Abstract
Xanthomonas oryzae pv. oryzicola, the cause of bacterial leaf streak in rice, possesses clusters of hrp genes that determine its ability to elicit a hypersensitive response (HR) in nonhost tobacco and pathogenicity in host rice. A 27-kb region of the genome of X. oryzae pv. oryzicola (RS105) was identified and sequenced, revealing 10 hrp, 9 hrc (hrp conserved), and 8 hpa (hrp-associated) genes and 7 regulatory plant-inducible promoter boxes. While the region from hpa2 to hpaB and the hrpF operon resembled the corresponding genes of other xanthomonads, the hpaB-hrpF region incorporated an hrpE3 gene that was not present in X. oryzae pv. oryzae. We found that an hrpF mutant had lost the ability to elicit the HR in tobacco and pathogenicity in adult rice plants but still caused water-soaking symptoms in rice seedlings and that Hpa1 is an HR elicitor in nonhost tobacco whose expression is controlled by an hrp regulator, HrpX. Using an Hrp phenotype complementation test, we identified a small hrp cluster containing the hrpG and hrpX regulatory genes, which is separated from the core hrp cluster. In addition, we identified a gene, prhA (plant-regulated hrp), that played a key role in the Hrp phenotype of X. oryzae pv. oryzicola but was neither in the core hrp cluster nor in the hrp regulatory cluster. A prhA mutant failed to reduce the HR in tobacco and pathogenicity in rice but caused water-soaking symptoms in rice. This is the first report that X. oryzae pv. oryzicola possesses three separate DNA regions for HR induction in nonhost tobacco and pathogenicity in host rice, which will provide a fundamental base to understand pathogenicity determinants of X. oryzae pv. oryzicola compared with those of X. oryzae pv. oryzae.
The interaction of many gram-negative plant-pathogenic bacteria with plants is modulated by two sets of genes: the avirulence (avr) or virulence (vir) genes determine host specificity via gene-for-gene interactions, and the hrp genes, which are usually clustered in a chromosomal region that spans 20 to 30 kb, are involved in induction of the hypersensitive response (HR) in resistant host and nonhost plants and pathogenicity in susceptible host plants (2, 7). HR is a rapid, local, programmed cell death that is induced upon recognition of the pathogen and concomitant with the inhibition of pathogen growth within the infected plants. Commonly, the nine hrp genes, which are highly conserved in plant and animal bacterial pathogens, are known as the hrc (hrp conserved) genes (6). The hrc genes encode the type III secretion system (TTSS) and are critical for pathogenicity and initiation of disease (2, 34). Recently, it was established that the hrpF gene in plant-pathogenic bacteria encodes a translocon that is a component of the TTSS (13, 31, 49). These hrc genes are also found in animal pathogens such as Salmonella and Shigella and are thought to be evolutionarily related to the flagellar apparatus (18, 19, 20). The hpa (hrp-associated) genes contribute to pathogenicity and to the induction of the HR in nonhost plants but are not essential for the pathogenic interactions of bacteria with plants (24, 28, 32).
Our knowledge of hrp genes in Xanthomonas arises mainly from studies of X. campestris species. The best characterized systems are the hrp gene clusters of X. campestris pv. vesicatoria (3, 12, 54), the causal agent of bacterial spot on pepper and tomato; X. axonopodis pv. glycines (31), the pathogen of bacterial pustule on soybean; X. axonopodis pv. citri (16), a pathogen of citrus canker; X. campestris pv. campestris (16), a pathogen of black rot of crucifers; and X. oryzae pv. oryzae (41, 49, 61), the causal agent of bacterial blight in rice. The core hrp cluster of xanthomonads contains six operons from hrpA to hrpF. However, little is known about the structure and function of the hrp cluster in X. oryzae pv. oryzicola.
The expression of hrp genes is highly regulated, being induced only in plants or certain nutrient-poor synthetic media (3, 27, 35, 46, 47, 50, 55). There are two types of hrp regulatory systems in plant-pathogenic bacteria (2, 23). In group I systems, which are found in Pseudomonas syringae, Erwinia amylovora, and Pantoea stewartii, a member of the extracytoplasmic function family of alternative sigma factors, called HrpL, functions as the regulator for the other hrp genes (36, 59). On the other hand, in group II systems, which are found in Xanthomonas species or pathovars and Ralstonia solanacearum, either the AraC-type transcriptional activator HrpX (Xanthomonas) or HrpB (R. solanacearum) regulates expression of the hrpB-to-hrpF operon along with some effector proteins (5, 22, 30, 40, 45, 54). Commonly, the HrpX regulons in xanthomonads are preceded by a consensus sequence motif, called the plant-inducible promoter (PIP) box (TTCGC-N15-TTCGC) (18, 40, 41, 51, 54). The expression of hrpA and hrpX is activated by the product of hrpG, which belongs to the OmpR family of two-component regulatory systems (56, 57). Interestingly, the existence of plant factors that specifically regulate hrp genes has been proposed. In R. solanacearum, the outer membrane protein PrhA (plant-regulated hrp) controls the plant-responsive regulatory cascade composed of PrhR, PrhI, PrhJ, HrpG, and HrpB, the final activator of hrp transcription units 1 to 4 and 7 (1, 11, 35). In contrast, our knowledge of the hrp regulation system in X. oryzae pv. oryzicola is rudimentary.
The harpins, i.e., HrpN of E. amylovora (52), HrpZ of P. syringae (25), HpaG of X. axonopodis pv. glycines (31), Hpa1 of X. oryzae pv. oryzae (53), and XopA of X. campestris pv. vesicatoria (40), have been characterized as HR elicitors in nonhost plants, but their individual contributions to pathogenicity in host plants vary greatly. The hpa1 gene of X. oryzae pv. oryzae encodes a 13-kDa glycine-rich protein with a composition similar to those of harpins in xanthomonads and PopA in R. solanacearum (62). To date, no harpin-like proteins with elicitor activity have been reported to exist in X. oryzae pv. oryzicola.
X. oryzae pv. oryzicola colonizes the intercellular spaces, the apoplast, of the mesophyll to cause bacterial leaf streak in rice. This disease is of increasing importance throughout Asia, and especially in China, where many high-yield hybrids are very susceptible (15, 60). Scientifically, X. oryzae pv. oryzicola is considered an ideal pathogen to understand molecular mechanisms of rice-Xanthomonas interactions (44). The ability of the bacteria to induce the HR on nonhost plants and cause disease on host plants is controlled by hrp genes (14). Considering the progress in determining the rice genome (44), it is expected that knowledge of the hrp genes of X. oryzae pv. oryzicola will be key to understanding rice-bacterium interactions. However, our knowledge of the hrp clusters in this bacterium is still rudimentary. Consequently, we have sought to identify and clone the hrp clusters of X. oryzae pv. oryzicola, which we report upon here. In starting to characterize the individual hrp genes, we confirmed that the hpa1 gene product is a TTSS-dependent HR elicitor in nonhost tobacco and that the expression of the hpa1 gene is controlled by an hrp regulator gene, hrpX. In addition, we identified the prhA locus, which contributes to the hypersensitive response in nonhost tobacco and to pathogenicity in host rice.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 1. All of the X. oryzae pv. oryzicola strains used in these experiments were derivatives of the parent strain RS105. Escherichia coli cells were cultivated at 37°C in Luria broth (LB) or on LB agar plates. The Xanthomonas strains were grown at 28°C on nutrient agar (NA) (0.5% yeast extract, 1% polypeptone, and 1% NaCl) plates or in nutrient broth without agar. Antibiotics were used at the following concentrations: 100 μg/ml ampicillin, 15 μg/ml chloramphenicol, 25 μg/ml kanamycin, and 25 μg/ml spectinomycin for E. coli and 50 μg/ml kanamycin, 50 μg/ml spectinomycin, and 50 μg/ml rifampin for X. oryzae pv. oryzicola. Tetracycline was used at 10 μg/ml for E. coli and at 2 μg/ml for X. oryzae pv. oryzicola.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Reference or source |
|---|---|---|
| Strains | ||
| X. oryzae pv. oryzicola | ||
| RS105 | Wild type, Rifr | This lab |
| RABC | prhA::Cm, prhA mutant, Rifr Cmr | This study |
| RCX | hrpX::Cm, hrpX mutant, Rifr Cmr | This study |
| RCG | hrpG::Cm, hrpG mutant, Rifr Cmr | This study |
| RFBC | hrpF::Cm, hrpF mutant, Rifr Cmr | This study |
| M55 | hrpX mutant generated by chemical mutagenesis | 14 |
| M1005 | hrpG mutant generated by chemical mutagenesis | 14 |
| Escherichia coli | ||
| DH5α | F− φ80dlacZΔM15 Δ(lacZYA-argF)U169 endA1 deoR recA1 hsdR17(rK− mK+) phoA supE44 λ−thi-1 gyrA96 relA1 | Clontech |
| S17-1 | Tra+recA Spr | 48 |
| BL21(DE3) | F−ompT hsdB(rB− mB−) gal dcm (DES) | Novagene |
| Bhpa1 | Conjugant of BL21(DE3) with plasmid pETHpa1 | This study |
| Plasmids | ||
| pUFR034 | Kmr, IncW, Mob(p), Mob+, LacZa+, PK2 replicon, cosmid | 17 |
| pBlueScript II KS(−) | Phagemid, pUC derivative, Apr | Stratagene |
| pBC SK | pUC ori, Cmr | Stratagene |
| pUC18 | pUC ori, Apr | This lab |
| pMD-18-T | Apr, T-easy vector for PCR products | TaKaRa |
| p6 | Kmr, positive hrp gene clone screened from X. oryzae pv. oryzicola genomic library | This study |
| pPK12.3 | Kmr, a 12.3-kb fragment containing the hrpG and hrpX operons screened from X. oryzae pv. oryzicola genomic library | 14 |
| pA1 | Kmr, positive hrp gene clone screened from X. oryzae pv. oryzicola genomic library | This study |
| pA1-E2 | Apr, a 5.1-kb EcoRI fragment from pA1 in pUC18 | This study |
| pHA1.5 | Apr, a 1.5-kb hrpA of X. oryzae pv. oryzae ligated into pUC18 as a probe for screening the genomic library of X. oryzae pv. oryzicola | This study |
| pHF1.62 | Apr, a 1.62-kb hrpF fragment of X. oryzae pv. oryzae ligated into pBluescript KS(−) as a probe for screening the genomic library of X. oryzae pv. oryzicola | This study |
| p6-28 | Apr, a 9,611-bp EcoRI fragment from p6 ligated into pBluescript KS(−) | This study |
| p6-7 | Apr, a 5,071-bp EcoRI fragment from p6 ligated into pBluescript KS(−) | This study |
| p6-8 | Apr, a 7,808-bp EcoRI fragment from p6 ligated into pBluescript KS(−) | This study |
| p685 | Apr, a 685-bp EcoRI fragment from p6 ligated into pMD-18-T vector | This study |
| p6-3 | Apr, a 1,996-bp EcoRI fragment from p6 ligated into pBluescript KS(−) | This study |
| pBSK579 | Apr, a 579-bp EcoRI-KpnI fragment from p6 ligated into pBluescript KS(−) | This study |
| p6121-2 | Apr, a 6,381-bp KpnI fragment from p6 ligated into pBluescript KS(−) | This study |
| pHpa21 | Apr, a 1,356-bp fragment containing the hap2 and hpa1 genes ligated into pMD-18-T | This study |
| p61 | Kmr, a 2,163-bp prhA gene from pHA1.5 ligated in pUFR034 | This study |
| phrA2.1 | Apr, PCR product of a 2,163-bp prhA gene ligated into pBluescript | This study |
| pABC | Cmr, a 112-bp PstI-SacII fragment from phrA2.1 ligated in pBC SK(−) | This study |
| pEThpa1 | Apr, a 414-bp hpa1 gene of X. oryzae pv. oryzicola cloned into pET21a | This study |
| pUhpa1 | Kmr, a 614-bp hpa1 gene plus the promoter region together with the fragment encoding a His6 tag cloned into pUFR034 | This study |
| pBCX147 | Cmr, a 147-bp PstI-KpnI fragment from the hrpX gene ligated in pBC SK(−) | This study |
| pBCG121 | Cmr, a 121-bp EcoRI-SacI fragment from the hrpG gene into pBC SK(−) | This study |
| pFBC | Cmr, a 351-bp BamHI-PstI fragment from p6-3 linked into pBC SK(-) | This study |
| pHrpF | Kmr, a 2,409-bp hrpF gene by PCR linked into pUFR034 | This study |
Apr, ampicillin resistance; Cmr, chloramphenicol resistance; Kmr, kanamycin resistance; Rifr, rifampin resistance; Spr, spectinomycin resistance.
To obtain an hrp-inducing medium for X. oryzae pv. oryzicola, we modified XVM2 [20 mM NaC1, 10 mM (NH4)2SO4, 5 mM MgSO4, 1 mM CaCl, 0.16 mM KH2PO4, 0.32 mM K2HPO4, 0.01 mM FeSO4, 10 mM fructose, 10 mM sucrose, 0.03% Casamino Acids (pH 6.7)] (46, 54) and developed a synthetic medium (termed XOM3) containing 1% sugar source, 650 μM dl-methionine, 10 mM sodium l-(+)-glutamate, 15.0 mM KH2PO4, 40 μM MnSO4, 240 μM Fe2-EDTA, and 5 mM MgCl2, pH 6.5. Antibiotics were added to XOM3 for the growth of Xanthomonas bacteria as required.
Bacterial conjugation and transformation.
For construction of genomic DNA (gDNA) libraries, the total genomic DNA from strain RS105 was partially digested by EcoRI, and the resulting 30- to 40-kb fragments were ligated into the vector pUFR034 (17). Subclones were first introduced into E. coli strain DH5α for lacZ selection by transformation and then into S17-1 (48) for later biparental conjugation.
As recipients, each hrp mutant of X. oryzae pv. oryzicola (108 CFU/ml) was harvested at 4,000 rpm for 10 min and resuspended in 200 μl of LB broth. A portion of a single colony of the donor S17-1, harboring a corresponding cosmid with a matching hrp gene, was transferred onto a nylon film (2 cm2) and then mixed with 30 μl of the recipients. The mixture on the film was incubated on NA at 28°C for 48 h and then plated on NA complemented with kanamycin and rifampin. For Hrp phenotype tests, single-colony transfers were used to purify the transconjugants after culture at 28°C for 4 to 6 days. The transformation of X. oryzae pv. oryzicola strains, which led to homologous recombination of the incoming DNA with marker exchange, was performed according to the method of Boucher et al. (9).
Hpa1 protein expression and purification in E. coli.
The hpa1 gene (accession no. AY875714) was amplified by PCR from genomic DNA of the X. oryzae pv. oryzicola strain RS105 with the primers hpa1-F1 and hpa1-R1 (Table 2) and ligated into pGEM-T Easy vector (Promega). After restriction enzyme digestion of the vector with NdeI and XhoI, the resulting hpa1 fragment was ligated into pET21a (Novagen), generating a construct (pEThpa1) to express the hpa1 gene with a C-terminal hexahistidine tag. The construct was transformed into E. coli strain BL21(DE3) (Invitrogen). Bhpa1 cells were grown in an orbital shaker at 37°C and 220 rpm until the culture reached an absorbance, at 600 nm, of 0.5 to 0.6 and then were induced by the addition of 1 mM isopropyl-β-d-thiogalactopyranoside at 37 °C and 200 rpm for 3 h. The cells were harvested by centrifugation at 7,000 × g for 8 min at 4°C, resuspended in buffer A (20 mM Tris [pH 7.4], 300 mM NaCl, 20% glycerol, 5 U/ml DNase I, one 100-ml protease inhibitor mixture tablet), and disrupted by three passages through a Constant System cell disrupter (15 kpsi, model Z-plus 1.1 kW; Constant Systems). Disrupted cells were subjected to ultracentrifugation at 220,000 × g for 90 min at 4°C. The supernatant fraction was purified by affinity chromatography using a 1-ml HiTrap chelating column (Amersham Bioscience) immobilized with Ni2+ equilibrated with buffer C (20 mM Tris [pH 7.8], 100 mM NaCl, 10% glycerol). The column was washed with 50 mM imidazole added to the buffer C, and the extracted harpin was eluted with 500 mM imidazole in buffer B.
TABLE 2.
Primers used in this study
| Primer | Sequence (5′→3′)a | Purpose |
|---|---|---|
| prhA-F1 | CTGAATTCATGTCTGCGACCTTCCC | To amplify the prhA gene from X. oryzae pv. oryzicola |
| prhA-R1 | AGGAATTCCTAGAAATCCACCTGC | |
| hpa1-F1 | CATATGAATTCTTTGAACACACAATTC | To express the hpa1 gene in E. coli |
| hpa1-R1 | CTCGAGCTGCATCGATCCGCTGTCGTTCG | |
| hpa1-F2 | GGATCCGATCTGTTATCGATCCTAAAAAATTTTCCAC | To express the hpa1 gene together with a hexahistidine tag in X. oryzae pv. oryzicola |
| hpa1-R2 | GGTACCTCAGTGGTGGTGGTGGTGGTGCTGCATCGATCCGCTGTCGTTCG | |
| hrpA-F1 | CGGATCCACTTAACGGGCAAGAAAAAAG | To amplify the hrpA gene as a probe for screening for hrp clones from an X. oryzae pv. oryzicola genomic library |
| hrpA-R1 | TGGATCCGGTACCGGGTCTGTCAAAGATTC | |
| hrpF-F1 | GAAGGATCCGCGGTTGTTCTTCGCCATC | As a probe for screening for hrp clones from an X. oryzae pv. oryzicola genomic library |
| hrpF-R1 | TGCAAGCTTGGAGGCACGTTCATACGAACG | |
| hrpF-F2 | GGTACCATGTCGCTCAACATGCTTTCTAC | To express the hrpF gene in the hrpF mutant of X. oryzae pv. oryzicola |
| hrpF-R2 | GGTACCTTATCTGCGACGTATCCTGACATTG | |
| hrpX-F | CTGGATCCATGATCCTTTCGACCTACTTTG | To amplify the hrpX gene from X. oryzae pv. oryzicola |
| hrpX-R | ATGGATCCTTACCGTTGCAAGGTT | |
| hrpG-F | GGATCCATGAACATCCCTTGCCCCCTTG | To amplify the hrpG gene from X. oryzae pv. oryzicola |
| hrpG-R | GGATCCTCAGCAGGCGGCTGTGCGATG | |
| hpa2-F | CTATTCACCAATCACACCAC | For cloning the region incorporating the hpa2 to hpa1 genes from X. oryzae pv. oryzicola |
| hpa1-R | TTACTGCATCGATGCGCT |
Restriction sites are underlined.
Detection of Hpa1 in the supernatant of the Xanthomonas cell culture.
The promoter and coding regions of the hpa1 gene were amplified by PCR with the primers hpa1-F2 and hpa1-R2, which incorporated the sequence encoding the His tag (Table 2). The resulting PCR product was cloned into the BamHI/KpnI sites of pUFR034, giving pUhpa1. The cosmid was transconjugated into the hrpX mutant RCX and the wild-type RS105, as described elsewhere in this paper, producing conjugants of RCX and RS105 with the pUhpa1 plasmid. The conjugants were grown at 28°C for 16 h in 500 ml of XOM3 medium. After two successive centrifugations (8,000 × g, 15 min), the culture supernatant was concentrated 20-fold by ultrafiltration with an Amicon Centricon 10 miniconcentrator (10,000-Mr cutoff) (37). This concentrated supernatant preparation was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Meanwhile, harvested cells were resuspended in 0.05× the original volume of buffer A. Xanthomonas cells were disrupted and Hpa1 protein purified as described above.
Electrophoresis of proteins by 12.5% SDS-PAGE was performed as described by Laemmli (33). Before electrophoresis, the purified protein was treated with 0.5 M Tris(hydroxypropyl)phosphine solution (Novagen) in order to dissociate any dimers. After electrophoresis, proteins were either stained using the Stain Plus kit (Bio-Rad) or transferred onto a nitrocellulose membrane. Immunoblotting experiments were performed with mouse anti-His6 monoclonal antibody (Roche Molecular Biochemical), which allows the detection of His6-tagged recombinant proteins, according to the manufacturer's protocol.
Hrp phenotype tests in planta.
Hypersensitive response and pathogenicity assays were performed as described by Hopkins et al. (26). Xanthomonas bacteria grown in NA broth with appropriate antibiotics to 108 CFU/ml were infiltrated into tobacco leaves (Nicotiana tobacum cultivar NC89) by using needleless syringes and inoculated into leaves of adult rice plants (IR24, susceptible to the pathogen) by using leaf needling for lesion length measurement or needleless syringes for detection of water soaking in rice seedlings. Plant responses were scored at 24 h (for HR), 3 days (for water soaking), and 14 days (for pathogenicity) after inoculation. Scores are the means of those for three leaves. All plants were grown in growth chambers at 28°C with a 12-h photoperiod. Experiments were repeated at least three times.
RT-PCR assay.
The expression of the hpa1 gene of X. oryzae pv. oryzicola was assayed by reverse transcription-PCR (RT-PCR) with the primers hpa1-F1 and hpa1-R1 (Table 2). First, the xanthomonad bacteria were preincubated in NA medium for 16 h, suspended at an optical density of 600 nm of 2.0 in sterilized water, and washed twice. Then, 40 μl of this bacterial suspension was inoculated into 1 ml of the modified XOM3. As a template, total RNA from the bacteria was prepared using the RNeasy plant minikit (QIAGEN). Reverse transcription and PCR for the hpa1 gene with the primers were performed using ReverTra Ace (TaKaRa, China) according to the manufacturer's directions.
Generation of hrpG, hrpX, prhA, and hrpF mutants of X. oryzae pv. oryzicola.
The hrpG, hrpX, prhA, and hrpF mutants of X. oryzae pv. oryzicola were constructed using the methodology described by Mongkolsuk et al. (38, 39). To create a polar insertion of hrpX, the pBCX147 plasmid was constructed in the following manner. A 1,430-bp fragment of the hrpX gene was amplified (with primers described in Table 2) using gDNA of strain RS105 as a template. The product was ligated into the vector pMD-18-T and digested with PstI and KpnI. The excised 147-bp fragment was inserted into the PstI and KpnI sites of pBC SK(−), giving pBCX147. Using the same strategy, a 121-bp fragment from the hrpG gene (primers are described in Table 2) was cloned into the EcoRI and SacI sites of pBC SK(−), a 112-bp fragment from the prhA gene (primers are described in Table 2) was cloned into the PstI and SacII sites of pBC SK(−), and a 351-bp fragment from p6-3 was ligated into the BamHI and PstI sites of pBC SK(−), giving pBCG121, pABC, and pFBC, respectively. Subsequently, the reconstructed plasmids were electroporated into strain RS105, using the methodology described by Mongkolsuk et al. (39), and single transformants were selected on chloramphenicol-NA plates after a 4-day incubation. Mutants at a single crossing with marker exchange were identified by Southern blotting with corresponding probes (unpublished data), and these strains were termed RABC for the prhA mutant, RCX for the hrpX mutant, RCG for the hrpG mutant, and RFBC for the hrpF mutant, respectively.
Nucleotide sequencing and data analysis.
The inserted DNA in p6 digested with single EcoRI or KpnI enzymes was mapped physically first and then subcloned into pBluescript II SK(−) prior to sequencing. Universal and reverse primers were used for the primary reactions, and synthesized primers were then used to sequence both strands completely. DNA sequencing was performed using ABI PRISM dideoxy terminator kits and analyzed on an ABI model 373A automated sequencer in TaKaRa (Dalian, China). The sequence data were analyzed with the BLAST program at the National Center for Biotechnology Information, MEGALIGN software (DNASTAR), and Vector NTI software (Invitrogen). Since there are no regions containing the hpa2 gene and the hpa1 promoter in p6, we PCR amplified a 1,356-bp fragment containing the hpa2 and hpa1 genes with the primers hpa2-F and hpa1-R (Table 2), using X. oryzae pv. oryzicola gDNA as the template. The fragment was ligated into pMD-18-T vector to give pHpa21. Using primers outside the fragment, with X. oryzae pv. oryzicola gDNA as the template, we were able to confirm that the amplified DNA was native to X. oryzae pv. oryzicola (data not shown).
Nucleotide sequence accession numbers.
The core hrp cluster, the cluster of the hrpG and hrpX genes, and the prhA gene have been assigned GenBank accession numbers AY875714, AY272885, and AY129230, respectively.
RESULTS
DNA sequence and putative ORFs of the core hrp region.
Based on the sequence of the hrp cluster of X. oryzae pv. oryzae (accession no. AB115081), we PCR amplified a 1.5-kb hrpA operon with the primers hrpA-F1 and hrpA-R1 (Table 2) and a 1.62-kb hrpF gene fragment with the primers hrpF-F1 and hrpF-R1 (Table 2) from the gDNA of X. oryzae pv. oryzae and used these as probes for screening a genomic library for the hrp cluster of X. oryzae pv. oryzicola. In situ hybridization with the probes used for screening the genomic library established that p6 was an hrp-positive clone (data not shown). Furthermore, a Southern hybridization with the two probes and the p6 clone, digested with EcoRI, indicated that the clone contained the hrp region from the hrpA to the hrpF operons (Fig. 1). Subsequent subcloning and sequencing demonstrated that clone p6 contained a 26,243-bp hrp cluster of X. oryzae pv. oryzicola, comprising 26 open reading frames (ORFs) that included the region from the partial hpa1 gene to the hpaF gene (Fig. 1 and 2; Table 3). There were two genes for a transposase and an IS1748 sited at the right border of hpaF (Fig. 1). In contrast to the case for X. campestris pv. vesicatoria (12), X. axonopodis pv. glycines (31), and X. axonopodis pv. citri (16), there are no tRNAs bordering after the hpaF gene (data not shown). The hpa2 and hpa1 genes were directly adjacent to the left of the hrpA (Fig. 1). Therefore, the length of the core hrp cluster from hpa2 to hpaF is about 27 kb.
FIG. 1.
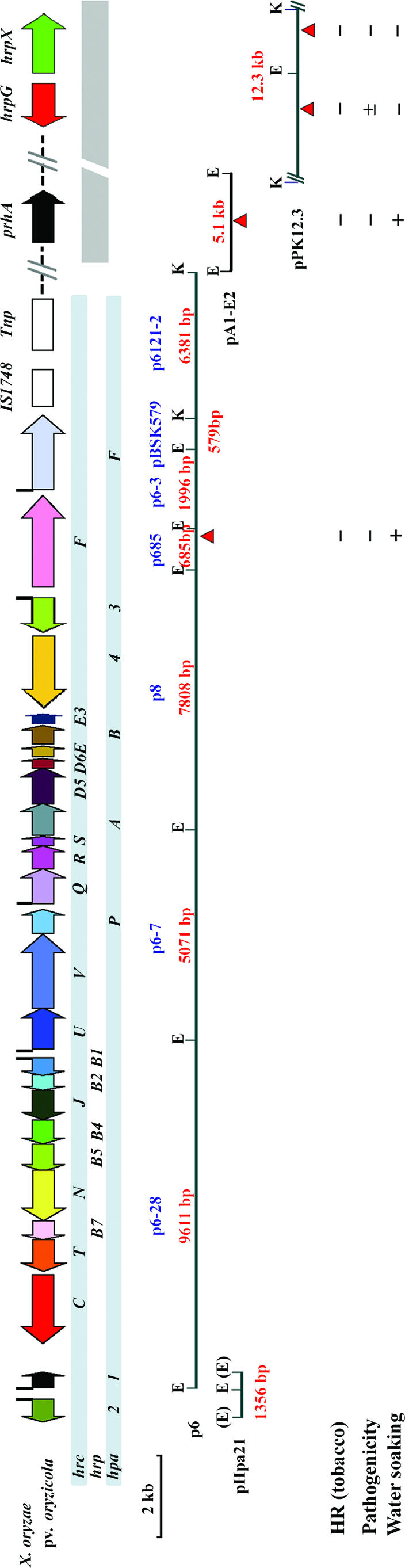
Genetic organization and restriction maps of the Hrp clusters of X. oryzae pv. oryzicola, which were cloned in p6, pPha21, pA1-E2, and pPK12.3. Colored open arrows indicate the positions and orientations of the hrp, hrc, and hpa genes. Black rectangles above open arrows indicate the positions of the PIP boxes. Red arrows below the plasmid maps indicate the positions and orientations of the marker-exchanged insertions, and the major phenotypes of the mutants are represented below the corresponding restriction maps. −, either no HR in tobacco or no pathogenicity in adult rice plants and no water-soaking symptoms in rice seedlings. ±, weak pathogenicity in formation of short lesion length in adult rice plants but no water-soaking symptoms in rice seedlings (data not shown). Tnp, transposase. Enzyme sites from the vector are shown in parentheses. E, EcoRI; K, KpnI.
FIG. 2.
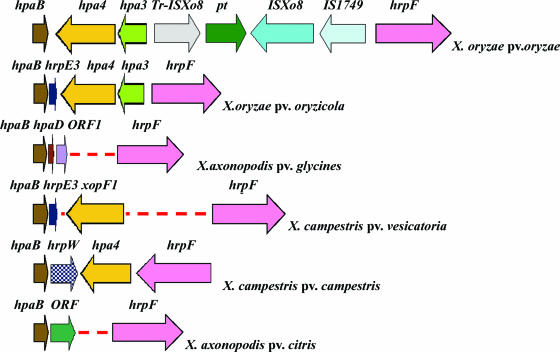
Comparisons of the hpaB-hrpF interspace region in the core hrp clusters of six Xanthomonas species or pathovars. Colored arrows display the orientations and genetic organizations of genes in the hpaB-hrpF region. The gene size and organization are based on the sequence from the GenBank database, with accession numbers AB115081 for X. oryzae pv. oryzae, AY875714 for X. oryzae pv. oryzicola, AF499777 for X. axonopodis pv. glycines, AF056246 for X. campestris pv. vesicatoria, AE008922 for X. campestris pv. campestris, and AE008923 for X. axonopodis pv. citri. pt, putative transposase.
TABLE 3.
Nucleotide sequence homology between clustered hrp genes of Xanthomonas oryzae pv. oryzicola and the hrp genes of X. oryzae pv. oryzae
| Gene | Size (bp) in:
|
% Identical nucleotides compared with X. oryzae pv. oryzae | Gap (bp) | PIP boxa
|
||
|---|---|---|---|---|---|---|
| X. oryzae pv. oryzicola | X. oryzae pv. oryzae | Sequence | Position | |||
| hpa2 | 564 | 474 | 98 | 90 | I | 147 |
| hpa1 | 414 | 420 | 86 | 12 | P | 160 |
| hrcC (hrpA) | 1,824 | 1,818 | 97 | 6 | ||
| hrcT (hrpB8) | 831 | 831 | 98 | 0 | ||
| hrpB7 | 510 | 510 | 97 | 0 | ||
| hrcN (hrpB6) | 1,329 | 1,329 | 98 | 0 | ||
| hrpB5 | 702 | 702 | 98 | 0 | ||
| hrpB4 | 630 | 630 | 97 | 0 | ||
| hrcJ (hrpB3) | 765 | 762 | 94 | 3 | ||
| hrpB2 | 393 | 393 | 96 | 0 | ||
| hrpB1 | 456 | 456 | 98 | 0 | P | 121 |
| hrcU (hrpC1) | 1,074 | 1,080 | 97 | 6 | P | 96 |
| hrcV (hrpC2) | 1,938 | 1,938 | 99 | 0 | ||
| hpaP (hrpC3) | 618 | 618 | 97 | |||
| hrcQ (hrpD1) | 915 | 915 | 97 | P | 273 | |
| hrcR (hrpD2) | 645 | 645 | 98 | |||
| hrcS (hrpD3) | 261 | 261 | 98 | |||
| hpaA (hrpD4) | 828 | 828 | 96 | |||
| hrpD5 | 939 | 939 | 96 | |||
| hrpD6 | 243 | 243 | 97 | |||
| hrpE | 282 | 282 | 97 | |||
| hpaB | 471 | 471 | 97 | |||
| hrpE3 | 260 | |||||
| hpa4 | 1,932 | 1,986 | 95 | 56 | ||
| hpa3 | 447 | 447 | 96 | 0 | II | 121 |
| hrpF | 2,409 | 2,409 | 98 | 0 | ||
| hpaF | 2,409 | 2,040 | 97 | 9 | III | 239 |
| IS1748 | 1,062 | |||||
| Putative transposase gene | 1,059 | 1,059 | 97 | 0 | ||
A PIP box or similar sequence upstream of the putative translational start site of the gene. I, imperfect PIP box TTCGC-N15-TTCGT; P, PIP box TTCGC-N15-TTCGC; II, imperfect PIP box TTCGT-N15-TTCGC; III, imperfect PIP box TTCGC-N9-TTCGC.
The complete DNA sequence of the core hrp cluster of X. oryzae pv. oryzicola revealed that there were 10 hrp genes, 9 hrc genes, and 8 hpa genes (Fig. 1; Table 3). For most genes, there was 86% to 99% identity in the nucleotide sequences for each ORF between the two pathovars of X. oryzae (Table 3). However, significant differences were found in the sequences of four genes: the ORF for the hpa2 genes was 90 bp larger at the 5′ end than that of X. oryzae pv. oryzae, the hpa1 gene had only 86% similarity with that of X. oryzae pv. oryzae, the hpa4 gene lacked a 63-bp fragment that existed in hpa4 of X. oryzae pv. oryzae, and the 5′ end of hpaF was 96 bp longer than that in X. oryzae pv. oryzae (accession numbers AY536514 and AB115081) (Table 3). In total, there were seven predicted PIP boxes in the hrp cluster. Four genes (hpa1, hrpB1, hrcU, and hrcQ) had perfect PIP boxes (TTCGC-N15-TTCGC), while three genes (hpa2, hpa3, and hpaF) had imperfect PIP boxes (hpa2, TTCGC-N15-TTCGT; hpa3, TTCGT-N15-TTCGC; hpaF, TTCGC-N9-TTCGC) in their putative promoter regions (Fig. 1; Table 3). The imperfect PIP box in the hpaF locus was found only in X. oryzae pv. oryzicola among Xanthomonas species or pathovars, including X. oryzae pv. oryzae. Additionally, Hpa1 of X. oryzae pv. oryzicola had 23.1% identity to PopA1 of R. solanacearum and 42% to 72.1% identity to Hpa1 proteins of other Xanthomonas species or pathovars (see Fig. 6), indicating that the hpa1 locus is variable in xanthomonads.
FIG. 6.
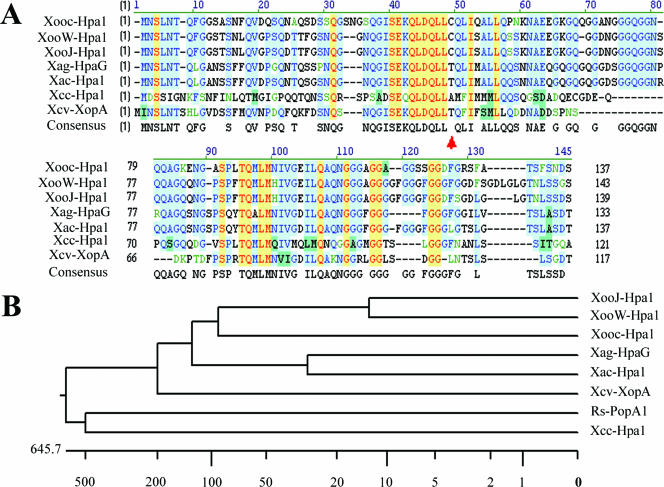
Comparison of Hpa1 proteins among xanthomonads and Ralstonia solanacearum. A neighbor-joining bootstrap tree is derived from the amino acid sequences of all the Hpa1 proteins in plant-pathogenic bacteria by using the Vector NTI align program. The protein names are indicated as the abbreviated names of the corresponding plant-pathogenic bacteria. The abbreviations are as follows, with accession numbers in GenBank or Gene ID in parentheses: Xooc, X. oryzae pv. oryzicola (AY875714); XooJ, in strain MAFF301237 of X. oryzae pv. oryzae (NC006834); XooW, in strain PXO99A of X. oryzae pv. oryzae (AB115081); Xag, X. axonopodis pv. glycines (AF4997777); Xac, X. axonopodis pv. citri (Xac0416); Xcv, X. campestris pv. vesicatoria (U33548); Rs, Ralstonia solanacearum (AB026629); and Xcc, X. campestris pv. campestris (Xcc1240). A. Sequence alignment of xanthomonad Hpa1 proteins. The red arrow indicates the position of the cysteine residue in Hpa1 of X. oryzae. B. Phylogenetic relationship of Hpa1 protein among xathomonads and R. solanacearum.
The interspace region from hpaB to hrpF is variable in the highly conserved hrp cluster of xanthomonads.
We used a combination of BLAST, PSI-BLAST, and FASTA programs to search for homology in the deduced amino acid sequence of each Hrp protein of X. oryzae pv. oryzicola and to predict its site of localization in the bacterial or plant cell (data not shown). The 20 genes from hrcC to hpaB, which were the main genes for the TTSS, of the core region were present in all six Xanthomonas species tested and were highly conserved with the corresponding genes from X. oryzae pv. oryzae (Table 3). In contrast to the corresponding region in X. oryzae pv. oryzae, we could not find an insertion sequence or a transposase in the interspace region between hpaB and hrpF, which encodes the hrpE3, hpa4, and hpa3 genes (Fig. 2). The hrpE3 gene, which encodes an 86-amino-acid protein, is highly homologous to hrpE3 of X. campestris pv. vesicatoria. The hpa4 gene, which encodes a 643-amino-acid protein that is predicted to be soluble and located in the cytoplasm, was homologous to hpa4 in X. oryzae pv. oryzae (accession no. AB115081) (49), which is also located in the interspace region between hpaB and hrpF of the hrp gene cluster (Fig. 2). A homology search revealed a 254-residue protein with homology to the protein XopF1 in X. axonopodis pv. citri (AE011919-5) (16) (Fig. 2). The hpa3 gene, which is predicted to encode a 148-residue lipoprotein that is anchored to the inner and/or outer membrane, is similar to hpa3 of X. oryzae pv. oryzae but differs by the presence of an imperfect PIP box (TTCGT-N15-TTCGC) that is not present in the promoter region in X. oryzae pv. oryzae (49). However, the hrpW homolog from P. syringae pathovars and Erwinia amylovora were present only in the interspace region of X. campestris pv. campestris and not in X. oryzae pv. oryzicola (Fig. 2). In contrast to the case for other xanthomonads, there is an IS1748 and a gene for a putative transposase following the hpaF gene (Fig. 1) in X. oryzae pv. oryzicola. There are no tRNAs after the hpaF gene in X. oryzae pv. oryzicola, in contrast to those found in X. campestris pv. vesicatoria, X. axonopodis pv. glycines, and X. axonopodis pv. citri (data not shown).
Hrp regulatory genes, hrpG, hrpX, and prhA, are outside of the core hrp cluster.
To isolate the Hrp regulatory genes of X. oryzae pv. oryzicola, we transferred individual clones of the RS105 genomic library into the hrp mutants M55 and M1005, which were previously confirmed as hrpX and hrpG mutants (14), and isolated a cosmid clone (pPK12.3) with the function of restoring the hrp mutants to HR induction in tobacco and pathogenicity in rice. A restriction enzyme digestion analysis established that pPK12.3 had an insert of approximately 12.3 kb (Fig. 1), while a Southern blot confirmed that the inserted fragment was colinear with the RS105 genome (data not shown). The complete DNA sequence of the 12.3-kb insert in pPK12.3 was determined and found to contain two clustered hrp regulatory genes, hrpG and hrpX, which were highly conserved with those in other xanthomonads (data not shown). However, the sequences on either side of the hrpG and hrpX locus had no homology to hrp genes in other xanthomonads, indicating that the hrpG and hrpX genes are clustered but located outside the core hrp cluster, elsewhere in the chromosome (Fig. 1).
We were interested in identifying a gene encoding a putative siderophore receptor similar to PrhA in Ralstonia solanacearum (35), which acts as a sensor that detects the plant cell wall, triggering the transcriptional activation of bacterial virulence genes (1). Using the phrA gene in a BLAST search, we identified a homolog that encodes a putative siderophore receptor in X. oryzae pv. oryzae (accession number AF325732). Based upon this sequence, we designed primers (Table 2) and PCR amplified the gene from X. oryzae pv. oryzae, which was used to construct a prhA mutant of X. oryzae pv. oryzicola that was conjugated with each clone of the RS105 genomic library (unpublished data) in order to identify the adjacent genes and to determine whether they are related to any genes involved in Hrp phenotypes, HR induction in nonhost plants, and pathogenicity in host plants. The prhA mutant lost HR induction in tobacco and pathogenicity in rice (Fig. 3). The subsequent determination of the phenotypes of the conjugants in tobacco and rice led to the isolation of a cosmid clone (pA1) that restored the ability of the mutant to cause HR induction in tobacco and pathogenicity in rice (Fig. 3). A subclone, pA1-E2, which harbored a 5.1-kb DNA fragment was rescued as the smallest DNA fragment with the ability to restore the Hrp phenotype to the mutant (Table 1; Fig. 1). The complete sequence of this fragment showed that it contained a gene, which we term prhA, encoding a putative siderophore receptor which had 29% identity to PrhA of R. solanacearum (35). The sequences adjacent to the prhA locus had no homology with hrp genes in other xanthomonads (data not shown).
FIG. 3.
Phenotypic analysis of a prhA mutant derived from X. oryzae pv. oryzicola. The mutation in the prhA gene resulted in the loss of the hypersensitive response in tobacco (Nc89) (C) and pathogenicity in adult rice (A), but the ability to cause water-soaking symptoms in rice seedlings (3 weeks old, IR24) was retained (B). 1, wild-type strain RS105 with an empty vector pUFR034; 2, prhA mutant RFBC produced by marker exchange via a single crossover event when the reconstructed plasmid pFBC was electroporated into the wild-type strain RS105; 3, the conjugate of the prhA mutant complemented with plasmid pA1 harboring the prhA gene of X. oryzae pv. oryzicola.
Phenotypes of the hrpF, hrpG, hrpX, and prhA mutants.
Most of the hrp, hrc, and hpa genes of Xanthomonas oryzae have been mutated in an attempt to understand their roles in the hypersensitive response in nonhosts and pathogenicity in rice (41, 49, 62). To elucidate the hrp regulatory cassette for the core hrp cluster of X. oryzae pv. oryzicola, we constructed hrpF, prhA, hrpG, and hrpX mutants by using the homologous recombination and marker exchange methodology described by Rabibhadana et al. (42). Each mutant was confirmed by Southern hybridization (data not shown) and assayed for HR induction in tobacco and pathogenicity in rice. Aside from the fact that the hrpF, hrpX, and prhA mutants had lost their ability to induce the HR in tobacco and pathogenicity in adult rice, interestingly, the hrpF and prhA mutants, when infiltrated into leaves of rice seedlings (21 days old, cultivar IR24), retained the ability to trigger water soaking comparable to that caused by the wild-type strain RS105 (Fig. 3, 4, and 5). In contrast, the hrpG mutant had completely lost its ability to cause water soaking in IR24 seedling leaves and HR induction in tobacco but retained weak pathogenicity in the form of short lesion length when inoculated into leaves of adult rice plants by the leaf-needling method (Fig. 4). All of the mutant strains were complemented in HR induction and pathogenicity with the clones that incorporated the corresponding genes: p6 complemented the hrpF mutant (Fig. 1 and 5), pA1-E2 complemented the prhA mutant (Fig. 1), and pPK12.3 complemented the hrpG and hrpX mutants (Fig. 1). Since the three clones p6, pA1-E2, and pPK12.3 were nonoverlapping clones, derived from the RS105 genomic library, this confirms that the Hrp phenotype is controlled by three separate DNA regions: a core hrp cluster, an hrpG and hrpX cluster, and a prhA locus. The identification of genes in addition to prhA in pA1-E2 is ongoing.
FIG. 4.
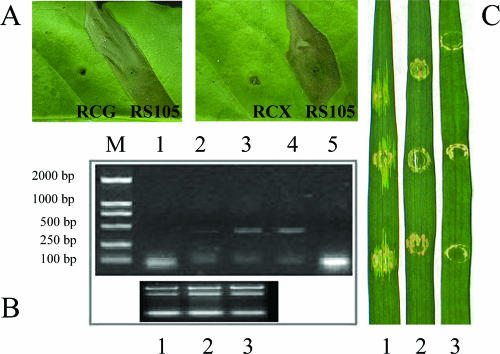
The hrpX gene of X. oryzae pv. oryzicola controls expression of the hpa1 gene in XOM3 medium. Xanthomonas strains were incubated in XOM3, and the phenotypes of the hrp regulatory gene mutants, hrpX and hrpG, were tested in tobacco leaves (cultivar Nc89) (A) and in rice (IR24) (C), respectively. The induction of hpa1 expression in XOM3 medium was identified by RT-PCR (B), which was performed using total RNA as the template. The hrpX mutant RCX (1), the hrpG mutant RCG (2), and the wild-type strain RS105 (3) were grown at 28°C in XOM3 (pH 6.5). Total RNA was extracted from each bacterium after incubation for 16 h. Specific primers that amplified a 414-bp DNA fragment corresponding to the hpa1 gene were used for RT-PCR. PCR products (B, top) and rRNA (B, bottom) were separated by agarose gel electrophoresis and stained with ethidium bromide. A positive PCR control using RS105 gDNA as the template (4) and a negative PCR control with no RT (5) are also indicated.
FIG. 5.
Phenotypic analysis of the hrpF mutant derived from X. oryzae pv. oryzicola. The hrpF mutant had lost pathogenicity in adult rice (IR24) when inoculated into leaves by using leaf needling for lesion length measurements (B) and had lost the hypersensitive response in tobacco (Nc89) when infiltrated into leaves with needleless syringes (C), but it retained to the ability to cause water-soaking symptoms in rice seedlings (A). A. Water-soaking symptoms caused by the hrpF mutant. The third leaf of 14-day-old IR24 seedlings was infiltrated using needleless syringes individually with the wild-type strain RS105 harboring pUFR034 (empty plasmid) (1), RFBC (hrpF mutant) (2), RFBC harboring p6 (the core hrp cluster) (3), and RFBC harboring pHrpF (the hrpF gene) (4). The water-soaking symptoms after 3 days of infiltration are shown. B. Measurements of lesion length caused by hrpF mutants. The third leaf of IR24 adult plants was inoculated with corresponding bacteria as described above, using leaf needling. The lesion lengths after 14 days are shown. C. Hypersensitive response in tobacco induced by hrpF mutants. The leaves were infiltrated using needleless syringes with the bacteria as described above, and the reaction was recorded within 24 h. Three replicates were conducted for identification of the phenotypes.
Hpa1 is a TTSS-dependent HR elicitor.
Hpa1 is a harpin-like protein encoded by the hpa1 gene of X. oryzae pv. oryzae (61) and is considered to be an Hrp type III-secreted elicitor (31). The sequence of the putative Hpa1 protein from X. oryzae pv. oryzicola had quite high similarities with Hpa1 of X. oryzae pv. oryzae (71.3% to 72.1% identity), Hpa1 of X. axonopodis pv. citri (67.4% identity), HpaG of X. axonopodis pv. glycines (64.4% identity), XopA of X. campestris pv. vesicatoria (53.0% identity), Hpa1 of X. campestris pv. campestris (42.0% identity), and PopA1 of R. solanacearum (23.1% identity) (Fig. 6). In contrast to XopA (8% glycine) of X. campestris pv. vesicatoria and Hpa1 (13% glycine) of X. campestris pv. campestris, Hpa1 (21% glycine) of X. oryzae pv. oryzicola, HpaG (21% glycine) of X. axonopodis pv. glycines, Hpa1 (22% glycine) of X. axonopodis pv. citri, and Hpa1 (26% glycine) of X. oryzae pv. oryzae had relatively high glycine contents (Fig. 6). Another significant difference was that both Hpa1 of X. oryzae pv. oryzicola and Hpa1 of X. oryzae pv. oryzae had a cysteine residue that is not present in Hpa1 or HpaG from other xanthomonad species (Fig. 6). Indeed, a feature of the harpins found in P. syringae pathovars, Erwinia species, R. solanacearum, and other xanthomonads is that they do not possess cysteine residues. A phylogenetic analysis of the relationship of Hpa1 with others of xathomonads and R. solanacearum demonstrated that Hpa1of X. oryzae pv. oryzicola is grouped with Hpa1 of X. oryzae pv. oryzae (Fig. 6).
To explore how the expression of the hpa1 gene is modulated in X. oryzae pv. oryzicola, an hrp-inducing medium, XOM3, was developed based on XOM2, which was used for X. oryzae pv. oryzae (50). Total RNAs were extracted from the Xanthomonas strain RS105 and the hrpG and hrpX mutants growing in NA and XOM3 media, respectively, and used as templates for RT-PCR assays of the expression of the hpa1 gene. This analysis indicated that hpa1 expression is induced by the nutrient-poor medium XOM3 (Fig. 4) but not by a nutrient-rich medium, NA (data not shown), and that the expression of hpa1 in X. oryzae pv. oryzicola is under the control of the hrpX gene, because the hpa1 gene was not expressed when the hrpX gene was mutated (Fig. 4). The expression level of the hpa1 gene in the wild type was higher than that in the hrpG mutant of X. oryzae pv. oryzicola (Fig. 4), suggesting that the hrpG gene participated in regulating the expression of hpa1 when the bacterium was grown in the hrp-inducing medium for 16 h.
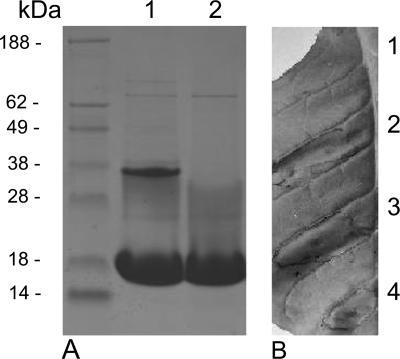
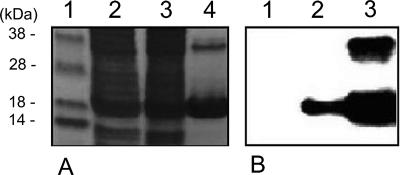
In order to investigate whether Hpa1 of X. oryzae pv. oryzicola had HR elicitor activity in the nonhost tobacco, Hpa1 that was expressed and purified from E. coli (Fig. 7A) was injected (at, e.g., 1 μg/ml) into tobacco leaves, where it was found to elicit the HR (Fig. 7B). Interestingly, the purified Hpa1 existed both as a monomer and a dimer (Fig. 7A), possibly due to cross-linking of the cysteine residue in Hpa1 (Fig. 6A). An immunoblotting analysis was performed to determine if Hpa1 secretion was via the type III secretion system; Hpa1 was not detected in the disrupted cells of the hrpX mutant harboring the pUhpa1 plasmid that carries the hpa1 gene, whereas it was detected both in the disrupted cells and in the culture supernatant of the wild-type strain (Fig. 8), thus indicating that Hpa1 is a TTSS effector that triggers the HR in nonhost tobacco.
FIG. 7.
Hypersensitive reaction of the purified Hpa1 protein of X. oryzae pv. oryzicola. A. A 12.5% SDS-polyacrylamide gel showing Hpa1 that was expressed and purified from E. coli (BL21). Two bands are apparent in lane 1, while only one band is apparent in lane 2 after treatment of the protein with 0.5 M Tris(hydroxypropyl)phosphine (Novagen), suggesting that the higher-Mr band is a dimer due to cysteine cross-linking. B. Comparison of Hpa1 activity with that of well-known harpin HrpN. 1, BL21 harboring empty vector pET21a; 2, RS105 (108 CFU/ml); 3, Hpa1 (1 μg/ml); 4, HrpN (5 μg/ml) (kindly provided by Z. Wei, Eden Biotech). The tobacco leaves were photographed 24 h after injection using needleless syringes. Three replicates of the assay were conducted. The first lane in panel A is the protein marker.
FIG. 8.
In vitro secretion analysis of X. oryzae pv. oryzicola expressing Hpa1. Purified proteins from disrupted cells and the culture supernatant were analyzed by 12.5% SDS-PAGE (A) and immunoblotted with the monoclonal antihexahistidine antibody (B). The plasmid pUhpa1 was used to express the His-tagged Hpa1 in bacteria that were cultured in XOM3 for 16 h. Lanes 1, disrupted cells of the hrpX mutant RCX harboring pUhpa1; lanes 2, disrupted cells of the wild-type RS105 harboring pUhpa1; lanes 3, supernatant of the wild-type RS105 with plasmid pUhpa1. The Hpa1 monomer and dimer are shown as the lower and upper bands in lanes 3. The first lane in panel A is the protein marker.
DISCUSSION
Here we report on the isolation of a core cluster of hrp genes, a clustered pair of hrp regulatory genes (hrpX and hrpG), and a prhA gene from the Chinese strain RS105 of X. oryzae pv. oryzicola. Mutations in the isolated hrp genes, hrp regulatory genes, and a prhA gene of X. oryzae pv. oryzicola resulted in a loss of pleiotropic phenotypes for both the ability to induce the HR in nonhost tobacco and pathogenicity in host rice. When the nucleotide sequence for the core hrp cluster from X. oryzae pv. oryzicola was compared with those from X. oryzae pv. oryzae (41, 49), X. campestris pv. campestris and X. axonopodis pv. citri (17), X. campestris pv. vesicatoria (8, 28, 40), and X. axonopodis pv. glycines (31), we found that, in common with other xanthomonads, the hrc genes encoding the TTSS apparatus were completely conserved. A comparison of the Hrp pathogenicity islands of six Xanthomonas species demonstrated variabilities in the hpaB-hrpF interspace region (Fig. 2). Only the hrpE3, hpa4, and hpa3 genes were present in the hpaB-hrpF region of X. oryzae pv. oryzicola, which differed greatly from the regions of the other five Xanthomonas species. The hpa4 and hpa3 genes appear to be linearly placed within an operon that is preceded by a PIP box (TTCGT-N15-TTCGC) located 121 bp upstream of the putative operon (Table 3). This indicates that the interspace region is part of the core hrp cluster of X. oryzae pv. oryzicola and that the expression of the hpa4 and hpa3 genes is regulated by HrpX. However, direct evidence for the involvement of hrpE3, hpa4, and hpa3 in the pathogenicity of X. oryzae pv. oryzicola is required. Previous studies have shown that mutations in either hpa4 or hpa3 of X. oryzae pv. oryzae did not affect the virulence (49) and that the product of xopF1, an homolog of hpa4, is a TTSS effector of X. campestris pv. vesicatoria (43). The variability in the hpaB-hrpF region revealed in this report suggests that, in comparison to the hrp clusters of other xanthomonads, the core hrp cluster of X. oryzae pv. oryzicola is novel.
The core hrp cluster of gram-negative plant pathogenic bacteria forms a TTSS apparatus to deliver virulence effectors into the host cytosol that interfere with and alter host processes. The proposed function of HrpF as a translocon and as a secreted protein places the protein at the plant-bacterium interface (13, 49). Interestingly, mutation of the hrpF locus of X. oryzae pv. oryzicola resulted in the loss of pathogenicity in rice and the ability to induce HR in nonhost tobacco. This was consistent with previous reports that mutations in hrpF of X. campestris pv. vesicatoria or X. axonopodis pv. glycines resulted in strains that were nonpathogenic in host plants and unable to elicit race-specific HRs (13, 31). This contrasts with the behavior of the hrpF mutant of X. oryzae pv. oryzae, which retained its pathogenicity but displayed both a reduced ability to grow within rice and a reduced ability to cause lesions (49). However, in common with the hrpF mutant of X. oryzae pv. oryzae, our investigation revealed that the hrpF mutant of X. oryzae pv. oryzicola still caused water-soaking symptoms in susceptible rice seedlings (Fig. 5). Importantly, the mutation in the hrpF gene of X. oryzae pv. oryzae had no effect on HR induction in rice when there was an avr gene in the pathogen that correspondingly matched an R gene in rice (49). The production of water-soaking symptoms in susceptible rice seedlings is one of the functions of avrBs3 family members in X. oryzae pv. oryzae (58). In this study, we found that mutation of the hrpX gene led to a complete loss of pathogenicity, not only as measured by lesion length in adult rice plants but also in a lack of ability to cause water-soaking symptoms in rice seedlings (Fig. 1 and 4). However, the hrpG mutant retained weak pathogenicity in lesion length only at the inoculation site and had completely lost its ability to cause water soaking in rice seedlings (Fig. 4). Recently, it was reported that there are diverse members of the avrBs3 family in X. oryzae pv. oryzicola (15, 62). Considering the fact that the expression of hrpA and hrpX is activated by HrpG and the activation of hrpB-hrpF operons by HrpX established in other xanthomonads (51, 54, 56, 57), we postulate that AvrBs3 family members, the critical TTSS effectors, are not delivered through the HrpF translocon of X. oryzae pv. oryzicola into rice cells, but further evidence is needed to support this postulate. Importantly, the prhA mutant of X. oryzae pv. oryzicola retained the ability to cause water-soaking symptom in rice seedlings, indicating that the prhA gene had no effect on the roles of avrBs3 family members in X. oryzae pv. oryzicola. Therefore, modulating the expression of avrBs3 family genes in xanthomonads at the hrpF and prhA loci should elucidate the process of secretion when X. oryzae pv. oryzicola interacts with rice.
Our knowledge and understanding of the hrp regulatory cascade in X. oryzae pv. oryzicola are still rudimentary. Previous studies revealed that the hrp genes in xanthomonads, which are induced in plants, are not expressed when bacteria are grown in rich media but are strongly expressed in media that mimic the plant apoplastic medium (35, 47, 48, 50, 55). In this report, we developed an XOM2-based hrp-inducing medium, XOM3, that is suitable for X. oryzae pv. oryzicola (50). The hpa1 gene, as an indicator detected by RT-PCR, was expressed strongly in the wild-type strain and constitutively in the hrpG mutant but was not expressed in the hrpX mutant when the bacteria were grown on XOM3 (Fig. 4). The His-tagged Hap1 was detected by immunoblotting in the supernatant and disrupted cells of the wild-type strain that had been transformed with the pUhpa1 plasmid and grown in XOM3, while it was not detected in disrupted cells of the hrpX mutant (Fig. 8). However, a prhA mutant of X. oryzae pv. oryzicola, which had lost the ability to trigger HR in tobacco and pathogenicity in adult rice but retained the ability to cause water-soaking symptoms in rice seedlings, had no effect on hpa1 expression in XOM3 (data not shown). The phenotypes in tobacco and rice caused by the prhA mutant were not comparable to those caused by the prhA mutant of R. solanacearum. In R. solanacearum, PrhA perceived plant cell wall-derived signals during the bacterium-plant interaction and activated hrp gene expression through a six-gene regulatory cascade (e.g., prhA, prhR, prhI, prhJ, hrpG and hrpB) (1, 10). Recently, Genin et al. (21) reported that the repression of hrp genes in nutrient-rich medium was relieved in a phcA mutant of R. solanacearum and that the Prh plant-responsive pathway and an unidentified minimal medium pathway connected hrp gene regulation to the global virulence regulator PhcA at the branching point of HrpG. Therefore, further genetic evidence is required in order to elucidate the hrp regulatory network in xanthomonads, especially in X. oryzae pv. oryzicola.
The expression of some TTSS effector genes that have a PIP box with the consensus nucleotide sequence TTCGC-N15-TTCGC in their promoter region is HrpX dependent (7, 18, 51). In analogy to hpa1 in X. oryzae pv. oryzae, we found that there was a PIP box in the promoter region of the hpa1 gene of X. oryzae pv. oryzicola. The hpa1 gene was not expressed in the hrpX mutant when the bacterium was grown in the hrp-inducing medium XOM3 but was expressed in the hrpG mutant at a much lower level. Moreover, immunoblotting failed to detect the Hpa1 protein in the hrpX mutant of X. oryzae pv. oryzicola, consistent with the proposal that Hpa1 is a TTSS effector and that the transcriptional expression of the hpa1 gene was directly controlled by HrpX.
Harpins constitute a family of secreted effector proteins which are translocated via the type III pathway in plant-pathogenic bacteria, triggering disease resistance-associated responses, such as hypersensitive cell death, and thus activating the plant's surveillance system (20, 32). Genes encoding such proteins have been identified in E. amylovora and Erwinia chrysanthemi (hrpN) (52); P. syringae pv. syringae, tomato, and glycinea (hrpZ) (25); R. solanacearum (popA) (4); X. axonopodis pv. glycines (hpaG) (31); X. campestris pv. vesicatoria (xopA) (40); and X. oryzae pv. oryzae (hpa1) (53, 61). Prior to the present study, no harpin-like proteins had been isolated in X. oryzae pv. oryzicola. Zhao et al. (60) demonstrated that avrRxo1 of X. oryzae pv. oryzicola induced an HR on maize with Rxo1 but not on maize without Rxo1. In our study, the putative translation product of the hpa1 gene of X. oryzae pv. oryzicola was similar to the harpins mentioned above: it is a heat-stable, glycine-rich protein. The purified product of the hpa1 gene, which elicited an HR in tobacco at 1.0 μM, is the first HR-eliciting protein identified in X. oryzae pv. oryzicola.
Among the Hpa1 homologs of xanthomonads, only Hpa1 from X. oryzae pv. oryzicola, Hpa1 from X. oryzae pv. oryzae (53), and HpaG from X. axonopodis pv. glycines (31) have demonstrated harpin-like elicitor activity. Although all of the homologs exhibited high levels of identity at the amino acid level, two interesting differences were found in the amino acid alignment of Hpa1, HpaG, and XopA. There were two regions containing more glycine residues in Hpa1 of X. oryzae pv. oryzicola (Fig. 6A). HpaG lacked the residues GFGGG that corresponded to positions 114 to 117 in Hpa1 of X. oryzae pv. oryzicola. XopA lacked two glycine-rich regions that were present in Hpa1, suggesting that this region is critical for Hpa1 homologs to act as elicitors on nonhost plants. Currently, we are investigating which amino acid residues in Hpa1 are critical for conferring HR activity. Another notable difference is that only Hpa1 of X. oryzae pv. oryzicola and X. oryzae pv. oryzae possess a cysteine residue. The purified Hpa1 of X. oryzae pv. oryzicola, expressed both in X. oryzae pv. oryzicola and in E. coli, displayed two bands when run on SDS-polyacrylamide gels, which we have putatively assigned as the monomer and dimer. We postulated that these oligomers are due to cross-linking of the monomer due to disulfide bond formation, but we do not know if the dimer has any functional significance. In order to gain further insight into the functional role of Hpa1, we are using biophysical methods to determine if it undergoes structural changes upon exposure to artificial membranes, and we have succeeded in its crystallization. Such studies may lead us to determine if the harpin-induced HR is mediated by direct insertion of harpin into plant membranes or by a specific interaction with a proteinaceous or nonproteinaceous receptor.
Acknowledgments
We are grateful to Adan Bogdanov for critical discussions.
This study was supported by funds from the National Natural Science Foundation of China (grant 30370926), the Key Basic Science Research Project from Jiangsu Province (grant BK2001207), and the Key Science and Technology Project from the Ministry of Education of China (grant 106093).
REFERENCES
- 1.Aldon, D., B. Brito, C. Boucher, and S. Genin. 2000. A bacterial sensor of plant cell contact controls the transcriptional induction of Ralstonia solanacearum pathogenicity genes. EMBO J. 19:2304-2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alfano, J. R., and A. Collmer. 1997. The type III (Hrp) secretion pathway of plant pathogenic bacteria: trafficking harpins, Avr proteins, and death. J. Bacteriol. 179:5655-5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arlat, M., C. Gough, C. E. Barber, C. Boucher, and M. Daniels. 1991. Xanthomonas campestris contains a cluster of hrp genes related to the larger hrp cluster of Pseudomonas solanacearum. Mol. Plant-Microbe Interact. 4:593-601. [DOI] [PubMed] [Google Scholar]
- 4.Arlat, M., F. Van Gijsegem, J. C. Huet, J. C. Pernollet, and C. A. Boucher. 1994. PopA1, a protein which induces a hypersensitivity-like response on specific Petunia genotypes, is secreted via the Hrp pathway of Pseudomonas solanacearum. EMBO J. 13:543-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Astua-Monge, G., G. V. Minsavage, R. E. Stall, M. J. Davis, U. Bonas, and J. B. Jones. 2000. Resistance of tomato and pepper to T3 strains of Xanthomonas campestris pv. vesicatoria is specified by a plant-inducible avirulence gene. Mol. Plant-Microbe Interact. 13:911-921. [DOI] [PubMed] [Google Scholar]
- 6.Bogdanove, A. J., S. V. Beer, U. Bonas, C. A. Boucher, A. Collmer, D. L. Coplin, G. R. Cornelis, H. C. Huang, S. W. Hutcheson, N. J. Panopoulos, and F. van Gijsegem. 1996. Unified nomenclature for broadly conserved hrp genes of phytopathogenic bacteria. Mol. Microbiol. 20:681-683. [DOI] [PubMed] [Google Scholar]
- 7.Bonas, U. 1994. hrp genes of phytopathogenic bacteria. Curr. Top. Microbiol. Immunol. 192:79-98. [DOI] [PubMed] [Google Scholar]
- 8.Bonas, U., R. Schulete, S. Fenselau, G. V. Minsavage, B. J. Staskawicz, and R. E. Stall. 1991. Isolation of a gene cluster from Xanthomonas campestris pv. vesicatoria that determines pathogenicity and the hypersensitive response on pepper and tomato. Mol. Plant-Microbe Interact. 4:81-88. [Google Scholar]
- 9.Boucher, C. A., P. A. Bareris, A. Trigalet, and D. A. Demery. 1985. Transposon mutagenesis of Pseudomonas solanacearum: isolation of Tn5-induced avirulent mutants. J. Gen. Microbiol. 131:2449-2457. [Google Scholar]
- 10.Brito, B., D. Aldon, P. Barberis, C. Boucher, and S. Genin. 2002. A signal transfer system through three compartments transduces the plant cell contact-dependent signal controlling Ralstonia solanacearum hrp genes. Mol. Plant-Microbe Interact. 15:109-119. [DOI] [PubMed] [Google Scholar]
- 11.Brito, B., M. Marenda, P. Barberis, C. Boucher, and S. Genin. 1999. prhJ and hrpG, two new components of the plant signal dependent regulatory cascade controlled by PrhA in Ralstonia solanacearum. Mol. Microbiol. 31:237-251. [DOI] [PubMed] [Google Scholar]
- 12.Buttner, D., and U. Bonas. 2003. Common infection strategies of plant and animal pathogenic bacteria. Curr. Opin. Plant Biol. 6:312-319. [DOI] [PubMed] [Google Scholar]
- 13.Buttner, D., D. Nennstiel, B. Klusener, and U. Bonas. 2002. Functional analysis of HrpF, a putative type III translocon protein from Xanthomonas campestris pv. vesicatoria. J. Bacteriol. 184:2389-2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen, G., X. Zhang, X. Zhang, and J. Wang. 2000. Characterization of hrp mutants and their conjugates with hrp gene clones of Xanthomonas oryzae pv. oryzicola. J. Agric. Biotechnol. 8:117-122. [Google Scholar]
- 15.Chen, G., L. Zou, X. Wu, and J. Wang. 2005. avr/pth13 gene of Xanthomonas oryzae pv. oryzicola, a novel virulence member of avrBs3/PthA family, strengthening virulence of Xanthomonas oryzae pv. oryzae on rice. Chin. J. Rice Sci. 19:291-296. [Google Scholar]
- 16.da Silva, A. C. R., J. A. Ferro, F. C. Reinach, C. S. Farah, L. R. Furlan, R. B. Quaggio, C. B. Monteiro-Vitorello, M. A. Van Sluys, N. F. Almeida, L. M. C. Alves, A. M. do Amaral, M. C. Bertolini, L. E. Camargo, G. Camarotte, F. Cannavan, J. Cardozo, F. Chambergo, L. P. Ciapina, R. M. B. Cicarelli, L. L. Coutinho, J. R. Cursino-Santos, H. El-Dorry, J. B. Faria, A. J. S. Ferreira, R. C. C. Ferreira, M. I. T. Ferro, E. F. Formighieri, M. C. Franco, C. C. Greggio, A. Gruber, A. M. Katsuyama, L. T. Kishi, R. P. Leite, E. G. M. Lemos, M. V. F. Lemos, E. C. Locali, M. A. Machado, A. M. B. Madeira, N. M. Martinez-Rossi, E. C. Martins, J. Meidanis, C. F. M. Menck, C. Y. Miyaki, D. H. Moon, L. M. Moreira, M. T. M. Novo, V. K. Okura, M. C. Oliveira, V. R. Oliveira, H. A. Pereira, A. Rossi, J. A. D. Sena, C. Silva, R. F. de Souza, L. A. F. Spinola, M. A. Takita, R. E. Tamura, E. C. Teixeira, R. I. D. Tezza, M. Trindade dos Santos, D. Truffi, S. M. Tsai, F. F. White, J. C. Setubal, and J. P. Kitajima. 2002. Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature 417:459-463. [DOI] [PubMed] [Google Scholar]
- 17.de Feyter, R., C. I. Kado, and D. W. Gabriel. 1990. Small, stable shuttle vectors for use in Xanthomonas. Gene 88:65-72. [DOI] [PubMed] [Google Scholar]
- 18.Fenselau, S., and U. Bonas. 1995. Sequence and expression analysis of the hrpB pathogenicity operon of Xanthomonas campestris pv. vesicatoria which encodes eight proteins with similarity to components of the Hrp, Ysc, Spa and Fli secretion systems. Mol. Plant-Microbe Interact. 8:845-854. [DOI] [PubMed] [Google Scholar]
- 19.Fenselau, S., I. Balbo, and U. Bonas. 1992. Determinants of pathogenicity in Xanthomonas campestris pv. vesicatoria are related to proteins involved in secretion in bacterial pathogens of animals. Mol. Plant-Microbe Interact. 5:390-396. [DOI] [PubMed] [Google Scholar]
- 20.Galán, J. E., and A. Collmer. 1999. Type III secretion machines: bacterial devices for protein delivery into host cells. Science 284:1322-1328. [DOI] [PubMed] [Google Scholar]
- 21.Genin, S., B. Brito, P. T. Denny, and C. Boucher. 2005. Control of the Ralstonia solanacearum type III secretion system (Hrp) genes by the global virulence regulator PhcA. FEBS Lett. 579:2077-2081. [DOI] [PubMed] [Google Scholar]
- 22.Genin, S., C. L. Gough, C. Zischek, and C. A. Boucher. 1992. Evidence that the hrpB gene encodes a positive regulator of pathogenicity genes from Pseudomonas solanacearum. Mol. Microbiol. 6:3065-3076. [DOI] [PubMed] [Google Scholar]
- 23.Gophna, U., E. Z. Ron, and D. Graur. 2003. Bacterial type III secretion systems are ancient and evolved by multiple horizontal-transfer events. Gene 312:151-163. [DOI] [PubMed] [Google Scholar]
- 24.Gough, C. L., S. Genin, V. Lopes, and C. A. Boucher. 1993. Homology between the HrpO protein of Pseudomonas solanacearum and bacterial proteins implicated in a signal peptide-independent secretion mechanism. Mol. Gen. Genet. 239:378-392. [DOI] [PubMed] [Google Scholar]
- 25.He, S. Y., H. C. Huang, and A. Collmer. 1993. Pseudomonas syringae pv. syringae HarpinPss: a protein that is secreted via the Hrp pathway and elicits the hypersensitive response in plants. Cell 73:1255-1266. [DOI] [PubMed] [Google Scholar]
- 26.Hopkins, C. M., F. F. White, S. H. Choi, A. Guo, and J. E. Leach. 1992. A family of avirulence genes from Xanthomonas oryzae pv. oryzae. Mol. Plant-Microbe Interact. 5:451-459. [DOI] [PubMed] [Google Scholar]
- 27.Huang, H., S. W. Hutcheson, and A. Collmer. 1991. Characterization of the hrp cluster from Pseudomonas syringae pv. syringae 61 and TnphoA tagging of genes encoding export of membrane spanning Hrp proteins. Mol. Plant-Microbe Interact. 4:469-476. [Google Scholar]
- 28.Huguet, E., K. Hahn, K. Wengelnik, and U. Bonas. 1998. hpaA mutants of Xanthomonas campestris pv. vesicatoria are affected in pathogenicity but retain the ability to induce host-specific hypersensitive reaction. Mol. Microbiol. 29:1379-1390. [DOI] [PubMed] [Google Scholar]
- 29.Reference deleted.
- 30.Kamdar, H. V., S. Kamoun, and C. I. Kado. 1993. Restoration of pathogenicity of avirulent Xanthomonas oryzae pv. oryzae and X. campestris pathovars by reciprocal complementation with the hrpXo and hrpXc genes and identification of HrpX function by sequence analyses. J. Bacteriol. 175:2017-2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim, J.-G., B. K. Park, C.-H. Yoo, E. Jeon, J. Oh, and I. Hwang. 2003. Characterization of the Xanthomonas axonopodis pv. glycines Hrp pathogenicity island. J. Bacteriol. 185:3155-3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kjemtrup, S., Z. Nimchuk, and J. L. Dangl. 2000. Effector proteins of phytopathogenic bacteria: bifunctional signals in virulence and host recognition. Curr. Opin. Microbiol. 3:73-78. [DOI] [PubMed] [Google Scholar]
- 33.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 34.Lahaye, T., and U. Bonas. 2001. Molecular secrets of bacterial type III effector proteins. Trends Plant Sci. 6:479-485. [DOI] [PubMed] [Google Scholar]
- 35.Marenda, M., B. Brito, D. Callard, S. Genin, P. Barberis, C. Boucher, and M. Arlat. 1998. PrhA controls a novel regulatory pathway required for the specific induction of Ralstonia solanacearum hrp genes in the presence of plant cells. Mol. Microbiol. 27:437-453. [DOI] [PubMed] [Google Scholar]
- 36.Merighi, M., D. R. Majerczak, E. H. Stover, and D. L. Coplin. 2003. The HrpX/HrpY two-component system activates hrpS expression, the first step in the regulatory cascade controlling the Hrp regulon in Pantoea stewartii subsp. stewartii. Mol. Plant-Microbe Interact. 16:238-248. [DOI] [PubMed] [Google Scholar]
- 37.Mithöfer, A., F. Lottspeich, and J. Ebel. 1996. One-step purification of the beta-glucan elicitor-binding protein from soybean (Glycine max L.) roots and characterization of an anti-peptide antiserum. FEBS Lett. 381:203-207. [DOI] [PubMed] [Google Scholar]
- 38.Mongkolsuk, S., S. Loprasert, P. Vattanaviboon, C. Chanvanichayachai, S. Chamnongpol, and N. Supsamran. 1996. Heterologous growth phase- and temperature-dependent expression and H2O2 toxicity protection of a superoxide-inducible monofunctional catalase gene from Xanthomonas oryzae pv. oryzae. J. Bacteriol. 178:3578-3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mongkolsuk, S., S. Rabibibhadana, R. Sukchavalit, and G. Vaughn. 1998. Construction and physiological analysis of a Xanthomonas oryzae pv. oryzae recA mutant. FEMS Microbiol. Lett. 169:269-275. [DOI] [PubMed] [Google Scholar]
- 40.Noël, L., F. Thieme, D. Nennstiel, and U. Bonas. 2002. Two novel type III- secreted proteins of Xanthomonas campestris pv. vesicatoria are encoded within the hrp pathogenicity island. J. Bacteriol. 184:1340-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oku, T., K. Tanaka, M. Iwamoto, Y. Inoue, H. Ochiai, H. Kaku, S. Tsuge, and K. Tsuno. 2004. Structural conservation of hrp gene cluster in Xanthomonas oryzae pv. oryzae. J. Gen. Plant Pathol. 70:159-167. [Google Scholar]
- 42.Rabibhadana, S., S. Chamnongpol, J. E. Trempy, N. P. Ambulos, and S. Mongkolsuk. 1993. Isolation and expression in Escherichia coli of a Xanthomonas oryzae recA-like gene. Gene 132:113-118. [DOI] [PubMed] [Google Scholar]
- 43.Roden, J. A., B. Belt, J. B. Ross, T. Tachibana, J. Vargas, and M. B. Mudgett. 2004. A genetic screen to isolate type III effectors translocated into pepper cells during Xanthomonas infection. Proc. Natl. Acad. Sci. USA 101:16624-16629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ronald, P., and H. Leung. 2002. The rice genome: the most precious things are not jade and pearls. Science 296:58-59. [DOI] [PubMed] [Google Scholar]
- 45.Schell, M. A. 2000. Regulation of virulence and pathogenicity genes in Ralstonia solanacearum by a complex network. Annu. Rev. Phytopathol. 38:263-292. [DOI] [PubMed] [Google Scholar]
- 46.Schulte, R., and U. Bonas. 1992. A Xanthomonas pathogenicity locus is induced by sucrose and sulfur-containing amino acid. Plant Cell 1992:79-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schulte, R., and U. Bonas. 1992. Expression of the Xanthomonas campestris pv. vesicatoria hrp gene cluster, which determines pathogenicity and hypersensitivity on pepper and tomato, is plant inducible. J. Bacteriol. 174:815-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for genetic engineering: transposon mutagenesis in gram negative bacteria. Biotechnology 1:784-791. [Google Scholar]
- 49.Sugio, A., B. Yang, and F. F. White. 2005. Characterization of the hrpF pathogenicity peninsula of Xanthomonas oryzae pv. oryzae. Mol. Plant-Microbe Interact. 18:546-554. [DOI] [PubMed] [Google Scholar]
- 50.Tsuge, S., A. Furutani, R. Fukunaka, T. Oku, K. Tsuno, H. Ochiai, Y. Inoue, H. Kaku, and Y. Kubo. 2002. Expression of Xanthomonas oryzae pv. oryzae hrp genes in XOM2, a novel synthetic medium. J. Gen. Plant Pathol. 68:363-371. [Google Scholar]
- 51.Tsuge, S., T. Terashima, A. Furutani, H. Ochiai, T. Oku, K. Tsuno, H. Kaku, and Y. Kubo. 2005. Effects on promoter activity of base substitutions in the cis-acting regulatory element of HrpXo regulons in Xanthomonas oryzae pv. oryzae. J. Bacteriol. 187:2308-2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wei, Z. M., R. J. Laby, C. H. Zumoff, D. W. Bauer, S. Y. He, A. Collmer, and S. V. Beer. 1992. Harpin, elicitor of the hypersensitive response produced by the plant pathogen Erwinia amylovora. Science 257:85-88. [DOI] [PubMed] [Google Scholar]
- 53.Wen, W., M. Shao, G. Chen, and J. Wang. 2003. Defense response in plants induced by harpinXoo, an elicitor of hypersensitive response from Xanthomonas oryzae pv. oryzae. J. Agric. Biotechnol. 11:192-197. [Google Scholar]
- 54.Wengelnik, K., and U. Bonas. 1996. HrpXv, an AraC-type regulator, activates expression of five of the six loci in the hrp cluster of Xanthomonas campestris pv. vesicatoria. J. Bacteriol. 178:3462-3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wengelnik, K., C. Marie, M. Russel, and U. Bonas. 1996. Expression and localization of HrpA1, a protein of Xanthomonas campestris pv. vesicatoria essential for pathogenicity and induction of the hypersensitive reaction. J. Bacteriol. 178:1061-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wengelnik, K., O. Rossier, and U. Bonas. 1999. Mutations in the regulatory gene hrpG of Xanthomonas campestris pv. vesicatoria result in constitutive expression of all hrp genes. J. Bacteriol. 181:6828-6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wengelnik, K., G. Van den Ackerveken, and U. Bonas. 1996. HrpG, a key hrp regulatory protein of Xanthomonas campestris pv. vesicatoria, is homologous to two-component response regulators. Mol. Plant-Microbe Interact. 9:704-712. [DOI] [PubMed] [Google Scholar]
- 58.White, F. F., B. Yang, and L. B. Johnson. 2000. Prospect for understanding avirulence gene function. Curr. Opin. Plant Biol. 3:291-298. [DOI] [PubMed] [Google Scholar]
- 59.Xiao, Y., and S. W. Hutcheson. 1994. A single promoter sequence recognized by a newly identified alternate sigma factor directs expression of pathogenicity and host range determinants in Pseudomonas syringae. J. Bacteriol. 176:3089-3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhao, B., E. Y. Ardales, A. Raymundo, J. Bai, H. N. Trick, and J. E. Leach. 2004. The avrRxo1 gene from the rice pathogen Xanthomonas oryzae pv. oryzicola confers a non-host defense reaction on maize with resistance gene Rxo1. Mol. Plant-Microbe Interact. 17:771-779. [DOI] [PubMed] [Google Scholar]
- 61.Zhu, W., M. M. Magbanua, and F. F. White. 2000. Identification of two novel hrp-associated genes in the hrp gene cluster of Xanthomonas oryzae pv. oryzae. J. Bacteriol. 182:1844-1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zou, L., G. Chen, X. Wu, and J. Wang. 2005. Cloning and sequence analysis of diverse members of avrBs3/PthA family of Xanthomonas oryzae pv. oryzicola. Sci. Agric. Sin. 38:929-935. [Google Scholar]