Abstract
Bacterial communities reside in basal ice, sediment, and meltwater in the supra-, sub-, and proglacial environments of John Evans Glacier, Nunavut, Canada. We examined whether the subglacial bacterial community shares common members with the pro- and supraglacial communities, and by inference, whether it could be derived from communities in either of these environments (e.g., by ice overriding proglacial sediments or by in-wash of surface meltwaters). Terminal restriction fragment length polymorphism analysis of bacterial 16S rRNA genes amplified from these environments revealed that the subglacial water, basal ice, and sediment communities were distinct from those detected in supraglacial meltwater and proglacial sediments, with 60 of 142 unique terminal restriction fragments (T-RFs) detected exclusively in subglacial samples and only 8 T-RFs detected in all three environments. Supraglacial waters shared some T-RFs with subglacial water and ice, likely reflecting the seasonal flow of surface meltwater into the subglacial drainage system, whereas supraglacial and proglacial communities shared the fewest T-RFs. Thus, the subglacial community at John Evans Glacier appears to be predominantly autochthonous rather than allochthonous, and it may be adapted to subglacial conditions. Chemical analysis of water and melted ice also revealed differences between the supraglacial and proglacial environments, particularly regarding electrical conductivity and nitrate, sulfate, and dissolved organic carbon concentrations. Whereas the potential exists for common bacterial types to be broadly distributed throughout the glacial system, we have observed distinct bacterial communities in physically and chemically different glacial environments.
The subglacial environment, comprising sediment and water beneath glaciers and debris-rich ice accreted to the glacier sole, was historically considered to be devoid of life, and subglacial geochemical activity was explained exclusively in terms of abiotic processes (28). However, this view has been superseded with the recent discovery of microbial communities in the refrozen lake water above an Antarctic subglacial lake (27) and at the beds of alpine (14, 29) and high Arctic glaciers (30, 32). The microbiota may facilitate redox reactions and chemical weathering at the glacier bed (30), and their existence has potentially important implications for the global carbon cycle on glacial-interglacial timescales (32). By characterizing the source, composition, distribution, and biogeochemical function of subglacial microbiota in these cold, dark, oligotrophic environments, we extend our understanding of the limitations to life on Earth.
Whereas previous studies established the presence and viability of subglacial bacteria and evidence of their redox reaction products in the laboratory at near in situ conditions (29, 32), neither the source nor the relatedness of communities inhabiting different regions of glacier environments has been studied. Comparison of supra-, sub-, and proglacial bacterial communities present, respectively, on the surface of, beneath, and adjacent to glaciers could establish whether the subglacial microbiota are allochthonous (originating from adjacent environments) or autochthonous (indigenous communities unique and potentially adapted to the subglacial environment). If the subglacial community is distinct from the proglacial and supraglacial communities, it implies the existence of a discrete subglacial ecosystem.
We therefore hypothesized that different glacial environments (e.g., subglacial sediments versus supraglacial meltwaters) would harbor unique assemblages of microbes because they arose from different sources and/or diverged over time in response to specific selective pressures in situ, yet they could share some members due to cross-inoculation. To test this hypothesis, we used terminal restriction fragment length polymorphism (T-RFLP) analysis of bacterial 16S rRNA genes, a cultivation-independent technique that is especially useful for analyzing communities with low to intermediate levels of species diversity (1, 4, 10, 12, 13). Although T-RFLP analysis currently does not readily permit identification of community members, it does enable comparison of whole microbial assemblages across spatial and temporal scales without the same biases inherent in cultivation-dependent analyses (11, 21). Therefore, the supra-, sub-, and proglacial bacterial community T-RFLP patterns derived from a high Arctic glacier were used to study the relatedness between the communities and infer the sources of the subglacial microbiota.
MATERIALS AND METHODS
Field sites and sample collection.
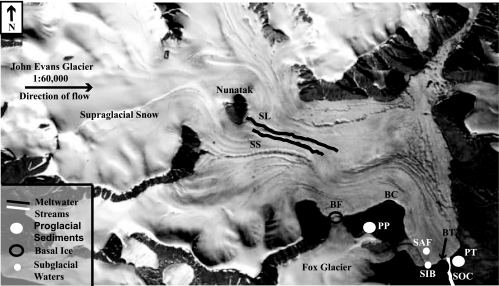
John Evans Glacier (79°40′N, 74°00′W) is a high Arctic polythermal valley glacier located on the eastern coast of Ellesmere Island, Nunavut, Canada. The glacier is about 20 km long, reaches a maximum thickness of ∼400 m at the equilibrium line, and terminates on land. The underlying bedrock is an Ordovician/Silurian carbonate/evaporite sequence with a small clastic component (17). The glacier consists mainly of ice at subfreezing temperatures, but there is a thin basal layer of temperate ice at the pressure melting point in its ablation area (9). Melting in the temperate core of the glacier and seasonal inputs of meltwater from the glacier surface provide liquid water at the base (32). The unfrozen sediments beneath the glacier are inaccessible except by drilling, but these sediments are entrained into basal ice where meltwaters refreeze beneath the cold-based marginal regions of the glacier. Ice flow then transports them to the glacier margins, enabling sampling of the basal ice as an analog of the unfrozen sediments and their associated microbiota (32). During the 2002 summer melt season, samples from the supra-, sub-, and proglacial environments (Fig. 1) were collected aseptically for molecular and hydrochemical analyses as previously described (32).
FIG. 1.
Locations of subglacial, supraglacial, and proglacial sampling sites on John Evans Glacier, Ellesmere Island, Nunavut, Canada (79°40′N, 74°00′W). Sample sites are indicated using site abbreviations defined in Materials and Methods.
The supraglacial samples comprised dry snow and seasonal meltwater on the glacier's surface. Dry snow (snow at subfreezing temperatures that has not yet undergone seasonal melting) was collected aseptically using ethanol-flamed tools and transferred to sterile plastic bags (WhirlPak; Nasco Products, New Hambury, Ontario, Canada). The snow samples were melted on-site in the bags and vacuum filtered through a sterile analytical filter unit with a removable 0.2-μm cellulose nitrate membrane (Nalgene, Rochester, New York). The filter membrane was stored in a new sterile plastic bag (WhirlPak) on ice in situ and during transport for 33 days and then frozen at −80°C upon arrival in the laboratory until analyzed.
Meltwater samples were collected from two different streams. One supraglacial stream drained a catchment composed entirely of glacier ice (“surface stream” [SS]). The second stream (“supraglacial lake” [SL]) drained onto the glacier from an ice-marginal lake. The runoff in this stream had contact with ice-marginal sediments and bedrock before draining onto the glacier, whereas the surface stream runoff did not. Water samples were collected aseptically in twice-autoclaved screw-cap bottles (Nalgene) that had been rinsed in deionized water and the respective sample stream (three times each) prior to sampling. Subsequently, the samples were stored on ice in situ and during transport for up to 22 days and then frozen at −20°C in the original bottles upon arrival at the laboratory until they were thawed for processing.
The subglacial samples included basal ice, frozen sediment, and liquid water representing different regions beneath the glacier. Basal ice containing sediment entrained from the bed at the core of the glacier was collected from three locations: the glacier terminus (“basal ice, terminus” [BT]), an ice cave exposed by a stream cutting into the glacier bed at the glacier's west margin (“basal ice, cave” [BC]), and the junction between John Evans Glacier and the tributary Fox Glacier (“basal ice, Fox Glacier” [BF]). Samples were collected aseptically (14, 32) in doubled Whirl-Pak bags and maintained frozen in situ for up to 23 days and during transport and storage at −20°C in the laboratory.
Subglacial water samples represented three different sources. Samples were collected from the first meltwaters to emerge from the glacier snout during the melt season on 30 June 2002 at 19:00 h (“subglacial initial burst” [SIB]). These solute-rich samples consist of water stored over the winter in contact with the glacier bed (3). The initial outburst persisted until 10:00 h on 1 July 2002, during which time the outflow shifted to an artesian fountain that emerged on the glacier surface, approximately 100 m from the snout (“subglacial artesian fountain” [SAF]). The initial burst outflow ceased shortly after the artesian fountain outflow started at 9:00 h. The artesian fountain persisted for 1 week (1 to 7 July 2002), during which time the outflow of subglacial waters at the glacier front resumed via a major channel (“subglacial outburst channel” [SOC]) (4 July to 1 August). The SIB samples are considered “early” season waters and the SOC samples “late” season waters, based on the progression of outflow and on chemical criteria (Table 1). The SAF samples represent a transition period during which waters that had been stored at the glacier bed over the winter were progressively diluted by the arrival at the bed of the new season's supraglacial meltwater. Samples were collected, frozen, and handled as described for supraglacial waters; they were stored on ice in situ and during transport for up to 8 days. No salt precipitates were observed in the bottles, indicating that the freezing process did not substantially affect the water chemistry.
TABLE 1.
Summary of pH, EC, and DOC measured in samples from John Evans Glacier
| Sample type | Median pH (range) (n) | Mean EC (μS/cm) (range) (n) | Mean DOC (ppm) (range) (n) |
|---|---|---|---|
| Supraglacial snow | 8.6 (6.8-8.7) (3) | 11.8 (3.4-21) (3) | 1.0 (0.38-1.35) (3) |
| Supraglacial waters (SS, SL) | 7.5 (6.4-8.1) (14) | 20.3 (4.3-49) (15) | 1.2 (0.3-3.4) (11) |
| Early subglacial waters (SIB) (30 June to 1 July 2002) | 8.0 (7.9-8.3) (16) | 558 (436-694) (16) | 0.4 (1) |
| Transitional subglacial waters (SAF) (1 to 7 July 2002) | 8.3 (7.4-8.5) (8) | 389 (210-492) (9) | 0.3 (0.2-0.3) (2) |
| Late subglacial waters (SOC) (4 July to 1 August 2002) | 7.9 (7.4-8.8) (5) | 282 (185-361) (6) | 1.7 (0.3-3.7) (6) |
| Basal ice (melted) (BT, BC, BF) | 7.3 (6.4-8.2) (5) | 140 (62-235) (5) | 62 (0.6-244) (5) |
Proglacial samples were sediments collected from sites in front of and adjacent to John Evans Glacier that were either exposed by retreat of the glacier or deposited by proglacial melt streams after deglaciation. Samples comprised fine sediments and gravels from exposed stream bank cuts directly in front of the glacier terminus (“proglacial terminus” [PT]) and from sorted stone polygons adjacent to the glacier (“proglacial polygons” [PP]) on which there was an algal crust and a few plants (e.g., Arctic poppy, Papaver radicatum; purple saxifrage, Saxifraga oppositifolia). Proglacial sediments were collected aseptically from several sites at 20- to 40-cm depths after removing the surface sediments and stored on ice in situ and during transport for up to 16 days until frozen at −20°C in the laboratory.
Hydrochemical analyses.
Replicate samples of supraglacial and subglacial waters were processed on-site for measurements of electrical conductivity (EC) and pH. Major anions (nitrate and sulfate) and dissolved organic carbon (DOC) were measured in the laboratory using the protocols described by Skidmore and Sharp (31) and Lafreniere and Sharp (18, 19). Samples for EC, pH, and ion chromatography were collected from streams in clean 1-liter polyethylene wide-mouth bottles (Nalgene) that were rinsed three times each in deionized water and the sample stream prior to sampling. Separate bottles were used for supraglacial and subglacial sampling to avoid cross-contamination. Subglacial ice samples were filtered and the filtrate analyzed chemically (18, 19, 31) when samples were thawed for T-RFLP analysis.
Sample processing.
Typically, two or more samples were collected from each site, and two or more aliquots of each sample were carried through separate amplification, digestion, and polyacrylamide gel electrophoresis steps to provide 141 DNA preparations for T-RFLP analyses.
In the laboratory, frozen supra- and subglacial water samples were thawed at 4°C in their original bottles for 1 to 2 days and processed for microbial analyses. The thawed 1-liter water samples were vacuum filtered through a sterile 0.2-μm-pore-size analytical filter unit (Nalgene). The filtrates for hydrochemical analyses (above) were collected in clean plastic bottles for EC, pH, and ion analysis and in combusted glassware for DOC analysis. The filters were used for T-RFLP analysis as follows. The filter chamber was disassembled, and the membrane was cut into sections (halves, quarters, or eighths) using a fresh sterile disposable scalpel blade. Each section was transferred with twice-autoclaved, bleach-rinsed forceps into a sterile 2.0-ml polypropylene screw-cap microcentrifuge tube with an o-ring seal and conical bottom (Biospec Products, Bartlesville, OK). Each tube contained approximately 0.5 g (each) of twice-sterilized 0.1- and 2.5-mm-diameter zirconium-silica beads (3.7 g/ml) (Biospec Products). Filled tubes were stored at −70°C prior to cell lysis and nucleic acid extraction.
Basal ice samples were thawed in twice-sterilized covered glass beakers at 4°C until just melted (1 to 3 days) and then filtered and processed as described above. Sediment or sediment-water slurry remaining in the beaker after filtration was measured into bead-filled tubes, using twice-autoclaved, bleach-rinsed spatulas, in 0.50-g or 500-μl aliquots and stored at −70°C until extraction.
Small replicate subsamples (0.5 g) of proglacial sediment were weighed into sterile bead-filled tubes and stored at −70°C until cell lysis and nucleic acid extraction.
Cell lysis, nucleic acid extraction, and PCR.
DNA was extracted from samples using a Mini-BeadBeater (Biospec Products) as previously described (14). The duration of bead beating (40 to 60 s) was selected through preliminary optimization trials with John Evans Glacier subglacial sediment. PCR was used to amplify near-full-length bacterial 16S rRNA genes (ca. 1,500 bp) from the extracted DNA using the universal bacterial primers PB36 (5′-AG[AG]GTTTGATC[AC]TGGCTCAG-3′) and PB38 (5′-G[GT]TACCTTGTTACGACTT-3′) (14), corresponding to the Escherichia coli 16S rRNA gene positions 8 to 27 and 1509 to 1492, respectively (numbering per Brosius et al. [5]). The forward primer, PB36, was synthesized with the 5′ end labeled with a fluorescein phosphoramidite dye (FAM; MWG Biotech, High Point, North Carolina), and all subsequent manipulations were protected from light. Generally, the reaction mixtures for PCR comprised 5 μl of template DNA, 200 μM concentrations of each deoxynucleotide, 10 μM concentrations of each primer, and 5 U/μl of Taq polymerase and buffer (Roche Diagnostics, Laval, Quebec, Canada) in a final volume of 50 μl. Some reactions were performed with either 5% or 2.5% dimethyl sulfoxide to obtain better PCR products. PCR was performed using a Techne Flexigene thermal cycler (Techne Flexigene, Princeton, New Jersey) and the following program: 94°C for 3 min, then 30 cycles of 94°C for 45 s, 53°C for 30 s, 72°C for 90 s, and finally, 72°C for 7 min. The concentration of the amplified product was estimated on a 1.5% agarose gel by comparing the intensity of amplicon bands to a 100-bp ladder (DNA molecular weight marker XIV; Roche) of known mass.
16S rRNA gene T-RFLP analysis.
Aliquots (60 ng) of the FAM-labeled PCR products were digested with HaeIII or HhaI (Roche), and 6 ng of each digested product was mixed with 1 μl of formamide in loading buffer (Applied Biosystems, Foster City, Calif.) plus 0.5 μl of DNA fragment length internal standard (TAMRA 2500; Applied Biosystems). After denaturation at 94°C for 5 min and chilling on ice, 2-μl aliquots were loaded onto a 36-cm 5% denaturing polyacrylamide gel, and the fragments were separated by electrophoresis for 6 h on a model ABI 377 XL automated sequencer (Applied Biosystems). The lengths of the 5′-terminal restriction fragments (T-RFs) were determined by comparison with the internal standard using GeneScan software (version 3.1; Applied Biosystems). If a peak was split, the right-hand peak was always selected. If the standard baseline in any lane was above approximately 15 relative fluorescence units (RFU), that lane was omitted from subsequent analyses. A sample T-RFLP gel and corresponding electropherogram are shown in Fig. S1 in the supplemental material.
A minimum peak amplitude of 50 RFU and an absolute minimum RFU threshold level (peak height) of 100 RFU were selected to eliminate background peaks and to identify valid peaks. Each valid peak represents a base pair category that theoretically corresponds to a different organism. The fluorescence threshold level is necessarily more rigorous than the minimum peak amplitude because the former defines the basic criterion of a valid base pair category, whereas the latter refers to the minimum height for a peak to be recognized and placed into the appropriate category. These criteria were determined after preliminary analysis of T-RF data and then applied to all electropherograms. Each peak exceeding 100 RFU and 80 bp in length was analyzed, and the T-RF category tolerances (i.e., acceptable variation in a calculated base pair size within a category) were established by analyzing replicate aliquots on the same gel and on independent gels to account for intra- and intergel variation, respectively (13, 20). Comparative analysis revealed that the variation resulting between replicated peaks run on different gels (intergel variation) was greater than the variation resulting from different FAM-PCR and restriction enzyme preparations (intragel variation). Thus, the intergel variation was used to calculate the category tolerance levels and establish a more conservative guideline for category differentiation. The average variations among 341 peaks on six different gels from identical samples (same extraction, amplification, and digestion) per base pair range were plotted (not shown), and a function was fit to the resulting logarithmic curve (y = 0.0487e0.0034x[r] and R2 = 0.8371, where x is the size in base pairs and y is the average variation in bp between replicated T-RFs on different gels). A master category list was created comprising all the valid T-RF base pair categories and edited to remove some redundancy, resulting in a list of categories with discrete base pair sizes. The accuracy of the category tolerances predicted by the function was established by visual inspection of the minimum and maximum base pair values within a gel and found to satisfactorily predict the appropriate breaks in the category list between discrete peaks.
Each gel data file was imported separately into GenoTyper (version 2.0; Applied Biosystems), and peak information (base pair values of valid T-RFs in each category for each sample site) was exported to Microsoft Excel (Microsoft Corp.), where it was converted to a binary data table and used in statistical analyses.
Precautions and controls.
In addition to standard precautions to reduce PCR contamination, additional steps were taken to ensure that extracted and amplified DNA originated from the environmental samples rather than from contaminating DNA. All sample manipulations were conducted in a UV-sterilized biohazard safety cabinet with HEPA-filtered airflow. All tools and containers (forceps, spatulas, and beakers) were autoclaved twice on successive days before use. Forceps and spatulas were also rinsed in 5% bleach prior to use. Reagent solutions were made from chemical stocks used only for PCR and that had not been touched with spatulas. DNA-free filter-barrier micropipette tips (Fisher Scientific) and heat-sterilized microcentrifuge tubes (Rose Scientific, Edmonton, Alberta) were used.
Extraction controls (bead-filled tubes containing only extraction reagents) were included with each extraction procedure conducted and subsequently amplified to certify that the reagents and beads were DNA free. A control consisting of autoclaved deionized water, filtered and processed in the same manner as the subglacial and supraglacial water samples, was used to confirm that the Nalgene filter unit, cellulose nitrate membrane (Nalgene), and water were DNA free. Negative (template-free) controls were included in every PCR set to certify that the reagents were DNA free. All controls were found to be negative by agarose gel electrophoresis. The extraction and PCR-negative controls were digested, loaded onto the polyacrylamide gel, and analyzed on T-RFLP gels with authentic samples to check for contamination by extraneous DNA. The few T-RF peaks greater than the minimum peak amplitude for import (50 RFU, see above) that appeared in some control lanes were eliminated as categories from the master T-RF category list. Thus, any potential contamination derived from the laboratory procedures was identified and eliminated from subsequent analysis.
Statistical analyses.
Principal component analysis (PCA) and cluster analysis were applied to T-RF peak information using Statistica (version 5; StatSoft, Inc., Tulsa, OK). The HaeIII and HhaI datasets were analyzed separately, generating two independent sets of statistical results (25). PCA identified 15 factors each for the HaeIII and HhaI samples, which explained >70% of the total variance in each data set. These factors represented groups of samples with a common complement of T-RFs, identified by their strong loadings (>0.50) on the factor. The percentage of total variance explained by each factor decreased from 24% (factor 1) to 1.5% (factor 15). Further detail on the factors is provided in Table S1 in the supplemental material. Cluster analysis was conducted using the PCA factor loadings of each DNA preparation in the HaeIII and HhaI datasets to group the samples into clusters so that samples sharing the same T-RFs or groups of T-RFs were placed in the same class. Samples were clustered using a single linkage agglomerative hierarchical method with generalized Euclidean distances as a measure of similarity between samples.
RESULTS AND DISCUSSION
Hydrochemical analyses of water and snow samples.
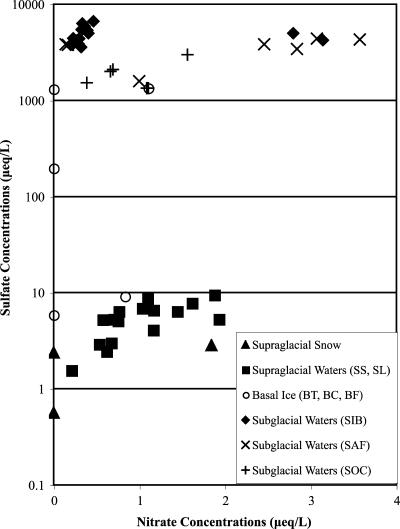
The pH values of all waters and melted snow samples were similar, being neutral to slightly alkaline (Table 1). Other hydrochemical signatures distinguish the sub- and supraglacial waters: snow and supraglacial meltwater samples were characterized by lower EC values (range, 3.4 to 49 μS/cm) and sulfate concentrations (range, 0.6 to 9 μeq/liter) than the basal ice and subglacial water samples (EC range, 62 to 694 μS/cm; sulfate range, 1,339 to 6,674 μeq/liter) (Table 1; Fig. 2), in accordance with previous data (31). These values imply that the initially dilute meltwaters acquired solutes as a result of rock-water interaction either on the glacier surface or during transit beneath the glacier (28).
FIG. 2.
Concentrations of nitrate and sulfate in 3 supraglacial snow samples, 16 supraglacial water samples (SS, SL), 5 basal ice samples (BT, BC, and BF), 17 early subglacial water samples (SIB; 30 June to 1 July 2002), 7 transitional subglacial water samples (SAF; 1 to 7 July 2002), and 6 late subglacial water samples (SOC; 4 July to 1 August 2002) corresponding to samples in Table 1.
At John Evans Glacier, sulfate is derived primarily from the dissolution of gypsum and/or anhydrite, which are known to outcrop beneath the glacier, so both EC and solute content provide an index of the extent and duration of water-rock contact (31). The EC of the subglacial water samples (mean, 458 μS/cm) was generally much higher than that of the basal ice samples (mean, 140 μS/cm) (Table 1). This is expected because solutes are rejected from ice as it forms from water. Among the subglacial waters, the early-season SIB waters (representing meltwaters stored at the glacier bed over winter) were solute rich (mean, 558 μS/cm), followed in order of appearance by the SAF (mean, 389 μS/cm) and the late-season SOC waters (mean, 282 μS/cm) that were progressively diluted by supraglacial meltwaters which probably drained rapidly through the glacier with limited contact with subglacial sediments (3).
The depleted DOC concentrations in the limited number of samples of SIB (0.4 ppm) and SAF (average, 0.3 ppm) analyzed compared with supraglacial snow and waters (means, 1.0 ppm and 1.2 ppm, respectively) and the SOC waters (mean, 1.7 ppm) suggest that the DOC was used subglacially as a microbial substrate over the winter (Table 1). However, more subglacial water samples are required to confirm significant differences.
Chemical analyses of basal ice samples.
Samples from the three basal ice sites (BT, BC, and BF) had varying DOC (Table 1), sulfate, and nitrate concentrations (Fig. 2). The mean DOC concentration in the melted basal ice samples was higher than in all other samples collected at John Evans Glacier, due to two extremely high DOC values (63 ppm and 244 ppm) at the BC site, whereas the single BT sample and two BF samples had DOC concentrations of ≤1.0 ppm, comparable to those of the subglacial waters (Table 1). This variability in basal ice DOC concentrations suggests that the distribution of organic carbon in the subglacial environment at John Evans Glacier is highly heterogeneous (2). Basal ice is chemically and physically distinct from the overlying glacier ice because it contains substantial amounts of sediment and is formed by processes that occur at the glacier bed (16, 32). The degree of contact between source waters, rock material, and organic carbon prior to formation of the basal ice layer influences the chemistry and heterogeneity of the basal ice samples (32).
The sulfate concentrations of the basal ice varied over several orders of magnitude (from 5.9 μeq/liter to 1,343 μeq/liter) (Fig. 2), illustrating the varying extent and duration of water-rock contact among the waters that form the basal ice sampled at the different sites. The low mean nitrate concentration in the basal ice (0.4 μeq/liter) may reflect microbial activity in anoxic microenvironments, either within the basal ice layer or in areas of the subglacial drainage system that supply the water that forms the basal ice (6). Skidmore et al. (32) observed the presence and activity of nitrate-reducing microbes in cultured water and ice from John Evans Glacier, demonstrating that such activity is possible in situ.
T-RFLP analysis.
T-RFLP analysis of 141 DNA preparations digested with HaeIII and 126 DNA preparations digested with HhaI resolved totals of 142 and 102 unique T-RFs, respectively. The same conclusions were drawn from both data sets, so only the HaeIII results are discussed in detail here.
Considering first the broad interenvironmental distribution of T-RFs among the subglacial (basal ice and subglacial waters), supraglacial (surface streams), and proglacial (sediment) environments, only 8 of the total 142 unique T-RFs were detected in all three environments (Table 2). This represents a small minority (8 to 18%) of the T-RFs found in each environment. In contrast, many T-RFs were unique to one of the three environments, and a minority was shared between two environments. For example, 60 (58%) of the 104 T-RFs detected in subglacial waters and ice were found only in subglacial samples. Likewise, 23 (46%) of the T-RFs detected in proglacial samples were unique to those samples. Only 16 T-RFs were common to both the subglacial and proglacial samples, indicating limited interaction between these sources of microbiota. The supraglacial waters had fewer unique T-RFs (12 of 43 total; 28%), but shared a large proportion (47%) with the subglacial samples. This pattern is consistent with the observed flow of supraglacial water into the subglacial drainage system, which likely provides seasonal “inoculation” of the subglacial system. The supraglacial and proglacial communities shared the lowest absolute and relative numbers of overlapping T-RFs (6 to 7%), consistent with limited wind-borne deposition being the most likely source of shared microbiota between these environments. Although T-RFs were detected in the supraglacial streams arising from melted surface snow and ice, no T-RFs were detected in the dry snow samples nor in samples from the BT basal ice site. It is possible that the use of filters with 0.2-μm pore diameters did not retain ultrasmall cells such as those recently detected in glacier ice cores (23). Thus, although some T-RFs were shared between and among environments, the data indicate that in-wash of supraglacial microbes and overriding of proglacial sediments cannot fully account for the composition of the subglacial community. Therefore, many T-RFs might represent autochthonous bacteria adapted to the subglacial environment.
TABLE 2.
Summary of the presence and distribution of 142 unique T-RFs generated by HaeIII digestion of DNA amplified from samples collected at John Evans Glacier
| Distribution of T-RFs | No. (%) of T-RFs in environmenta:
|
||
|---|---|---|---|
| Subglacial | Supraglacial | Proglacial | |
| Total detected in each environment | 104 (100) | 43 (100) | 50 (100) |
| Unique to subglacial samples | 60 (58) | ||
| Unique to supraglacial samples | 12 (28) | ||
| Unique to proglacial samples | 23 (46) | ||
| Common to sub- and supraglacial samples | 20 (19) | 20 (47) | |
| Common to sub- and proglacial samples | 16 (15) | 16 (32) | |
| Common to supra- and proglacial samples | 3 (7) | 3 (6) | |
| Common to sub-, supra-, and proglacial samples | 8 (8) | 8 (18) | 8 (16) |
Sample sites have been grouped into three broad categories to simplify comparison. “Subglacial” comprises samples from SIB, SAF, SOC waters and BT, BC and BF basal ice; “Supraglacial” comprises SS and SL stream waters; “Proglacial” includes PT and PP sediments.
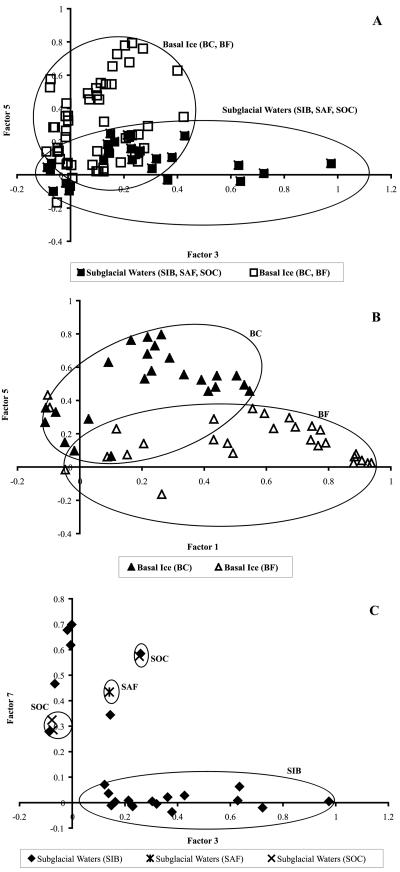
Multivariate analysis of the HaeIII data set (presented as a dendrogram in Fig. S2 in the supplemental material) confirmed that the bacterial communities from the sub-, supra-, and proglacial environments are distinct (i.e., showed interenvironmental differences). PCA presented as two-dimensional scatter plots of PCA factor loadings revealed intraenvironmental differences (Fig. 3). For example, within the subglacial environment, the waters and basal ice samples shared some T-RFs, as would be expected with refreezing of subglacial water onto the sole of the glacier, but the T-RF compositions of the subglacial waters and basal ice also varied as a result of the presence of unique T-RFs in each subenvironment (Fig. 3A). The two basal ice samples also overlapped, suggesting a common source for a core group of T-RFs as well as site-specific T-RFs that allowed the BC and BF samples to be distinguished using scatterplots of factor loadings (Fig. 3B). There were also some similarities among the T-RF compositions of six of the SIB waters and the SAF and SOC waters, but the majority of the SIB waters had distinctive T-RF compositions (Fig. 3C). This suggests that the T-RF composition of the SIB waters represents that portion of the bacterial community that is indigenous to regions of the glacier bed with limited hydrological activity. The SIB waters were also hydrochemically distinct from the SAF and SOC waters, having a higher mean EC value and depleted nitrate concentrations (Table 1 and Fig. 2). The chemical signatures of six SIB outliers (Fig. 3C) did not differ significantly from the majority of SIB samples that clustered together, so chemistry cannot explain the position of the six SIB outliers, and the reason for their variance is unknown. Additional interenvironmental distinctions are shown in Fig. S3 in the supplemental material for the supraglacial (SS and SL) and proglacial samples (PT and PP).
FIG. 3.
Scatterplots based on PCA of T-RFs detected in samples from different glacial environments showing intraenvironmental differences in T-RF complements. (A) Two types of subglacial samples (basal ice and subglacial waters), based on principal components (factors) 3 and 5. (B) Basal ice samples from two sites (BC and BF), based on factors 1 and 5. (C) Subglacial water samples (SIB, SAF, and SOC), based on factors 3 and 7. In each case, the factors were selected for presentation because the samples loaded heavily on those factors (see Table S1 in the supplemental material).
The subglacial microbial environment: hot spots and hot moments.
Physical and chemical differences among the different glacial environments and subenvironments likely result in a heterogeneous distribution of energy sources, nutrients, and bacteria at both the micro- and macroscale that produces the inter- and intraenvironmental differences in T-RF distributions. Thus, microbial niches may be defined temporally (“hot moments”), i.e., during over-winter storage of meltwater at the glacier bed, and/or spatially (“hot spots”), i.e., as a result of localization of sources of organic carbon and/or nutrients (2).
Features that distinguish the subglacial environment from the supra- and proglacial environments also make it habitable for microbes, including the presence of liquid water, nutrients, and terminal electron acceptors, as well as insulation from UV radiation and the large temperature fluctuations that occur at the surface (29). Because John Evans Glacier is polythermal, liquid water persists at the ice-bed interface in some regions year-round (29, 32), whereas only small quantities may be present at the glacier surface in winter (as a quasi-liquid water layer on the exterior of ice crystals) (7, 26). Nutrients, such as sulfate and nitrate, and DOC are released during subglacial weathering of the bedrock and sediments and are more readily available in subglacial environments than in supraglacial streams where atmospheric deposition is a primary source of nutrients (22, 33, 34). Furthermore, a potential supply of particulate and dissolved organic carbon in the subglacial environment exists in the bedrock, soils, and plants overridden during glacier advances (32). Legacy carbon from these sources could sustain and be recycled within dark subglacial communities, even in the absence of new primary production (2). Alternatively, subglacial organic carbon may be derived from chemoautotrophic metabolism or supraglacial in-wash of particulates. Finally, the subglacial environment likely includes anaerobic microhabitats and alternative electron acceptors like nitrate and sulfate (36). For example, Skidmore et al. (32) observed nitrate and sulfate reduction as well as methanogenesis following anaerobic incubation of basal ice samples and suggested that, although the subglacial meltwaters were primarily aerobic (from input of aerated supraglacial waters or release of dissolved O2 from melting glacier ice), the debris-rich basal ice and subglacial sediments probably contain anaerobic microenvironments. Only bacterial 16S rRNA gene primers were used for the current analysis, so it is possible that Archaea-specific primers would have detected sequences corresponding to the methanogenic activity detected by Skidmore et al. (32). The waters draining through major channels at the glacier bed (e.g., SOC) may not effectively sample those parts of the bed drained by a more distributed drainage system (e.g., SIB). The distributed system likely affords a greater opportunity for nutrient acquisition via subglacial weathering and is characterized by low rates of water flow that aid retention of fine sediments and associated bacteria (15, 35). Furthermore, Tranter et al. (35) suggested that, as oxygen levels decrease within meltwaters in the distributed system and associated sediments over the winter, possibly due to microbial activity, a different array of bacteria (e.g., nitrate and sulfate reducers and methanogens) capable of withstanding seasonal anoxia could become established.
Currently, T-RFLP analysis does not readily lend itself to identifying T-RFs from environmental samples due to a limited database of identified fragments. Furthermore, T-RFLP analysis can underestimate the true diversity of a microbial community, because of lack of sensitivity and resolution of similar T-RFs, or overestimate diversity by falsely identifying T-RFs that represent microheterogeneities or PCR errors rather than truly unique 16S rRNA genes. However, complementary cultivation-dependent (S. M. Cheng and J. Foght, unpublished data) and independent (30) studies at John Evans Glacier are beginning to reveal the identities of its subglacial microbiota. Skidmore et al. (30) have determined that about half (56%) of the 16S rRNA gene clones representing the subglacial bacteria at John Evans were related to the β-Proteobacteria and 25% were related to the Cytophaga/Flavobacterium/Bacteroides group, with a diversity of other phyla detected at a lower frequency. The subglacial microbiota of Bench Glacier in Alaska, in contrast, was less diverse but shared some sequences with John Evans. The basal ice from two temperate New Zealand glaciers was also characterized by cultivable β-Proteobacteria and Cytophaga/Flavobacterium/Bacteroides isolates (14). Further study is needed to determine whether there exists a widely distributed core group of bacteria adapted to the subglacial environment and to what extent each glacier's geography, geochemistry, and history make its microbiota unique.
Potential sources of subglacial microbiota.
Although recent studies of subsurface microbial consortia in cold environments (e.g., temperate alpine subglacial environments, Greenland glacier ice cores, and accreted ice from Lake Vostok) have discerned the microbial community composition through culture techniques and DNA sequencing analysis (8, 14, 24, 27), none has identified the origin of uniquely subglacial communities. Possible mechanisms for establishing a subglacial microbial community include (i) transport to the glacier bed by supraglacial meltwaters flowing down crevasses and moulins, (ii) vertical transport of viable cells from the surface by the flow of glacier ice, and (iii) basal incorporation of soils and sediments overridden during glacier advances. If the subglacial bacterial communities were established and replenished by either the first or second mechanism, the subglacial microbiota should comprise primarily supraglacial T-RFs, yet our analysis shows <20% overlap of supraglacial T-RFs in subglacial samples (Table 2). If the subglacial communities were established by the third mechanism, the majority of the subglacial T-RFs should be shared with the proglacial environment. Instead, T-RFLP analysis indicates that the subglacial microbiota can be distinguished from that detected in both the supra- and proglacial environments. It is possible that the origin of the subglacial microbes was one or both of these external environments and that the relative isolation of the subglacial environment from its surroundings resulted in the subsequent selection and establishment of a unique microbial community. However, continued inputs from both of these potential sources has not resulted in a simple amalgam of communities, suggesting that physical and chemical conditions at the bed exclude or select against certain components of the potential source communities, allow selected minor components to survive, and/or support T-RFs that cannot thrive in the surface environments. Further research is needed to elucidate the in situ effects of hydrochemistry on the microbiota and vice versa.
It is now clear that the subglacial environment at John Evans Glacier harbors microbes warranting further study and requiring examination of the subglacial material specifically, rather than surrogate materials such as surficial glacier ice and water or proglacial sediments. It is also clear that such samples must be collected from multiple subglacial sources to take into account subsurface heterogeneity.
Supplementary Material
Acknowledgments
This research was supported by the Canadian Circumpolar Institute and Northern Scientific Training Program (M.B.) and by NSERC (M.S., J.F.). Logistical support was provided by the Polar Continental Shelf Project (PCSP), Canada. This is PCSP contribution no. 00406.
Fieldwork was carried out with permission from the Nunavut Research Institute and the hamlets of Grise Fjord and Resolute Bay. S. Boon, M. Hanson, and D. Lewis assisted in collection of field samples. We are grateful to S. Cheng for assistance with sample processing for molecular analyses and to J. Barker for anion and DOC analyses. T-RFLP analysis was conducted in the Molecular Biology Services Unit (Biological Sciences Department, University of Alberta, Edmonton, Canada). We are grateful to C. Davis and H. Rai for helpful discussions.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Avaniss-Aghajani, E., K. Jones, D. Chapman, and C. Brunk. 1994. A molecular technique for identification of bacteria using small-subunit ribosomal-RNA sequences. BioTechniques 17:144-146, 148-149. [PubMed] [Google Scholar]
- 2.Barker, J. D., M. J. Sharp, S. J. Fitzsimons, and R. J. Turner. 2006. Abundance and dynamics of organic carbon in glacier systems. Arct. Antarc. Alp. Res. 38:163-172. [Google Scholar]
- 3.Bingham, R. G., P. W. Nienow, M. J. Sharp, and S. Boon. 2005. Subglacial drainage processes at a High Arctic polythermal valley glacier. J. Glaciol. 51:15-24. [Google Scholar]
- 4.Blackwood, C. B., T. Marsh, S.-H. Kim, and E. A. Paul. 2003. Terminal restriction fragment length polymorphism data analysis for quantitative comparison of microbial communities. Appl. Environ. Microbiol. 69:926-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brosius, J., T. J. Dull, D. D. Sleeter, and H. F. Noller. 1981. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J. Mol. Biol. 148:107-127. [DOI] [PubMed] [Google Scholar]
- 6.Campen, R. K., T. Sowers, and R. B. Alley. 2003. Evidence of microbial consortia metabolizing within a low latitude mountain glacier. Geology 31:231-234. [Google Scholar]
- 7.Carpenter, E. J., S. Lin, and D. G. Capone. 2000. Bacterial activity in South Pole snow. Appl. Environ. Microbiol. 66:4514-4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christner, B. C., E. Mosley-Thompson, L. G. Thompson, and J. N. Reeve. 2001. Isolation of bacteria and 16S rDNAs from Lake Vostok accretion ice. Environ. Microbiol. 3:570-577. [DOI] [PubMed] [Google Scholar]
- 9.Copland, L., and M. Sharp. 2001. Mapping thermal and hydrological conditions beneath a polythermal glacier with radio-echo sounding. J. Glaciol. 47:232-242. [Google Scholar]
- 10.Denaro, R., G. D'Auria, G. Di Marco, M. Genovese, M. Troussellier, M. M. Yakimov, and L. Giuliano. 2005. Assessing terminal restriction fragment length polymorphism suitability for the description of bacterial community structure and dynamics in hydrocarbon-polluted marine environments. Environ. Microbiol. 7:78-87. [DOI] [PubMed] [Google Scholar]
- 11.Dunbar, J., L. O. Ticknor, and C. R. Kuske. 2000. Assessment of microbial diversity in four southwestern United States soils by 16S rRNA gene terminal restriction fragment analysis. Appl. Environ. Microbiol. 66:2943-2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dunbar, J., L. O. Ticknor, and C. R. Kuske. 2001. Phylogenetic specificity and reproducibility and new method for analysis of terminal restriction fragment profiles of 16S rRNA genes from bacterial communities. Appl. Environ. Microbiol. 67:190-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Engebretson, J. J., and C. L. Moyer. 2003. Fidelity of select restriction endonucleases in determining microbial diversity by terminal restriction fragment length polymorphism. Appl. Environ. Microbiol. 69:4823-4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foght, J., J. Aislabie, S. Turner, C. E. Brown, J. Ryburn, D. J. Saul, and W. Lawson. 2004. Culturable bacteria in subglacial sediments and ice from two Southern Hemisphere glaciers. Microb. Ecol. 47:329-340. [DOI] [PubMed] [Google Scholar]
- 15.Hodson, A. J., P. N. Mumford, J. Kohler, and P. M. Wynn. 2005. The High Arctic glacial ecosystem: new insights from nutrient budgets. Biogeochemistry 72:233-256. [Google Scholar]
- 16.Hubbard, B., and M. Sharp. 1989. Basal ice formation and deformation: a review. Prog. Phys. Geogr. 13:529-558. [Google Scholar]
- 17.Kerr, J. W. 1972. Map 1358A geology: Dobbin Bay, District of Franklin. Geological Survey of Canada, Ottawa, Ontario.
- 18.Lafreniere, M., and M. Sharp. 2005. A comparison of solute fluxes and sources from glacial and non-glacial catchments over contrasting melt season. Hydrol. Process. 19:2991-3012. [Google Scholar]
- 19.Lafreniere, M., and M. Sharp. 2004. The concentration and spectrofluorometric properties of dissolved organic carbon (DOC) in surface waters: interpreting the flow routing of runoff and DOC sources in glacial and non-glacial catchments. Arct. Antarc. Alp. Res. 36:156-165. [Google Scholar]
- 20.Liu, W., T. L. Marsh, H. Cheng, and L. J. Forney. 1997. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl. Environ. Microbiol. 63:4516-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lukow, T., P. F. Dunfield, and W. Liesack. 2000. Use of T-RFLP technique to assess spatial and temporal changes in the bacterial community structure within an agricultural soil planted with transgenic and non-transgenic potato plants. FEMS Microbiol. Ecol. 32:241-247. [DOI] [PubMed] [Google Scholar]
- 22.Lyons, W. B., K. A. Welch, A. G. Fountain, G. L. Dana, B. H. Vaughn, and D. M. McKnight. 2003. Surface glaciochemistry of Taylor Valley, southern Victoria Land, Antarctica, and its relationship to stream chemistry. Hydrol. Process. 17:115-130. [Google Scholar]
- 23.Miteva, V. I., and J. E. Brenchley. 2005. Detection and isolation of ultrasmall microorganisms from a 120,000-year-old Greenland glacier ice core. Appl. Environ. Microbiol. 71:7806-7818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miteva, V. I., P. P. Sheridan, and J. E. Brenchley. 2004. Phylogenetic and physiological diversity of microorganisms isolated from a deep Greenland glacier ice core. Appl. Environ. Microbiol. 70:202-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Osborne, C. A., G. N. Rees, Y. Bernstein, and P. H. Janssen. 2006. New threshold confidence estimates for terminal restriction fragment length polymorphism analysis of complex bacterial communities. Appl. Environ. Microbiol. 72:1270-1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Price, P. B. 2000. A habitat for psychrophiles in deep Antarctica ice. Proc. Natl. Acad. Sci. USA 97:1247-1251. [DOI] [PMC free article] [PubMed]
- 27.Priscu, J. C., E. E. Adams, W. B. Lyons, M. A. Voytek, D. W. Mogk, R. L. Brown, C. P. McKay, C. D. Takacs, K. A. Welch, C. F. Wolf, J. D. Krishtein, and R. Avci. 1999. Geomicrobiology of subglacial ice above Lake Vostok, Antarctica. Science 286:2141-2144. [DOI] [PubMed] [Google Scholar]
- 28.Raiswell, R. 1984. Chemical models of solute acquisition in glacial meltwaters. J. Glaciol. 30:49-57. [Google Scholar]
- 29.Sharp, M., J. Parkes, B. Cragg, I. J. Fairchild, and M. Tranter. 1999. Widespread bacterial populations at glacier beds and their relationship to rock weathering and carbon cycling. Geology 27:107-110. [Google Scholar]
- 30.Skidmore, M., S. P. Anderson, M. Sharp, J. Foght, and B. D. Lanoil. 2005. Comparison of microbial community composition in two subglacial environments reveals a possible role for microbes in chemical weathering processes. Appl. Environ. Microbiol. 71:6986-6997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Skidmore, M. L., and M. J. Sharp. 1999. Drainage behaviour of a high Arctic polythermal glacier. Ann. Glaciol. 28:209-215. [Google Scholar]
- 32.Skidmore, M. L., J. M. Foght, and M. J. Sharp. 2000. Microbial life beneath a high Arctic glacier. Appl. Environ. Microbiol. 66:3214-3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Toom-Sauntry, D., and L. A. Barrie. 2002. Chemical composition of snowfall in the high Arctic 1990-1994. Atmos. Environ. 36:2683-2693. [Google Scholar]
- 34.Tranter, M. 1982. Controls on the chemical composition of Alpine glacial meltwaters. Ph.D. thesis. University of East Anglia, East Anglia, United Kingdom.
- 35.Tranter, M., M. Skidmore, and J. L. Wadham. 2005. Hydrological controls on microbial communities in subglacial environments. Hydrol. Process. 19:995-998. [Google Scholar]
- 36.Tranter, M., M. J. Sharp, H. R. Lamb, G. H. Brown, B. P. Hubbard, and I. C. Willis. 2002. Geochemical weathering at the bed of Haut Glacier d'Arolla, Switzerland-a new model. Hydrol. Process. 16:959-993. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.