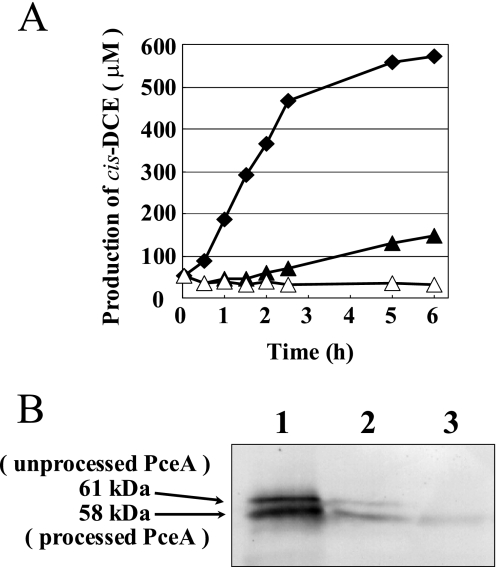
Because CF significantly affected the growth of strain Y51, we determined PCE dehalogenation activity when the cells were grown in MMYPF medium with or without CF (Fig. 2A). The cell extracts were subjected to a dechlorination assay. The cell extracts from strain Y51 grown without CF exhibited a significant dechlorination activity toward PCE. On the other hand, the cell extracts from strain Y51 grown with 0.1 μM CF showed reduced PCE dechlorination. The cell extract from strain Y51 grown with 1 μM CF had no PCE-dechlorinating ability. These results agreed with the results of the immunoblot analysis using the anti-PceA antiserum (Fig. 2B). The PceA protein signal became significantly weaker in the cells grown with 0.1 μM CF and was dramatically weaker with 1 μM CF.
FIG. 2.
(A) PCE dechlorination of cell extracts from wt strain Y51 after growth in MMYPF medium alone (⧫) or supplemented with 0.1 μM CF (▴) or 1 μM CF (▵). (B) Immunoblot analysis using the anti-PceA antiserum of cell extracts from wt strain Y51. Lanes 1 to 3 depict strain Y51 grown in MMYPF medium, MMYPF with 0.1 μM CF, and MMYPF with 1 μM CF, respectively.
High frequency of emergence of pceA deletion variants in the presence of CF.
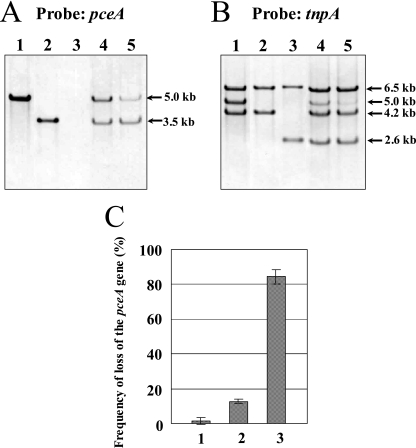
Because the PCE dehalogenase activity of cells grown with CF was significantly reduced, we investigated how CF affects the pce genes. Southern blot analysis, using pceA as a probe, was done for the genomic DNA from strain Y51 cultivated in MMYPF medium with or without CF (Fig. 3A). The 5.0-kb and 3.5-kb EcoRI fragments originating from wt Y51 and SD variant were detected, respectively, and the signal was not detected from the LD variant. On the other hand, in wt Y51 grown with 0.1 and 1 μM CF, the intensity of the 5.0-kb band became faint, and the thin 3.5-kb band appeared. These results suggested that CF promotes the high frequency of emergence of the pceA deletion variant and slight emergence of SD variant at less than 1 μM. In addition, we used the tnpA gene as a probe to investigate the emergence of the LD variant, which had lost the pce genes (Fig. 3B). The genome analysis revealed that three tnpA genes are present in strain Y51 (15); three EcoRI fragments of 6.5, 5.0, and 4.2 kb were detected from wt Y51, in which the 6.5-kb EcoRI fragment is derived from tnpA located far from the pce gene cluster, the 5.0-kb fragment contains tnpAa, and the 4.2-kb fragment contains tnpAb. Because the SD variant lost ISDesp1, including the tnpAa gene, the 5.0-kb band disappeared (Fig. 4A). From the LD variant, the 2.6-kb band was detected, which contains a hybrid tnpAab gene generated by homologous recombination between tnpAa and tnpAb (8). The 6.5-kb band was detected in both SD and LD variants. In the cells grown with 0.1 and 1 μM CF, the intensity of the 5.0-kb band (in wt Y51) became faint and the 2.6-kb band (in the LD variant) appeared. These results indicate that CF promotes the high frequency of emergence of the LD variant.
FIG. 3.
(A) Southern blot analysis of the genomic DNA from strain Y51 grown with or without CF using pceA as a probe. Genomic DNA was digested by EcoRI. Strains and culture conditions for the lanes are as follows: lane 1, wt Y51, MMYPF medium; lane 2, SD variant, MMYPF; lane 3, LD variant, MMYPF; lane 4, wt Y51, MMYPF with CF at 0.1 μM; lane 5, wt Y51, MMYPF with CF at 1 μM. (B) Southern blot analysis of the genomic DNA from strain Y51 grown with or without CF using tnpA as a probe. Genomic DNA was digested by EcoRI. Strains and culture conditions were the same as in panel A. (C) Effect of CF on the frequency of loss of the pceA gene in wt strain Y51. Two hundred colonies were subjected to colony hybridization using pceA as a probe. Culture conditions for the bars are as follows: 1, MMYPF medium; 2, MMYPF with CF at 0.1 μM; 3, MMYPF with CF at 1 μM.
We also confirmed the pceA deletion variant by colony hybridization using pceA as a probe (Fig. 3C). Even after growth in the presence of 1 μM CF, 84% ± 4% of the cells lost pceA. In addition, we confirmed that the 200 μM biogenic cis-DCE did not affect the frequency of emergence of the pceA deletion variants (data not shown).
Effects of CF on the growth of SD and LD variants.
We were interested in investigating the reason why the cells grew normally after an extended lag phase when wt strain Y51 was inoculated with CF. For this purpose, we investigated the growth of SD and LD variants in the presence or absence of CF. The SD and LD variants grew normally in the MMYPF medium using fumarate as an electron acceptor (data not shown). Surprisingly, no significant growth inhibition of these nondechlorinating variants was observed in the presence of CF at concentrations of 0.01, 0.1, 1, 10, 100, and 1,000 μM (data not shown). Thus, such SD and LD variants seem to be selectively enriched in the presence of CF.
Effects of CD and CT on the growth of the strain and deletion of the pceA gene.
We next investigated whether the negative effects caused by CF are also true for other chloromethanes, such as carbon dichloride (CD) and carbon tetrachloride (CT). First, we confirmed that strain Y51 could not dechlorinate CT and CD as well as it could dechlorinate CF (data not shown). The addition of CD at 1,000 μM did not affect the growth of wt Y51 and SD and LD variants (data not shown). However, the addition of CT caused a significant growth inhibition of wt Y51 even at 0.1 μM. The prolonged lag period of the growth of wt Y51 was observed with the increased concentrations of CT up to 100 μM as follows: 0.1 μM, ca. 6 h; 1 μM, ca. 21 h; 10 μM, ca. 24 h; and 100 μM, ca. 90 h. On the other hand, the growth of the SD and LD variants was not affected by CT up to 1 μM; however, at 10 μM and 100 μM, an extension of the lag phase was observed as follows: 10 μM, ca. 6 h; and 100 μM, ca. 79 h. At 1,000 μM CT, the growth of wt Y51 and SD and LD variants was completely inhibited. Thus, CT exhibited growth inhibition not only for wt Y51 but also for the SD and LD variants, but to a much lesser extent for the latter two.
Subsequently, we investigated the effects of CT and CD on the stability of the pceA gene. Colony hybridization using pceA as a probe demonstrated that CT promoted enrichment of the LD variant, as did CF. The growth of strain Y51 with 1 μM CT resulted in the generation of 94% ± 6% pceA deletion variants. However, CD did not generate such deletion variants even at 1,000 μM.
Transcriptional analyses of the pce gene cluster.
We were interested in investigating the reason why growth inhibition was different for the wt strain Y51 and the SD and LD variants. Since the difference in the genotype between wt Y51 and the deletion variants is the regions around the pceABCT gene cluster, it is likely that the difference in the sensitivities to CF and CT between wt Y51 and its deletion variants is due to the difference in the expression of pceABCT genes. Therefore, RT-PCR and Northern blot analyses were done to investigate the transcription of the pce gene cluster (Fig. 4). RNA isolated from the cells grown with pyruvate (as an electron donor) and fumarate (as an electron acceptor) revealed that PceA is significantly produced in the absence of chloroethenes. RT-PCR toward the total RNA from wt Y51 with respective primer pairs designed to amplify the intergenic regions between pceA and pceB (using primers 1 and 2), pceB and pceC (primers 3 and 4), pceC and pceT (primers 5 and 6), and pceA and pceC (primers 1 and 4) yielded products with the expected sizes (Fig. 4B). Northern blot analyses revealed that ca. 3.5-kb mRNA was detected, indicating that the pceA, pceB, and pceC genes were transcribed in a polycistronic manner in wt Y51 (Fig. 4C). Also, ca. 1.1-kb mRNA revealed that pceT was transcribed in a monocistronic manner (Fig. 4C). On the other hand, in the SD variant, pceA, pceB, and pceC mRNA were not detected, but the pceT mRNA was transcribed as in wt Y51. These results indicated that pceA, pceB, and pceC were cotranscribed and that pceT was transcribed alone. However, the RT-PCR result (using primers 5 and 6) indicated that the pceT gene is also transcribed along with pceABC to some extent. Thus, the growth inhibition of wt strain Y51 by CF or CT seems to be caused by the interaction of PceA, PceB, or PceC with CF or CT.
DISCUSSION
In a previous study, the stability of pceA in D. hafniense strain Y51 was investigated after serial subculturing with or without chloroethenes (7). Initially, we found that the growth of strain Y51 was inhibited and more than 80% of the cells in a stationary phase lost the pceA gene after a single subculture in the presence of 5 mM cis-DCE. Afterward it turned out that the cis-DCE used in the previous study contained 0.62% (mol/mol) CF, which results in the contamination of 31 μM CF in the culture medium containing 5 mM cis-DCE. This concentration of CF is more than enough to inhibit the growth of wt strain Y51, but not SD and LD variants. We also confirmed that biogenic cis-DCE did not cause such growth inhibition. These results indicate that cis-DCE does not affect the growth of wt Y51 and enrichment of LD variant. PCE used in the previous study also stimulated the appearance of the LD variant at 5 mM but to a lesser extent than does commercial cis-DCE. This is simply because the addition of 5 mM PCE results in the contamination of 0.085 μM CF; this amount is much lower than that in 5 mM cis-DCE (31 μM CF). On the other hand, 5 mM TCE results in 0.013 μM CF, so that no significant growth inhibition or enrichment of LD variant occurred at this concentration. In the present study, we found that CF significantly affected dehalogenation activity and prevalence of pce gene deletion mutants at a concentration of 0.1 μM, but not at 0.01 μM. This is in perfect agreement with the effects observed for commercial chloroethene preparations.
CT also caused a growth delay in wt strain Y51. Unlike CF and CT, no negative effects were observed with CD. Therefore, the effects of chloromethanes on the growth inhibition of wt strain Y51 seem to be strictly dependent on the chlorine substitution on the methane structure. CT seems to have an additional toxic effect different from CF because the growth of SD and LD variants were also inhibited by CT at higher concentrations. Although wt Y51 did not dechlorinate these chloromethanes, Desulfitobacterium hafniense TCE1 did, forming a small amount of CD (9). Because strain TCE1 dechlorinates PCE to cis-DCE by PCE dehalogenase, which exhibits a 99.8% similarity to that of strain Y51, strain TCE1 may have other enzymes responsible for the reductive dechlorination of chlorinated methanes.
In the previous study, it was found that the LD variant was generated 6.5 times more frequently than the SD variant when wt strain Y51 was subcultured in MMYPF medium (8). This is comparable to the high emergence of LD variant when wt strain Y51 was cultured in the presence of CF and CT. These SD and LD variants were desensitized to CF even at 1,000 μM. The pceA, pceB, and pceC genes failed to be expressed in the SD variant because of the deletion of the promoter, and the same genes were completely deleted in the LD variant. In this context, the difference in the growth inhibition by CF between wt Y51 and the SD and LD variants seems to be caused by the presence or absence of three proteins encoded by pceA, pceB, and pceC. Among these proteins, PceA is a reductive dehalogenase, PceB is believed to be a membrane anchor protein associated with PceA, and PceC is an unknown protein similar to NosR that is involved in nitrous oxide (N2O) respiration (3, 23). Our hypothesis is that one of these three proteins may interact with CF or CT to inhibit the growth of the wt strain. Since SD and LD variants do not produce these proteins, CF does not inhibit the growth of SD and LD variants. Our efforts are currently focusing on which protein, if any, interacts with CF and what the mechanism is.
Maymo-Gatell et al. found that the commercially synthesized cis-DCE contained approximately 0.4% (mol/mol) CF, which was inhibitory for chloroethene utilization by Dehalococcoides ethenogenes strain 195 (12). They biologically produced cis-DCE from PCE using a Desulfitobacterium culture. Such a biogenic cis-DCE was readily utilized by strain 195 (12). Chloromethanes were reported to be inhibitory to a PCE dehalogenase of Sulfurospirillum multivorans (14). A 50% inhibition of the PCE dehalogenase reaction was observed at the following concentrations: 50 μM for CD, 25 μM for CF, and 100 μM for CT. In the chloroethene-dechlorinating consortium, it was also reported that CF inhibited the dechlorination reaction, accumulating vinyl chloride (6). The complete inhibition of dechlorination of vinyl chloride by CF was observed at 2.5 μM. Furthermore, CF is reported to be a potent inhibitor of methanogenesis (1, 2). However, the biochemical and genetic bases of these inhibitory effects by chloroethenes remain to be elucidated. In this study, we suggested for the first time that the growth inhibition by CF and CT could take place with interaction between a certain protein(s) (PceA, PceB, or PceC) and CF or CT in the halorespiring D. hafniense strain Y51.
The results obtained in this study should be important from the viewpoints of toxicology and bioremediation including natural attenuation. It should be also important to investigate how CF affects various halorespiring anaerobes in the environment because CF and CT are widely used and known as environmental contaminants all over the world. We are currently investigating how such anaerobes dechlorinate when chloromethanes and chloroethenes coexist in the environment.
Acknowledgments
This work was supported by a grant-in-aid (Hazardous Chemicals) from the Ministry of Agriculture, Forestry, and Fisheries of Japan (HC-04-2321-1) and by a grant-in-aid (Mechanism of Biodegrading and Processing) from New Energy and Industrial Technology Development Organization (NEDO). T.F. was supported by a research fellowship of the Japan Society for the Promotion of Science for Young Scientists.
REFERENCES
- 1.Bagley, D. M., and J. M. Gossett. 1995. Chloroform degradation in methanogenic methanol enrichment cultures and by Methanosarcina barkeri 227. Appl. Environ. Microbiol. 61:3195-3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bauchop, T. 1967. Inhibition of rumen methanogenesis by methane analogues. J. Bacteriol. 94:171-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cuypers, H., A. Viebrock-Sambale, and W. G. Zumft. 1992. NosR, a membrane-bound regulatory component necessary for expression of nitrous oxide reductase in denitrifying Pseudomonas stutzeri. J. Bacteriol. 174:5332-5339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Damborsky, J. 1999. Tetrachloroethene-dehalogenating bacteria. Folia Microbiol. 44:247-262. [DOI] [PubMed] [Google Scholar]
- 5.DeWeerd, K. A., L. Mandelco, R. S. Tanner, C. T. Woese, and J. M. Suflita. 1990. Desulfomonile tiedjei gen. nov. and sp. nov., a novel anaerobic, dehalogenating, sulfate-reducing bacterium. Arch. Microbiol. 154:23-30. [Google Scholar]
- 6.Duhamel, M., S. D. Wehr, L. Yu, H. Rizvi, D. Seepersad, S. Dworatzek, E. E. Cox, and E. A. Edwards. 2002. Comparison of anaerobic dechlorinating enrichment cultures maintained on tetrachloroethene, trichloroethene, cis-dichloroethene and vinyl chloride. Water Res. 36:4193-4202. [DOI] [PubMed] [Google Scholar]
- 7.Furukawa, K., A. Suyama, Y. Tsuboi, T. Futagami, and M. Goto. 2005. Biochemical and molecular characterization of a tetrachloroethene dechlorinating Desulfitobacterium sp. strain Y51: a review. J. Ind. Microbiol. Biotechnol. 32:534-541. [DOI] [PubMed] [Google Scholar]
- 8.Futagami, T., Y. Tsuboi, A. Suyama, M. Goto, and K. Furukawa. 2006. Emergence of two types of nondechlorinating variants in the tetrachloroethene-halorespiring Desulfitobacterium sp. strain Y51. Appl. Microbiol. Biotechnol. 70:720-728. [DOI] [PubMed] [Google Scholar]
- 9.Gerritse, J., O. Drzyzga, G. Kloetstra, M. Keijmel, L. P. Wiersum, R. Hutson, M. D. Collins, and J. C. Gottschal. 1999. Influence of different electron donors and acceptors on dehalorespiration of tetrachloroethene by Desulfitobacterium frappieri TCE1. Appl. Environ. Microbiol. 65:5212-5221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holliger, C., and W. Schumacher. 1994. Reductive dehalogenation as a respiratory process. Antonie Leeuwenhoek 66:239-246. [DOI] [PubMed] [Google Scholar]
- 11.Holliger, C., G. Wohlfarth, and G. Diekert. 1998. Reductive dechlorination in the energy metabolism of anaerobic bacteria. FEMS Microbiol. Rev. 22:383-398. [Google Scholar]
- 12.Maymo-Gatell, X., I. Nijenhuis, and S. H. Zinder. 2001. Reductive dechlorination of cis-1,2-dichloroethene and vinyl chloride by “Dehalococcoides ethenogenes.” Environ. Sci. Technol. 35:516-521. [DOI] [PubMed] [Google Scholar]
- 13.Mohn, W. W., and J. M. Tiedje. 1992. Microbial reductive dechlorination. Microbiol. Rev. 56:482-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neumann, A., G. Wohlfarth, and G. Diekert. 1996. Purification and characterization of tetrachloroethene reductive dehalogenase from Dehalospirillum multivorans. J. Biol. Chem. 271:16515-16519. [DOI] [PubMed] [Google Scholar]
- 15.Nonaka, H., G. Keresztes, Y. Shinoda, Y. Ikenaga, M. Abe, K. Naito, K. Inatomi, K. Furukawa, M. Inui, and H. Yukawa. 2006. Complete genome sequence of the dehalorespiring bacterium Desulfitobacterium hafniense Y51 and comparison with Dehalococcoides ethenogenes 195. J. Bacteriol. 188:2262-2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shelton, D. R., and J. M. Tiedje. 1984. Isolation and partial characterization of bacteria in an anaerobic consortium that mineralizes 3-chlorobenzoic acid. Appl. Environ. Microbiol. 48:840-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smidt, H., A. D. L. Akkermans, J. van der Oost, and W. M. de Vos. 2000. Halorespiring bacteria—molecular characterization and detection. Enzyme Microb. Technol. 27:812-820. [DOI] [PubMed] [Google Scholar]
- 18.Smidt, H., and W. M. de Vos. 2004. Anaerobic microbial dehalogenation. Annu. Rev. Microbiol. 58:43-73. [DOI] [PubMed] [Google Scholar]
- 19.Suyama, A., R. Iwakiri, K. Kai, T. Tokunaga, N. Sera, and K. Furukawa. 2001. Isolation and characterization of Desulfitobacterium sp. strain Y51 capable of efficient dehalogenation of tetrachloroethene and polychloroethanes. Biosci. Biotechnol. Biochem. 65:1474-1481. [DOI] [PubMed] [Google Scholar]
- 20.Suyama, A., M. Yamashita, S. Yoshino, and K. Furukawa. 2002. Molecular characterization of the PceA reductive dehalogenase of Desulfitobacterium sp. strain Y51. J. Bacteriol. 184:3419-3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilson, K. 1987. Preparation of genomic DNA from bacteria, p. 241-245. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. Greene Publishing Associates and Wiley Interscience, New York, N.Y.
- 22.Wohlfarth, G., and G. Diekert. 1997. Anaerobic dehalogenases. Curr. Opin. Biotechnol. 8:290-295. [DOI] [PubMed] [Google Scholar]
- 23.Wunsch, P., and W. G. Zumft. 2005. Functional domains of NosR, a novel transmembrane iron-sulfur flavoprotein necessary for nitrous oxide respiration. J. Bacteriol. 187:1992-2001. [DOI] [PMC free article] [PubMed] [Google Scholar]