Abstract
Repetitive-element PCR (rep-PCR) is a method for genotyping bacteria based on the selective amplification of repetitive genetic elements dispersed throughout bacterial chromosomes. The method has great potential for large-scale epidemiological studies because of its speed and simplicity; however, objective guidelines for inferring relationships among bacterial isolates from rep-PCR data are lacking. We used multilocus sequence typing (MLST) as a “gold standard” to optimize the analytical parameters for inferring relationships among Escherichia coli isolates from rep-PCR data. We chose 12 isolates from a large database to represent a wide range of pairwise genetic distances, based on the initial evaluation of their rep-PCR fingerprints. We conducted MLST with these same isolates and systematically varied the analytical parameters to maximize the correspondence between the relationships inferred from rep-PCR and those inferred from MLST. Methods that compared the shapes of densitometric profiles (“curve-based” methods) yielded consistently higher correspondence values between data types than did methods that calculated indices of similarity based on shared and different bands (maximum correspondences of 84.5% and 80.3%, respectively). Curve-based methods were also markedly more robust in accommodating variations in user-specified analytical parameter values than were “band-sharing coefficient” methods, and they enhanced the reproducibility of rep-PCR. Phylogenetic analyses of rep-PCR data yielded trees with high topological correspondence to trees based on MLST and high statistical support for major clades. These results indicate that rep-PCR yields accurate information for inferring relationships among E. coli isolates and that accuracy can be enhanced with the use of analytical methods that consider the shapes of densitometric profiles.
Repetitive-element PCR (rep-PCR) is a method for genotyping bacteria that relies on the selective amplification of repetitive sequences dispersed throughout bacterial chromosomes (12). Because it can be performed quickly and cost-effectively and because amplified DNA can be electrophoresed on standard agarose gels, rep-PCR has great potential for epidemiological studies where large numbers of bacterial isolates must be analyzed efficiently. Nevertheless, analyses of rep-PCR data are complicated by subjectivity in the selection of analytical parameters for inferring genetic relationships among bacteria represented by complex electrophoretic patterns.
Multilocus sequence typing (MLST) is based on the nucleotide sequences of internal fragments of genomically dispersed housekeeping genes (7). The inherent objectivity and high discriminatory power of MLST make it favorable for studies of small numbers of bacterial isolates. However, MLST is still impractical for large-scale epidemiological studies, which can involve thousands of isolates. rep-PCR would be a more feasible choice in such situations if the accuracy of the technique for inferring genetic relationships among bacterial isolates were reasonably high. Because MLST data consist of nucleotide sequences of genes selected for their utility in inferring genetic relationships among isolates, the data are objective and provide a “gold standard” against which to evaluate data from rep-PCR.
To provide objective criteria for analyzing rep-PCR data and to investigate the accuracy of the method, we conducted both rep-PCR and MLST analyses with Escherichia coli isolates selected from a large database to span a wide range of pairwise genetic distances. Our goal was to define optimal analytical parameters that would maximize the correspondence between genetic relationships inferred from rep-PCR and those inferred from MLST, thereby also assessing the accuracy of rep-PCR relative to MLST for molecular epidemiological studies.
MATERIALS AND METHODS
Two hundred fifty E. coli isolates were collected from people and animals as part of a large-scale epidemiological study of interspecific bacterial transmission ecology. The isolates were not associated with clinical disease and were considered nonpathogenic. Bacteria from fresh fecal samples were isolated on MacConkey agar and confirmed to be E. coli by standard biochemical tests (6).
DNA was extracted from bacterial isolates using a DNeasy mini kit (QIAGEN, Valencia, CA) and eluted in a 100-μl volume. rep-PCR reactions were performed in 25-μl volumes containing 3.5 mM MgCl2, a 0.3 mM concentration of each deoxynucleoside triphosphate, 2 μM primer box AIR (5′-CTACGGCAAGGCGACGCTGACG-3′), and 1.25 U AmpliTaq Gold DNA polymerase (Applied Biosystems, Foster City, CA), with 1× buffer II (without MgCl2) and 2 μl template. Reaction mixtures were cycled in an iCycler thermocycler (Bio-Rad, Hercules, CA) at 95°C for 7 min and then for 30 cycles at 94°C for 1 min, 66°C for 8 min, and 71°C for 1 min, followed by a 71°C final extension step for 15 min and an indefinite 4°C soak. Immediately after cycling, 5 μl of gel loading dye (Amresco 6× agarose gel loading dye; Amresco, Solon, OH) was added, and reaction mixtures were stored at 4°C. These methods (modified from the method described by Johnson and O'Bryan [4]) were developed in our laboratory for maximum reliability and reproducibility, yield consistently high-quality fingerprints, and are robust enough to withstand pipetting errors of approximately 6% (T. L. Goldberg, unpublished data).
PCR products were electrophoresed in 20- by 30-cm gels of 200 ml 2.0% GenePure LE agarose (ISC BioExpress, Kaysville, UT) in 1× TAE (Tris-acetate-EDTA) buffer (model A2 gel electrophoresis system; Owl Separation Systems, Portsmouth, NH) at 4°C for 15 h at 62 V. Ten microliters of marker (GeneRuler 100-bp DNA ladder plus; Fermentas, Hanover, MD) was loaded after every sixth sample. Gels were stained for 20 min in 1× TAE containing 0.5 mg/liter ethidium bromide, destained for 60 min in 1× TAE, and photographed immediately under UV light with a Gel Doc XR system (Bio-Rad, Hercules, CA). To assess reproducibility, we performed all rep-PCRs twice, and we also performed 18 independent DNA extractions and rep-PCRs, each run on a separate electrophoretic gel, using a reference isolate from our collection over the course of the study.
Twelve E. coli isolates from among the 250 analyzed were chosen for MLST. These isolates were selected to span as wide a range as possible of pairwise genetic distances (from identical to maximally divergent), based on an initial evaluation of their rep-PCR genotypes using BioNumerics version 4.0 software (Applied Maths, Austin, TX) and default parameters. Seven loci (aspC, clpX, fadD, icdA, lysP, mdh, and uidA) were sequenced in each isolate, according to protocols available from the NIH/NIAID Shiga Toxin-Producing Escherichia coli Center website (www.shigatox.net). Amplicons were gel purified with a ZymoClean gel DNA recovery kit (Zymo Research, Orange, CA) and sequenced directly and in both directions with the same primers used for PCR. Sequencing was performed at the Roy J. Carver Biotechnology Center, University of Illinois. Genetic data from bacterial isolates (both rep-PCR fingerprints and MLST data) were stored in a database in BioNumerics.
The analytical parameters available from BioNumerics, both for identifying bands on electrophoretic gels and for calculating genetic distances among bacterial isolates from fingerprints, were varied systematically. The parameters that were varied during the identification of bands on electrophoretic gels were “minimum profiling” (a criterion for identifying bands based on the elevation of densitometric peaks relative to the highest peak in the pattern), “minimum area” (a criterion for identifying bands based on the area of densitometric features relative to the total area of the pattern), and “shoulder sensitivity” (an option that allows for the identification of doublet bands and of bands on the “shoulders” of other densitometric features). These parameters are commonly used to define objective criteria for identifying bands in complex electrophoretic patterns prior to genetic or phylogenetic analyses that rely on discrete character data.
The parameters that were varied during the calculation of genetic distances from fingerprints were optimization (the percentage of “shift” in total length allowed between two patterns being compared), position tolerance (the percentage of error in fragment length tolerated when similarly sized bands in two different patterns were identified as a “match”), and the method of calculating distances between patterns, which included both “curve-based” methods (which correlate intensities between densitometric curves) and “band-sharing coefficients” (which calculate indices of similarity based on shared and different bands). These parameters determine the degree of relative genetic distance inferred between pairs of isolates, which in turn affect inferences about their phylogenetic and epidemiological relationships.
The correspondence between the results of rep-PCR and MLST for different sets of parameters was measured as a Pearson's product-moment correlation coefficient between genetic similarity matrices, calculated with the “congruence of experiments” protocol in BioNumerics. Because of the large potential parameter space of this analysis, all combinations of parameters were not explored. Rather, initial parameter values were chosen based on defaults suggested in the BioNumerics user's manual and were varied independently in 1% increments, up to approximately 20% around these values, until local optima were found. Parameters were then covaried near these local optima in increments of 0.1% or 0.01% until the maximum correspondence between rep-PCR and MLST results was achieved.
RESULTS
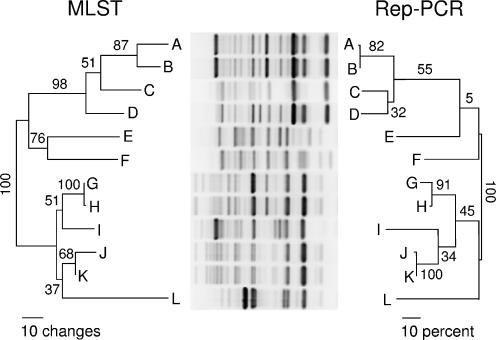
The 250 E. coli isolates analyzed contained 89 unique rep-PCR genotypes. From among these, 12 isolates were selected for MLST to represent the widest possible range of genetic distances, initially inferred using BioNumerics and default parameters. These included two pairs of isolates with visually indistinguishable fingerprint patterns (Fig. 1, isolates A and B and J and K).
FIG. 1.
Comparison of MLST and rep-PCR for inferring genetic relationships among 12 E. coli isolates. Isolates were initially chosen to represent the widest possible range of pairwise genetic distances inferred from rep-PCR genotypes using default parameters in the computer program BioNumerics, version 4.0 (Applied Maths, Inc.). The MLST tree was the likeliest tree (−ln likelihood score of 7730.2) found during a maximum likelihood search with the computer program PAUP* version 4.0b10 (11), with a model of molecular evolution selected using a hierarchical likelihood ratio test approach implemented with the computer program Modeltest, version 3.7 (8). Numbers beside branches are bootstrap values (percentages), based on an analysis of 1,000 maximum likelihood bootstrap replicates performed with PAUP* using the same parameters as those in the original search. The rep-PCR tree is a neighbor-joining tree (10) constructed from a matrix of distances between bacterial genotypes calculated as Pearson's product-moment correlation coefficients between densitometric curves generated from the fingerprint patterns shown using optimal curve-based analytical parameters (Table 2). Bootstrap values shown on the rep-PCR tree were generated by converting bacterial genotypes into a series of binary variables representing the presence/absence of bands at 41 band positions using the “bandmatch” procedure in BioNumerics and optimal parameters (Table 2); this data set was then imported into PAUP* for bootstrap analysis (1,000 replicates using the neighbor-joining algorithm), and resulting values were transcribed onto the corresponding clades of the tree as shown.
MLST revealed a high diversity of alleles among these 12 isolates (Table 1). Diversity was higher at the nucleic acid level for all loci analyzed than at the amino acid level, indicating a predominance of silent substitutions among the alleles sequenced. When all seven loci were combined, each of the 12 isolates had a unique sequence, including pairs of isolates with visually indistinguishable fingerprints.
TABLE 1.
Allelic diversity at seven MLST loci in 12 E. coli isolatesa
| Locus | Sequence length (nucleotides) | No. of alleles | % Nucleotide diversity
|
% Amino acid diversity
|
||
|---|---|---|---|---|---|---|
| Mean ± SE | Range | Mean ± SE | Range | |||
| aspC | 554 | 7 | 1.03 ± 0.27 | 2.17 | 0.00 ± 0.00 | 0.00 |
| clpX | 632 | 9 | 2.37 ± 0.36 | 5.06 | 0.00 ± 0.00 | 0.00 |
| fadD | 540 | 8 | 3.13 ± 0.37 | 9.63 | 0.09 ± 0.09 | 0.56 |
| icdA | 629 | 6 | 1.61 ± 0.27 | 3.18 | 0.22 ± 0.16 | 0.96 |
| lysP | 588 | 6 | 0.70 ± 0.24 | 1.53 | 0.00 ± 0.00 | 0.00 |
| mdh | 607 | 7 | 1.08 ± 0.24 | 1.81 | 0.23 ± 0.16 | 0.99 |
| uidA | 610 | 5 | 1.77 ± 0.36 | 3.11 | 0.62 ± 0.39 | 1.48 |
Nucleotide diversity, mean proportion of pairwise nucleotide sequence differences among isolates; amino acid diversity, mean proportion of pairwise amino acid sequence differences among isolates. Calculations include identical alleles. Standard errors were estimated from 1,000 replicates of bootstrap resampling. All calculations were performed with the computer program MEGA3 (5). Ranges represent nucleotide and amino acid sequence differences among alleles; values indicate upper ranges of pairwise differences.
The maximum correspondence between rep-PCR and MLST results achieved for any set of analytical parameters was 84.5% (Table 2). Curve-based methods and default parameters in BioNumerics yielded correspondences of approximately 80%. The correspondence between rep-PCR and MLST results using optimal parameters is shown in Fig. 1 as a comparison of phylogenetic trees. The topologies of the two trees are similar, and all clades in the MLST tree supported by bootstrap values of ≥80% are also present in the rep-PCR tree. The relationships between pairwise distances inferred from rep-PCR (R) and those inferred from MLST (M) were described by the linear equation M = 0.963 + 0.034R, indicating that a distance between isolates of 1% at the sequence level corresponds to a distance of approximately 3.4% at the rep-PCR level. This equation was derived from a least-squares regression line fitted to the pairwise distances determined by rep-PCR versus those determined by MLST.
TABLE 2.
Effects of varying analytical parameters on the correspondence between genetic distances inferred from rep-PCR and MLST
| Parameter | Optimal value (%) or coefficienta | Effect on correspondence (%)b | ||
|---|---|---|---|---|
| Band-sharing coefficient methods | ||||
| Minimum profiling | 0.0 | 0.0 | ||
| Minimum area | 1.8 | −4.5 | ||
| Shoulder sensitivity | 0.0 | 0.0 | ||
| Optimization | 1.00 | −0.7 | ||
| Tolerance | 0.52 | −2.7 | ||
| Algorithmc | (Dice) | −10.8 | ||
| Curve-based methods
|
||||
| Optimization | 8.0 | −0.7 | ||
| Algorithmd | (Pearson) | −0.4 | ||
Values are optimal for all parameters in combination, yielding maximum correspondences between rep-PCR and MLST of 80.3% for band-sharing coefficient methods and 84.5% for curve-based methods. Parameters are described in the text.
Values indicate maximum reductions in correspondence between rep-PCR and MLST data caused by varying each parameter independently up to 20% of its optimal value.
The band-sharing algorithms used were those available in BioNumerics, including the Jaccard, Dice, Jeffrey's X, Ochiai, and different-band coefficients.
Curve-based algorithms were those available in BioNumerics, including the Pearson's product-moment and cosine correlation coefficients between densiometric curves.
We also determined the optimal analytical parameters for rep-PCR data analyzed using band-sharing coefficients (Table 2). The type of binary data resulting from such analyses is amenable to bootstrapping and various other statistical procedures that require discrete character data. In this case, the optimal parameters yielded a maximum 80.3% correspondence between rep-PCR and MLST. Manual editing of band identifications (e.g., to remove “extraneous” bands assigned by the computer to particularly broad densitometric features or to flaws in the gel) altered congruence by only approximately ±3%. Band-sharing coefficient methods and default parameters in BioNumerics yielded correspondences between rep-PCR and MLST results of approximately 65%.
Analysis of electrophoretic patterns generated from the second, independent rep-PCR replicate experiment with our 12 E. coli isolates gave results very similar to those described above, yielding a correspondence value of 81.2% with MLST using optimal parameters derived from the first replicate (Table 2). Eighteen independent DNA extractions, PCRs, and electrophoreses performed with our reference isolate yielded reproducibility estimates of 96.7% ± 0.2% (mean ± standard error) similarity among patterns when data were analyzed using curve-based methods and 84.7% ± 1.0% similarity among patterns when data were analyzed using band-sharing coefficients, again with optimal parameters.
DISCUSSION
We identified analytical parameters in this study that yielded a high correspondence between rep-PCR and MLST results (up to 84.5%) and that accurately reconstructed phylogenetic relationships among E. coli isolates that were well supported statistically by MLST data. This high correspondence was achieved with the use of only a single rep-PCR primer. This is encouraging for epidemiological investigations in which large numbers of isolates must be genotyped. We suspect that the use of additional rep-PCR primers would increase the ability of the technique to discriminate among isolates. MLST has higher discriminatory power than does rep-PCR, as evidenced by the unique MLST sequences of pairs of isolates with visually indistinguishable genotypes resulting from rep-PCR.
A higher correspondence between rep-PCR and MLST data resulted from analytical methods that compared the shapes of densitometric profiles than from methods that scored the presence or absence of individual bands. Additionally, curve-based methods were markedly less sensitive to variations in user-specified parameter values (e.g., optimization) than were methods that relied on the scoring of bands. Curve-based methods, because they correlate densitometric intensities of fingerprints over the entire lengths of the patterns, avoid the subjectivity inherent in defining cutoff values for scoring bands. Even when we optimized parameters for band-sharing coefficient methods, the highest correspondence resulted from using a minimum-area cutoff to score bands, which considers the area of an entire densitometric feature, and not minimum profiling, which defines cutoffs based only on the heights of densitometric peaks.
Based on these results, we recommend the use of curve-based methods for inferring genetic relationships from rep-PCR data. This recommendation is in agreement with the results of Hassan et al., who found that curve-based methods were more accurate than band sharing indices for bacterial source tracking using rep-PCR data (3). We suspect that curve-based methods may also prove favorable for other types of data in which the intensities of bands within a pattern vary widely and where the intensities of different patterns also vary (e.g., most PCR-based methods, such as randomly amplified polymorphic DNA or amplified fragment length polymorphism). The disadvantage of curve-based methods is that they yield only a single distance metric and cannot be used for bootstrapping or other analyses in which discrete character data are needed. When discrete character data are required, we recommend the use of area-based algorithms for band scoring.
Although we did not conduct extensive tests of the reproducibility of rep-PCR, the technique proved to be consistent in our hands. Using curve-based methods and optimal analytical parameters, we achieved over 95% correspondence between independent rep-PCR replicates. This is in contrast to the results of Johnson and O'Bryan (4), who achieved maximum reproducibility values of only approximately 80%. We attribute the higher reproducibility of our results not only to our particular PCR method, which we optimized for reliability and reproducibility, but also to the use of curve-based methods for analyzing the resulting patterns. Our results, in combination with those of other studies (1, 2), suggest that the reputation of rep-PCR as a technique of inherently low reproducibility may not be warranted and that analytical parameters chosen to maximize the accuracy of the technique may also enhance its reproducibility. Automation may further enhance the reproducibility of rep-PCR; in the case of Staphylococcus aureus, automated rep-PCR performs favorably in comparison to the “industry standard” of pulsed-field gel electrophoresis (9).
Overall, our results suggest that rep-PCR yields accurate data for inferring genetic relationships among E. coli isolates and that it is a reasonable choice for molecular epidemiological studies involving large numbers of isolates. We would still recommend MLST for smaller-scale studies because of the inherent objectivity of nucleotide sequence data. We suspect that optimal parameter values for the analysis of rep-PCR data may differ for different sample sizes, for different populations of E. coli, and for different bacterial species. Ideally, comparisons of rep-PCR to more objective methods such as MLST should be performed to identify optimal analytical parameters specific to different laboratories and study systems. In the absence of such information, however, and because of the greater robustness of curve-based methods to accommodate variations in user-specified analytical parameter values, we suggest adopting such methods as analytical starting points.
Acknowledgments
We thank E. Estoff, J. Gibson, K. Knuffmann, K. Inendino, I. Rwego, and E. Wheeler for assistance with laboratory analyses. Three anonymous reviewers provided constructive comments on the manuscript.
This material is based upon work supported by The Morris Animal Foundation under award no. D04ZO-67.
REFERENCES
- 1.Baldy-Chudzik, K., J. Niedbach, and M. Stosik. 2001. Application of rep-PCR fingerprinting for genotyping of Escherichia coli strains in Wojnowskie Wschodnie and Wojnowskie Zachodnie lake. Acta Microbiol. Pol. 50:233-242. [PubMed] [Google Scholar]
- 2.Carson, C. A., B. L. Shear, M. R. Ellersieck, and J. D. Schnell. 2003. Comparison of ribotyping and repetitive extragenic palindromic-PCR for identification of fecal Escherichia coli from humans and animals. Appl. Environ. Microbiol. 69:1836-1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hassan, W. M., S. Y. Wang, and R. D. Ellender. 2005. Methods to increase fidelity of repetitive extragenic palindromic PCR fingerprint-based bacterial source tracking efforts. Appl. Environ. Microbiol. 71:512-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson, J. R., and T. T. O'Bryan. 2000. Improved repetitive-element PCR fingerprinting for resolving pathogenic and nonpathogenic phylogenetic groups within Escherichia coli. Clin. Diagn. Lab. Immunol. 7:265-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 6.MacFaddin, J. F. 1980. Enterobacteriaceae and other intestinal bacteria, p. 439-464. In J. F. MacFaddin (ed.), Biochemical tests for identification of medical bacteria. Williams & Wilkins, Baltimore, Md.
- 7.Maiden, M. C., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Posada, D., and K. A. Crandall. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 9.Ross, T. L., W. G. Merz, M. Farkosh, and K. C. Carroll. 2005. Comparison of an automated repetitive sequence-based PCR microbial typing system to pulsed-field gel electrophoresis for analysis of outbreaks of methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 43:5642-5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 11.Swofford, D. L. 2000. PAUP*. Phylogenetic analysis using parsimony (*and other methods), 4th ed. Sinauer Associates, Sunderland, Mass.
- 12.Versalovic, J., T. Koeuth, and J. R. Lupski. 1991. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 19:6823-6831. [DOI] [PMC free article] [PubMed] [Google Scholar]