Abstract
The marine epiphytic bacterium Pseudoalteromonas tunicata produces a range of extracellular secondary metabolites that inhibit an array of common fouling organisms, including fungi. In this study, we test the hypothesis that the ability to inhibit fungi provides P. tunicata with an advantage during colonization of a surface. Studies on a transposon-generated antifungal-deficient mutant of P. tunicata, FM3, indicated that a long-chain fatty acid-coenzyme A ligase is involved in the production of a broad-range antifungal compound by P. tunicata. Flow cell experiments demonstrated that production of an antifungal compound provided P. tunicata with a competitive advantage against a marine yeast isolate during surface colonization. This compound enabled P. tunicata to disrupt an already established fungal biofilm by decreasing the number of yeast cells attached to the surface by 66% ± 9%. For in vivo experiments, the wild-type and FM3 strains of P. tunicata were used to inoculate the surface of the green alga Ulva australis. Double-gradient denaturing gradient gel electrophoresis analysis revealed that after 48 h, the wild-type P. tunicata had outcompeted the surface-associated fungal community, whereas the antifungal-deficient mutant had no effect on the fungal community. Our data suggest that P. tunicata is an effective competitor against fungal surface communities in the marine environment.
Submerged surfaces within the marine environment are rapidly colonized by a range of microorganisms, sessile plants, and animals in a process known as biofouling. Biofouling is a highly dynamic process in which microfoulers, such as bacteria and fungi, can significantly influence the final composition of the mature fouling community through the production of secondary metabolites (34). The formation of mature biofouling communities can have detrimental effects on living surfaces, such as the loss of photosynthesis, the arrest of growth, reduced viability, and organism death (33). Also, fouling of marine surfaces can lead to decreases in the productivity of fish farms and reduction of shipping efficiency and, once established, is extremely difficult to eradicate (2).
Seaweeds have evolved a number of physical and chemical defense mechanisms to prevent fouling, such as the production of mucus, shedding of fouled tissue, and the production of inhibitory compounds (15). While remaining relatively unfouled in the marine environment, some seaweeds do not appear to produce physical or chemical defenses against fouling organisms. One such example is the green alga Ulva australis, which appears to rely on surface-associated microbes for defenses against fouling organisms (36).
Pseudoalteromonas species are marine bacteria commonly associated with surfaces. These bacteria produce a diverse range of biologically active compounds that specifically target a wide variety of marine fouling organisms (31). In a study of 10 Pseudoalteromonas species, Pseudoalteromonas tunicata was found to have the highest and broadest range of fouling inhibitory activities (31). P. tunicata colonizes a variety of eukaryotic surfaces in the marine environment, including Ulva australis, and produces antifouling compounds specifically targeting bacteria, protozoa, algal spores, invertebrate larvae, and fungi (32). The antibacterial activity exhibited by P. tunicata has been attributed to a 190-kDa multisubunit protein, designated AlpP, which is effective against both gram-negative and gram-positive bacteria from a range of environments (35). Production of AlpP by P. tunicata has been demonstrated to provide a competitive advantage when colonizing a surface in competition with other bacteria (50). In addition, P. tunicata is green due to the production of a yellow pigment, belonging to the tambjamine class of compounds (23), and a purple pigment.
Similar to bacterial biofilms, fungal biofilms are enclosed within an exopolysaccharide matrix and may occur on a wide variety of surfaces, including aquatic and terrestrial surfaces, metal, plastics, and plant and animal tissue (3, 16). The formation of fungal biofilms has been speculated to confer an ecological advantage including protection from environmental changes, increased antibiotic resistance, nutrient availability, metabolic cooperation, and the acquisition of new genetic traits (14, 17, 38, 49).
Complex interspecies and intraspecies interactions, such as coaggregation, have been found to occur in biofilms (21). For example, Candida albicans has been demonstrated to promote the attachment of different bacterial species, including Streptococcus spp., Escherichia coli, and Porphyromonas gingivalis (8, 9, 46, 47), and coadhesion between varieties of Streptococcus species promotes oral colonization by yeast cells (30). A more recent study demonstrated that biofilms of C. albicans are capable of entrapping other microorganisms and are more likely to be heterogeneous with other bacteria and fungi in the environment and on medical devices (20). Such interactions are also governed by production of bioactive compounds by the microorganisms. Miao et al. (43) reported that extracts from 32 out of 46 fungal strains isolated from seawater and marine surfaces exhibited antibacterial activity and that 17% of these isolates were in turn inhibited by at least one test bacterial species (43). Also, Gil-Turnes et al. (26) discovered that shrimp embryos are covered by a bacterium, Alteromonas sp., that produces the broad-spectrum antifungal compound isatin and that this compound protects the embryos from the pathogenic fungus Lagenidium callinectes (26). Likewise, embryos of Homarus americanus, a member of the lobster family, were protected from L. callinectes by a gram-negative bacterium that produces the antifungal metabolite tyrosol (25).
Interactions between fungi and bacteria in the marine environment are expected to be particularly prevalent on surfaces. However, while marine systems are currently being extensively explored for potential new biologically active compounds (7), especially against fungi, little is known as to whether such compounds provide an ecological advantage for fungal inhibitory bacteria when grown in competition with marine fungal species.
In this study, we tested the hypothesis that the production of the antifungal compound by P. tunicata provides a competitive advantage against marine fungi during surface colonization. A P. tunicata antifungal-deficient mutant was generated using transposon mutagenesis. The antifungal-deficient mutant and the wild type were used in colonization studies of glass flow cells in competition with a marine yeast isolated from U. australis. As these experiments demonstrated the ability of P. tunicata to disrupt an established fungal biofilm, the experiments were extended to the natural surface-associated fungal community of U. australis. Our data demonstrate that the production of the antifungal compound does provide P. tunicata a competitive advantage against fungi during surface colonization, an advantage not obtained through competition for colonization sites, physical space, or nutrients.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
All organisms used in this study are listed in Table 1. E. coli strains were maintained on Luria-Bertani agar (LB) (5) at 37°C. P. tunicata strains were maintained on Väätanen nine-salt solution agar (VNSS) (41) at room temperature. Marine minimal medium (48) supplemented with 0.5% trehalose as the carbon source (tMMM) was used for biofilm flow cell experiments. When required, streptomycin (100 μg/ml), ampicillin (100 μg/ml), and kanamycin (85 μg/ml) were added to media.
TABLE 1.
Organisms used in this study
| Species, strain, or plasmid | Relevant characteristic or genotype | Reference or source |
|---|---|---|
| Bacteria | ||
| P. tunicata | ||
| D2 | Wild type | 32 |
| D2SM | Spontaneous Smr | 19 |
| FM3 | Smr Kmr, afaA mutant | This study |
| E. coli | ||
| DH5α | 24 | |
| Sm10λ | mobRP4, π replicase (pir) | 24 |
| Yeast | ||
| Candida albicans | UNSW Culture Collectiona | |
| Rhototorula rubra | UNSW Culture Collection | |
| Y1 | Rhodosporidium sphaerocarpum isolated from Ulva australias | This study |
| Y2, Y3, Y4, Y5, Y6, Y7, Y8, and Y9 | Ulva australias surface yeast isolates | This study |
| Filamentous fungi | ||
| Aspergillus flavus | UNSW Culture Collection | |
| Aspergillus niger | UNSW Culture Collection | |
| Mucor generensis | UNSW Culture Collection | |
| Microsporon canis | UNSW Culture Collection | |
| Penicillium expansum | UNSW Culture Collection | |
| Rhizopus nigricans | UNSW Culture Collection | |
| Plasmid PLOF | Mini-Tn10 (Km), Kmr Ampr | 29 |
UNSW Culture Collection, School of Biotechnology and Biomolecular Science Culture Collection, University of New South Wales, Sydney, Australia.
Yeast and fungal strains were maintained on malt extract agar (Difco) supplemented with 10% NaCl for the marine yeast strains.
Transposon mutagenesis.
A mini-Tn10 transposon was used to generate mutants of the wild-type P. tunicata as previously described (19). To facilitate screening for mutants unable to inhibit fungal growth, nine-salt solution (NSS) (41) containing 104 fungal spores per ml was diluted 1:10 with the conjugation mixture before plating on selective media. Mutants were examined for zones of inhibition of fungal growth after 24 and 48 h. Mutants lacking a zone of inhibition after 48 h were selected.
Antifungal activity against yeast and fungal species was assessed using two agar plate-based methods. To test for inhibitory activity against yeasts, overnight cultures of yeast test strains were spread plated onto a VNSS agar plate and air dried. Filamentous fungi (Table 1) were streak inoculated in a single line. Thereafter, the wild-type P. tunicata and the antifungal-deficient mutant strain FM3 were stab inoculated from a fresh VNSS agar plate into the plate containing the fungal or yeast test strains. Plates were incubated for 48 h or until the fungi had created an even lawn of growth. At that time, zones of inhibition were visible surrounding the bacterial inoculations.
DNA extraction.
The XS buffer DNA extraction method was used to isolate genomic DNA as previously described (55). A bead beating method was used to isolate genomic DNA samples from yeast isolate Y1 as previously described (54).
Genotypic characterization of the antifungal-deficient mutant.
Genomic DNA surrounding the inserted transposon in the antifungal-deficient mutant was sequenced using the panhandle PCR method of Siebert et al. (52) as modified by Egan et al. (19). DNA sequencing was conducted using BigDye terminator cycle sequencing mix (Applied Biosystems) and analyzed on an ABI 377 DNA sequencing system at the Automated Sequencing Facility, University of New South Wales.
The completed DNA sequence was compared to known sequences in the GenBank database using the BLAST search algorithm (1), and open reading frames (ORFs) were defined using the ORF finder program (both programs made available through the National Center for Biotechnology Information [NCBI] website [http://www.ncbi.nlm.nih.gov]). Further analysis was performed using the appropriate programs in the GCG software package provided by the Australian National Genomic Information Service (ANGIS) (http://www.angis.org.au/WebANGIS/) and molecular biology analysis tools available through the ExPASy website (http://expasy.proteome.org.au/index.html).
Isolation and identification of a yeast isolate from Ulva australis.
Several yeast species were isolated from the surface of U. australis from the intertidal regions of Clovelly, Sydney, Australia. U. australis plants were harvested at low tide and stored in sterile NSS on ice during transport to the laboratory. The plants were rinsed three times with NSS and thereafter placed in 50-ml Falcon tubes filled with 10 ml of NSS. The samples were vortexed for 30 s, and the supernatants were spread plated onto malt extract agar. Individual colonies were isolated by streak plating and classified by using morphology. Nine yeast isolates, labeled Y1 to Y9, were assessed for their ability to attach to a glass surface. In brief, 10-ml samples of overnight cultures grown in tMMM were pelleted by centrifugation, resuspended in 10 ml of NSS, and added to the top of glass slides (Alltech) placed in sterile petri dishes. After 1 hour of incubation, the glass slides were examined microscopically. They were washed with 10 ml of NSS and placed in a sterile petri dish with 15 ml of tMMM and further incubated at room temperature with shaking at 30 rpm. The slides were examined microscopically after 24 h.
The yeast isolate Y1 was identified by sequencing of the internal transcribed spacer (ITS) region and partial sequences of the large 28 ribosomal DNA subunit (V3) regions of the chromosomal DNA to allow comparison with sequences in the EMBL fungal DNA database using Fasts3 sequence homology searches.
Primers used were as follows: ITS-1, TCCGTAGGTGAACCTGCGG (56); ITS-4, TCCTCCGCTTATTGATATGC (56); V3-1, GCATATCAATAAGCGGAGGAAAAG (22); and V3-2, GGTCGTGTTTCAAGACGG (22).
The cycle sequencing conditions using ITS-1 and ITS-4 primers were denaturing at 96°C for 10 s, annealing at 50°C for 5 s, and extension at 60°C for 4 min for 25 cycles, with a final extension at 60°C for 4 min. The annealing temperature was increased to 55°C for sequencing reactions using the V3-1 and V3-2 primers. Sequencing was conducted at the Automated Sequencing Facility, University of New South Wales.
Mixed glass flow cell biofilms.
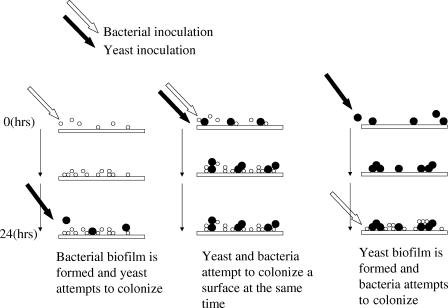
Mixed bacterial-yeast biofilms were studied using square glass flow cells (20 cm of 3-mm2 tubing; Friedrich & Dimmock Inc.). Flow cells were kept at a constant temperature by mounting on a heated brass plate connected to a solenoid. Starter cultures of the marine yeast isolate Y1, wild-type P. tunicata, and the P. tunicata antifungal-deficient mutant FM3 were grown overnight in tMMM to an optical density at 610 nm of 0.5 and washed twice by centrifugation and resuspension in sterile tMMM. The bacterial cultures were resuspended to 107 CFU/ml, and the marine yeast sample was resuspended to 106 CFU/ml. The flow cell chambers were prepared by allowing sterile media to flow through the system for 2 h at 4.8 ml/h. The flow cells were inoculated with the bacterial and yeast cultures by pumping the prepared cultures through the flow cell at 5 ml/h for 1 h. Three different inoculation strategies were conducted in the bacterial-fungal interactions (Fig. 1). During and after inoculation, the flow cells were maintained at 22°C. Sterile tMMM was pumped through the flow cells at 5 ml/h for 72 h. Biofilms were observed by transmitted bright-field microscopy using an Olympus BH2 microscope. A COHU 4612-5000 charge-coupled-device camera (Cohu, Inc., San Diego, Calif.) and a VG-5 PCI framestore board (Scion Inc., Frederick, Md.) were used to capture images. Image capture, processing, and analysis were done using NIH Image (rsb.info.nih.gov/nih-image/) on a Macintosh computer or Scion Image on a personal computer. Counts of attached yeast cells were calculated from five images taken in random locations of each flow cell. Each biofilm was conducted in triplicate in two separate experiments.
FIG. 1.
Bacterial and yeast inoculation methods for the mixed biofilm flow cell study. In the first instance (left), Rhodosporidium sphaerocarpum attachment to an established Pseudoalteromonas tunicata biofilm was investigated. In the second instance (middle), the P. tunicata and R. sphaerocarpum cultures competed in unison for an uncolonized surface, and in the third instance (right), the effect of P. tunicata attachment to an established R. sphaerocarpum biofilm was assessed.
Statistical analysis.
The mean and standard deviation were calculated from pooled counts and compared using one-way analysis of variance. Where appropriate for paired samples, t tests were used with all calculations carried out using SPSS (Statistical Package for the Social Sciences) 11 for Mac OS X software. Differences achieving a confidence level of 95% were considered significant.
Inoculation of Pseudoalteromonas tunicata wild-type and FM3 strains on surfaces of Ulva australis.
Samples of U. australis were harvested in July 2004 from the intertidal regions of Clovelly, Sydney, Australia. The plants were collected in sterile NSS and stored on ice for 2 h until processed. The U. australis samples were rinsed three times and separated at their base into four sections such that five round bore discs (25-mm diameter) could be sampled from each section. All four sections were attached to wire gauze and maintained aerated in sterile seawater using separate 1-liter beakers with a 14-h light-10-h dark cycle. After preparation, the algal samples were left for 24 h to stabilize to the experimental conditions. Overnight cultures of P. tunicata and the antifungal-deficient mutant FM3 were grown in VNSS to late logarithmic phase and harvested by centrifugation. The cells were washed in sterile seawater three times and resuspended to 108 CFU/ml in fresh sterile seawater. Samples (500 ml) of the resuspended bacterial cultures were added to the U. australis plants and allowed to attach to the algal surfaces for a period of 6 h. Control samples had sterile seawater added instead of bacterial culture. The algal discs were then removed and rinsed three times with sterile seawater and incubated with aeration at room temperature with the same light-dark cycle as described above. Each test condition was replicated five times, and the sterile seawater was changed every 24 h.
Extraction of DNA from the microbial surface community from Ulva australis.
Sections of the alga from the different treatments were collected after 0, 24, 48, and 120 h. Each sample was rinsed three times in NSS, and four discs (25-mm diameter) were taken from each section and combined for DNA extraction using the FastDNA SPIN kit for soil (BIO101 Systems, Q.BIOgene) per the manufacturer's instructions. DNA concentrations were standardized to 10 ng/μl for further PCRs.
PCR amplification and DG-DGGE.
DNA amplifications by PCR were carried out in a volume of 50 μl containing 5 μl of Sigma REDTaq buffer, 2.5 mM of each deoxynucleotide triphosphate, 25 pmol of each primer (nu-SSU-8175/nu-SSU-1196 [22]), 10 μg of bovine serum albumin, sterile filtered Milli-Q-water, 1 μl Taq polymerase (Sigma REDTaq), and 40 ng of template DNA. The thermocycling program was 3 min at 94°C, followed by 30 cycles of 1 min at 94°C, 1 min at 55°C, and 1.5 min at 72°C, and a final extension for 10 min at 72°C. PCR products were loaded on 6 to 8% polyacrylamide gradient gel (acrylamide-N,N′methylenebisacrylamide [60:1]) containing 0.13% (vol/vol) N,N,N′,N′-tetramethylethylenediamine (TEMED) and 0.06% (wt/vol) ammonium persulfate, parallel with 35 to 50% denaturant (100% denaturing solution is defined as 7 M urea and 40% formamide) using the Bio-Rad Dcode system. Fifty percent (wt/vol) glycerol was added to the 100% denaturing solution to generate the high denaturing solution to extend the density gradient in order to carry out double-gradient denaturing gradient gel electrophoresis (DG-DGGE) (13). Gels were run in 1× Tris-acetate-EDTA (TAE) buffer at 75 V for 16 h at a constant temperature of 60°C. The gels were stained with ethidium bromide (1 μg/ml), destained in 1× TAE buffer, and then photographed using a Gel-Doc 2000 imaging system (Bio-Rad).
Nucleotide sequence accession number.
The sequence has been placed in GenBank under accession number DQ385851.
RESULTS
Generation of antifungal-deficient Pseudoalteromonas tunicata mutant strains.
Mini-Tn10 transposon mutagenesis of P. tunicata produced a number of purple, yellow, and nonpigmented mutants. As pigment production has been linked to the broad range of antifouling activities of P. tunicata (19), these were not selected for study. A screen of 45,000 transposon mutants identified three mutants, designated FM1, FM2, and FM3, which produced both a yellow and purple pigment but were unable to inhibit the growth of the test Penicillium species. The mutants were unable to inhibit the growth of a range of both environmental (yeast isolates Y1 to Y9) and medical fungal strains (Penicillium expansum, Mucor generensis, Rhizopus nigricans, Aspergillus niger, Aspergillus flavus, Microsporon canis, Rhototorula rubra, and Candida albicans) that were inhibited by the wild-type strain. Analysis of the three mutants identified that the transposon insertion had occurred in the same location (data not shown); therefore, further studies were conducted using FM3 only.
Phenotypic analysis of the FM3 strain revealed the presence of all antifouling activities, apart from that of the fungal inhibitory compound associated with P. tunicata. The inhibition of bacterial growth, the settlement of invertebrate larvae (Balanus amphitrite and Hydroides elegans), and the germination of common algal spores (U. australis and Polysiphonia sp.) by the FM3 strain were similar to those of the wild type. Similarly, no differences were observed between the growth pattern of FM3 and the wild-type P. tunicata (see the supplemental material).
Genotypic characterization of the FM3 strain.
Sequence analysis was carried out via panhandle PCR and primer walking to generate sequence data flanking the transposon. A total of 3,736 bp of flanking sequence was obtained. Analysis indicated that the transposon had disrupted a 1,662-bp ORF, designated afaA (antifungal activity A), with 63% identity and 78% similarity (over 539 amino acid residues) to the E. coli long-chain-fatty-acid coenzyme A (CoA) ligase gene (fadD). The translational stop of afaA was followed by a GC-rich region of inverted repeats followed in turn by a series of six thymidine residues that may act as a ρ-independent terminator of transcription. An open reading frame designated afaB, which shares sequence similarity to a group of putative hydrolases was directly upstream of afaA. The close proximity of afaA and afaB and the lack of any putative terminator of transcription between them indicates that these two genes are cotranscribed.
Selection and molecular identification of a marine yeast isolate for use in colonization competition mixed glass flow cell experiments.
The marine yeast isolate Y1 was selected for the biofilm work as it both attached to the glass surface and maintained a consistent biofilm that could be easily monitored using microscopic techniques. Y1 was identified via partial sequencing and alignment of the ITS and V3 regions with the EMBL fungal DNA database using Fasts3 sequence homology searches. Both the ITS region (98.8% identity) and the V3 region (99.7% identity) were found to be closely matched to the ITS and V3 regions of the yeast Rhodosporidium sphaerocarpum.
Mixed bacterial-yeast biofilm experiments.
The wild-type P. tunicata, the P. tunicata antifungal-deficient mutant FM3, and the yeast isolate R. sphaerocarpum formed biofilms on the glass flow cell surface. The bacterial strains developed thicker biofilms and covered more of the surface than the yeast biofilm did. Microscopically, there was no difference between the formation of a biofilm by the wild-type P. tunicata and the antifungal-deficient mutant FM3 (Fig. 2). Both the wild type and the antifungal-deficient mutant rapidly covered the entire surface and formed a heterogeneous structure, consisting of dense cell clusters, within 16 to 24 h, similar to P. tunicata biofilms previously described (40). The yeast cells attached to the glass flow cell could be observed to bud and divide during the 72 h of the experiment.
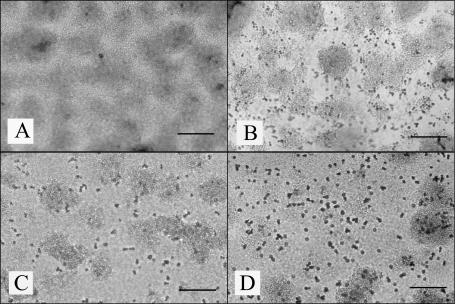
FIG. 2.
Biofilms on glass surfaces of the wild-type Pseudoalteromonas tunicata (A), P. tunicata antifungal-deficient mutant FM3 (B), and Rhodosporidium sphaerocarpum isolate (C). Images were taken 24 h after inoculation. Bars = 200 μm.
Yeast attachment to a bacterial biofilm.
The yeast R. sphaerocarpum was unable to attach to an established wild-type P. tunicata biofilm, whereas the antifungal-deficient mutant, FM3, did not inhibit yeast attachment (Fig. 3). In the wild-type P. tunicata biofilm samples, only free-living yeast cells were observed, while in the FM3 biofilm samples, the yeast cells were attached to the mutant biofilm. These attached yeast cells budded and divided in a similar fashion to the yeast monospecies biofilm (Fig. 4).
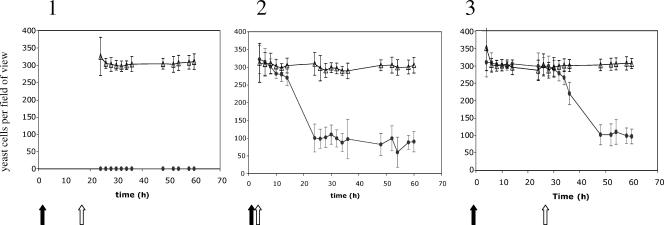
FIG. 3.
Counts of Rhodosporidium sphaerocarpum cells attached to the glass surface in the presence of the two bacterial species, Pseudoalteromonas tunicata (solid ovals) and P. tunicata antifungal-deficient mutant FM3 (open triangles), were conducted per field of view under ×40 magnification. The area of each field of view was 1.35 mm2. Three inoculation strategies were employed. First, P. tunicata strains were inoculated at time zero (open arrow) and allowed to establish a biofilm for 24 h before being inoculated with the R. sphaerocarpum (black arrow) (panel 1). Second, both the R. sphaerocarpum and P. tunicata strains were inoculated together (panel 2), and third, R. sphaerocarpum was allowed to establish a biofilm for 24 h before being inoculated with the P. tunicata strains (panel 3).
FIG. 4.
Effects of the wild-type strain and antifungal-deficient mutant FM3 strain of Pseudoalteromonas tunicata on surface attachment of Rhodosporidium sphaerocarpum. (A) A 24-h-old P. tunicata biofilm challenged with R. sphaerocarpum cells. (B) A 24-h-old P. tunicata antifungal-deficient mutant FM3 biofilm challenged with R. sphaerocarpum cells. (C). Wild-type P. tunicata was introduced to a 24-h-old R. sphaerocarpum biofilm. (D) The antifungal-deficient mutant FM3 was introduced to a 24-h-old R. sphaerocarpum biofilm. Images were captured 24 h after inoculation of the biofilm with R. sphaerocarpum (A and B), P. tunicata wild type (C), or P. tunicata FMS (D). Bars = 200 μm.
Bacterial and yeast competition for colonization of a clean glass surface.
The number of yeast cells attached to the surface did not vary significantly for the first 10 h after the dual attachment of bacterial and yeast cells of the clean surface (P = 0.36). However, there was a significant drop in the number of yeast cells attached to the surface in the mixed wild-type P. tunicata/R. sphaerocarpum biofilm after 10 to 12 h (P = 0.003) (Fig. 3). The number of yeast cells attached to the surface decreased by 66% ± 11% after 12 h (Fig. 3), and after 24 h, the yeast cells in the wild-type biofilm no longer appeared to be budding and dividing (Fig. 4). In contrast, the number of yeast cells attached to the surface was unaffected by the attachment of the antifungal-deficient mutant (P = 0.264), and budding and division could be observed throughout the experiment with the mixed antifungal-deficient mutant/yeast biofilm.
Bacterial colonization of a yeast biofilm.
The wild-type P. tunicata and P. tunicata antifungal-deficient mutant were both able to attach to an established R. sphaerocarpum biofilm. No difference in the number of yeast cells attached to the flow cell surface was observed for the first 10 h after bacterial inoculation. After an additional 10 h, the wild-type P. tunicata started to compete with the yeast cells and there was a significant decrease of 66% ± 5% (P < 0.001) of the yeast cell numbers attached to the surface between 32 h and 36 h. The number of yeast cells attached to the surface was unaffected by the attachment of the antifungal-deficient mutant (P = 0.114) (Fig. 3). After 48 h, the yeast biofilms colonized by the wild-type P. tunicata were not observed to be budding and dividing, whereas budding and division were not affected by the attachment of the P. tunicata antifungal-deficient mutant (Fig. 4).
The effect of the fungal inhibitory activity of Pseudoalteromonas tunicata on the fungal community associated with the green alga Ulva australis.
Between four and eight bands were separated from each U. australis control sample by DG-DGGE. Twenty hours after the inoculation of the U. australis with the wild-type P. tunicata, only two bands were amplified from five samples, whereas there was no difference in banding patterns produced by the control samples and the samples treated with P. tunicata antifungal-deficient mutant. After 48 and 120 h, no amplified bands were observed in the samples treated with wild-type P. tunicata, whereas the control samples and the samples treated with FM3 strain displayed similar banding patterns.
DISCUSSION
Members of the Pseudoalteromonas genus produce a diverse range of inhibitory metabolites targeting common fouling organisms. This has led to the hypothesis that the production of these compounds provides a competitive advantage during surface colonization (50). Previous studies have demonstrated that production of an antibacterial protein by P. tunicata provides a competitive advantage to the bacterium during surface colonization in the presence of other marine bacteria (50). P. tunicata is also able to inhibit the growth of a wide range of fungal species (40). To investigate whether the fungal inhibitory activity provides an ecological advantage to P. tunicata, a P. tunicata transposon mutant, FM3, unable to inhibit fungal growth was generated. This mutant was used in a series of glass flow cell experiments in competition with R. sphaerocarpum, a yeast species isolated from the surface of U. australis. The surface competition experiments were also extended to include a DG-DGGE analysis of the fungal community on the surface of U. australis inoculated with wild-type or FM3 P. tunicata strains.
Previous transposon studies of P. tunicata have indicated a correlation between the production of the yellow pigment and various antifouling activities including fungal inhibition (19). By using this screen, we avoided mutants with a loss of pigmentation to avoid pleiotropic effects. Analysis of the DNA sequence of the FM3 mutant revealed that the mini-Tn10 transposon had disrupted a gene (designated afaA for antifungal activity gene A) with a high sequence similarity to genes from various organisms coding for a long-chain-fatty-acid CoA synthetase (fatty acid CoA ligase, Amp-forming; EC 6.2.1.4). This long-chain-fatty-acid CoA synthetase catalyzes the formation of long-chain-fatty-acid CoA thioesters (fatty acyl-CoAs) from long-chain fatty acids, ATP, and CoA. Activated long-chain fatty acids, in the form of fatty acyl CoAs, are bioactive compounds involved in a variety of processes, such as intracellular protein transport (27), cell signaling (4, 18, 37), translocation of long-chain fatty acids across the plasma membrane (51), and transcriptional control, as well as being the first step in beta oxidation and phospholipid biosynthesis (6). The yellow pigment YP1 produced by P. tunicata contains a 2,2′-bipyrrole ring system with an unsaturated 12-carbon alkyl chain (23). YP1 belongs to the tambjamine class of compounds, which have previously been reported to exhibit antimicrobial activities and have been isolated from eukaryotic sources in the marine environment (39). This pigment is the first reported natural tambjamine with an unsaturated alkyl chain. It may be speculated that activation of long-chain fatty acids is required for the production of an active antifungal metabolite. Further studies into the biosynthesis of the antifungal compound will address the possibility that AfaA acts as a general activator of long-chain fatty acids or if it also has a more specific role in the production of the antifungal compound.
While the P. tunicata antifungal-deficient mutant FM3 did not inhibit any of the fungi or yeast included as test strains in this study, the broad spectrum of fungal inhibitory activity by P. tunicata was expanded beyond those already reported (19, 31, 32). These new test strains are of industrial, agricultural, and medical importance, and it was demonstrated that the growth of all these fungi was inhibited by wild-type P. tunicata. This further supports the observation that P. tunicata is a bacterium producing a powerful antifouling agent.
After 24 h of growth in the presence of P. tunicata strains, the P. tunicata wild-type biofilm was able to completely inhibit the attachment of R. sphaerocarpum to a glass flow cell surface. In contrast, a biofilm of the antifungal-deficient mutant FM3 did not affect the attachment of the yeast R. sphaerocarpum, demonstrating that production of the antifungal compound by P. tunicata enables the exclusion of R. sphaerocarpum from a biofilm. Studies by Gil-Turnes et al. (25, 26) found a similar effect with a fungal inhibitory bacterial strain isolated from the surface of embryos of two marine crustaceans. Palaemon marcodactylus and Homarus americanus embryos were covered exclusively by a single, gram-negative, bacterial population that was found to produce isatin and tyrsol, metabolites with antifungal activity. Removal of the bacterial population allowed the pathogenic fungi to colonize the crustacean embryos, resulting in death of the embryo. If the bacteria were reintroduced, the antifungal metabolites produced by the bacterial strain inhibited the attachment and growth of the pathogenic fungi and allowed the embryos to survive (25, 26).
When yeast and bacterial cells were coinoculated in competition for colonization of a clean surface, no difference in the number of yeast cells attached to the surface was observed for the first 10 h. After 10 h, there was a rapid decrease (66% ± 11%) in the number of yeast cells in the mixed wild-type P. tunicata/R. sphaerocarpum biofilm experiments. We suggest that the decrease in yeast numbers after 10 h correlates with the cessation of logarithmic growth, onset of stationary growth, and production of bacterial secondary metabolites. The yeast cells in the biofilm after 24 h were no longer observed to bud or divide, indicating that growth had been arrested. The FM3 strain, when attaching in competition for a clean surface, did not affect the attachment of yeast cells, as the yeast cells were observed to continue to bud and divide even when completely covered by FM3 cells.
It is well documented that fungal biofilms are persistent and have enhanced resistance to antimicrobial compounds. For example, Chandra et al. (10) found that biofilm formation by C. albicans increased drug resistance. In the current study, we found that a 24-h biofilm of R. sphaerocarpum did not provide any further protection against wild-type P. tunicata cells. Similar to the coattachment experiment, no decrease in yeast cell numbers was observed for 10 h after inoculation of the bacteria into the flow cell system. After 10 h, however, the number of yeast cells attached to the surface decreased rapidly. This is possibly due to the antifungal compound produced by P. tunicata. While the P. tunicata antifungal-deficient mutant FM3 attached to an established R. sphaerocarpum biofilm, it did not affect the number of yeast cells attached to the surface. The yeast cells continued to bud and divide in the presence of the P. tunicata antifungal-deficient mutant and displayed a slight increase in numbers. The results demonstrate that the antifungal compound produced by P. tunicata may offer an advantage during surface colonization in competition with yeast cells for a surface in the marine environment.
A study by Rao et al. (50) found that the production of the antibacterial protein AlpP gives P. tunicata a competitive advantage during surface colonization in the presence of other bacterial isolates from the surface of U. australis when single and mixed species bacterial biofilms containing P. tunicata were grown in glass flow cells. In pure culture, each of the marine bacterial isolates formed biofilms containing microcolony structures within 72 h. However, in mixed species biofilms, P. tunicata removed the competing strain unless its competitor was relatively insensitive to AlpP (for example, P. gracilis), or produced strong inhibitory activity against P. tunicata (for example, Roseobacter gallaeciensis). Moreover, biofilm studies conducted with an AlpP− mutant of P. tunicata indicated that the mutant was less competitive when introduced into preestablished biofilms, suggesting a role for AlpP during competitive bacterial biofilm formation. When single-species biofilms were allowed to form microcolonies before the introduction of a competitor, these microcolonies coexisted with P. tunicata for extended periods of time before being removed. In the glass flow cell experiments conducted by Rao et al. (50), the marine bacteria R. gallaeciencis and P. tunicata were found to be superior competitors; their dominance was attributed to their ability to both rapidly form microcolonies and produce extracellular antibacterial compounds (50).
To examine the interactions of the wild-type and FM3 strains of P. tunicata with a mixed natural microbial community, U. australis samples were collected and inoculated with these strains. DG-DGGE was used to study the effects of the bacteria on the diversity of the fungal community. This analysis is a fingerprint method that offers an overview of the microbial diversity in a community by separating species on the basis of differences in nucleotide sequences. There have been many ecological studies on bacterial populations using denaturing gradient gel electrophoresis (24, 44, 45). However, almost all DGGE fungal analyses have focused on terrestrial fungal communities, and very few have been directed at marine fungal communities.
Marine fungi are classified as an ecological group, rather than a taxonomic group, and grow on a wide variety of substrata (28). DG-DGGE analysis of the individual algal plants revealed a low but variable fungal diversity associated with the surface of U. australis.
Upon treatment with the wild-type P. tunicata, the fungi on the surface of U. australis were no longer detectable after 48 h. In contrast, the addition of the P. tunicata antifungal-deficient mutant to the algal plants did not affect the natural fungal community. The DG-DGGE banding patterns were similar to the banding patterns of the untreated communities, with four to eight bands being resolved in each sample. Similar to the flow cell experiments, P. tunicata does not seem to be able to outcompete fungi and yeasts without the expression of the antifungal compound. The natural fungal and yeast community on the surface of U. australis was unaffected by the likely increase in competition for nutrients due to the introduction of the P. tunicata fungal mutant. In contrast, the wild-type P. tunicata was able to disrupt the already established fungal community on the surfaces of the algal plants. This is interesting, as previous studies have suggested that the formation of biofilms increases the resistance of member species to antimicrobial compounds (10-12, 42, 53).
P. tunicata was found to be an effective colonizer of glass surfaces in flow cell systems. Mutation of afaA did not affect attachment or biofilm development but prevented the inhibition of fungal growth shown by the wild-type stain. Through the specific production of the antifungal compound, P. tunicata was able to exclude fungi from a biofilm and disrupt an already established fungal biofilm. Production of the antifungal compound is reliant on stationary-phase growth, being produced as part of secondary metabolite production. Moreover, the antifungal compound enabled P. tunicata to disrupt an established natural fungal community on the surface of the green alga U. australis. Thus, by the action of the antifungal compound and in combination with the production of the antibacterial protein, P. tunicata can greatly affect the composition of biofilm communities and therefore would also affect the process of biofouling and may provide protection for the host organisms in the marine environment.
Supplementary Material
Acknowledgments
This project was supported by funds from the Australian Research Council and the Centre for Marine Biofouling and Bio-Innovation, University of New South Wales, Sydney, Australia.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Armstrong, E., K. G. Boyd, and J. G. Burgess. 2000. Prevention of marine biofouling using natural compounds from marine organisms. Biotechnol. Annu. Rev. 6:221-241. [DOI] [PubMed] [Google Scholar]
- 3.Baillie, G. S., and L. J. Douglas. 2000. Matrix polymers of Candida biofilms and their possible role in biofilm resistance to antifungal agents. J. Antimicrob. Chemother. 46:397-403. [DOI] [PubMed] [Google Scholar]
- 4.Barber, C., J. Tang, J. Feng, M. Pan, T. Wilson, H. Slater, J. Dow, P. Williams, and M. Daniels. 1997. A novel regulatory system required for pathogenicity of Xanthomonas campestris is mediated by small diffusible signal molecules. Mol. Microbiol. 24:555-566. [DOI] [PubMed] [Google Scholar]
- 5.Bertani, G. 1951. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 62:293-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Black, P. N., and C. C. DiRusso. 1994. Molecular and biochemical analyses of fatty acid transport, metabolism, and gene regulation in Escherichia coli. Biochim. Biophys. Acta 1210:123-145. [DOI] [PubMed] [Google Scholar]
- 7.Blunt, J. W., B. R. Copp, M. H. Munro, P. T. Northcote, and M. R. Prinsep. 2004. Marine natural products. Nat. Prod. Rep. 21:1-49. [DOI] [PubMed] [Google Scholar]
- 8.Branting, C., M. L. Sund, and L. E. Linder. 1989. The influence of Streptococcus mutans on adhesion of Candida albicans to acrylic surfaces in vitro. Arch. Oral Biol. 34:347-353. [DOI] [PubMed] [Google Scholar]
- 9.Centeno, A., C. P. Davis, M. S. Cohen, and M. M. Warren. 1983. Modulation of Candida albicans attachment to human epithelial cells by bacteria and carbohydrates. Infect. Immun. 39:1354-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chandra, J., D. M. Kuhn, P. K. Mukherjee, L. L. Hoyer, T. McCormick, and M. A. Ghannoum. 2001. Biofilm formation by the fungal pathogen Candida albicans: development, architecture, and drug resistance. J. Bacteriol. 183:5385-5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Costerton, J. W., K.-J. Cheng, G. G. Geesey, T. I. Ladd, J. C. Nickel, M. Dasguputa, and T. J. Marrie. 1987. Bacterial biofilms in nature and disease. Annu. Rev. Microbiol. 41:435-464. [DOI] [PubMed] [Google Scholar]
- 12.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 13.Cremonesi, L., P. Carrera, A. Fumagalli, S. Lucchiari, E. Cardillo, M. Ferrari, S. C. Righetti, F. Zunino, P. G. Righetti, and C. Gelfi. 1999. Validation of double gradient denaturing gradient gel electrophoresis through multigenic retrospective analysis. Clin. Chem. 45:35-40. [PubMed] [Google Scholar]
- 14.Davey, M. E., and G. A. O'Toole. 2000. Microbial biofilms: from ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 64:847-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.deNys, R., P. D. Steinberg, P. Willemsen, S. A. Dworjanyn, C. L. Gabelish, and R. J. King. 1994. Broad spectrum effects of secondary metabolites from the red alga Delisea pulchra in antifouling assays. Biofouling 8:259-271. [Google Scholar]
- 16.Donlan, R. M. 2002. Biofilms: microbial life on surfaces. Emerg. Infect. Dis. 8:881-890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Douglas, L. J. 2003. Candida biofilms and their role in infection. Trends Microbiol. 11:30-36. [DOI] [PubMed] [Google Scholar]
- 18.Downard, J., and D. Toal. 1995. Branched-chain fatty acids: the case for a novel form of cell-cell signaling during Myxococcus xanthus development. Mol. Microbiol. 16:171-175. [DOI] [PubMed] [Google Scholar]
- 19.Egan, S., S. James, C. Holmström, and S. Kjelleberg. 2002. Correlation between pigmentation and antifouling compounds produced by Pseudoalteromonas tunicata. Environ. Microbiol. 4:433-442. [DOI] [PubMed] [Google Scholar]
- 20.El-Azizi, M. A., S. E. Starks, and N. Khardori. 2004. Interactions of Candida albicans with other Candida spp. and bacteria in the biofilms. J. Appl. Microbiol. 96:1067-1073. [DOI] [PubMed] [Google Scholar]
- 21.Elvers, K. T., K. Leeming, and H. M. Lappin-Scott. 2002. Binary and mixed population biofilms: time-lapse image analysis and disinfection with biocides. J. Ind. Microbiol. Biotechnol. 29:331-338. [DOI] [PubMed] [Google Scholar]
- 22.Fell, J. W. 1993. Rapid identification of yeast species using three primers in a polymerase chain reaction. Mol. Mar. Biol. Biotechnol. 2:174-180. [PubMed] [Google Scholar]
- 23.Franks, A., P. Haywood, C. Holmström, S. Egan, S. Kjelleberg, and N. Kumar. 2005. Isolation and structure elucidation of a novel yellow pigment from the marine bacterium Pseudoalteromonas tunicata. Molecules 10:1286-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fromin, N., J. Hamelin, S. Tarnawski, D. Roesti, K. Jourdain-Miserez, N. Forestier, S. Teyssier-Cuvelle, F. Gillet, M. Aragno, and P. Rossi. 2002. Statistical analysis of denaturing gel electrophoresis (DGE) fingerprinting patterns. Environ. Microbiol. 4:634-643. [DOI] [PubMed] [Google Scholar]
- 25.Gil-Turnes, M. S., and W. Fenical. 1992. Embryos of Homarus americanus are protected by epibiotic bacteria. Biol. Bull. 182:105-108. [DOI] [PubMed] [Google Scholar]
- 26.Gil-Turnes, M. S., M. E. Hay, and W. Fenical. 1989. Symbiotic marine bacteria defend crustacean embryos from a pathogenic fungus. Science 240:116-118. [DOI] [PubMed] [Google Scholar]
- 27.Glick, B. S., and J. E. Rothman. 1987. Possible role for fatty acyl-coenzyme A in intracellular protein transport. Nature 326:309-312. [DOI] [PubMed] [Google Scholar]
- 28.Hawksworth, D. L. 2001. The magnitude of fungal diversity: the 1.5 million species estimate revisited. Mycol. Res. 105:1422-1432. [Google Scholar]
- 29.Herrero, M., V. de Lorenzo, and K. N. Timmis. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172:6557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holmes, A. R., R. McNab, and H. F. Jenkinson. 1996. Candida albicans binding to the oral bacterium Streptococcus gordonii involves multiple adhesin-receptor interactions. Infect. Immun. 64:4680-4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holmström, C., S. Egan, A. Franks, S. McCloy, and S. Kjelleberg. 2002. Antifouling activities expressed by marine surface associated Pseudoalteromonas species. FEMS Microb. Ecol. 41:47-58. [DOI] [PubMed] [Google Scholar]
- 32.Holmström, C., S. James, B. Neilan, D. White, and S. Kjelleberg. 1998. Pseudoalteromonas tunicata sp. nov., a bacterium that produces antifouling agents. Int. J. Syst. Bacteriol. 48:1205-1212. [DOI] [PubMed] [Google Scholar]
- 33.Holmström, C., and S. Kjelleberg. 2000. Bacterial interactions with marine fouling organisms, p. 101-117. In L. V. Evans (ed.), Biofilms: recent advances in their study and control. Overseas Publishing Associates (UK), Amsterdam, The Netherlands.
- 34.Holmström, C., and S. Kjelleberg. 1994. The effect of external biological factors on settlement of marine invertebrate larvae and new antifouling technology. Biofouling 8:147-160. [Google Scholar]
- 35.James, S. G., C. Holmström, and S. Kjelleberg. 1996. Purification and characterization of a novel antibacterial protein from the marine bacterium D2. Appl. Environ. Microbiol. 62:2783-2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kjelleberg, S., and P. D. Steinberg. 2002. Defenses against bacterial colonisation of marine plants, p. 152-172. In S. E. Lindow, E. Hecht-Poinar, and V. Elliott (ed.), Phyllosphere microbiology. American Phytopathological Society Press, St. Paul, Minn.
- 37.Korchak, H. M., L. H. Kane, M. W. Rossi, and B. E. Corkey. 1994. Long chain acyl-coenzyme A and signaling in neutrophils. An inhibitor of acyl coenzyme A synthetase, triacsin C, inhibits superoxide anion generation and degranulation by human neutrophils. J. Biol. Chem. 269:30281-30287. [PubMed] [Google Scholar]
- 38.Lewis, K. 2001. Riddle of biofilm resistance. Antimicrob. Agents Chemother. 45:999-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lindquist, N., and W. Fenical. 1991. New tambjamine class alkaloids from the marine ascidian Atapozoa sp. and its nudibranch predators—origin of the tambjamines in Atapozoa. Experientia 47:504-506. [Google Scholar]
- 40.Mai-Prochnow, A., F. Evans, D. Dalisay-Saludes, S. Stelzer, S. Egan, S. James, J. S. Webb, and S. Kjelleberg. 2004. Biofilm development and cell death in the marine bacterium Pseudoalteromonas tunicata. Appl. Environ. Microbiol. 70:3232-3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mården, P., A. Tunlid, K. Malmcronafriberg, G. Odham, and S. Kjelleberg. 1985. Physiological and morphological changes during short-term starvation of marine bacterial isolates. Arch. Microbiol. 142:326-332. [Google Scholar]
- 42.Mercier-Bonin, M., K. Ouazzani, P. Schmitz, and S. Lorthois. 2004. Study of bioadhesion on a flat plate with a yeast/glass model system. J. Colloid Interface Sci. 271:342-350. [DOI] [PubMed] [Google Scholar]
- 43.Miao, L., and P. Qian. 2005. Antagonistic antimicrobial activity of marine fungi and bacteria isolated from marine biofilm and seawaters of Hong Kong. Aquat. Microbiol. Ecol. 38:231-238. [Google Scholar]
- 44.Muyzer, G., E. C. Dewaal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muyzer, G., and K. Smalla. 1998. Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) in microbial ecology. Antonie Leeuwenhoek 73:127-141. [DOI] [PubMed] [Google Scholar]
- 46.Nair, R. G., S. Anil, and L. P. Samaranayake. 2001. The effect of oral bacteria on Candida albicans germ-tube formation. APMIS 109:147-154. [DOI] [PubMed] [Google Scholar]
- 47.Nair, R. G., and L. P. Samaranayake. 1996. The effect of oral commensal bacteria on candidal adhesion to human buccal epithelial cells in vitro. J. Med. Microbiol. 45:179-185. [DOI] [PubMed] [Google Scholar]
- 48.Neidhardt, F. C., P. L. Bloch, and D. F. Smith. 1974. Culture medium for enterobacteria. J. Bacteriol. 119:736-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.O'Toole, G., H. B. Kaplan, and R. Kolter. 2000. Biofilm formation as microbial development. Annu. Rev. Microbiol. 54:49-79. [DOI] [PubMed] [Google Scholar]
- 50.Rao, D., J. S. Webb, and S. Kjelleberg. 2005. Competitive interactions in mixed-species biofilms containing the marine bacterium Pseudoalteromonas tunicata. Appl. Environ. Microbiol. 71:1729-1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schmelter, T., B. L. Trigatti, G. E. Gerber, and D. Mangroo. 2004. Biochemical demonstration of the involvement of fatty acyl-CoA synthetase in fatty acid translocation across the plasma membrane. J. Biol. Chem. 279:24163-24170. [DOI] [PubMed] [Google Scholar]
- 52.Siebert, P. D., A. Chenchik, D. E. Kellogg, K. A. Lukyanov, and S. A. Lukyanov. 1995. An improved PCR method for walking in uncloned genomic DNA. Nucleic Acids Res. 23:1087-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stoodley, P., S. Wilson, L. Hall-Stoodley, J. Boyle, H. M. Lappin-Scott, and J. W. Costerton. 2001. Growth and detachment of cell clusters from mature mixed-species biofilms. Appl. Environ. Microbiol. 67:5608-5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Taylor, M. W., P. J. Schupp, I. Dahllof, S. Kjelleberg, and P. D. Steinberg. 2004. Host specificity in marine sponge-associated bacteria, and potential implications for marine microbial diversity. Environ. Microbiol. 6:121-130. [DOI] [PubMed] [Google Scholar]
- 55.Tillett, D., and B. A. Neilan. 2000. Rapid nucleic acid isolation from cultured and environmental cyanobacteria: novel techniques based on xanthogenate. J. Phycol. 36:251-258. [Google Scholar]
- 56.White, T. J., T. D. Bruns, S. B. Lee, and J. W. Taylor. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p. 315-322. In N. Innis, D. Gelfand, J. Sninsky, and T. White (ed.), PCR: protocols and applications. A laboratory manual. Academic Press, New York, N.Y.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.