Abstract
Though a large fraction of primary production and organic matter cycling in the oceans occurs on continental shelves dominated by sandy deposits, the microbial communities associated with permeable shelf sediments remain poorly characterized. Therefore, in this study, we provide the first detailed characterization of microbial diversity in marine sands of the South Atlantic Bight through parallel analyses of small-subunit (SSU) rRNA gene (Bacteria), nosZ (denitrifying bacteria), and amoA (ammonia-oxidizing bacteria) sequences. Communities were analyzed by parallel DNA extractions and clone library construction from both sediment core material and manipulated sediment within column experiments designed for geochemical rate determinations. Rapid organic-matter degradation and coupled nitrification-denitrification were observed in column experiments at flow rates resembling in situ conditions over a range of oxygen concentrations. Numerous SSU rRNA phylotypes were affiliated with the phyla Proteobacteria (classes Alpha-, Delta-, and Gammaproteobacteria), Planctomycetes, Cyanobacteria, Chloroflexi, and Bacteroidetes. Detectable sequence diversity of nosZ and SSU rRNA genes increased in stratified redox-stabilized columns compared to in situ sediments, with the Alphaproteobacteria comprising the most frequently detected group. Alternatively, nitrifier communities showed a relatively low and stable diversity that did not covary with the other gene targets. Our results elucidate predominant phylotypes that are likely to catalyze carbon and nitrogen cycling in marine sands. Although overall diversity increased in response to redox stabilization and stratification in column experiments, the major phylotypes remained the same in all of our libraries, indicating that the columns sufficiently mimic in situ conditions.
Sandy sediments cover large areas of the shallow ocean, and recent technological developments in marine geochemistry have revealed that these sediments rapidly recycle organic matter and have the potential to play a large role in global biogeochemical cycles (22, 25, 34, 50). In fine-grained sediments that have been studied more extensively, molecular diffusion limits aerobic and suboxic microbial metabolism to a thin surface layer (26). In contrast, the high permeability of sands allows for rapid exchange of pore water with the overlying water column, thereby enhancing the transport of microbial substrates into and metabolic waste products out of the sediments. Hydrodynamic forces thus fuel high rates of microbial metabolism in permeable sands while supporting a low microbial abundance and organic matter content (8, 53, 54). In addition, such drastically different physicochemical parameters are likely to support a microbial community with a composition very different from that in fine-grained sediments. However, the community composition of microorganisms inhabiting permeable sediments is poorly known. A few studies have investigated community composition in permeable sands using lipid biomarkers or fluorescent in situ hybridization (FISH) approaches (8, 31, 54). To our knowledge, few or no previous studies have examined community composition using genetic cloning/sequencing in marine sands.
Nitrification and denitrification are critical microbially mediated processes in the nitrogen cycle that, when coupled, link the mineralization of nitrogenous compounds to the removal of nitrogen from the continental shelf (11, 28, 59). Nitrification is a two-step process resulting in the oxidation of NH3+ and the production of NO3−. The first step of the pathway, the oxidation of NH3+ to NO2− by ammonia-oxidizing bacteria (AOB), is considered to be the rate-limiting step of nitrification and is catalyzed by the ammonia monooxygenase (Amo) enzyme. The amoA gene, which codes for a subunit of ammonia monooxygenase, has been utilized as a gene target to explore AOB diversity in a variety of marine environments (3, 14, 27, 41, 44). However, previous studies of amoA in marine sediments have focused on highly impermeable marine muds, which generally contain much higher ammonium concentrations, are low in oxygen content, and are rich in organic matter (3, 9, 14, 41, 43). To our knowledge, nothing is known about the diversity of nitrifying bacteria in marine sands, and few studies of denitrifiers in comparable environments are available (55).
Numerous genes associated with denitrification have been identified and studied in detail (reviewed by Philippot [45]). Previous community-based studies have targeted the nirS and nirK genes, associated with the second step in the denitrification process (5, 6, 30). While nirS and nirK encode proteins earlier in the denitrification pathway, nosZ, encoding nitrous oxide reductase, is associated with the final step, thus representing the step directly related to the loss of biologically available nitrogen from the environment. Studies have frequently targeted nosZ by molecular techniques to characterize the denitrifying fraction of the microbial community; however, the database for marine sediments remains relatively small, and past studies have often focused on methodological development (42, 47, 55-57).
The overall goal of our study was to provide a detailed characterization of microbially diverse communities in an understudied marine sedimentary environment, permeable shelf sands, that plays a substantial role in global biogeochemical cycles. Our work was conducted in combination with a geochemical study, using packed sediment column experiments, of N cycling in permeable sediments (49). Here we present results from the investigation of diversity in these column experiments and unmanipulated sediment cores collected from the same site. We hypothesized that the stabilization and stratification of redox conditions in the columns would act to increase detectable community diversity and that we would observe a change in community composition across redox boundaries within the columns. Our results have revealed predominant phylotypes that are likely to catalyze carbon and nitrogen cycling in marine sands. Although overall diversity increased in response to redox stabilization and stratification in column experiments, the major phylotypes remained the same in all of our libraries. Our results provide an initial sequence database for the development of improved probes and primer sets to be used in quantifying the metabolically active members of microbial communities in permeable shelf sediments.
MATERIALS AND METHODS
Site and sample description.
Sediments were sampled from the South Atlantic Bight (SAB) off the coast of Savannah, Ga. The SAB shelf seafloor is a high-energy, nonaccumulating environment consisting of medium and coarse sands (51). Sediment and water column parameters at the time of sampling were characterized as follows: porosity, 0.5; median sediment grain size, 500 μm; permeability, 4.7 × 10−11 m2; range of pore water exchange with surface water, 0 to 4 cm; near-bottom current speed, 30 cm s−1; surface water temperature, 27.5°C; salinity, 36.1 ppt; percent surface PAR (photosynthetically available radiation) at the seafloor, 2 to 10%. The average water depth of the SAB sampling area was 27.9 m.
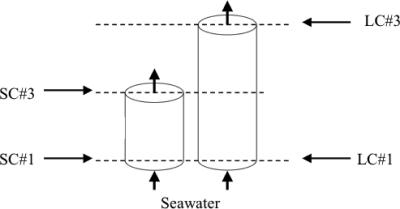
Sediment cores and grab samples were collected from the surface (depth interval, 0 to 5 cm) at the W27 site of the SAB shelf (31o29′N, 80o26′W) in June 2004. A detailed description of the sediment column experiments is presented by Rao and Jahnke (49). In brief, sediment was homogenized and packed into short (8-cm) and long (32-cm) columns with seawater pumped through the columns from the bottom upward at a constant rate (Fig. 1). Rates of metabolic activity were quantified from the difference in chemical constituents between the column inflow and outflow according to the continuous-flow method (1). Columns were constructed on the day of sampling and equilibrated for 6 days to reach steady state. Triplicate inflow and outflow samples were collected once daily over several days for rate determinations. Then 20 μM of labeled 15NO3− was spiked into the column inflow to track microbial nitrogen transformation pathways, and constituents were again sampled. Column pore waters were analyzed for ammonium and nitrate (62), oxygen (Winkler titration), and total carbon dioxide (alkalinity titration) using standard methods. Aqueous nitrogen gas and nitrogen transformation pathways were quantified as described by An et al. (1). Total incubation time was 2 weeks prior to sacrificing of sediments for nucleic acid extraction. Sediment was collected adjacent to the inflow and outflow of the short column (SC#1 and SC#3) and the long column (LC#1 and LC#3) (Fig. 1), as well as from the top 5 cm of duplicate pooled in situ SAB core samples.
FIG. 1.
Experimental design of column incubations. Column heights were 8 and 32 cm, respectively, with a diameter of 7.6 cm. SAB samples from the sediment surface (depth, 0 to 5 cm) were packed into these columns.
DNA extraction and gene analysis.
Microbial community DNA was extracted directly from the sediment using the RNA/DNA extraction protocol described by Hurt et al. (23) and then cleaned using a QIAGEN (Valencia, CA) RNA/DNA Midi kit. Bacterial nosZ, amoA, and SSU rRNA genes were amplified by PCR in an Eppendorf (Westbury, NY) Mastercycler EP Gradient PCR machine. The standard PCR mix included 1× PCR buffer containing 1.5 mM MgCl2 (Takara Bio Inc., Japan), 250 μM of each deoxynucleoside triphosphate (Takara Bio Inc., Japan), 1 pmol each of forward and reverse primers, 0.025 U μl−1 rTaq enzyme (Takara Bio Inc., Japan), and distilled H2O. To each reaction, 10 to 20 μg of DNA was added as a template. Primer sequences and PCR conditions are listed in Table S1 in the supplemental material. PCR products were cleaned using a QIAGEN (Valencia, CA) PCR purification kit and cloned using a TOPO TA cloning kit (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Cloned inserts were amplified using the PCR conditions described above. However, for the SSU rRNA gene clones, the vector-specific primers M13F and M13R (37) were used to avoid amplification of host Escherichia coli SSU rRNA genes. The M13 primer sequences and PCR conditions are reported in Table S1 in the supplemental material. Clones were screened and grouped into phylotypes using restriction fragment length polymorphism (RFLP) as described by Mills et al. (38), except that restriction enzymes HaeIII (New England Biolabs, Inc., Beverly, MA) and MspI (Promega, Madison, WI) were used in this study. Select clones were sequenced using an Applied Biosystems 3100 genetic analyzer at the Florida State University sequencing facility. For the percentage of similarity to previously reported sequences as determined by BLAST analysis and frequency of detection for each phylotype, see Tables S2 to S4 in the supplemental material.
Phylogenetic and statistical analyses.
Clone sequences were checked for chimeras using Chimera Check from Ribosomal Database Project II (33). Sequences from this study and reference sequences, as determined by BLAST analysis, were subsequently aligned using the Fast Aligner algorithm in the ARB package (63). All alignments were then visually verified and adjusted by hand according to the E. coli SSU rRNA secondary structure. Neighbor-joining trees incorporating a Jukes-Cantor distance correction were created from the alignments using the ARB software package (63). An average of 500 (i.e., amoA) to 1,000 (i.e., SSU rRNA and nosZ clones) nucleotides were included in the phylogenetic analyses. Bootstrap data represented 1,000 samplings. Rarefaction analysis, based on equations described by Heck et al. (17), was included as Fig. S1 in the supplemental material. Sorensen's (1/D) and Shannon-Wiener indices were calculated using standard equations. Species richness was determined by EstimateS (10, 12, 13). Additional statistical estimators, including gene (40) and nucleotide (40, 64) diversity and θ(π) (64), were calculated using Arlequin (58).
Nucleotide sequence accession numbers.
The 88 nucleotide sequences were submitted to the GenBank database under accession numbers DQ289896 to DQ289983.
RESULTS
Sediment sampling and geochemical determinations.
The composition of the community of Bacteria in the SAB was determined by SSU rRNA gene, nosZ, and amoA phylogenetic analyses of DNA-derived clone libraries extracted from sediments in two flowthrough columns and two sediment cores. Extracted DNA concentrations were comparable for all four column samples, i.e., SC#1, SC#3, LC#1, and LC#3, and the homogenized core sediment, designated SAB core (data not shown).
Rapid microbial mineralization of organic matter in SAB sediments was indicated by oxygen consumption and the production of CO2 in column experiments (Table 1). The short- and long-column experiments had pore water residence times of 3 and 12 h, respectively. Flow rates in the columns were adjusted to mimic pore water exchange rates measured at the site. Oxygen consumption in the columns increased with residence time of the column, resulting in O2 concentrations at the outflow of 54.5 μM and 8.3 μM for the short and long columns, respectively. Oxygen concentration decreased linearly with column height between the inflow and outflow. Therefore, strong oxygen gradients were observed in all columns, and the long-column outflow was nearly anoxic. A hydrogen sulfide odor was detected in the outflow from the long columns, and a black precipitate, indicative of iron sulfide, was observed on the outflow tubing.
TABLE 1.
Summary of concentrations of dissolved gases and nutrients in column experiments
| Concn (μM)a of:
|
|||||||
|---|---|---|---|---|---|---|---|
| O2 | 28N2 | 29N2 | 30N2 | NO3− | NH4+ | Total CO2 | |
| SC#1 (inflow) | |||||||
| Unamended | 214.8 ± 4.12b | 415.94 ± 3.02 | BDL | BDL | BDL | 0.33 ± 0.45 | 2,071.65 ± 23.43 |
| Amended | 219.55 | 415.16 | BDL | BDL | 17.95 | 0.31 | 2,024.17 |
| SC#3 (outflow) | |||||||
| Unamended | 101.14 ± 1.029 | 423.99 ± 0.478 | BDL | BDL | 2.78 | 0.40 ± 0.19 | 2,206.77 ± 19.26 |
| Amended | 54.53 | 422.95 | 0.13 | 10.15 | 16.57 | 0.82 | 2,282.00 |
| LC#1 (inflow) | |||||||
| Unamended | 214.8 ± 4.12 | 415.94 ± 3.021 | BDL | BDL | BDL | 0.33 ± 0.45 | 2,071.65 ± 23.43 |
| Amended | 219.55 | 415.16 | BDL | BDL | 17.95 | 0.31 | 2,024.17 |
| LC#3 (outflow) | |||||||
| Unamended | 7.31 ± 1.26 | 413.72 ± 0.208 | BDL | BDL | BDL | 0.12 ± 0.08 | 2,637.49 ± 2.69 |
| Amended | 8.26 | 418.28 | 3.47 | 2.88 | BDL | 1.05 | 2,464.63 |
Values for unamended columns are means ± standard deviations. Single samples were analyzed for all amended columns. BDL, below detection limit.
Nitrate release in the short column indicated net regeneration or ammonification followed by nitrification, while no nitrate was detected in the outflow of the long column (Table 1). Nearly all nitrogen in the outflow was in the form of 28N2, indicating that the majority of regenerated N was being denitrified. Further, little or no NH4+, N2O, 29N2, or 30N2 accumulated in the column outflow (Table 1), ruling out other N transformation pathways (dissimilatory nitrate reduction to ammonium or anammox [1]). Upon enrichment with 15NO3−, nearly all of the labeled nitrate was consumed in the long columns while only slight consumption was observed in the short columns. Little to no change in the ammonium concentration was observed in all column treatments. Production of 28N2 indicated that nitrification and denitrification were actively occurring and tightly coupled in the short columns, and 28N2 flux was not impacted by 15NO3− addition. Thus, coupled nitrification-denitrification was confirmed as a critical component of the marine N cycle in marine sands, representing a primary pathway for N removal from these ecosystems.
Geochemical constituents in SAB sediments at the W27 sampling site more closely resembled those observed in our short-column experiments. Pore water nitrate and ammonium concentrations in the surface sediments (depth, 0 to 5 cm) were in the low micromolar range over a seasonal cycle (34). Pore water oxygen concentrations ranged from 200 μM to near-anoxia, decreasing with sediment depth (34).
RFLP and statistical analyses of clone libraries.
Fifteen clone libraries constructed from SC#1, SC#3, LC#1, LC#3, and SAB core sediments resulted in a total of 309 SSU rRNA gene clones, 291 nosZ clones, and 202 amoA clones belonging to Bacteria. Clones were analyzed and grouped into phylotypes according to observed RFLP patterns. The total percent coverage of all five SSU rRNA gene clone libraries was nearly 88%, with individual library coverages ranging from 58 to 89%, with the exception of SC#1 (41%) (Table 2). The total percentages of coverage for the nosZ and amoA clone libraries were 93 and 99%, respectively (data not shown), with individual clone library percentages of coverage ranging from 66 to 89% for nosZ and 98 to 100% for amoA (Table 2). Rarefaction curves for the combined SSU rRNA gene and nosZ libraries, and the combined and individual amoA gene libraries, suggested that a sufficient number of clones were sampled to represent the diversity of these particular libraries. The rarefaction curves for the SSU rRNA and nosZ genes in SC#1, SC#3, LC#1, LC#3, and SAB core libraries did not indicate saturation; however, numerically dominant RFLP groups were observed. Estimation of species richness indicated that all of the column-derived SSU rRNA and nosZ gene libraries were more diverse than libraries from the SAB cores (Table 2). Shannon-Wiener and 1/D indices indicated a significant difference between the four column samples and the SAB core for both the SSU rRNA gene (P ≤ 0.05) and the nosZ gene (P < 0.01). Similar nucleotide and gene diversity, evenness, and θ(π) were calculated for all SSU rRNA and nosZ gene clone libraries (Table 2). All statistical estimators, including rarefaction and species richness, indicated low diversity in the five amoA gene clone libraries (Table 2). Shannon-Wiener and 1/D indices did not indicate a significant difference in diversity between the five amoA-derived clone libraries (P > 0.10).
TABLE 2.
Statistical analyses of SSU rRNA, nosZ, and amoA gene clone libraries using standard ecological and molecular estimates of sequence diversity
| PCR target and sample | No. of clones (phylotypes) | % Coverage | Species richness (95% confidence interval) | Evenness | Shannon-Wiener index | 1/D | Gene diversity (mean ± SD) | Nucleotide diversity (mean ± SD) | θ(π) (mean ± SD) |
|---|---|---|---|---|---|---|---|---|---|
| SSU rRNA gene | |||||||||
| SC#1 | 61 (48) | 41.0 | 120 (87, 195) | 0.84 | 4.01 | 158 | 0.98 ± 0.01 | 0.22 ± 0.11 | 279.4 ± 135.4 |
| SC#3 | 62 (40) | 58.1 | 88 (67, 138) | 0.86 | 3.84 | 86.1 | 0.98 ± 0.01 | 0.20 ± 0.10 | 248.7 ± 119.9 |
| LC#1 | 48 (29) | 60.4 | 65 (40, 147) | 0.76 | 3.18 | 32.2 | 0.95 ± 0.02 | 0.18 ± 0.09 | 217.8 ± 105.6 |
| LC#3 | 70 (37) | 71.4 | 57 (44, 96) | 0.85 | 3.42 | 39.0 | 0.97 ± 0.01 | 0.18 ± 0.09 | 229.3 ± 110.2 |
| SAB | 75 (23) | 89.3 | 32 (29, 45) | 0.92 | 3.18 | 29.5 | 0.96 ± 0.01 | 0.19 ± 0.09 | 230.9 ± 110.9 |
| nosZ | |||||||||
| SC#1 | 73 (39) | 69.2 | 67 (48, 120) | 0.83 | 3.49 | 44.5 | 0.96 ± 0.01 | 0.23 ± 0.11 | 213.3 ± 102.6 |
| SC#3 | 52 (29) | 65.9 | 46 (35, 82) | 0.85 | 3.27 | 40.2 | 0.97 ± 0.01 | 0.22 ± 0.11 | 207.4 ± 100.6 |
| LC#1 | 65 (31) | 78.5 | 43 (35, 73) | 0.86 | 3.25 | 32.5 | 0.95 ± 0.01 | 0.24 ± 0.12 | 216.3 ± 104.4 |
| LC#3 | 44 (24) | 71.2 | 52 (32, 127) | 0.74 | 2.95 | 22.0 | 0.93 ± 0.02 | 0.22 ± 0.11 | 199.3 ± 97.1 |
| SAB | 57 (21) | 89.5 | 23 (21, 33) | 0.90 | 2.82 | 17.9 | 0.93 ± 0.02 | 0.22 ± 0.11 | 208.8 ± 100.9 |
| amoA | |||||||||
| SC#1 | 48 (3) | 100.0 | 3 (3, 3) | 0.60 | 0.66 | 1.71 | 0.41 ± 0.07 | 0.007 ± 0.004 | 2.94 ± 1.74 |
| SC#3 | 32 (2) | 100.0 | 2 (2, 2) | 0.81 | 0.56 | 1.63 | 0.39 ± 0.08 | 0.006 ± 0.004 | 2.32 ± 1.45 |
| LC#1 | 48 (3) | 97.9 | 3 (3, 3) | 0.53 | 0.58 | 1.53 | 0.32 ± 0.07 | 0.005 ± 0.003 | 1.90 ± 1.22 |
| LC#3 | 49 (2) | 97.9 | 2 (2, 2) | 1.00 | 0.69 | 2.03 | 0.51 ± 0.02 | 0.008 ± 0.004 | 3.05 ± 1.79 |
| SAB | 25 (2) | 100.0 | 2 (2, 2) | 0.85 | 0.59 | 1.72 | 0.42 ± 0.08 | 0.006 ± 0.004 | 2.52 ± 1.56 |
Phylogenetic analysis based on the SSU rRNA gene.
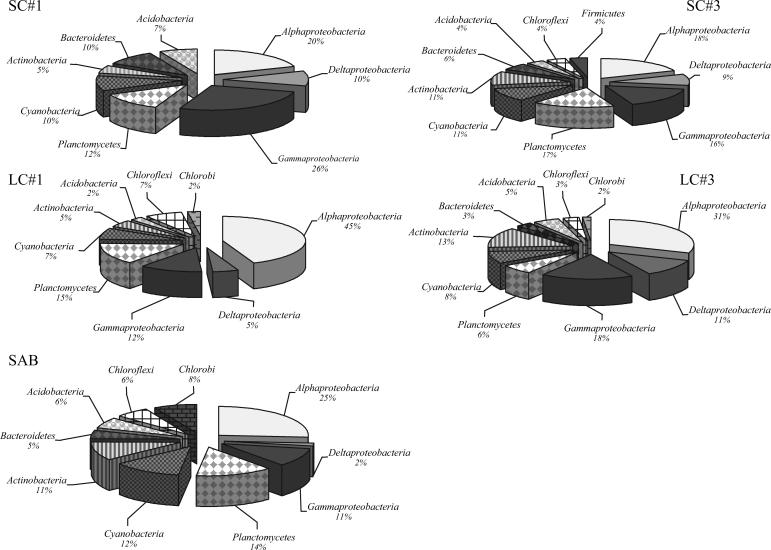
Sequence analysis of 48 of the 88 SSU rRNA gene phylotypes (phylotypes comprising more than a single clone; 257 total clones) indicated 10 distinct phyla, with a majority of the sequences most closely related to uncultured lineages from marine environments. The most frequently detected phylum was Proteobacteria, comprising 42% of the total SSU rRNA gene clones, with Alphaproteobacteria-related clones alone comprising 23% (Fig. 2). Planctomycetes (11%), Actinobacteria (7%), and Cyanobacteria (8%) comprised approximately one-third of all SSU rRNA gene clones (Fig. 2). The other four lineages detected (Acidobacteria, Bacteroidetes/Chlorobi, Chloroflexi, and Firmicutes) comprised between 1 and 6% of all SSU rRNA gene clones (Fig. 2). Interestingly, clone sequences most closely related to the class Betaproteobacteria were not detected.
FIG. 2.
Frequencies of bacterial phylogenetic lineages detected in SSU rRNA gene clone libraries derived from SC#1, SC#3, LC#1, LC#3, and SAB samples. Calculations were made based on the total number of clones associated with phylotypes from which a representative clone had been sequenced.
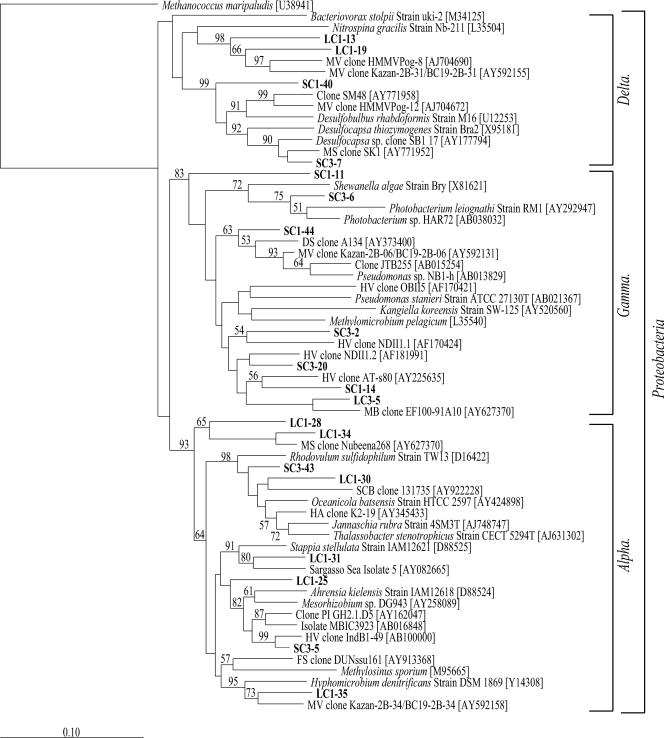
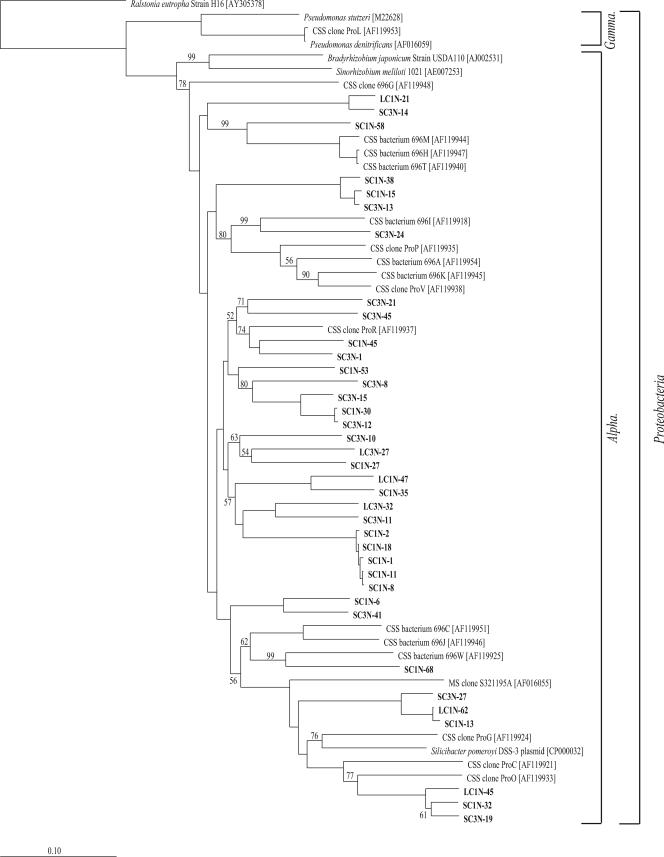
Two separate distance-based neighbor-joining trees were constructed with the 19 Proteobacteria- and 27 non-Proteobacteria-related sequences from this study and reference sequences from the GenBank database (Fig. 3 and 4). The Proteobacteria-related clones grouped into three classes, i.e., Alpha-, Delta-, and Gammaproteobacteria. Of the 19 phylotypes, 4 were most closely related to Deltaproteobacteria and, with the exception of phylotype SC1-40, grouped within two families, Desulfarculaceae and Desulfobulbaceae, of the order Desulfobacterales (Fig. 3). No similar taxonomic level could be determined for phylotype SC1-40; however, it was the most frequently detected Deltaproteobacteria-related phylotype and the only one to be detected in the SAB core sediment. Only 31% of the deltaproteobacterial clones were found in the SC#1 and LC#1 samples, compared to 63% found in the SC#3 and LC#3 samples, with no single sediment-related phylotype found in all five clone libraries. Phylotype SC3-7, the second most frequently detected of the phylotypes, was detected only in the two outflow samples, SC#3 and LC#3 (Fig. 3).
FIG. 3.
Phylum Proteobacteria neighbor-joining phylogenetic tree, incorporating a Jukes-Cantor distance correction, of SSU rRNA genes from SC#1, SC#3, LC#1, LC#3, and SAB samples. Sequences from this study and close relatives were aligned using the Fast Aligner algorithm, verified by hand, and compared to the E. coli SSU rRNA secondary structure using the ARB software package. Bootstrap analyses were conducted on 1,000 samples, and percentages greater than 50% are indicated at the nodes. Methanococcus maripaludis was used as the outgroup. Bar, 0.10 change per nucleotide position.
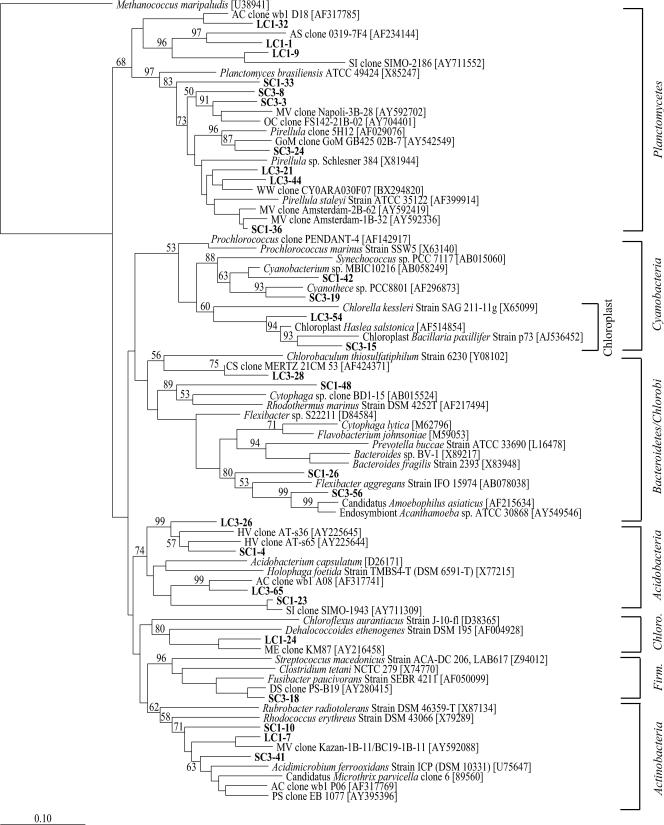
FIG. 4.
Nonproteobacterial neighbor-joining phylogenetic tree, incorporating a Jukes-Cantor distance correction, of SSU rRNA genes from SC#1, SC#3, LC#1, LC#3, and SAB samples. Sequences from this study and close relatives were aligned using the Fast Aligner algorithm, verified by hand, and compared to the E. coli SSU rRNA secondary structure using the ARB software package. Bootstrap analyses were conducted on 1,000 samples, and percentages greater than 50% are indicated at the nodes. Methanococcus maripaludis was used as the outgroup. Bar, 0.10 change per nucleotide position.
A total of 7 of the 19 Proteobacteria-related phylotypes grouped within the class Gammaproteobacteria (Fig. 3). Compared to the deltaproteobacterial phylotypes, the Gammaproteobacteria-related phylotypes were more diverse, representing multiple families and unclassified lineages. Although four phylotypes, represented by clones SC3-2, SC3-20, SC1-14, and LC3-5, grouped into a clade with no sequences from cultured isolates (Fig. 3), phylotypes SC3-2 and SC1-14 were most closely related to clone sequences associated with sulfur-oxidizing symbionts (32) and SC3-20 was most closely related to a nitrogen-fixing symbiont (67). Phylotypes SC3-2 and SC3-20 were two of the three most frequently detected phylotypes and were the only Gammaproteobacteria-related phylotypes to be detected in the SAB core.
The remaining eight Proteobacteria-related phylotypes were most closely related to the class Alphaproteobacteria and represented 23% of the total SSU rRNA gene clones. As with the Delta- and Gammaproteobacteria-related clones, the phylotypes that were most frequently detected overall were detected in the SAB cores. A total of 5 of the 8 phylotypes grouped within the order Rhodobacterales and the family Rhodobacteraceae, including the two most numerically dominant phylotypes, represented by clones LC1-31 and LC1-30. Phylotype LC1-35 clustered with Hyphomicrobium denitrificans strain DSM 1869 within the order Rhizobiales and family Hyphomicrobiaceae (Fig. 3). Hyphomicrobium denitrificans strain DSM 1869 was previously characterized as a facultative methylotroph capable of denitrification (48). Phylotypes LC1-34 and LC1-28 clustered with Nubeena 268, an uncultured marine sediment bacterium (unpublished data), in a clade distinct from the other orders of Alphaproteobacteria on the tree, as supported by strong bootstrap values (Fig. 3).
A second distance-based neighbor-joining tree was constructed with 27 non-Proteobacteria-related phylotypes grouping into seven phyla (Fig. 4). As with the Proteobacteria-related phylotypes, a majority of sequences collected were closely related to noncultured environmental clones from numerous marine habitats. Of the 27 non-Proteobacteria-related phylotypes, 10 grouped within the phylum Planctomycetes. With the exception of phylotypes SC3-3 and SC1-36, all of the phylotypes related to Planctomycetes were <97% similar to any previously identified SSU rRNA gene sequence. Although the phylum Planctomycetes has a single class, order, and family currently identified (15), three phylotypes, represented by LC1-32, LC1-1, and LC1-9 (42% of sequences related to Planctomycetes), clustered apart from the cultured Planctomycetes and were supported by good bootstrap values (Fig. 4). Phylotype LC1-32, the most frequently detected phylotype related to Planctomycetes, was detected in both inflow sediment samples and the SAB core sediment. The remaining seven phylotypes clustered with members of the genera Planctomyces and Pirellula.
The four cyanobacterium-related phylotypes (8% of the total clones) grouped into two clades (Fig. 4). Phylotypes SC1-42 and SC3-19 clustered within the order Chlorococcales and were most closely related to the nitrogen-fixing Cyanobacterium sp. strain MBIC10216 (94%) (unpublished data) and Cyanothece sp. strain PCC 8801 (97%) (66). The remaining two phylotypes, represented by clones LC3-54 and SC3-15, were most similar (both 97%) to chloroplast DNA sequences from the diatoms Haslea salstonica and Bacillaria paxillifer, respectively (unpublished data).
Four additional non-Proteobacteria-related phylotypes grouped within the two gram-positive phyla, Actinobacteria and Firmicutes. Although all three phylotypes related to Actinobacteria (7% of the total SSU rRNA clones) were detected in the SC#3 and SAB core clone libraries, only phylotype SC1-10, the most frequently detected phylotype related to Actinobacteria, was detected in all five clone libraries. The only phylotype related to Firmicutes, SC3-18, had limited similarity to Fusibacter paucivorans (90%) (Fig. 4).
The remaining nine non-Proteobacteria-related phylotypes represented the phyla Acidobacteria and Chloroflexi and the superphyla Bacteroidetes/Chlorobi (<17% of total clones). The four phylotypes related to Acidobacteria grouped within the class Acidobacteria, order Acidobacteriales, and family Acidobacteriaceae. Phylotype LC3-65, the most frequently detected phylotype related to Acidobacteria, was also the only phylotype related to Acidobacteria that was detected in the SAB core clone library. The only phylotype related to Chloroflexi, LC1-24 (4% of the total SSU rRNA clones), branched into a clade with the nitrogen-fixing Dehalococcoides ethenogenes strain 195 (36, 60). A total of four phylotypes grouped into the superphyla Bacteroidetes/Chlorobi. The most numerically dominant phylotype related to Bacteroidetes, SC3-56, was 92% similar to an uncharacterized endosymbiont (unpublished data). The only phylotype related to Chlorobi, LC3-28, branched into a clade with a green sulfur bacteria, Chlorobaculum thiosulfatiphilum strain 6230 (Fig. 4).
Phylogenetic analysis based on the nosZ gene.
Representatives of 37 of the 65 total phylotypes, i.e., the phylotypes comprising more than a single clone, were sequenced and analyzed (238 total nosZ-derived clones). The percentage of similarity for all phylotypes to previously identified sequences ranged from 79 to 88%; however, intralibrary sequence similarity ranged from 48 to 99%. Interestingly, 6 of the 10 most frequently detected phylotypes were identified in each of the five clone libraries. Only six phylotypes were detected in a single clone library, and none of these phylotypes contained more than three clones.
A distance-based neighbor-joining tree was constructed with the 37 nosZ sequences collected in this study (Fig. 5). Although nosZ has been detected in Beta- and Gammaproteobacteria, all sequences from this study were most closely related to nosZ genes from the class Alphaproteobacteria (Fig. 5). Several deep-branching clades were composed solely of clones from this study (Fig. 5). The largest clade incorporated 21 of the 37 phylotypes (57% of the total nosZ clones) and was most similar to a nosZ clone, ProR, identified from San Clemente Island, California (55) (Fig. 5).
FIG. 5.
Neighbor-joining phylogenetic tree, incorporating a Jukes-Cantor distance correction, of nosZ genes from SC#1, SC#3, LC#1, LC#3, and SAB samples. Sequences from this study and close relatives were aligned using the Fast Aligner algorithm and verified by hand using the ARB software package. Bootstrap analyses were conducted on 1,000 samples, and percentages greater than 50% are indicated at the nodes. Ralstonia eutropha strain H16 was used as the outgroup. Bar, 0.10 change per nucleotide position.
Phylogenetic analysis based on the amoA gene.
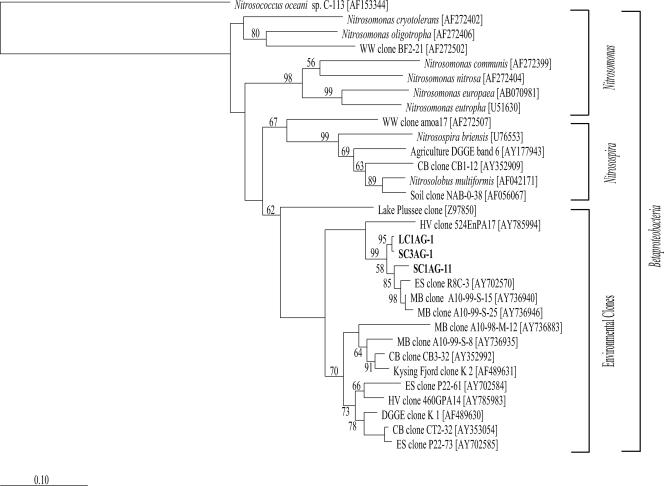
Analysis of the 202 amoA clones obtained from the four column samples and the in situ core indicated relatively low phylogenetic diversity. Representatives from all three phylotypes were sequenced and analyzed. Phylotypes LC1AG-1 and SC3AG-1 were detected in all five clone libraries and represented 99% of the total amoA clone sequences. Phylotype SC1AG-11 was found only in SC#1 and represented one clone.
A distance-based neighbor-joining tree was constructed with the three amoA sequences from this study (Fig. 6). All three sequenced clones clustered within the betaproteobacterial amoA group and branched into a large clade of environmental clones (Fig. 6), distinct from previously identified Nitrosomonas and Nitrosospira amoA clades. All three sequenced clones were 96 to 97% similar to clone A10-99-S-15 from Monterey Bay (44). Interestingly, all three phylotypes branch into their own clades, with an intralibrary sequence similarity of 96 to 98% (Fig. 6).
FIG. 6.
Neighbor-joining phylogenetic tree, incorporating a Jukes-Cantor distance correction, of amoA genes from SC#1, SC#3, LC#1, LC#3, and SAB samples. Sequences from this study and close relatives were aligned using the Fast Aligner algorithm and verified by hand using the ARB software package. Bootstrap analyses were conducted on 1,000 samples, and percentages greater than 50% are indicated at the nodes. Nitrosococcus oceani C-113 was used as the outgroup. Bar, 0.10 change per nucleotide position.
DISCUSSION
Past studies have hypothesized that severe hydrodynamic conditions, lower specific surface area, lower organic-matter content, and higher predation pressure contribute to lower bacterial abundance and a fundamentally different microbial-community composition in marine sands (8, 31, 53). Permeable sands undergo rapid pore water exchange, resulting in physically unstable environments that tend to diminish the geochemical gradients, which are thought to stratify microbial communities in fine-grained muddy sediments. We examined microbial activity and diversity in understudied continental shelf sands from the South Atlantic Bight off the coast of Georgia. In corroboration of results from group-specific FISH probes (8, 31, 54) applied to similar sedimentary environments, we observed abundant phylotypes affiliated with the phylum Proteobacteria (classes Alpha-, Delta-, and Gammaproteobacteria). In addition, phylotypes related to Planctomycetes, Actinobacteria, Acidobacteria, Bacteroidetes/Chlorobi, and Firmicutes, all of which are commonly found in less permeable sediments of the deep sea and estuaries, were detected (2, 4, 29, 32, 35, 46, 65). As is often the case in environmental sequence analysis, the majority of phylotypes we detected were not closely related to any cultivated representatives, and the sequences of several microbial groups were detected which have not been observed in past studies of permeable sands. We conclude that the microbial diversity of permeable sands has just begun to be revealed, and thus our clonal analysis provides a solid sequence database for the development of improved genetic probes designed to quantify the metabolically active microbial groups in these poorly studied but biogeochemically significant ecosystems. In addition, the data presented here suggest that previous reports may have underestimated sandy-sediment microbial diversity due to the fact that numerous lineages are below detection limits.
Phylogenetic diversity was shown to be relatively constant and consistent with geochemical determinations in column experiments designed to mimic in situ carbon and nitrogen flow. Sands in column experiments were exposed to a stable, redox-stratified environment for a 2-week incubation period. Changes in pore water geochemistry were observed by consumption of oxygen and production of N2, TCO2, Mn(II), Fe(II), and sulfide. In contrast, organic-matter degradation and coupled nitrification-denitrification, as indicated by total CO2 and N2 production, did not differ substantially between the column experiments (Table 1) (49). Similarly, microbial community diversity remained relatively constant between column experiments and across geochemical gradients within the columns. Abundant phylotypes of all gene targets remained relatively the same across large gradients in oxygen content and nitrogen species, with the exception of an increase in deltaproteobacterial sequences affiliated with S-transforming microorganisms from the inflow to the outflow of the long columns. From these results, it appears that column experiments provide a fairly accurate representation of the in situ microbial diversity of permeable shelf sediments.
The observed stable diversity may be a result of microbial community adaptations to large fluctuations in physicochemical parameters in permeable sands. Alternatively, due to the low organic-matter content and potentially low growth rates, cloning/sequencing targeted to DNA may not be sensitive enough to detect community change even after a 2-week incubation period. Further analysis using redesigned probes from our sequence database will be required in order to confirm the ecological significance of our observations.
A surprising observation was that denitrification occurred under largely oxic conditions in the bulk phase of the short columns (49). Such “aerobic denitrification” has been demonstrated in pure cultures of classic denitrifiers and nitrifiers (68). Denitrification of nitrite or nitrate to N2 can occur at near-atmospheric concentrations of O2 (52), and some strains show enhanced rates of aerobic denitrification at low oxygen concentrations (16). Column experiment results show that this process could be significant in permeable shelf sands. Therefore, as suggested by previous geochemical studies (7), denitrification, typically an anoxic process, may occur aerobically or within anaerobic microniches of otherwise aerobic sediment within the columns. Such microniches may have provided an environment suitable for denitrifiers that were below detection limits in the in situ SAB core to be identified in clone libraries from the sediment columns.
Microbial diversity was lower in the SAB core clone libraries than in the libraries constructed from column inflow and outflow sediment DNA extracts, as determined by species richness, the Shannon-Wiener index, and the reciprocal of Simpson's index (Table 2). Although less diverse, clones from the SAB core-derived library grouped into 10 of the 11 total phyla detected, including the most frequently detected phylotype in each of those phyla. Thus, the lack of SAB core library diversity was in the detection of multiple phylotypes within each phylum, not in a lack of phyla, as was supported by similar gene and nucleotide diversity indices for all libraries.
The lower diversity of the SAB core clone libraries may be explained by the geophysical properties of the shelf sediments. The highly permeable sediments of the South Atlantic Bight experience rapid, tidally driven bottom currents and migrating sediment wave forms (24). Tidal currents across migrating wave forms create variable advective flow rates into the sediments over short periods (20, 21, 61). Such frequently changing environmental conditions have been shown to restrict bacterial population growth (53), keeping some lineages below detection limits by standard cloning and sequencing techniques. Therefore, we hypothesized that, within the microbial community, the detectable diversity would expand in response to the creation of a stable redox interface in the columns. Interestingly, only one Deltaproteobacteria-related phylotype was detected in the SAB core clone library. However, in stable, stratified columns, numerous Deltaproteobacteria-related phylotypes were detected and were related to both aerobic sulfur oxidizers and anaerobic sulfate reducers. Therefore, a more complete description of the metabolic potential of the SAB permeable sediments was achieved by examining the microbial diversity across redox-stabilized geochemical gradients within the columns. By quantitative techniques with lower detection limits, future analysis of in situ core sediments from specific redox zones will be used to support our conclusions indicating additional diversity within the column libraries.
While the column experiments provided a means to detect a wider range of phylogenetic groups, some groups may have been overrepresented or, conversely, remained below detection limits. For example, although nitrification activity was demonstrated in geochemical determinations, nitrifying Betaproteobacteria-related phylotypes were absent in the SSU rRNA gene libraries from either the SAB core or column sediments. The presence of nitrifiers within the SAB sediments was indicated initially by amoA clonal analysis and then confirmed by amplification and cloning of SSU rRNA gene amplicons using primer sets specific for known nitrifiers (data not shown). As in other studies of marine environments (2, 4, 19, 35, 38, 39, 65), Betaproteobacteria-related phylotypes, a group that plays a critical role in ecosystem function, were in low abundance. In our study, the lack of Betaproteobacteria-related phylotypes is believed to be the result of these taxa being below our detection limits rather than being absent, and thus these taxa are underrepresented in our SSU rRNA-derived clone libraries. This conclusion is supported by FISH studies that show a decreased abundance and detectability of major microbial taxa in marine sands in comparison to muds (8, 31, 53).
Using amoA sequence analysis, we determined that the AOB in permeable shelf sediments were most similar to sequences retrieved during past studies of marine environments, primarily the water column of Monterey Bay and a variety of estuarine sediments (3, 44). Although our amoA sequences were not closely related to any amoA genes from cultured AOB, all phylotypes were affiliated with Nitrosospira spp., providing further evidence that this AOB group is ubiquitous in marine sediments (3, 14, 41). A number of physicochemical factors, including porosity, salinity, and ammonium and oxygen concentrations, have been implicated in the control of AOB species distribution in sediments (18). Salinity, in particular, was shown to correlate with changes in amoA sequence diversity in estuarine sediments (3, 14). In corroboration of the work of Bernhard et al. (3), we observed an AOB community that exhibited relatively low diversity at a high-salinity site. In addition, the most abundant amoA phylotypes in our study were most closely affiliated with environmental clones from sites where the salinity did not deviate from that of full-strength seawater (3, 44).
Interestingly, sediment manipulation and redox stabilization in the column experiments did not increase AOB community diversity, and our results suggest that other environmental parameters beyond salinity warrant further study. All previous studies of AOB diversity focused on marine muds that contained high porosity, low oxygen tension, and much higher ammonium concentrations relative to the sands we studied (3, 14, 41). The majority of phylotypes from these muddy sediments formed a separate sister clade in comparison to the sequences we retrieved. The detection of nearly identical phylotypes from physicochemically different marine environments emphasizes the need for further cultivation of AOB that are well represented in clone libraries but underrepresented in culture collections. Physiological characterization of new marine isolates would greatly aid in predicting niche differentiation and resource exploitation.
In contrast to the lack of diversity observed in the amoA-derived clone libraries, the diversity estimations for denitrifiers in the nosZ-derived clone libraries approached the levels of diversity established for the SSU rRNA gene-derived clone libraries. Although SSU rRNA gene libraries contained clones related to Gammaproteobacteria, those clones were <93% similar to any known denitrifiers. Therefore, it is not surprising that no nosZ-derived clones were related to Gammaproteobacteria and all were related to previously identified sequences from the Alphaproteobacteria. Based on this analysis, both SSU rRNA and nosZ gene sequence data indicated that Alphaproteobacteria are an important microbial group in permeable shelf sediments. Although the intralibrary nosZ sequence similarity was as low as 52% between some clone sequences, all contained characteristic histidine residues (69). Therefore, these sequences may be assumed accurate, although the limited relatedness to previously identified alphaproteobacterial sequences will require characterization of nosZ genes from cultured isolates to determine the appropriate host taxa. Additionally, some sequences within our libraries were 99% similar; however, we interpreted these sequences as separate phylotypes based on RFLP phylotyping. Regardless, this study represents a significant contribution of nosZ sequences to a relatively small database from marine sediments. Similar high levels of nosZ diversity were previously reported by Scala and Kerkhof (55) when characterizing denitrifiers in the coastal sediments of New Jersey and San Clemente Island. However, as indicated by the statistical estimators in this study, increased nosZ diversity was detected within the column sediments. Thus, by stabilizing the geochemical gradients within the sediment columns, allowing community stratification, increased diversity was identified not only in the total microbial community but also in the fraction associated with denitrification.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Science Foundation (OCE-0424967) and Florida State University (PEG 513680014).
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.An, S., W. S. Gardner, and T. Kana. 2001. Simultaneous measurement of denitrification and nitrogen fixation using isotope pairing with membrane inlet mass spectrometry analysis. Appl. Environ. Microbiol. 67:1171-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asami, H., M. Aida, and K. Watanabe. 2005. Accelerated sulfur cycle in coastal marine sediment beneath areas of intensive shellfish aquaculture. Appl. Environ. Microbiol. 71:2925-2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernhard, A. E., T. Donn, A. E. Giblin, and D. A. Stahl. 2005. Loss of diversity of ammonia-oxidizing bacteria correlates with increasing salinity in an estuary system. Environ. Microbiol. 7:1289-1297. [DOI] [PubMed] [Google Scholar]
- 4.Bowman, J. P., and R. D. McCuaig. 2003. Biodiversity, community structural shifts, and biogeography of prokaryotes within Antarctic continental shelf sediment. Appl. Environ. Microbiol. 69:2463-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braker, G., H. L. Ayala-del-Rio, A. H. Devol, A. Fesefeldt, and J. M. Tiedje. 2001. Community structure of denitrifiers, Bacteria, and Archaea along redox gradients in Pacific Northwest marine sediments by terminal restriction fragment length polymorphism analysis of amplified nitrite reductase (nirS) and 16S rRNA genes. Appl. Environ. Microbiol. 67:1893-1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braker, G., J. Z. Zhou, L. Y. Wu, A. H. Devol, and J. M. Tiedje. 2000. Nitrite reductase genes (nirK and nirS) as functional markers to investigate diversity of denitrifying bacteria in Pacific Northwest marine sediment communities. Appl. Environ. Microbiol. 66:2096-2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brandes, J. A., and A. H. Devol. 1995. Simultaneous nitrate and oxygen respiration in coastal sediments—evidence for discrete diagenesis. J. Mar. Res. 53:771-797. [Google Scholar]
- 8.Buhring, S. I., M. Elvert, and U. Witte. 2005. The microbial community structure of different permeable sandy sediments characterized by the investigation of bacterial fatty acids and fluorescence in situ hybridization. Environ. Microbiol. 7:281-293. [DOI] [PubMed] [Google Scholar]
- 9.Caffrey, J. M., N. Harrington, I. Solem, and B. B. Ward. 2003. Biogeochemical processes in a small California estuary. 2. Nitrification activity, community structure and role in nitrogen budgets. Mar. Ecol. Prog. Ser. 248:27-40. [Google Scholar]
- 10.Chao, A. 1987. Estimating the population size for capture-recapture data with unequal catchability. Biometrics 43:783-791. [PubMed] [Google Scholar]
- 11.Christensen, J. P. 1994. Carbon export from continental shelves, denitrification and atmospheric carbon dioxide. Cont. Shelf Res. 14:547-576. [Google Scholar]
- 12.Colwell, R. K. 1997. EstimateS: statistical estimation of species richness and shared species from samples, version 5. User's guide and application. [Online.] http://viceroy.eeb.uconn.edu/estimates.
- 13.Colwell, R. K., and J. A. Coddington. 1994. Estimating terrestrial biodiversity through extrapolation. Philos. Trans. R. Soc. Lond. B 345:101-118. [DOI] [PubMed] [Google Scholar]
- 14.Francis, C. A., G. D. O'Mullan, and B. B. Ward. 2003. Diversity of ammonia monooxygenase (amoA) genes across environmental gradients in Chesapeake Bay sediments. Geobiology 1:129-140. [Google Scholar]
- 15.Garrity, C., R. O. Ramseier, R. Peinert, S. Kern, and G. Fischer. 2005. Water column particulate organic carbon modeled fluxes in the ice-frequented Southern Ocean. J. Mar. Syst. 56:133-149. [Google Scholar]
- 16.Goreau, T. J., W. A. Kaplan, S. C. Wofsy, M. B. McElroy, F. W. Valois, and S. W. Watson. 1980. Production of NO2− and N2O by nitrifying bacteria at reduced concentrations of oxygen. Appl. Environ. Microbiol. 40:526-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heck, K. L., G. van Belle, and D. Simberloff. 1975. Explicit calculation of the rarefaction diversity measurement and the determination of sufficient sample size. Ecology 56:1459-1461. [Google Scholar]
- 18.Herbert, R. A. 1999. Nitrogen cycling in coastal marine ecosystems. FEMS Microbiol. Rev. 23:563-590. [DOI] [PubMed] [Google Scholar]
- 19.Holmes, A. J., N. A. Tujula, M. Holley, A. Contos, J. M. James, P. Rogers, and M. R. Gillings. 2001. Phylogenetic structure of unusual aquatic microbial formations in Nullarbor caves, Australia. Environ. Microbiol. 3:256-264. [DOI] [PubMed] [Google Scholar]
- 20.Huettel, M., and G. Gust. 1992. Impact of bioroughness on interfacial solute exchange in permeable sediments. Mar. Ecol. Prog. Ser. 89:253-267. [Google Scholar]
- 21.Huettel, M., W. Ziebis, and S. Forster. 1996. Flow-induced uptake of particulate matter in permeable sediments. Limnol. Oceanogr. 41:309-322. [Google Scholar]
- 22.Huettel, M., W. Ziebis, S. Forster, and G. W. Luther. 1998. Advective transport affecting metal and nutrient distributions and interfacial fluxes in permeable sediments. Geochim. Cosmochim. Acta 62:613-631. [Google Scholar]
- 23.Hurt, R. A., X. Y. Qiu, L. Y. Wu, Y. Roh, A. V. Palumbo, J. M. Tiedje, and J. H. Zhou. 2001. Simultaneous recovery of RNA and DNA from soils and sediments. Appl. Environ. Microbiol. 67:4495-4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jahnke, R., M. Richards, J. Nelson, C. Robertson, A. Rao, and D. Jahnke. 2005. Organic matter remineralization and porewater exchange rates in permeable South Atlantic Bight continental shelf sediments. Cont. Shelf. Res. 25:1433-1452. [Google Scholar]
- 25.Jahnke, R. A., J. R. Nelson, R. L. Marinelli, and J. E. Eckman. 2000. Benthic flux of biogenic elements on the Southeastern US continental shelf: influence of pore water advective transport and benthic microalgae. Cont. Shelf Res. 20:109-127. [Google Scholar]
- 26.Joergensen, B. B. 2000. Bacteria and marine biogeochemistry, p. 173-207. In H. D. Schultz and M. Zabel (ed.), Marine geochemistry. Springer, Berlin, Germany.
- 27.Lam, P., J. P. Cowen, and R. D. Jones. 2004. Autotrophic ammonia oxidation in a deep-sea hydrothermal plume. FEMS Microbiol. Ecol. 47:191-206. [DOI] [PubMed] [Google Scholar]
- 28.Laursen, A. E., and S. P. Seitzinger. 2002. The role of denitrification in nitrogen removal and carbon mineralization in Mid-Atlantic Bight sediments. Cont. Shelf Res. 22:1397-1416. [Google Scholar]
- 29.Li, L. N., C. Kato, and K. Horikoshi. 1999. Bacterial diversity in deep-sea sediments from different depths. Biodiversity Conserv. 8:659-677. [Google Scholar]
- 30.Liu, X. D., S. M. Tiquia, G. Holguin, L. Y. Wu, S. C. Nold, A. H. Devol, K. Luo, A. V. Palumbo, J. M. Tiedje, and J. Z. Zhou. 2003. Molecular diversity of denitrifying genes in continental margin sediments within the oxygen-deficient zone off the Pacific coast of Mexico. Appl. Environ. Microbiol. 69:3549-3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Llobet-Brossa, E., R. Rossello-Mora, and R. Amann. 1998. Microbial community composition of Wadden Sea sediments as revealed by fluorescence in situ hybridization. Appl. Environ. Microbiol. 64:2691-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lopez-Garcia, P., S. Duperron, P. Philippot, J. Foriel, J. Susini, and D. Moreira. 2003. Bacterial diversity in hydrothermal sediment and epsilonproteobacterial dominance in experimental microcolonizers at the Mid-Atlantic Ridge. Environ. Microbiol. 5:961-976. [DOI] [PubMed] [Google Scholar]
- 33.Maidak, B. L., J. R. Cole, C. T. Parker, Jr., G. M. Garrity, N. Larsen, B. Li, T. G. Lilburn, M. J. McCaughey, G. J. Olsen, R. Overbeek, S. Pramanik, T. M. Schmidt, J. M. Tiedje, and C. R. Woese. 1999. A new version of the RDP (Ribosomal Database Project). Nucleic Acids Res. 27:171-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marinelli, R. L., R. A. Jahnke, D. B. Craven, J. R. Nelson, and J. E. Eckman. 1998. Sediment nutrient dynamics on the South Atlantic Bight continental shelf. Limnol. Oceanogr. 43:1305-1320. [Google Scholar]
- 35.Matsui, G. Y., D. B. Ringelberg, and C. R. Lovell. 2004. Sulfate-reducing bacteria in tubes constructed by the marine infaunal polychaete Diopatra cuprea. Appl. Environ. Microbiol. 70:7053-7065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maymo-Gatell, X., Y. T. Chien, J. M. Gossett, and S. H. Zinder. 1997. Isolation of a bacterium that reductively dechlorinates tetrachloroethene to ethene. Science 276:1568-1571. [DOI] [PubMed] [Google Scholar]
- 37.Messing, J. 1983. New M13 vectors for cloning. Methods Enzymol. 101: 20-79. [DOI] [PubMed] [Google Scholar]
- 38.Mills, H. J., C. Hodges, K. Wilson, I. R. MacDonald, and P. A. Sobecky. 2003. Microbial diversity in sediments associated with surface-breaching gas hydrate mounds in the Gulf of Mexico. FEMS Microbiol. Ecol. 46:39-52. [DOI] [PubMed] [Google Scholar]
- 39.Mussmann, M., K. Ishii, R. Rabus, and R. Amann. 2005. Diversity and vertical distribution of cultured and uncultured Deltaproteobacteria in an intertidal mud flat of the Wadden Sea. Environ. Microbiol. 7:405-418. [DOI] [PubMed] [Google Scholar]
- 40.Nei, M. 1987. Molecular evolutionary genetics. Columbia University Press, New York, N.Y.
- 41.Nicolaisen, M. H., and N. B. Ramsing. 2002. Denaturing gradient gel electrophoresis (DGGE) approaches to study the diversity of ammonia-oxidizing bacteria. J. Microbiol. Methods 50:189-203. [DOI] [PubMed] [Google Scholar]
- 42.Nogales, B., K. N. Timmis, D. B. Nedwell, and A. M. Osborn. 2002. Detection and diversity of expressed denitrification genes in estuarine sediments after reverse transcription-PCR amplification from mRNA. Appl. Environ. Microbiol. 68:5017-5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nold, S. C., J. Z. Zhou, A. H. Devol, and J. M. Tiedje. 2000. Pacific Northwest marine sediments contain ammonia-oxidizing bacteria in the beta subdivision of the Proteobacteria. Appl. Environ. Microbiol. 66:4532-4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O'Mullan, G. D., and B. B. Ward. 2005. Relationship of temporal and spatial variabilities of ammonia-oxidizing bacteria to nitrification rates in Monterey Bay, California. Appl. Environ. Microbiol. 71:697-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Philippot, L. 2002. Denitrifying genes in bacterial and Archaeal genomes. Biochim. Biophys. Acta 1577:355-376. [DOI] [PubMed] [Google Scholar]
- 46.Piza, F. F., P. I. Prado, and G. P. Manfio. 2004. Investigation of bacterial diversity in Brazilian tropical estuarine sediments reveals high actinobacterial diversity. Antonie Leeuwenhoek 86:317-328. [DOI] [PubMed] [Google Scholar]
- 47.Purkhold, U., A. Pommerening-Roser, S. Juretschko, M. C. Schmid, H. P. Koops, and M. Wagner. 2000. Phylogeny of all recognized species of ammonia oxidizers based on comparative 16S rRNA and amoA sequence analysis: implications for molecular diversity surveys. Appl. Environ. Microbiol. 66:5368-5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rainey, F. A., N. Ward-Rainey, C. G. Gliesche, and E. Stackebrandt. 1998. Phylogenetic analysis and intrageneric structure of the genus Hyphomicrobium and the related genus Filomicrobium. Int. J. Syst. Bacteriol. 48:635-639. [DOI] [PubMed] [Google Scholar]
- 49.Rao, A. F., and R. A. Jahnke. Nitrogen cycling in permeable continental shelf sediments on the South Atlantic Bight. Submitted for publication.
- 50.Reimers, C. E., H. A. Stecher, G. L. Taghon, C. M. Fuller, M. Huettel, A. Rusch, N. Ryckelynck, and C. Wild. 2004. In situ measurements of advective solute transport in permeable shelf sands. Cont. Shelf Res. 24:183-201. [Google Scholar]
- 51.Riggs, S. R., S. W. Snyder, A. C. Hine, and D. L. Mearns. 1996. Hardbottom morphology and relationship to the geologic framework: Mid Atlantic continental shelf. J. Sed. Res. 66:830-846. [Google Scholar]
- 52.Robertson, L. A., T. Dalsgaard, N. P. Revsbech, and J. G. Kuenen. 1995. Confirmation of aerobic denitrification in batch cultures, using gas-chromatography and N-15 mass-spectrometry. FEMS Microbiol. Ecol. 18:113-119. [Google Scholar]
- 53.Rusch, A., S. Forster, and M. Huettel. 2001. Bacteria, diatoms and detritus in an intertidal sandflat subject to advective transport across the water-sediment interface. Biogeochemistry 55:1-27. [Google Scholar]
- 54.Rusch, A., M. Huettel, C. E. Reimers, G. L. Taghon, and C. M. Fuller. 2003. Activity and distribution of bacterial populations in Middle Atlantic Bight shelf sands. FEMS Microbiol. Ecol. 44:89-100. [DOI] [PubMed] [Google Scholar]
- 55.Scala, D. J., and L. J. Kerkhof. 1999. Diversity of nitrous oxide reductase (nosZ) genes in continental shelf sediments. Appl. Environ. Microbiol. 65:1681-1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Scala, D. J., and L. J. Kerkhof. 2000. Horizontal heterogeneity of denitrifying bacterial communities in marine sediments by terminal restriction fragment length polymorphism analysis. Appl. Environ. Microbiol. 66:1980-1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Scala, D. J., and L. J. Kerkhof. 1998. Nitrous oxide reductase (nosZ) gene-specific PCR primers for detection of denitrifiers and three nosZ genes from marine sediments. FEMS Microbiol. Lett. 162:61-68. [DOI] [PubMed] [Google Scholar]
- 58.Schneider, S., D. Roessli, and L. Excoffier. 2000. Arlequin ver. 2.000: a software for population genetics data analysis. Genetics and Biometry Laboratory, University of Geneva, Geneva, Switzerland.
- 59.Seitzinger, S. P., and A. E. Giblin. 1996. Estimating denitrification in North Atlantic continental shelf sediments. Biogeochemistry 35:235-260. [Google Scholar]
- 60.Seshadri, R., L. Adrian, D. E. Fouts, J. A. Eisen, A. M. Phillippy, B. A. Methe, N. L. Ward, W. C. Nelson, R. T. Deboy, H. M. Khouri, J. F. Kolonay, R. J. Dodson, S. C. Daugherty, L. M. Brinkac, S. A. Sullivan, R. Madupu, K. T. Nelson, K. H. Kang, M. Impraim, K. Tran, J. M. Robinson, H. A. Forberger, C. M. Fraser, S. H. Zinder, and J. F. Heidelberg. 2005. Genome sequence of the PCE-dechlorinating bacterium Dehalococcoides ethenogenes. Science 307:105-108. [DOI] [PubMed] [Google Scholar]
- 61.Shum, K. T. 1992. Wave-induced advective transport below a rippled water-sediment interface. J. Geophys. Res. 97:789-808. [Google Scholar]
- 62.Strickland, J. D. H., and T. R. Parsons. 1972. A practical handbook of seawater analysis. Fisheries Research Board of Canada, Ottawa, Ontario, Canada.
- 63.Strunk, O., and W. Ludwig. 1997. ARB: software for phylogenetic analysis. Distributed by the Technical University of Munich, Munich, Germany.
- 64.Tajima, F. 1983. Evolutionary relationship of DNA sequences in finite populations. Genetics 105:437-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Teske, A., K. U. Hinrichs, V. Edgcomb, A. D. Gomez, D. Kysela, S. P. Sylva, M. L. Sogin, and H. W. Jannasch. 2002. Microbial diversity of hydrothermal sediments in the Guaymas Basin: evidence for anaerobic methanotrophic communities. Appl. Environ. Microbiol. 68:1994-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Turner, S., T. C. Huang, and S. M. Chaw. 2001. Molecular phylogeny of nitrogen-fixing unicellular cyanobacteria. Bot. Bull. Acad. Sinica 42:181-186. [Google Scholar]
- 67.Wang, E. T., P. van Berkum, X. H. Sui, D. Beyene, W. X. Chen, and E. Martinez-Romero. 1999. Diversity of rhizobia associated with Amorpha fruticosa isolated from Chinese soils and description of Mesorhizobium amorphae sp. nov. Int. J. Syst. Bacteriol. 49:51-65. [DOI] [PubMed] [Google Scholar]
- 68.Zehr, J. P., and B. B. Ward. 2002. Nitrogen cycling in the ocean: new perspectives on processes and paradigms. Appl. Environ. Microbiol. 68:1015-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zumft, W. G. 1997. Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61:533-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.