Abstract
Xenorhabdus strains from entomopathogenic nematodes isolated from United Kingdom soils by using the insect bait entrapment method were characterized by partial sequencing of the 16S rRNA gene, four housekeeping genes (asd, ompR, recA, and serC) and the flagellin gene (fliC). Most strains (191/197) were found to have genes with greatest similarity to those of Xenorhabdus bovienii, and the remaining six strains had genes most similar to those of Xenorhabdus nematophila. Generally, 16S rRNA sequences and the sequence types based on housekeeping genes were in agreement, with a few notable exceptions. Statistical analysis implied that recombination had occurred at the serC locus and that moderate amounts of interallele recombination had also taken place. Surprisingly, the fliC locus contained a highly variable central region, even though insects lack an adaptive immune response, which is thought to drive flagellar variation in pathogens of higher organisms. All the X. nematophila strains exhibited a consistent pattern of insecticidal activity, and all contained the insecticidal toxin genes xptA1A2B1C1, which were present on a pathogenicity island (PAI). The PAIs were similar among the X. nematophila strains, except for partial deletions of a peptide synthetase gene and the presence of insertion sequences. Comparison of the PAI locus with that of X. bovienii suggested that the PAI integrated into the genome first and then acquired the xpt genes. The independent mobility of xpt genes was further supported by the presence of xpt genes in X. bovienii strain I73 on a type 2 transposon structure and by the variable patterns of insecticidal activity in X. bovienii isolates, even among closely related strains.
Xenorhabdus spp. are symbionts of entomopathogenic nematodes (Steinernematidae), where they reside in monoculture in a special vesicle in infective juveniles. When a nematode enters an insect, bacterial cells are released into the hemocel and replicate rapidly, destroying the insect and releasing a nutrient soup for the reproducing nematodes (1). A high level of specialization occurs in this mutualism, with a nematode carrying a single strain of Xenorhabdus. Recent evidence suggests that horizontal transmission of Xenorhabdus strains to other nematodes is effectively prevented by this specialization (26). Indeed, experiments showed that some nonnative strains were actually pathogenic to the nematode to which they were introduced (26). In addition to this, upon infection of the insect, Xenorhabdus strains produce potent antibiotics and other antimicrobial substances (31), leading to competitive exclusion of other strains and ensuring a virtual monoculture, which provides ideal conditions for the reproducing nematodes (5). Therefore, contact with other bacteria and scope for gene transfer and genetic recombination in Xenorhabdus should, in theory, be extremely limited in both the nematode and the infected insect. This would lead to clonal population structures emerging independently in each strain of nematode. However, genetic analysis of the closely related genus Photorhabdus, which is associated with nematodes in the same way as Xenorhabdus, seems to disprove this theory. The genome sequence of Photorhabdus luminescens TTO1 contains a large number of mobile genetic elements, suggesting continuously ongoing gene transfer. Phage remnants account for 4% of the genome, and there are 195 insertion sequences (ISs) or IS fragments present on the chromosome (4). Photorhabdus strains also contain many genomic islands, presumably acquired via horizontal transmission (34).
Multilocus sequence typing (MLST) types bacteria based on the sequence of a number of housekeeping genes and is thought to reflect the phylogeny of strains more accurately than other methods (11). Also, because a number of genes at different locations on the chromosome are sequenced, the amount of recombination occurring in a population can be calculated. The method has been applied almost exclusively to important bacterial pathogens and not to other types of bacteria. Xenorhabdus strains have the potential to become commercially important genera, producing oral insecticidal toxins for use in biocontrol and novel antibiotics with the potential to control clinically important microorganisms. Although the type strains have been well studied, and both an Xenorhabdus nematophila strain and an Xenorhabdus bovienii strain have been sequenced (http://xenorhabdus.danforthcenter.org), the extent of diversity within the species is largely unknown. Therefore, whether populations exhibit enough diversity to warrant screening for novel toxins and/or antibiotics, not present in the type strains, is not yet known.
The xpt genes of X. nematophila PMFI296 are responsible for oral insecticidal activity against a range of Lepidopteran insects (24). The four xpt genes involved in this activity, xptA1, xptA2, xptB1, and xptC1, are tightly linked on the chromosome of X. nematophila PMFI296 (13). Analogous genes have been found in P. luminescens (3), Yersinia pestis (33) and Serratia entomophilia (8). To agree with current nomenclature for the toxin genes, two of the original names have been changed: the xptB1 gene has been renamed xptC1, and xptC1 has been named xptB1 (24). To date, most xpt-like genes have been found associated with lysogenic phage-like elements. In Photorhabdus strain W14, tcdA and tcdB are associated with the lysR phage gene and are expressed from phage-like promoters induced by mitomycin C (33). It has been postulated that tc genes in Photorhabdus may be part of a cryptic prophage (33). Indeed, tcdA and tcdB from P. luminescens W14 have been shown to produce phage-like particles when expressed in Escherichia coli (33). In Y. pestis, the xpt-like genes are interspersed with the lysogenic phage-like genes gp13 and gp19, which code for a porin and a lysine, respectively. In Serratia, although the sep genes are located on a plasmid (8), they are associated with a gp19-like gene. In X. nematophila PMFI296, the xpt genes are not associated with lysogenic phage genes and were perhaps horizontally acquired by other means. The aim of this work was to characterize United Kingdom Xenorhabdus isolates by sequencing key housekeeping genes and to determine whether typing schemes correlated to patterns of insecticidal activity. For selected strains the aim was to examine the context of xpt genes and recombination at housekeeping loci and fliC in order to give an indication of the amount of potential gene exchange in the populations.
MATERIALS AND METHODS
Bacterial strains and media.
The type strain of X. nematophila used was NCIMB9965, which is referred to in this paper as strain 9965. The other type strains used were X. bovienii UQM2872, Xenorhabdus poinarii UQM2211, and Xenorhabdus beddingii (ATCC 4921). These type strains (referred to here as X. bovienii TS, X. poinarii TS, and X. beddingii TS) were gifts from Noel Boemare, INRA-CNRS, Paris, France. Sequence data for the fully sequenced X. bovienii strain (referred to as X. bovienii GS), isolated from a new species of nematode (29), was obtained from http://xenorhabdus.danforthcenter.org. The strains were grown and maintained on NA (Merck, Darmstadt, Germany). Isolation medium (NBTA) consisted of NA supplemented with bromothymol blue (25 mg ml−1) and triphenyl-2,3,5-tetrazolium chloride (4 mg ml−1).
Bacterial strain isolation.
Xenorhabdus strains were isolated from nematodes obtained from over 20 locations throughout the United Kingdom by an entrapment method using Galleria melonella larvae. Soil samples were collected and stored at 18°C for a maximum of 21 days. Leaf litter and large invertebrates were removed from the soil by hand, and approximately 30-g samples were placed in vented plastic sandwich boxes and baited with four G. melonella larvae. After 3, 7, and 14 days, the G. melonella larvae were observed for mortality and hemolymph from dead larvae was examined under a microscope. If rod-shaped bacteria were identified, the hemolymph sample was plated onto NBTA medium and incubated at 25°C for 72 h. To obtain pure cultures, dark blue and green colonies were selected and subcultured onto fresh NBTA plates, from which single colonies were picked and plated onto NA. In order to differentiate between the presumptive Xenorhabdus and Photorhabdus strains, they were grown for 16 h in 5 ml LB at 28°C. One milliliter of this culture was then placed in a luminometer (Wallac), and those that gave a positive signal (emitted light) were presumed to be Photorhabdus spp. and were not included in the study.
DNA manipulation techniques.
Chromosomal DNA was extracted from 1.5 ml of overnight X. nematophila culture by using a genomic DNA isolation kit (QIAGEN, Crawley, United Kingdom). PCR products were gel purified using a QIAquick PCR purification kit (QIAGEN) and products cloned using a pGEM Easy TA kit (Promega) according to the manufacturer's instructions. Cosmids and PCR products were sequenced using PCR, subcloning, and primer walking.
Sequencing of housekeeping genes and flagellin gene.
An internal fragment of the 16S rRNA, asd (accession no. AY317151.1), ompR (U07746.1), recA (U87924.1), serC (AY077462.1), fliC (X91047.1), and xptA3 genes was amplified by PCR. Each reaction was carried out in a 50-μl volume containing 1 μM of each primer (Table 1), 0.2 mM deoxynucleoside triphosphates, 200 ng chromosomal DNA, and 1 U Dynazyme (Flowgen). PCR cycles consisted of 94°C for 2 min and 25 cycles of 94oC for 15 s, 50 to 55oC (see Table 1 for value for each primer set) for 30 s, and 72°C for 45 s. The PCR products were purified and 200 ng of purified DNA used for sequencing with the same primers that had been used for the PCR amplification. Sequences of individual genes were edited using EditSeq (DNA*). Multiple-sequence alignments were generated using ClustalX and reedited to ensure correct alignment within Megalign. Sequences were compared using the packages Dnadist, Dnapars, Seqboot, Consense, and Neighbor within the Phylip suite of packages. Trees were viewed using Treeview.
TABLE 1.
Primer sequences, expected product sizes, and annealing temperatures used in PCR amplification of the 16S rRNA gene, housekeeping genes, fliC, and xptA3
| Gene | Forward primer | Reverse primer | Approx size (bp) | Annealing temp (oC) |
|---|---|---|---|---|
| 16S rRNA | GATGGAGGGGGATAACCACT | TTGTCCAGGGGGCCGCCT | 720 | 54 |
| asd | GAAGAGCGTCGCATTGTTGATCA | CGTTTTGGTGGTGCACTGATGGC | 335 | 51 |
| ompR | AATGCAGAACAGATGGATCG | TCCCCGGGCCAGGCTCATCAAT | 470 | 51.5 |
| recA | CCAATGGGCCGTATTGTTGA | TCATACGGATCTGGTTGATGAA | 420 | 54 |
| rpoS | CGCACAATTCGTCTGCCTAT | CTTGAATCTGACGCACTCTC | 430 | 53 |
| serC | CCACCAGCAACTTTGTGTCCTTTC | AAAGAAGCAGAAAAATATTGCAC | 670 | 53 |
| fliC | CCGCGTAGTCAGCATCTTCG | GAAGACAGACGCTCAATAGC | 755 | 52 |
| xptA3 | CTCCGCCACTTAATGGAC | CCGTTATAGACGGTATCC | 520 | 55 |
Analysis of housekeeping alleles.
For each locus the sequences obtained for all isolates were compared, and different sequences were assigned arbitrary allele numbers. For each isolate, the combination of alleles obtained at each locus defined its allelic profile. Those isolates with the same allelic profile were assigned to the same sequence type (ST), which was again identified by an arbitrary number. Allelic profiles and allele sequences were imported into the START program (http://outbreak.ceid.ox.ac.uk/software.html), where the dN/dS ratios and the homoplasy test were performed. The standardized index of association, ISA, was calculated using LAIN 3.1 (http://adenine.biz.fh-weihenstephan.de/lian). Sawyer's test was performed using GeneConv 1.81 (http://www.math.wustl.edu/∼sawyer/geneconv) with silent polymorphisms only (r option). SplitsTrees were constructed using SplitsTree4 (http://www-ab.informatik.uni-tuebingen.de/software/jsplits) with the default settings. Bootscanning was implemented using Simplot 2.5 (http://sray.med.som.jhmi.edu/RaySoft/SimPlot) with the default settings, with the exception of a window size of 120 bp and the Dnapars algorithm.
Insect bioassays.
The activity of each strain against Pieris brassicae and Phaedon cochleariae was measured as previously described (24). The activity of each strain against Aedes aegypti was measured as described by Thomas et al. (32). All bioassays were carried out in duplicate, and positive insecticidal activity was determined as greater than 30% insect mortality within 3 days. Negative control samples had less than 5% mortality in repeat assays.
Southern blotting.
Chromosomal DNA (1 μg) was restricted and the fragments separated on a 0.7% agarose gel. The gel was blotted onto a positively charged nylon membrane (Roche) using 20× SSC (3 M NaCl, 300 mM Na-citrate, pH 7.0). Membranes were prehybridized in hybridization solution (5× SSC, 0.5% Roche blocking agent,1% N-lauryl sarcosine, 0.02% sodium dodecyl sulfate [SDS]) for 2 h at 68°C and hybridized in the same solution containing 20 to 50 ng μl−1 of gel-purified labeled probe. Probes (approximately 500 bp in length) were generated by PCR using 3 μl digoxigenin DNA-labeling mix, 1 μM of each primer, 1 U Dynazyme (Flowgen), and 0.5 μg template. To create probes, a template consisting of the cloned PCR product containing the IS element was used. After hybridization, membranes were washed in 2× SSC containing 0.1% (wt/vol) SDS for 5 min at room temperature and then twice with 0.1× SSC containing 0.1% (wt/vol) SDS at 68°C for 15 min. Membranes were blocked in Tris (100 mM, pH 7.0) containing NaCl (150 mM) and 1% (wt/vol) blocking agent (Roche) for 1 h, before addition of antidigoxin-AP (Roche) as described by the manufacturers. Membranes were developed using nitroblue tetrazolium/5-bromo-4-chloro-3-indolylphosphate (Sigma, Gillingham, United Kingdom).
PCR mapping of xpt loci.
PCR was carried out using 1.7 U of the enzyme Expand HiFi (Roche) with 1 μM primer, 200 μM deoxynucleoside triphosphates, 0.5 μg DNA, and the recommended buffer. Cycling conditions consisted of 2 min at 94°C; 10 cycles of 94°C for 15 s, 60°C for 30 s, and 68°C for 8 min; 20 cycles of 94°C for 15 s, 60°C for 30 s, and 68°C for 8 min with a 5-s increment for each subsequent cycle; and 68°C for 10 min. PCR products were analyzed by gel electrophoresis on 0.7% agarose gels and characterized by restriction digestion and sequencing where appropriate.
RESULTS
Characterization of the culture collection by partial 16S rRNA gene sequencing.
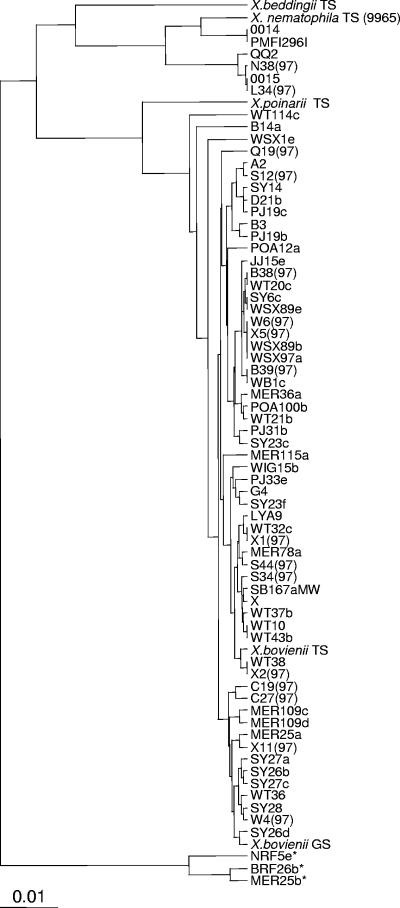
The presumptive Xenorhabdus isolates were initially compared to each other and characterized strains (X. bovienii, X. poinarii, X. beddingii, and X. nematophila type strains and the fully sequenced X. bovienii GS) by sequence analysis of the 16S rRNA gene. The results from the analysis of the 16S rRNA gene can be seen in Fig. 1. Most of the strains showed greatest sequence similarity to sequences from X. bovienii, with only a few isolated strains showing sequence similarity to X. nematophila. Within strains with similarity to the X. bovienii group, a number of subdivisions were observed. Thus, these strains could represent subspecies of X. bovienii. Overall, the results confirmed that the strains isolated by the nematode insect bait entrapment method, from United Kingdom soils, were strains from within the genus Xenorhabdus. Three strains, NRF5e, BRF26b, and MER25b, possessed 16S rRNA sequences that showed greatest identity to Providencia stuartii and uncharacterized bacteria isolated from insect-pathogenic nematodes infecting Drosophila (12). However, phenotypically these strains were indistinguishable from the other X. bovienii strains, producing a yellow pigment when grown on LB agar and exhibiting exactly the same morphology when viewed microscopically. Sequences from the housekeeping genes also suggested that they were X. bovienii strains, and they therefore are referred to as such throughout this paper.
FIG. 1.
Phylogenetic tree showing distances based on the 16S rRNA sequences obtained from isolates within insect-pathogenic nematodes that infected G. melonella. The analysis was carried out using the programs Dnadist (Jukes-Cantor maximum likelihood) and Neighbor (unweighted-pair group method using average linkages). The scale bar indicates 0.1% estimated sequence divergence. Sequences homologous to that of P. stuartii are indicated by asterisks. The strain shown is one representative of the group whose sequences differ by less than 2 bp.
Diversity of alleles and sequence types.
The percentage of polymorphic sites in the X. bovienii loci examined ranged from 7.5 to 10.7% of the total sequence obtained (Table 2). This high diversity, particularly at the asd and ompR loci, was largely due to divergent alleles from strains X. bovienii GS and WSX97a. When alleles from these two strains were removed from the analysis, the percentage of polymorphic sites fell to around 5% for all loci, which is comparable to that for loci used in other MLST analyses. The variable sites were used to calculate the dN/dS (15) ratio for each locus (Table 2), which gives an indication of the selection pressure on this locus. A low dN/dS ratio (substantially lower than 1) indicates that most changes are silent and therefore there is no selective pressure for mutation of the protein. This is expected for housekeeping genes and is ideal for MLST analysis, which requires variation that accumulates slowly and is expected to be selectively neutral. All four loci fulfilled these criteria, with dN/dS ratios substantially less than 1 (Table 2).
TABLE 2.
Genetic diversity at the four housekeeping loci in X. bovienii strains
| Gene | Fragment size (bp) | No. of allelesa | No. of polymorphic sitesa | % Polymorphic sitesa | dN/dSa |
|---|---|---|---|---|---|
| asd | 323 | 22 (20) | 34 (16) | 10.5 (5.0) | 0.048 (0.032) |
| ompR | 468 | 26 (24) | 48 (21) | 10.3 (4.5) | 0.031 (0.025) |
| recA | 424 | 16 (14) | 32 (25) | 7.5 (5.9) | 0.042 (0.048) |
| serC | 391 | 16 (14) | 42 (33) | 10.7 (8.4) | 0.121 (0.126) |
Numbers in parentheses show the values obtained when alleles from strain WSX97a and the fully sequenced strain, X. bovienii GS, were excluded.
Strains with identical alleles at all four loci were putatively assigned to a unique sequence type. Out of 193 strains, there were 94 STs; ST 22 contained the most strains (12), including the type strain X. bovienii (Table 3). The majority of STs (66%) were comprised of only one strain. For the seven X. nematophila strains, there were three STs. The two strains WSX97e and X. bovienii GS each contained unique alleles at four loci (Table 3), yet their 16S rRNA sequences (Fig. 1) placed them in groups with other X. bovienii strains. Conversely, the strains NRF5e, BRF26b, and MER25b possessed common housekeeping alleles, yet their 16S rRNA sequences were more similar to those of P. stuartii than to those of X. bovienii.
TABLE 3.
Allele type and insecticidal activity for Xenorhabdus strains
| ST | No.a | Reference strain | Allele |
No. active/no. testedb for: |
ST | No. | Reference strain | Allele |
No. active/no. tested for: |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| asd | ompR | recA | serC | P. brassicae | A. aegyptae | P. cochleriae | asd | ompR | recA | serC | P. brassicae | A. aegyptae | P. cochleriae | |||||||
| X. bovienii | 50 | 1 | POA14a | 2 | 1 | 11 | 2 | 0/1 | 0/1 | 0/0 | ||||||||||
| 1 | 1 | 5MER | 1 | 1 | 1 | 1 | 0/1 | 0/0 | 0/1 | 51 | 1 | POA15 | 1 | 1 | 7 | 10 | 0/1 | 0/1 | 0/1 | |
| 2 | 4 | WSX89c | 2 | 2 | 2 | 2 | 0/2 | 0/2 | 1/2 | 52 | 1 | POA2 | 1 | 1 | 11 | 2 | 0/1 | 0/1 | 0/1 | |
| 3 | 1 | A17(97) | 3 | 3 | 3 | 1 | 0/1 | 0/1 | 1/1 | 53 | 1 | POA23 | 14 | 2 | 2 | 2 | 0/1 | 0/1 | 0/1 | |
| 4 | 1 | A2 | 4 | 3 | 4 | 3 | 0/1 | 1/1 | 0/0 | 54 | 2 | POA59c | 3 | 3 | 3 | 11 | 0/2 | 0/2 | 0/0 | |
| 5 | 7 | SB121G | 5 | 2 | 1 | 2 | 0/5 | 2/5 | 5/5 | 55 | 1 | POA73 | 1 | 6 | 3 | 3 | 0/1 | 0/1 | 0/0 | |
| 6 | 2 | A53(97) | 1 | 2 | 5 | 2 | 0/2 | 0/2 | 1/1 | 56 | 1 | POA78b | 11 | 19 | 2 | 5 | 0/1 | 0/1 | 0/0 | |
| 7 | 6 | SY27c | 3 | 3 | 3 | 3 | 2/4 | 1/4 | 1/2 | 57 | 1 | POA84a | 10 | 10 | 1 | 5 | 0/1 | 0/1 | 0/1 | |
| 8 | 3 | SY28 | 3 | 3 | 4 | 3 | 1/2 | 1/2 | 0/0 | 58 | 1 | Q19(97) | 5 | 16 | 2 | 5 | 0/1 | 0/1 | 0/0 | |
| 9 | 2 | C19(97) | 4 | 3 | 3 | 3 | 0/1 | 1/1 | 0/0 | 59 | 1 | Q24(97) | 1 | 20 | 2 | 5 | 0/0 | 0/0 | 0/0 | |
| 10 | 2 | SB250aG | 3 | 3 | 4 | 1 | 0/1 | 0/1 | 1/1 | 60 | 1 | Q25(97) | 1 | 2 | 1 | 3 | 0/1 | 0/1 | 0/0 | |
| 11 | 2 | POA26 | 1 | 2 | 2 | 4 | 1/2 | 0/2 | 1/2 | 61 | 1 | S12(97) | 3 | 6 | 4 | 1 | 0/1 | 0/1 | 1/1 | |
| 12 | 1 | B39(97) | 1 | 4 | 2 | 3 | 0/1 | 0/0 | 1/1 | 62 | 3 | X1(97) | 5 | 2 | 5 | 5 | 0/3 | 0/3 | 1/2 | |
| 13 | 4 | D2e | 6 | 3 | 4 | 1 | 1/2 | 1/2 | 2/2 | 63 | 1 | SB102bG | 8 | 21 | 12 | 5 | 0/1 | 1/1 | 0/1 | |
| 14 | 10 | BRF26b | 1 | 2 | 2 | 2 | 0/5 | 0/6 | 2/3 | 64 | 4 | SY27d | 3 | 6 | 3 | 11 | 2/3 | 0/3 | 0/0 | |
| 15 | 8 | S46 | 1 | 5 | 1 | 5 | 1/7 | 0/7 | 2/3 | 65 | 3 | SB167fG | 8 | 5 | 1 | 5 | 0/3 | 0/3 | 1/3 | |
| 16 | 1 | CDN41 | 3 | 6 | 3 | 6 | 0/1 | 0/1 | 1/1 | 66 | 1 | SB205bG | 15 | 7 | 3 | 3 | 0/1 | 0/1 | 0/1 | |
| 17 | 9 | WT37b | 1 | 2 | 1 | 5 | 1/7 | 0/7 | 3/3 | 67 | 1 | SB79dG | 8 | 5 | 2 | 5 | 0/1 | 0/1 | 0/1 | |
| 18 | 1 | H31(97) | 3 | 7 | 4 | 3 | 1/1 | 0/1 | 1/1 | 68 | 1 | SB9aMW | 1 | 5 | 1 | 3 | 1/1 | 1/1 | 0/1 | |
| 19 | 1 | H51(97) | 4 | 7 | 3 | 3 | 0/1 | 0/1 | 1/1 | 69 | 2 | SY1b | 1 | 3 | 3 | 3 | 0/2 | 0/2 | 1/1 | |
| 20 | 2 | W6(97) | 3 | 8 | 3 | 3 | 0/1 | 0/1 | 1/1 | 70 | 1 | SY23f | 2 | 2 | 2 | 5 | 0/1 | 0/1 | 0/0 | |
| 21 | 1 | I73 | 3 | 9 | 6 | 7 | 1/1 | 1/1 | 1/1 | 71 | 1 | SY26a | 1 | 3 | 3 | 12 | 0/1 | 1/1 | 0/0 | |
| 22 | 12 | X. bovienii TS | 1 | 2 | 1 | 2 | 0/9 | 0/9 | 4/5 | 72 | 1 | SY26b | 16 | 3 | 3 | 12 | 0/1 | 0/1 | 0/0 | |
| 23 | 10 | SY23c | 1 | 2 | 2 | 5 | 0/8 | 0/8 | 1/2 | 73 | 1 | SY26d | 3 | 22 | 3 | 12 | 0/2 | 0/2 | 0/0 | |
| 24 | 1 | LYA10b | 1 | 5 | 2 | 5 | 0/0 | 0/0 | 0/0 | 74 | 1 | SY29a | 1 | 5 | 13 | 2 | 0/3 | 0/3 | 0/0 | |
| 25 | 2 | P39(97) | 2 | 2 | 1 | 2 | 0/0 | 0/0 | 0/0 | 75 | 1 | SY29b | 3 | 5 | 13 | 2 | 0/4 | 0/4 | 0/0 | |
| 26 | 3 | POA84c | 5 | 10 | 1 | 5 | 0/2 | 0/2 | 0/1 | 76 | 1 | SY6c | 1 | 23 | 2 | 3 | 0/5 | 0/5 | 1/1 | |
| 27 | 1 | MER109c | 4 | 11 | 3 | 3 | 0/1 | 0/0 | 0/1 | 77 | 1 | W4(97) | 3 | 3 | 4 | 13 | 0/6 | 0/6 | 1/1 | |
| 28 | 1 | MER109d | 4 | 12 | 3 | 3 | 0/1 | 0/0 | 0/1 | 78 | 1 | WB1c | 11 | 2 | 2 | 2 | 0/7 | 0/7 | 1/1 | |
| 29 | 3 | MER99a | 1 | 1 | 7 | 1 | 0/3 | 0/0 | 1/3 | 79 | 1 | WIG15b | 17 | 16 | 2 | 3 | 0/1 | 0/0 | 1/1 | |
| 30 | 1 | MER11b | 7 | 13 | 8 | 8 | 0/1 | 0/0 | 0/1 | 80 | 1 | WIG32b | 1 | 2 | 2 | 3 | 0/1 | 0/0 | 0/1 | |
| 31 | 1 | MER122c | 7 | 13 | 9 | 8 | 0/1 | 0/0 | 1/1 | 81 | 1 | WSX1d | 2 | 2 | 2 | 3 | 0/0 | 0/0 | 0/0 | |
| 32 | 1 | MER16b | 4 | 8 | 3 | 3 | 0/1 | 0/0 | 1/1 | 82 | 1 | WSX1e | 11 | 2 | 2 | 3 | 0/0 | 0/0 | 0/0 | |
| 33 | 3 | S40b(97) | 8 | 2 | 2 | 5 | 0/3 | 0/2 | 0/1 | 83 | 1 | WSX97a | 18 | 24 | 14 | 14 | 0/0 | 0/0 | 0/0 | |
| 34 | 1 | MER25a | 9 | 14 | 9 | 8 | 1/1 | 0/0 | 0/1 | 84 | 3 | WT32c | 1 | 16 | 2 | 3 | 0/1 | 0/1 | 0/0 | |
| 35 | 1 | MER25b | 10 | 14 | 9 | 8 | 0/1 | 0/0 | 1/1 | 85 | 1 | WT114c | 19 | 13 | 9 | 8 | 0/0 | 0/0 | 0/0 | |
| 36 | 1 | MER35c | 11 | 1 | 7 | 1 | 1/1 | 0/0 | 1/1 | 86 | 1 | WT20b | 10 | 2 | 15 | 3 | 0/1 | 0/1 | 0/0 | |
| 37 | 1 | MER36a | 12 | 15 | 6 | 9 | 0/0 | 0/0 | 0/0 | 87 | 1 | WT20c | 20 | 2 | 15 | 3 | 0/1 | 0/1 | 0/0 | |
| 38 | 1 | MER38b | 13 | 2 | 1 | 2 | 0/1 | 0/0 | 0/1 | 88 | 1 | WT21b | 1 | 16 | 1 | 15 | 0/0 | 0/0 | 0/0 | |
| 39 | 1 | MER3a | 11 | 5 | 1 | 2 | 1/1 | 0/0 | 0/1 | 89 | 1 | WT23 | 2 | 16 | 1 | 15 | 0/1 | 0/1 | 0/0 | |
| 40 | 3 | NRF5e | 1 | 16 | 2 | 2 | 1/3 | 0/2 | 0/3 | 90 | 1 | WT36 | 21 | 13 | 9 | 8 | 0/1 | 0/1 | 0/0 | |
| 41 | 1 | MER66a | 1 | 17 | 2 | 5 | 0/1 | 0/0 | 0/1 | 91 | 2 | WT43c | 1 | 25 | 1 | 2 | 0/2 | 0/2 | 0/0 | |
| 42 | 1 | MER78a | 1 | 18 | 1 | 2 | 0/1 | 0/0 | 0/1 | 92 | 1 | X. bov GS | 22 | 26 | 16 | 16 | 0/0 | 0/0 | 0/0 | |
| 43 | 1 | NRF40b | 11 | 2 | 2 | 5 | 0/1 | 0/1 | 1/1 | 93 | 1 | X11(97) | 1 | 7 | 3 | 3 | 0/1 | 0/1 | 1/1 | |
| 44 | 2 | SY6b | 1 | 5 | 5 | 2 | 0/2 | 0/2 | 1/1 | 94 | 1 | X2(97) | 2 | 5 | 1 | 3 | 0/1 | 0/1 | 1/1 | |
| 45 | 1 | PJ23 | 5 | 16 | 1 | 5 | 0/1 | 0/1 | 0/0 | |||||||||||
| 46 | 2 | PJ31b | 5 | 2 | 1 | 5 | 0/2 | 0/1 | 2/2 | X. nematophila | ||||||||||
| 47 | 6 | SB225aG | 1 | 15 | 10 | 2 | 0/4 | 0/4 | 2/4 | 1 | 2 | MFI | 1 | 1 | 1 | 1 | 2/2 | 2/2 | 2/2 | |
| 48 | 4 | POA78a | 1 | 19 | 2 | 5 | 0/4 | 0/4 | 1/2 | 2 | 4 | 0015 | 2 | 1 | 2 | 1 | 4/4 | 4/4 | 4/4 | |
| 49 | 1 | POA100c | 2 | 19 | 2 | 5 | 0/1 | 0/1 | 0/0 | 3 | 1 | 9965 | 3 | 1 | 2 | 2 | 1/1 | 1/1 | 1/1 | |
Number of strains within a sequence type.
Positive insecticidal activity was determined as greater than 30% insect mortality within 3 days. Negative control samples had less than 5% mortality in repeat assays.
Evidence for recombination.
Evidence of intralocus recombination of the four housekeeping genes was examined by performing two statistical tests. The first was the homoplasy test (27), which was shown to perform well with low levels of sequence divergence (18); the results are shown in Table 4. The test requires a minimum of six sequences containing at least 10 homoplasies at which the rarer of two alternative bases is present at least twice. This was not the case with the asd locus, and therefore no ratio could be calculated. The ompR and recA loci gave nonsignificant values (P > 0.2), indicating many fewer homoplasies than would be expected under free recombination. However, serC gave a significant value (P = 0.001) of 0.342, indicating frequent recombination at this locus. This finding was supported by Sawyer's test (23), which is based on pairwise examination of silent polymorphic sites between all alleles. Again this test showed significant recombination at the serC locus (Table 4) but not at any of the other loci.
TABLE 4.
Measure of recombination with homoplasy and Sawyer's test
| Gene | Homoplasy test |
Sawyer's test |
||
|---|---|---|---|---|
| H ratio | P value | Maximum score | P value | |
| asd | NDa | ND | 2.336 | 0.223 |
| ompR | 0.091 | 0.354 | 2.372 | 0.193 |
| recA | 0.078 | 0.371 | 2.134 | 0.214 |
| serC | 0.345 | 0.006 | 4.176 | 0.018 |
ND, not determined. The homoplasy ratio could not be determined because of too few informative sites in the alleles.
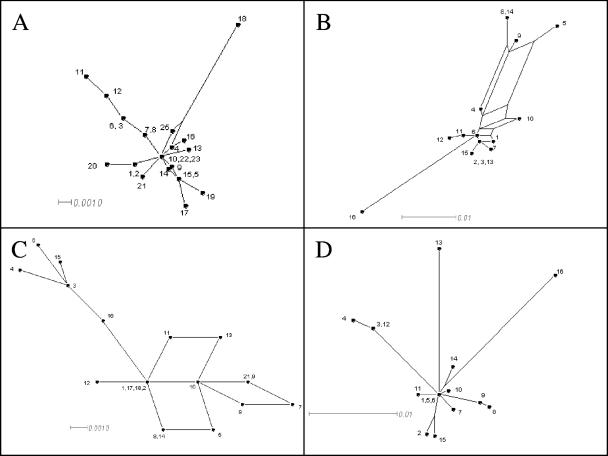
Split decomposition analysis (9) was used on the alleles at each locus to construct phylogenetic trees, whose patterns give some indication of recombination at that locus. Split decomposition analysis does not make the assumption of a branching or tree-like process and is therefore able to display conflicting results in the phylogenetic descent of sequences. The SplitsTree graphs of recA (Fig. 2D) and ompR (Fig. 2A) show essentially a star-like structure, consisting of a single origin in the center from which single branches radiate. This is typical of genes that have evolved from a single ancestor by mutation, with no or little recombination, and supports the results obtained with the two recombination tests. The graph for serC (Fig. 2B), on the other hand, shows a complex web pattern, implying extensive intraspecific recombination leading to the assembly of genes from different ancestral alleles. The graph for asd also shows a web-like pattern, albeit less complex than that for serC, suggesting a moderate amount of recombination. This is in contrast to the results obtained from the homoplasy and Sawyer tests.
FIG. 2.
Split decomposition analysis of housekeeping genes with SplitsTree. By this method, evolutionary data are canonically decomposed into a sum of weakly compatible splits and are represented by a splits graph. A, ompR; B, serC; C, asd; D, recA. Each scale bar represents the percent difference among the sequence types. The divergent asd and ompR alleles from strains X. bovienii GS and WSX97a were omitted from the analysis.
To measure the amount of inter allele recombination, the level of linkage disequilibrium was examined by analyzing the allele combinations of all the strains. The standardized index of association, ISA, is a measure of the level of linkage disequilibrium and is zero for a population in linkage equilibrium (free recombination). The value of ISA for all isolates using the four loci was 0.194, which suggests a moderate amount of recombination on a par with that for Campylobacter jejeni (ISA = 0.256) (30). In comparison, with Neisseria meningitidis, in which recombination is frequent, an ISA of 0.059 was observed (7), whereas the highly clonal plant pathogen Pseudomonas syringae showed an ISA of 0.61, based on an IA of 3.69 (22). In this study the difference from linkage equilibrium (P = 0.01 by Monte Carlo simulation with 1,000 repetitions) was significant, implying that the linkage between alleles is still reasonably strong.
fliC analysis.
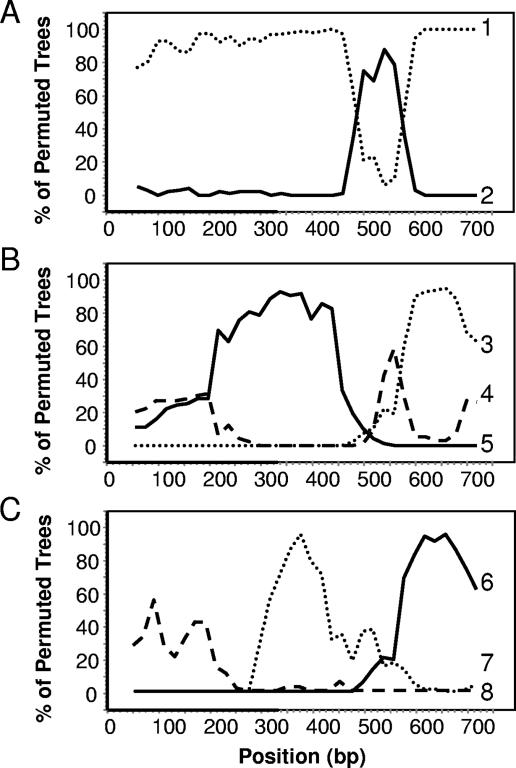
From the 193 strains sequenced, there were 41 unique fliC sequences (alleles), which were aligned using the ClustalW algorithm. The fliC gene showed a typical predicted flagellin protein structure, with fairly conserved NH2- and COOH-terminal sections and a highly variable internal section. The aligned sequences were analyzed by Sawyer's test and gave a maximum score of 75.8 (P = 0.0001), implying extensive recombination. Indeed, a total of 216 inner fragments (recombination events) were identified as being significant, 93 of which were still significant when Bonferroni corrected. The recombination events that were detected were throughout the fragment and were not localized to the internal highly variable region. Although it is unlikely that all putative recombination events are real, the actual number is still likely to be large, leading to a highly mosaic structure for this gene. This mosaic structure was further supported by analyzing the fliC alleles by using the bootscanning method (21) that is employed by the program Simplot. For such analysis, the 42 aligned fliC alleles were split into 12 groups, with alleles in each group showing no greater than 5% divergence. Using the bootscanning procedure, the nucleotide similarity of the consensus for each group was compared against the consensus of all the other groups (reference sequences) across the entire length of the gene. The resulting graph shows similarity between the sequence being studied and the reference sequences. Figure 3 shows examples of alleles that are composed of fragments from two (Fig. 3A) or three (Fig. 3B and C) other alleles. Despite this frequent recombination leading to complex mosaic genes, four alleles dominated, contributing to over 55% of the 193 strains. Presumably, despite frequent recombination, only the successful combinations survive and predominate. In order to study the origin of the highly variable central regions, the internal regions from position 377 to 722 of the aligned sequences were analyzed separately. There were 26 unique sequences, which could be split into 11 groups consisting of sequences showing less than 5% divergence. Only two groups showed any significant identity (e < 0.01) to any protein sequence in the database by BLASTX searching. Group 5 showed 32% identity to phase 1 flagellin from Salmonella, and group 9 showed 39% identity to a flagellin from Photorhabdus.
FIG. 3.
Bootscanning of nucleotide similarity of different fliC alleles, highlighting their mosaic structure. The graphs show examples of the similarity between the consensus sequence being studied (query sequence) and all other consensus sequences (reference sequences) across the entire length of the fliC gene PCR product. The query consensus sequences for each graph were composed of the following alleles: A, fliC27 and -40; (B) fliC4,-15, -16, and -29; and (C), fliC10 and -39. The reference sequences shown were composed of the following alleles: 1, fliC20, -21, -31, -33, -34, and -41; 2, fliC22 and -30; 3, fliC10 and -39; 4, fliC3, -8, -11, -12, -32, -35, and -36; 5, fliC1, -7, -13, -18, -19, and -23; 6, fliC4, -15, -16, and -29; 7, fliC3, -8, -11, -12, -32, -35, and -36; and 8, fliC5.
Insecticidal activity.
The oral insecticidal activity of most strains was tested by performing bioassays against members of three orders of insect pests: the cabbage white caterpillar Pieris brassicae, the mosquito larva Aedes aegypti, and the mustard beetle Phaedon cochleariae. The insecticidal activity for the X. bovienii strains was variable (Table 3), with 46 out of the 94 STs exhibiting no activity where tested. Only one ST (ST 21), containing the single strain I73, was active against all three insects. Even strains within a single ST exhibited different patterns of insecticidal activity (e.g., STs 7, 12, and 40) (Table 3). This was in contrast to the case for the X. nematophila strains, which all showed insecticidal activity towards all three test insects.
Characterization of the xpt PAI.
The xpt genes present in cHRIM1 (13) were detected by PCR in all seven X. nematophila strains. The area surrounding these genes was mapped in all seven strains by carrying out a combination of PCR and sequencing. The region to the right of the xpt loci in all strains shows the classical features of a pathogenicity island (PAI), with an integrase gene adjacent to the leucine (leu) tRNA gene and the core chromosomal genes uvrC and pgsA. This region is a common site for the insertion of PAIs and bacteriophages in many bacteria.
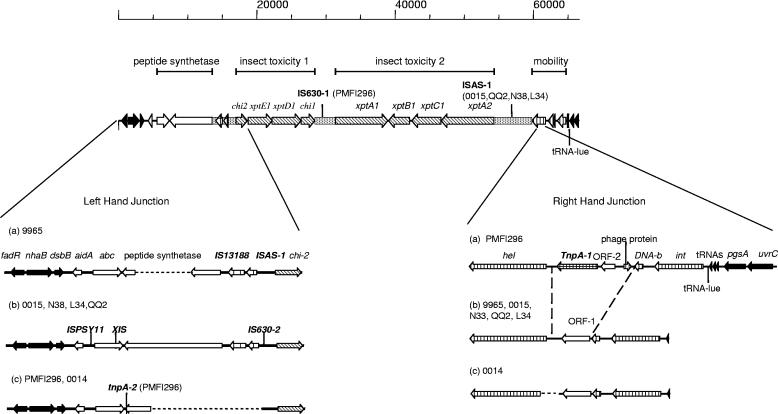
Upstream of the xptA to -C genes were the potential insect toxin genes xptD1 and xptE1 and a second exochitinase gene, chi2 (Fig. 4). Further upstream of this was a region of DNA that when translated showed homology to a peptide synthetase fragment and associated ATP binding cassette (ABC transporter) gene, homologues of which have been postulated to pump out peptide antibiotics from the cell (16). Beyond this were the core chromosomal genes fadR, nhaB, and dsbB, suggesting that this marked the right-hand junction (RHJ) of the PAI. The region containing the xpt genes and chitinase genes appeared, by PCR analysis, to be conserved in all seven X. nematophila strains, but small rearrangements or point mutations could not be discounted. Other regions, however, showed differences; at the RHJ, strain 0014 contained an approximately 500-bp deletion around the start of the helicase gene, and PMFI296 contained a region not present in the other strains, which was associated with a tnpA-like transposon. Deletions of parts of the peptide synthetase gene had occurred in both the 9965 and PMFI296/0014 strains (Fig. 4).
FIG. 4.
Structures of the xpt presumptive pathogenicity island in X. nematophila isolates, highlighting differences between the strains. Variations (a to c) in the left- and right-hand junctions of strains 9965, 0014, 0015, N38, L34, QQ2, and PMFI296 are highlighted. The positions of IS630-1 in strain PMFI296 and ISAS-1 in strains 0015, QQ2, N38, and L34 are indicated on the map line. The scale is in bp. ░⃞, AT-rich regions; ▪, core chromosomal genes; ▧, insecticidal toxin genes; ▥, phage-like genes; ⊔, transposon-like genes; □, other genes. Dotted lines show areas of deletion.
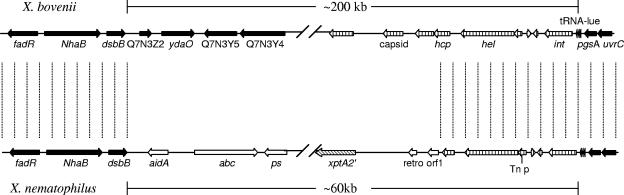
Comparison of this region with that of X. bovienii showed that the homology with X. nematophila extended beyond that of the core chromosomal and tRNA genes and extended into the PAI integrase and helicase genes (Fig. 5). Downstream of this in X. bovienii were phage-like genes, in contrast to the xpt-like genes in X. nematophila. Thus, a possible explanation may be that a phage-like structure was first inserted into this region in a common ancestor and that in X. nematophila, the phage genes were replaced by xpt-like genes.
FIG. 5.
Comparison of the xpt loci of X. nematophila PMFI296 to those of X. bovienii GS. Areas of colinearity are marked with a dotted line. ▪, core chromosomal genes; ▧, insecticidal toxin genes; ▥, phage-like genes; □, other genes. Note that the homology between the strains extends beyond the core chromosomal genes into the phage-like genes at the RHJ of the putative xpt PAI.
Insertion sequences and copy number.
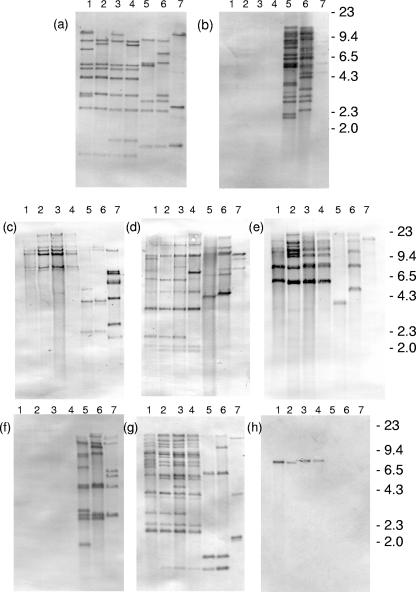
The main difference between the xpt PAIs of the seven X. nematophila strains was the presence or absence of IS elements (Fig. 6; Table 5). IS630-1, IS630-2, ISPSY-11, and IS1328 show homology to IS elements from closely related Enterobacteriaceae such as E. coli, Salmonella, and Shigella. IS630-1 was present only in strains 0014 and PMFI296 but was present in about 15 copies (Fig. 6). Its distribution in the two strains was substantially different, even though the 16S rRNA and housekeeping gene sequences of the two strains gave identical results. This indicates that transposition of this IS occurs at a high frequency. In contrast IS1328 and IS630-2 occurred in all seven strains (Fig. 6c and d), and their distribution pattern confirmed the relatedness of the seven strains as observed by 16S rRNA and housekeeping gene sequences. tnpA-1 and tnpA-2, present in the PAI of PMFI296, showed limited identity to tnpA from Alcaligenes eutrophus (54% and 58%, respectively) and about 70% identity to each other. Although tnpA-1 was present in all seven strains, tnpA-2 was present only in PMFI296, 0014, and 9965 (Fig. 6e and f). There was one complete copy and one partial copy of an ISAS1-like IS from Aeromonas salmonicida, a fish pathogen and a member of the Vibrionaceae, present in the PAI. Probing against the genome revealed from 3 copies in 9965 to 10 copies in 0015 (Fig. 6a). Finally, two open reading frames (ORFs) similar to two contiguous ORFs in P. luminescens disrupted the ABC transporter gene in L34-type strains. Clearly, by comparison with other strains, this element is capable of insertion, but its single copy number in all four L34-like strains suggests that it may not be a promiscuous IS or that it is a recent acquisition.
FIG. 6.
Southern hybridization of restriction-digested bacterial chromosomal DNA with digoxigenin-labeled IS probes. (a) HindIII-digested DNA probed with ISAS-1; (b) EcoRI-digested DNA probed with IS630-1; (c) HindIII-digested DNA probed with IS1388; (d) HindIII-digested DNA probed with IS630-2; (e) HindIII-digested DNA probed with TnpA-1; (f) HindIII-digested DNA probed with TnpA-2; (g) HindIII-digested DNA probed with ISPSY-11; (h) HindIII-digested DNA probed with Xis. X. nematophila isolates: 1, L34; 2, N38; 3, QQ2; 4, 0015; 5, 0014; 6, PMFI296; 7, 9965. Markers are λ DNA cut with HindIII.
TABLE 5.
Insertion sequences present in the xpt PAI of X. nematophilus strains
| Insertion sequence (ORF length, bp) | Homologous insertion sequence, organism (accession no.) | % Identity | Inverted repeata | Direct repeat | Strain(s) in which insertion sequence is present |
|---|---|---|---|---|---|
| IS630-1 (343) | IS630, Shigella flexneri (X05955) | 58 | AAATAAGATTTCAAAATAGT | None | PMFI296 |
| ACAATTATGAAAACTTATTT | |||||
| ISAS-1 (365) | ISAS1, Aeromonas salmonicida (Q44285) | 73 | CAGGGCGCGAGCATACTTT | ACCCTCACCC | L34, QQ2, N38, 0015 |
| ISAS-1p, partial (213) | ISAS1, Aeromonas salmonicida (Q44285) | 70 | CAGGGCGCGAGCATACTTT | NAb | L34, QQ2, N38, 0015, 9965 |
| IS630-2 (343) | IS630, Shigella sonnei (Q8VSK9) | 75 | AATA | TA | 0015, L34, QQ2, N38 |
| IS1328 (327) | IS1328, Shigella flexneri 2a (Q93EZ2) | 79 | None | None | L34, QQ2, N38, 0015, 9965 |
| TnpA-1 (351) | TnpA, Alcaligenes eutrophus (Q7WWT0) | 54 | ATGAACGCGTCCC | None | PMFI296 |
| GGGGCGCGTTCAT | |||||
| TnpA-2 (339) | TnpA, Alcaligenes eutrophus (Q7WWT0) | 58 | ATGAACGCATCCC | None | PMFI296 |
| ISPSY-11 | ISPSY-11, Salmonella enterica serovar Typhimurium | TGTCCCGTCCTAA | ATT | L34, QQ2, N38, 0015 | |
| TCAGGACTAGACA | |||||
| orfA (129) | OrfA (Q8ZQE1) | 66 | |||
| orfB (282) | OrfB (Q8ZQE2) | 64 | |||
| Xis | Photorhabdus luminescens | None | None | L34, QQ2, N38, 0015 | |
| orfA (113) | OrfA (Q7N0B2) | 56 | |||
| orfB (98) | OrfB (Q7NOB1) | 50 |
Perfect repeats are only shown once, or both imperfect repeats are shown with differences underlined.
NA, not applicable.
xpt genes in X. bovienii.
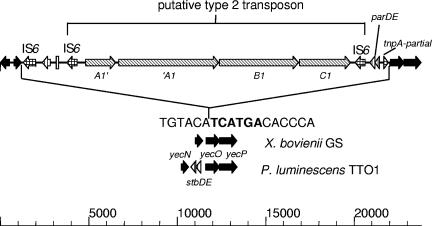
While sequencing a nematode-active cosmid from a genomic library of the X. bovienii strain I73, an xptC1-like gene was found on one extremity of the insert. The entire locus was sequenced (Fig. 7) and shown to contain three xpt-like genes in the typical arrangement xptA1, -B1, -C1 (in the current nomenclature; these genes were formerly called xptA, -C1, and -B1, respectively). The xptA1-like gene (xptA3) contained a 7-bp duplication leading to a frameshift mutation, which resulted in two potential open reading frames (xpt′A3 and xptA3′). Three IS elements of the IS6 family, showing 70% identity to IS327 from the pathogenicity-associated plasmid of the plant pathogen Erwinia herbicola (10), were present at this locus, as well as a partial tnpA-like IS. Two degenerate copies of the IS6 element flanked the xpt genes, in a manner analogous to that of a type 2 transposon. When this region was compared with that of the sequenced X. bovienii GS, the exact insertion point of the xpt element could be ascertained. The element had inserted in the yec operon, between the yecN and yecO genes, leading to duplication of a 6-bp palindromic region (TCATGA) of the target sequence. The left-hand junction of the element corresponded exactly to the indirect repeat of the intact IS6 element and the RHJ to the truncated tnpA-like IS.
FIG. 7.
Structure of the xpt genes in X. bovienii isolate I73, highlighting classical features of a type 2 transposon. The same loci on the X. bovienii and P. luminescens chromosomes are also shown for comparison. The sequence of the predicted point of insertion (based on the X. bovienii GS sequence) is shown, with the duplicated target sequence highlighted in boldface. The scale is in bp. ▪, core chromosomal genes; ▧, insecticidal toxin genes; ▥, plasmid-like genes; ⊔, transposon-like genes.
The presence of the xptA3 gene in the other strains in the culture collection was determined by PCR of the 500-bp region containing the 7-bp duplication. A PCR product was obtained only with the X. bovienii strains WSX1d and wig32b, which are closely related to each other but not to I73. For WSX1d, a larger PCR fragment corresponded to the presence of an IS1-like IS. Therefore, the only potentially active xptA3 gene in all 193 strains examined was in strain wig32b.
DISCUSSION
The first major finding of this study was that the vast majority of strains obtained from nematodes in United Kingdom soils by using the insect bait entrapment method were X. bovienii strains. No such study has been carried out previously, and therefore it is uncertain whether this predominance is common in other countries. It is possible that X. bovienii/nematode complexes exhibit better fitness traits in all the United Kingdom soils tested. However, the possibility that this finding is an artifact due to the bait entrapment method employed in these studies that favors isolation of X. bovienii above other Xenorhabdus strains cannot be discounted. The second major finding was the poor oral insecticidal activity of the X. bovienii isolates, with only strain I73 being active against all three insects tested. Although toxins that are active by injection are presumably essential for bacteria to rapidly kill infected insects, the purpose of orally active toxins remains obscure. One theory is that these toxins are produced to deter scavenging of the insect cadaver while the nematodes are reproducing. The fact that the vast majority of X. bovienii strains did not exhibit oral toxicity suggests that it is not essential for the bacterium/nematode complex. In contrast, all the X. nematophila strains exhibited oral insecticidal activity against all three test insects, and all xpt genes appeared to be intact. This is despite the presence of large numbers of IS elements in their genomes, which had inserted elsewhere on the xpt PAI. Although only seven strains were tested, the conserved oral insecticidal activity of X. nematophila may reflect its importance in the life cycle of the bacteria, perhaps infecting insects that are more likely to be scavenged while the nematodes are reproducing.
The four housekeeping loci examined in this study proved to be ideal candidates for use in MLST analysis, having low dN/dS ratios and only moderate variation (around 5% polymorphic sites). Even with analysis of only four loci, STs of the strains appeared to be more informative than analysis of 16S rRNA sequences alone. For example, the ST of X. bovienii strain GS, obtained from a novel nematode species in the American midwest (29), showed, as expected, that it was distinct from the strains isolated in the United Kingdom. However, analysis of the 16S rRNA of this strain could not differentiate it from the United Kingdom isolates. The United Kingdom isolate strain WSX97a also had a common 16S rRNA sequence but unique alleles at all four loci and therefore probably represents a rare United Kingdom strain, which would be interesting for further study. Surprisingly, the 16S rRNA sequences of strains NRF5e, BRF26b, and MER25b were distinct from those of all the other X. bovienii strains and showed closest homology to the 16S rRNA gene of P. stuartii. In contrast to this, housekeeping alleles of these three strains were common among the X. bovienii strains examined. Indeed, strain BRF26b shared an ST with nine other strains, and even the highly variable fliC gene in the three strains was identical to those in other X. bovienii strains. Therefore, it seems likely that the 16S rRNA gene had been subject to recombination with an external source, and this highlights the problems of typing bacteria based on a single gene.
Statistical evidence indicated that recombination had occurred at one of the four loci (serC), and the ISA of 0.256 suggested that only moderate interallele recombination had taken place. This is consistent with the fact that opportunity for genetic exchange would be limited due to the life cycle of the bacterium, which resides in monoculture in its host nematode and produces potent antimicrobial substances during insect infection, again limiting contact with other bacteria. Despite this, however, extensive recombination had taken place at the fliC locus, resulting in a highly variable central region, which corresponds to the exposed region of the flagella. This is similar to the situation with the fliC genes of bacterial pathogens which infect higher organisms and is thought to be a result of an “arms race” between the bacteria and the adaptive immune response of the host. However, for an insect pathogen such as X. bovienii, this is surprising, since insects possess only an innate immune system similar to the innate systems of higher organisms. The innate immune response is usually triggered by specific conserved patterns in bacteria. Indeed, Toll-like receptor 5, a protein responsible for triggering the innate response to bacterial infection, interacts with a conserved 13-amino-acid motif in the bacterial flagella (28). Therefore, the recognition of conserved domains in proteins is unlikely to lead to the diversity exhibited by the fliC genes of the X. bovienii isolates. Hence, other pressures must be at work in order to create this diversity. Similar diversity is seen in the fliC genes of Pseudomonas spp. (2). Like Xenorhabdus, most Pseudomonas species are not pathogens of higher organisms, and therefore it was hypothesized that the variation was a result of evading infection by bacteriophages. This could also be a possible explanation for the diversity seen in Xenorhabdus. All of the highly variable regions of the fliC genes examined showed little homology to any sequence in the database, implying that they were acquired from as-yet-undiscovered sequences or that frequent recombination had led to sequences that were no longer discernible. Acquisition of DNA from diverse sources in Xenorhabdus was also implied by the presence of IS elements that were similar to those from A. eutrophus and A. salmonicida in X. nematophila strains and a 16S rRNA gene similar to that of P. stuartii in three X. bovienii strains.
This work has also shown that the xpt genes are present on a PAI associated with a leu tRNA. The homologous tca genes in Photorhabdus have also recently been assigned to a putative PAI by virtue of the facts that it is unstable, contains phage-like genes, and is associated with aspV tRNA (25). However, the xpt PAI represents a more classical PAI with similarities to the high PAI of Yersinia spp. (20). Both the xpt PAI and the high PAI share a P4-like integrase at the right-hand junction, a core region containing pathogenicity genes, and an AT-rich left-hand junction containing many IS elements that has a tendency to delete (20). The left-hand junction of the PAI is likely to be between the ABC transporter gene and the dsbB gene. However, there is no direct repeat corresponding to the 3′ end of the leu tRNA present in this region, which usually marks the end of a PAI. It is most likely that deletions have occurred in this area, removing the repeat, as seen with the Yersinia high PAI in E. coli (20). It has been postulated that PAIs are evolutionary relics, where, after acquisition and over time, functions such as mobility are gradually lost as the island becomes incorporated into the chromosome (6). Comparing the PAI locus with that of X. bovienii GS appears to support this, as phage-like genes in X. bovienii appear to have been replaced by xpt genes in X. nematophila as the locus is converted from a phage to a PAI. The presence of the peptide synthetase and associated ABC transporter in the PAI would give the bacterium and presumably its nematode host a competitive advantage by killing competing bacteria in the insect cadaver. It is interesting that novel colicin and immunity proteins are also found associated with insecticidal tc genes in Photorhabdus spp. (25).
In the PAI all genes are tightly grouped into two insect-pathogenic regions, a mobility region and a region putatively involved in peptide antibiotic synthesis and resistance, which are all separated by AT-rich junctions (Fig. 4). In Photorhabdus, the tca genes are associated with lysogenic phage genes, and therefore it is conceivable that these genes originated from an integrated phage. This work has shown that the xpt genes in X. nematophila are not associated with phage genes but show close linkage to IS elements and therefore may have originated on transposons. Thus, unlike the tc genes of Photorhabdus, the xpt genes are unlikely to be expressed from phage-like promoters or form phage-like particles. The xpt loci found in X. bovienii I73 would appear to support the hypothesis that the xpt genes originated from transposons. The IS elements that flank the I73 xpt genes are homologous to members of the IS6 family, which have been found to constitute composite transposons containing antibiotic resistance (14). Interestingly, the locus includes accessory regions outside of this putative transposon. A possible theory to explain the left-hand accessory region is that “local hopping” of an IS element (19) from the original transposon to a position further downstream would generate a larger transposon, capturing the intervening region. The original IS, now internal in the transposon, would no longer be required for mobility and could lead to degeneracy, as is the case with the two internal IS6 elements at this locus. The extra plasmid-like region to the right of the putative transposon could also have been acquired in a similar manner. Although no IS6-like element exists at the RHJ, a partial tnpA-like element at this location may contribute the necessary transpositional functions. To date IS elements in Xenorhabdus have not been studied. This work has shown that X. nematophila strains, as well as containing common Enterobacteriaceae IS elements such as IS630 and IS1388, also contain elements that are rarer in Enterobacteriaceae, such as ISAS1 which shows over 75% homology to IS elements in A. salmonicida yet only 33% to any IS currently identified in Photorhabdus. The many different types of IS elements present in X. nematophila strains and their high copy number would indicate a very fluid genome, perhaps playing a significant role in the evolution of strains. This is surprising for a bacterium that resides in the relatively sheltered environment of the nematode gut.
In the xpt PAI, most IS elements detected were present in the AT-rich junctions that flanked the groups of insect virulence and mobility genes. These areas may represent hot spots for insertion, thus allowing the PAI to evolve with the acquisition of new genes without the disruption of existing loci. Indeed, the presence of IS elements in these regions may enhance expression of the xpt loci. Strain PMFI296 had the highest insecticidal activity against P. brassicae, and expression of the xptA1 gene in this strain was shown to be approximately 10-fold greater than that of strain 9965 (data not shown). Since PMFI296 was the only strain to contain an IS630-like element downstream of the xptA1 gene, it seems likely that the higher insecticidal activity and increased expression of xptA1 is likely to be due to the presence of the IS630-like element. It is also possible that the presence of the ISAS1-like element in strains 0015, N38, QQ2, and L34 may increase expression of the upstream xptA2 operon. Since xptA1 and xptA2 have been shown to be involved in toxicity to different insects (24), the integration of IS elements in the xpt PAI may modulate pathogenicity towards different insect species and thus tailor the bacterium/nematode complex to inhabit slightly different niches.
The differences, even among closely related strains, in insecticidal activity of the X. bovienii strains observed in this study can be explained not only by the presence of these genes on mobile genetic elements but also by frequent loss of gene function, as indicated by the frameshift and IS insertion in the xptA3 genes detected in X. bovienii strains. Frameshift mutations were also observed in analogous genes in Y. pestis (17). Therefore, the mobility of these genes and the spectrum of activity of different xpt genes (24) would imply that, in a manner analogous to that for B. thuringiensis cry genes, a whole raft of xpt genes exhibiting different biological activity exist in different Xenorhabdus strains. Therefore, further analysis of novel Xenorhabdus strains is required in order to isolate xpt genes that target other important insect pests.
Acknowledgments
This work was supported by a grant from the BBSRC.
REFERENCES
- 1.Akhurst, R. J., and G. B. Dunphy. 1993. Symbiotically associated entomopathogenic bacteria, nematodes, and their insect hosts, p. 1-23. In S. T. N. Beckage and B. Federici (ed.), Parasites and pathogens of insects, vol. 2. Academic Press, New York, N.Y. [Google Scholar]
- 2.Bellingham, N. F., J. A. W. Morgan, J. R. Saunders, and C. Winstanley. 2001. Flagellin gene sequence variation in the genus Pseudomonas. Syst. Appl. Microbiol. 24:157-165. [DOI] [PubMed] [Google Scholar]
- 3.Bowen, D., T. A. Rocheleau, M. Blackburn, O. Andreev, E. Golubeva, R. Bhartia, and R. H. Ffrench-Constant. 1998. Insecticidal toxins from the bacterium Photorhabdus luminescens. Science 280:2129-2132. [DOI] [PubMed] [Google Scholar]
- 4.Duchaud, E., C. Rusniok, L. Frangeul, C. Buchrieser, A. Givaudan, S. Taourit, S. Bocs, C. Boursaux-Eude, M. Chandler, J. F. Charles, E. Dassa, R. Derose, S. Derzelle, G. Freyssinet, S. Gaudriault, C. Medigue, A. Lanois, K. Powell, P. Siguier, R. Vincent, V. Wingate, M. Zouine, P. Glaser, N. Boemare, A. Danchin, and F. Kunst. 2003. The genome sequence of the entomopathogenic bacterium Photorhabdus luminescens. Nat. Biotechnol. 21:1307-1313. [DOI] [PubMed] [Google Scholar]
- 5.Forst, S., B. Dowds, N. Boemare, and E. Stackebrandt. 1997. Xenorhabdus and Photorhabdus spp.: bugs that kill bugs. Annu. Rev. Microbiol. 51:47-72. [DOI] [PubMed] [Google Scholar]
- 6.Hacker, J., and J. B. Kaper. 2000. Pathogenicity islands and the evolution of microbes. Annu. Rev. Microbiol. 54:641-679. [DOI] [PubMed] [Google Scholar]
- 7.Haubold, B., and R. R. Hudson. 2000. LIAN 3.0: detecting linkage disequilibrium in multilocus data. Bioinformatics 16:847-848. [DOI] [PubMed] [Google Scholar]
- 8.Hurst, M. R. H., T. R. Glare, T. A. Jackson, and C. W. Ronson. 2000. Plasmid-located pathogenicity determinants of Serratia entomophila, the causal agent of amber disease of grass grub, show similarity to the insecticidal toxins of Photorhabdus luminescens. J. Bacteriol. 182:5127-5138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huson, D. H. 1998. SplitsTree: analyzing and visualizing evolutionary data. Bioinformatics 14:68-73. [DOI] [PubMed] [Google Scholar]
- 10.Lichter, A., S. Manulis, L. Valinsky, B. Karniol, and I. Barash. 1996. IS 1327, a new insertion-like element in the pathogenicity-associated plasmid of Erwinia herbicola pv gypsophilae. Mol. Plant-Microbe Interact. 9:98-104. [PubMed] [Google Scholar]
- 11.Maiden, M. C. J., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller, S. G., B. C. Campbell, J. Becnel, and L. Ehrman. 1995. Bacterial entomopathogens from the Drosophila-Paulistorum semispecies complex. J. Invertebr. Pathol. 65:125-131. [DOI] [PubMed] [Google Scholar]
- 13.Morgan, J. A. W., M. Sergeant, D. Ellis, M. Ousley, and P. Jarrett. 2001. Sequence analysis of insecticidal genes from Xenorhabdus nematophilus PMFI296. Appl. Environ. Microbiol. 67:2062-2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murphy, E. 1989. Transposable elements in gram-posotive bacteria, p. 269-288. In D. E. Berg and M. M. Howe (ed.), Mobile DNA. American Society for Microbiology, Washington, D.C.
- 15.Nei, M., and T. Gojobori. 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3:418-426. [DOI] [PubMed] [Google Scholar]
- 16.Ohki, R., K. Tateno, Y. Okada, H. Okajima, K. Asai, Y. Sadaie, M. Murata, and T. Aiso. 2003. A bacitracin-resistant Bacillus subtilis gene encodes a homologue of the membrane-spanning subunit of the Bacillus licheniformis ABC transporter. J. Bacteriol. 185:51-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parkhill, J., B. W. Wren, N. R. Thomson, R. W. Titball, M. T. G. Holden, M. B. Prentice, M. Sebaihia, K. D. James, C. Churcher, K. L. Mungall, S. Baker, D. Basham, S. D. Bentley, K. Brooks, A. M. Cerdeno-Tarraga, T. Chillingworth, A. Cronin, R. M. Davies, P. Davis, G. Dougan, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Leather, S. Moule, P. C. F. Oyston, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Genome sequence of Yersinia pestis, the causative agent of plague. Nature 413:523-527. [DOI] [PubMed] [Google Scholar]
- 18.Posada, D., and K. A. Crandall. 2001. Evaluation of methods for detecting recombination from DNA sequences: computer simulations. Proc. Natl. Acad. Sci. USA 98:13757-13762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rakin, A., C. Noelting, P. Schropp, and J. Heesemann. 2001. Integrative module of the high-pathogenicity island of Yersinia. Mol. Microbiol/ 39:407-415. [DOI] [PubMed] [Google Scholar]
- 20.Rakin, A., C. Noelting, S. Schubert, and J. Heesemann. 1999. Common and specific characteristics of the high-pathogenicity island of Yersinia enterocolitica. Infect. Immun. 67:5265-5274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salminen, M. O., J. K. Carr, D. S. Burke, and F. E. McCutchan. 1995. Identification of breakpoints in intergenotypic recombinants of HIV type-1 by bootscanning. AIDS Res. Hum. Retrovir. 11:1423-1425. [DOI] [PubMed] [Google Scholar]
- 22.Sarkar, S. F., and D. S. Guttman. 2004. Evolution of the core genome of Pseudomonas syringae, a highly clonal, endemic plant pathogen. Appl. Environ. Microbiol. 70:1999-2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sawyer, S. 1989. Statistical tests for detecting gene conversion. Mol. Biol. Evol. 6:526-538. [DOI] [PubMed] [Google Scholar]
- 24.Sergeant, M., P. Jarrett, M. Ousley, and J. A. W. Morgan. 2003. Interactions of insecticidal toxin gene products from Xenorhabdus nematophilus PMFI296. Appl. Environ. Microbiol. 69:3344-3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sharma, S., N. Waterfield, D. Bowen, T. Rocheleau, L. Holland, R. James, and R. Ffrench-Constant. 2002. The lumicins: novel bacteriocins from Photorhabdus luminescens with similarity to the uropathogenic-specific protein (USP) from uropathogenic Escherichia coli. FEMS Microbiol. Lett. 214:241-249. [DOI] [PubMed] [Google Scholar]
- 26.Sicard, M., J. B. Ferdy, S. Pages, N. Le Brun, B. Godelle, N. Boemare, and C. Moulia. 2004. When mutualists are pathogens: an experimental study of the symbioses between Steinernema (entomopathogenic nematodes) and Xenorhabdus (bacteria). J. Evol. Biol. 17:985-993. [DOI] [PubMed] [Google Scholar]
- 27.Smith, J. M., and N. H. Smith. 1998. Detecting recombination from gene trees. Mol. Biol. Evol. 15:590-599. [DOI] [PubMed] [Google Scholar]
- 28.Smith, K. D., E. Andersen-Nissen, F. Hayashi, K. Strobe, M. A. Bergman, S. L. R. Barrett, B. T. Cookson, and A. Aderem. 2003. Toll-like receptor 5 recognizes a conserved site on flagellin required for protofilament formation and bacterial motility. Nat. Immunol. 4:1247-1253. [DOI] [PubMed] [Google Scholar]
- 29.Spiridonov, S. E., K. Krasomil-Osterfeld, and M. Moens. 2004. Steinernema jollieti sp n. (Rhabditida: Steinernematidae), a new entomopathogenic nematode from the American midwest. Russian J. Nematol. 12:85-95. [Google Scholar]
- 30.Suerbaum, S., M. Lohrengel, A. Sonnevend, F. Ruberg, and M. Kist. 2001. Allelic diversity and recombination in Campylobacter jejuni. J. Bacteriol. 183:2553-2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thaler, J. O., M. H. BoyerGiglio, and N. E. Boemare. 1997. New antimicrobial barriers produced by Xenorhabdus spp. and Photorhabdus spp. to secure the monoxenic development of entomopathogenic nematodes. Symbiosis 22:205-215. [Google Scholar]
- 32.Thomas, D. J. I., J. A. W. Morgan, J. M. Whipps, and J. R. Saunders. 2001. Plasmid transfer between Bacillus thuringiensis subsp. israelensis strains in laboratory culture, river water, and dipteran larvae. Appl. Environ. Microbiol. 67:330-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Waterfield, N., A. Dowling, S. Sharma, P. J. Daborn, U. Potter, and R. H. Ffrench-Constant. 2001. Oral toxicity of Photorhabdus luminescens W14 toxin complexes in Escherichia coli. Appl. Environ. Microbiol. 67:5017-5024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Waterfield, N. R., P. J. Daborn, and R. H. Ffrench-Constant. 2002. Genomic islands in Photorhabdus. Trends Microbiol. 10:541-545. [DOI] [PubMed] [Google Scholar]