Abstract
Several microbes and chemicals have been considered as potential tracers to identify fecal sources in the environment. However, to date, no one approach has been shown to accurately identify the origins of fecal pollution in aquatic environments. In this multilaboratory study, different microbial and chemical indicators were analyzed in order to distinguish human fecal sources from nonhuman fecal sources using wastewaters and slurries from diverse geographical areas within Europe. Twenty-six parameters, which were later combined to form derived variables for statistical analyses, were obtained by performing methods that were achievable in all the participant laboratories: enumeration of fecal coliform bacteria, enterococci, clostridia, somatic coliphages, F-specific RNA phages, bacteriophages infecting Bacteroides fragilis RYC2056 and Bacteroides thetaiotaomicron GA17, and total and sorbitol-fermenting bifidobacteria; genotyping of F-specific RNA phages; biochemical phenotyping of fecal coliform bacteria and enterococci using miniaturized tests; specific detection of Bifidobacterium adolescentis and Bifidobacterium dentium; and measurement of four fecal sterols. A number of potentially useful source indicators were detected (bacteriophages infecting B. thetaiotaomicron, certain genotypes of F-specific bacteriophages, sorbitol-fermenting bifidobacteria, 24-ethylcoprostanol, and epycoprostanol), although no one source identifier alone provided 100% correct classification of the fecal source. Subsequently, 38 variables (both single and derived) were defined from the measured microbial and chemical parameters in order to find the best subset of variables to develop predictive models using the lowest possible number of measured parameters. To this end, several statistical or machine learning methods were evaluated and provided two successful predictive models based on just two variables, giving 100% correct classification: the ratio of the densities of somatic coliphages and phages infecting Bacteroides thetaiotaomicron to the density of somatic coliphages and the ratio of the densities of fecal coliform bacteria and phages infecting Bacteroides thetaiotaomicron to the density of fecal coliform bacteria. Other models with high rates of correct classification were developed, but in these cases, higher numbers of variables were required.
Determining the source of fecal contamination in aquatic environments is essential for estimating the health risks associated with pollution, facilitating measures to remediate polluted waterways, and resolving legal responsibility for remediation. Source tracking methods should enable investigators to uncover the sources of fecal pollution in a particular water body (40). Candidate microbes and chemicals have been investigated and reviewed (15, 54, 55) as potential tools for the identification of human fecal sources. More recently, new approaches using eukaryotic mitochondrial DNA to differentiate fecal sources in feces-contaminated surface waters have been explored (43). However, field studies using most of the numerous chemical and microbiological methods available to track sources of fecal contamination have shown that existing methods are limited (15, 40, 54, 55, 56). These limitations to source identification approaches could be inferred from the reviews cited above. These include the assay of complex samples (such as those that are highly diluted or that are from an undetermined mixed origin or those containing pollution that is not recent), the use of approaches that are not spatially stable, overemphasis on limited improvement of technical aspects of methods rather than on the identification of appropriate source indicators (tracers), or trying to determine an appropriate tracer and source tracking method at the same time.
In our opinion, it is necessary to first identify tracers or combinations of tracers demonstrating high discrimination and then adapt these methods to the needs of source tracking studies. Consequently, both new conceptual and methodological approaches are needed in order to develop models for microbial source tracking. These new approaches should address, step by step, the factors that could influence the successful determination of the source of fecal pollution. These factors include the nature of the dominant fecal pollution contributions (anthropogenic or nonanthropogenic pollution), dilution, the persistence of indicators and parameters, the presence of complex mixtures from several distinct animal species, and the selection of appropriate and consistent numerical methods for the development of models.
The study described herein was initially designed to focus on five key components that were selected after analyzing the results of several previous investigations reported in recent reviews (15, 54, 55). Initially, the study focused only on the differentiation between human and nonhuman sources. Secondly, examination of highly polluted wastewaters or slurries was included because failures previously reported in the literature were often related to the use of dilute samples that give values under the threshold of the method investigated (46, 47). The third key element was to study widely different geographical areas, as clear geographical variations in results have been reported for several approaches described in the literature (50, 57, 59). The fourth key element was to include several indicators of fecal pollution from both human and nonhuman sources throughout the study, since this is needed for defining ratios between discriminant and nondiscriminant indicators and for defining the persistence of the values of fecal contaminants in the environment. Finally, the fifth key element was to identify statistical or machine learning methods to develop appropriate predictive models.
Recently, a multilaboratory study was undertaken in the following areas of Europe: northern Europe (Stockholm, Sweden), northwestern Europe (Brighton, United Kingdom), central Europe (Nancy, northeastern France), southeastern Europe (Nicosia, Cyprus), and southwestern Europe (Barcelona, Spain). During a first phase, quality control schemes were agreed upon and published (6). In order to fully acquaint all relevant laboratory personnel from the participating laboratories with the methods and the materials to be analyzed, collaborative training sessions, in which reference materials were used, were undertaken prior to an interlaboratory comparison study. Furthermore, the reference materials were used as standard samples for first-line quality control during the full study. In this first phase, nine methods achievable in all laboratories (enumeration of fecal coliforms, enterococci, Clostridium perfringens, somatic coliphages, F-specific RNA phages, and bacteriophages infecting Bacteroides fragilis RYC2056; genotyping of F-specific RNA phages; and biochemical phenotyping of fecal coliforms and enterococci by a miniaturized system) were applied to local samples in all the laboratories. The novel methods, or methods not achievable in some of the laboratories, were applied to local samples only in the laboratories that had the appropriate facilities and/or expertise. The novel methods were those related to the specific detection of Bifidobacterium species and bacteriophages infecting Bacteroides thetaiotaomicron GA17, detection of adenoviruses and enteroviruses by genomic methods (PCR and reverse transcription-PCR), genotyping of Giardia, and determination of fecal sterols. Following this first phase, several methods (detection of adenoviruses and enteroviruses by genomic methods, genotyping of Giardia, and analysis of antibiotic resistance profiles) were rejected either because of failures (suspected true or false negatives) or because the method gave unreliable results in some of the laboratories. Results of this first phase were reported elsewhere previously (6). Taking the results of the first phase of study into account, the second phase involved sampling of wastewaters and slurries of human and animal origin in the different geographical areas using the single and derived parameters presented in Table 1. Moreover, a number of statistical methods were tested to aid in the identification and classification of sources of fecal pollution in water based on microbial and/or chemical indicators, which have been proposed as discriminant tracers. These included discriminant analysis (17, 48, 58), the nearest-neighbor technique (maximum similarity) (10, 13, 51), and the use of artificial neural networks (9, 18). Until now, there has been no widespread consensus on when the use of each of the methods is the most appropriate, and to date, none of these methods have provided an interpretable model. Furthermore, there is no consensus on the most appropriate statistical analyses to determine the set of optimal variables for developing these predictive models (51). Other numerical methods should be assayed in order to develop predictive models for source tracking. Specifically, machine learning methods (45) have been used with a considerable degree of success in many disciplines. Their potential application to microbial source tracking should therefore be evaluated.
TABLE 1.
Distribution of samples from the geographical areas sampled by the research groups participating in this study
| Source | Geographic area
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Spain
|
France
|
Sweden
|
United Kingdom
|
Cyprus
|
||||||
| No. of samples | No. of sampling sites | No. of samples | No. of sampling sites | No. of samples | No. of sampling sites | No. of samples | No. of sampling sites | No. of samples | No. of sampling sites | |
| Human wastewater | ||||||||||
| Urban | 22 | 6 | 10 | 1 | 18 | 9 | 22 | 3 | 5 | 1 |
| Hospital | 16 | 1 | 5 | 5 | ||||||
| Military camp | 17 | 1 | ||||||||
| Animal wastewatera | ||||||||||
| Cow | 6 | 2 | 15 | 3 | 9 | 6 | 8 | 1 | ||
| Pig | 9 | 3 | 5 | 1 | 7 | 1 | 8 | 1 | ||
| Poultry | 8 | 2 | 4 | 1 | 7 | 1 | 9 | 1 | ||
| Horse | 4 | 3 | ||||||||
| Mixedb | 4 | 2 | 7 | 2 | 5 | 1 | ||||
| Total | 49 | 15 | 48 | 7 | 45 | 25 | 44 | 6 | 44 | 5 |
Slaughterhouses or farm slurries.
Cow, pig, and sheep.
The objectives of the present study were (i) to determine the most discriminant tracers showing wide and consistent geographical stability between all locations, (ii) to identify subsets of variables derived from tracers with the highest discriminant capacity, and (iii) to evaluate and compare statistical or machine learning methods to develop predictive models for source tracking using the minimum number of these variables.
MATERIALS AND METHODS
Samples and sampling.
A total of 230 samples were analyzed during a period of 2 years. Of these, 114 samples composed almost exclusively of feces of human origin were taken from municipal wastewater at the influent to treatment plants (77 samples), from hospital wastewaters (21 samples), and from wastewater from a military camp (17 samples). The remaining 116 samples, composed almost exclusively of feces of nonhuman (animal) origin, were taken from slaughterhouse wastewater effluents (57 samples) or farm slurries (59 samples) from different animals (cattle, sheep, pigs, horses, and poultry). The number of samples and the sampling sites for each geographical area are summarized in Table 1. Details for each sample are provided in the supplemental material. Similar proportions of samples of each type (human and nonhuman) were taken from each geographic area. Municipal wastewater came from communities with 1,000 to 1.5 million inhabitants. Hospital wastewater was taken from hospitals with at least 100 beds. The population contributing to the military camp wastewater was approximately 600. Slurry samples were derived from units of at least 10 animals. Wastewater was taken from different slaughterhouses processing between approximately 200 and 3,000 animals per day. Human and animal wastewater sampling occasions were randomly distributed along the 2-year period. No more than one sample was taken at each site on each occasion. For purposes of clarity, the first group of samples will be referred to here as human samples, and the second group will be referred to as animal samples. Sampling, transport, storage, and pretreatment of samples were performed according to standardized International Standardisation Organisation (ISO) protocols (20, 21, 22, 24, 26).
Detection and enumeration of bacterial indicators.
Three fecal indicators were measured: fecal coliforms, enterococci, and clostridia. Standardized methods (23, 25, 28) for the enumeration of these indicators were followed. Briefly, fecal coliforms were enumerated by membrane filtration on 0.45-μm-pore-size membranes followed by incubation for 24 h on mFC agar (Difco, Detroit, Mich.) at 44.5°C according to established procedures (28). Enterococci were also enumerated by membrane filtration according to standardized protocols by incubation on m-Enterococcus agar (Difco, Detroit, Mich.) at 37°C for 48 h. Membranes were then transferred to bile esculine agar (Difco) for 1 h at 44°C to confirm the enterococci colonies on the basis of the hydrolysis of esculine. Clostridia were enumerated by thermal shock of samples at 80°C for 10 min. Later, 10-fold dilutions were made in one-quarter-strength Ringer's solution, and 1 ml of each dilution was inoculated into 50 ml of liquid sulfite polymyxin sulfadiazine agar (Difco) followed by incubation at 44°C for 24 h.
Detection, enumeration, and typing of bacteriophages.
Somatic coliphages, F-specific RNA bacteriophages, and phages infecting Bacteroides fragilis RYC2056 were enumerated in accordance with ISO standardized methods (27, 29, 30). PFU of somatic coliphages were counted by the double-agar-layer technique using Escherichia coli strain WG5 according to ISO standard 10705-2 (29). Total numbers of F RNA and F-specific RNA bacteriophage PFU were determined using strain Salmonella enterica serovar Typhimurium strain WG49 (now classified as Salmonella enteritidis subsp. typhimurium) in accordance with ISO standard ISO 10705-1 (27). PFU of bacteriophages infecting Bacteroides fragilis strain RYC2056 were determined by the double agar layer method according to ISO standard 10705-4 (30). Phages infecting Bacteroides thetaiotaomicron GA17 were enumerated as described elsewhere previously (49) according to ISO standard 10705-4 (30); as stated in Results, plaques obtained on GA17 in the United Kingdom were very turbid. In this case, plaques were counted by researchers more experienced in the technique, and suspected plaques were verified by subculture (enrichment followed by spot test).
Genotyping of F-specific RNA phages.
The distribution of genotypes of F-specific RNA bacteriophages was carried out by plaque hybridization as previously described (52) using probes previously described (3).
Phenotyping of fecal coliforms and fecal streptococci.
From each sample, 24 fecal coliform colonies and 24 Enterococcus colonies were selected at random from selective agar plates (23, 28) containing between 30 and 100 colonies and were picked from these plates to obtain a pure culture for biochemical phenotyping. The number of bacterial isolates required in each sample for diversity analysis was previously determined by other authors (4). Biochemical phenotyping was performed using PhP-RE and PhP-RF microplates according to the manufacturer's instructions (PhP-Plate Microplates Techniques AB, Sweden) and previously described techniques (35). The basis of biochemical fingerprinting using these microplates has also been described previously (33). The biochemical profiles were calculated for each isolate as previously described (36) by using PhpWin software (PhP-Plate Microplates Techniques AB). Simpson's diversity index (Di) was used to calculate the diversity of bacterial populations in each group studied (2, 19), while similarity between populations was calculated by the population similarity coefficient (36). Calculations of diversity (Di), population similarity indices, and correlation coefficients and cluster analyses were also performed using PhpWin software (PhP-Plate Microplates Techniques) as previously described (36). In addition, the species distribution of Enterococcus species was analyzed using a previously described matrix (41) and a previously described procedure (5). The percentage of Enterococcus faecium plus Enterococcus faecalis isolates (variable FMFS) and the percentage of Enterococcus hirae isolates (variable HiR) were also calculated, because differences in their proportions in wastewaters of animal and human origin have been reported previously in other studies (34). Similarly, the percentage of E. coli within the fecal coliforms was determined by comparing isolates with E. coli PhP-Plate reference phenotypes. Additionally, the percentage of those fecal coliform isolates that did not demonstrate fermentation of cellobiose was also calculated. E. coli isolates are normally cellobiose negative (16), whereas other thermotolerant coliform species showing E. coli-like colonies on mFC agar are often cellobiose positive, and thus, this proportion is an estimation of the proportion of E. coli isolates among the E. coli-like isolates.
Bifidobacterium determinations.
Total bifidobacteria were counted on human bifido sorbitol agar as described previously by other authors (42). Yellow colonies on human bifido sorbitol agar were counted as sorbitol-fermenting bifidobacteria as described elsewhere previously (8). Additionally, the presence of Bifidobacterium dentium and Bifidobacterium adolescentis was determined by PCR amplification using specific primers of the 16S RNA genes as described elsewhere previously (7).
Determination of fecal sterols.
The procedure for analysis of sterols in wastewater with high concentrations of solid fraction was performed as previously described (37). First, separation of the solid fraction from 100-ml volumes of each sample was carried out by filtration through glass filters. The membranes were then weighed and frozen at −70°C until analysis. Gas chromatography with flame ionization detection analysis of four main fecal sterols (coprostanol [5β-cholestan-3β-ol], stigmastanol or 24-ethylcoprostanol [24-ethyl-5β-cholestan-3β-ol], epicoprostanol [5β-cholestan-3α-ol], and cholestanol [5-α-colestan-3β-ol]) was then performed.
Establishment of operating principles and quality assurance.
In order to establish a set of operating principles for data quality, a training session for operators from all the participant laboratories was undertaken. Noncertified reference materials (bacterial strains and bacteriophages) were prepared and used during the training session as previously described (39). These reference materials were provided to the partners at the end of an interlaboratory exercise session in order to evaluate the implementation of the methods in participating laboratories. Moreover, these reference materials were used in routine quality control practices at the participating laboratories. Taking into account the available facilities in the different laboratories and the results of the interlaboratory exercises, the following parameters were tested in each of the five laboratories: enumeration of fecal coliform bacteria, enterococci, clostridia, somatic coliphages, F-specific RNA phages, total bifidobacteria, sorbitol-fermenting bifidobacteria, bacteriophages infecting B. fragilis RYC2056, and bacteriophages infecting B. thetaiotaomicron GA17; genotyping of F-specific RNA phages; and phenotypic characterization of fecal coliforms and enterococci. Detection of Bifidobacterium dentium and Bifidobacterium adolescentis by PCR and fecal sterol analysis of all samples were performed in the laboratories of the University of Barcelona.
Data treatment and statistical analyses.
Raw data from the analyses performed provided 26 variables, as presented in Table 2. This initial group of variables used in the statistical analyses consisted of the 20 single variables and 6 derived variables from the phenotyping of fecal coliforms and enterococci (percentage of cellobiose-negative fecal coliforms [CNFC], diversity index for fecal coliforms [DiC], diversity index for enterococci [DiE], percentage of E. coli Ph-Plate phenotypes [ECP], FMFS, and HiR). Some values that were below the threshold value (lowest sensitivity) for the method were corrected to the threshold value.
TABLE 2.
Definition of terms used for single and derived variables in the statistical and machine learning methods of this study
| Variable | Label | Parameter |
|---|---|---|
| Single | BA | Detection of the presence (1) or absence (0) of Bifidobacterium adolescentis |
| BE | Detection of the presence (1) or absence (0) of Bifidobacterium dentium | |
| BTHPH | Enumeration of B. fragilis bacteriophages using the new host strain B. thetaiotaomicron GA17 | |
| CHOL | Concn of cholestanol or 5-α-colestan-3ß-ol | |
| CL | Enumeration of clostridia | |
| COP | Concn of coprostanol or 5ß-cholestan-3ß-ol | |
| EPICOP | Concn of epicoprostanol or 5ß-cholestan-3α-ol | |
| ETHYLCOP | Concn of stigmastanol or 24-ethylcoprostanol | |
| FC | Enumeration of fecal coliforms | |
| FE | Enumeration of fecal enterococci | |
| FRNAPH | Enumeration of F-specific RNA bacteriophages | |
| FRNAPH I | % of genotype I of F-specific RNA bacteriophages | |
| FRNAPH II | % of genotype II of F-specific RNA bacteriophages | |
| FRNAPH III | % of genotype III of F-specific RNA bacteriophages | |
| FRNAPH IV | % of genotype IV of F-specific RNA bacteriophages | |
| FTOTAL | Enumeration of F-specific bacteriophages | |
| RYC2056 | Enumeration of B. fragilis bacteriophages using the host strain RYC2056 | |
| SFBIF | Enumeration of sorbitol-fermenting bifidobacteria | |
| SOMCPH | Enumeration of somatic coliphages | |
| TBIF | Enumeration of total bifidobacteria | |
| Derived | CNFC | % of cellobiose-negative fecal coliforms |
| COP/EPICOP | Ratio of concn of coprostanol to that of epicoprostanol | |
| COP/ETHYLCOP | Ratio of concn of coprostanol to that of stigmastanol | |
| DA | Sum of values of BA and BE | |
| DiC | Simpson's diversity index for fecal coliforms | |
| DiE | Simpson's diversity index for enterococci | |
| ECP | % of E. coli Ph-Plate phenotypes | |
| FC/BTHPH | Ratio of the no. of fecal coliforms to that of the new host strain B. thetaiotaomicron GA17 | |
| FC/FE | Ratio of the no. of fecal coliforms to that of enterococci | |
| FC/RYC2056 | Ratio of the no. of fecal coliforms to that of phages on the host strain B. fragilis RYC2056 | |
| FC/SOMCPH | Ratio of the no. of fecal coliforms to that of coliphages | |
| FMFS | % of E. faecium and E. faecalis | |
| FRNAPH I + FRNAPH IV | Sum of the % of genotypes I and IV of F-specific RNA bacteriophages | |
| FRNAPH II + FRNAPH III | Sum of the % of genotypes II and III of F-specific RNA bacteriophages | |
| HiR | % of E. hirae | |
| SFBIF/TBIF | Ratio of the no. of sorbitol-fermenting bifidobacteria to that of total bifidobacteria | |
| SOMCPH/BTHPH | Ratio of the no. of somatic coliphages to that of the new host strain B. thetaiotaomicron GA17 | |
| SOMCPH/RYC2056 | Ratio of the no. of somatic coliphages to that of phages on the host strain B. fragilis RYC2056 |
Descriptive statistics (minimum and maximum values, mean, error, standard deviation, and median) were calculated for each of the single variables studied. First, Student's t test was performed in order to determine significant differences in microbial and chemical analytes between waste streams of human and nonhuman origin. Variables were analyzed by segregating them according to the measured parameters into several groups, namely, enumeration of bacterial and bacteriophage groups, genotypes of F-specific RNA phages, diversity index and percentage of certain species of enterococci and fecal coliforms, ratios between bifidobacteria populations, molecular detection of Bifidobacterium adolescentis and Bifidobacterium dentium, and concentration of fecal sterols (see Tables 3 to 7, respectively). Additionally, correlation, regression, and discriminant analyses were conducted. These data analyses were performed using Statgraphics statistical analysis software (Statgraphics plus 5.1; STSC Software Publishing Group, Rockville, MD) and aimed to assess the discriminatory power of the individual variables.
TABLE 3.
Bacterial indicators and bacteriophage densities
| Tracera | No. of samples | % Positive samples | Log10 CFU or PFU per 100 ml
|
P valueb | ||||
|---|---|---|---|---|---|---|---|---|
| Minimum | Maximum | Mean | SD | Median | ||||
| FC-H | 110 | 100 | 5.43 | 8.26 | 6.94 | 0.55 | 6.94 | <0.001 |
| FC-A | 111 | 100 | 3.90 | 10.24 | 7.43 | 0.94 | 7.39 | |
| FE-H | 108 | 100 | 4.01 | 7.20 | 6.01 | 0.53 | 6.04 | <0.001 |
| FE-A | 111 | 100 | 4.54 | 8.89 | 6.37 | 0.88 | 6.20 | |
| CL-H | 110 | 100 | 3.56 | 6.96 | 5.05 | 0.66 | 5.07 | 0.160 |
| CL-A | 111 | 100 | 3.28 | 8.30 | 5.24 | 1.25 | 4.89 | |
| TBIF-H | 56 | 100 | <5.60 | 8.24 | 7.01 | 0.57 | 6.98 | 0.119 |
| TBIF-A | 56 | 98.2 | <3.00 | 9.73 | 7.30 | 1.22 | 7.31 | |
| SFBIF-H | 54 | 100 | <5.00 | 8.15 | 6.40 | 0.59 | 6.41 | <0.001 |
| SFBIF-A | 56 | 21.4 | <3.00 | 9.00 | <5.27 | 1.39 | <5.08 | |
| SOMCPH-H | 110 | 100 | 4.04 | 7.66 | 6.03 | 0.80 | 6.07 | <0.001 |
| SOMCPH-A | 110 | 100 | 1.69 | 9.62 | 6.73 | 1.13 | 6.72 | |
| FRNAPH-H | 110 | 99 | <1.70 | 6.92 | 5.30 | 0.89 | 5.62 | 0.008 |
| FRNAPH-A | 110 | 80 | <1.70 | 8.40 | 4.76 | 1.84 | 5.21 | |
| RYC2056-H | 108 | 99 | <1.70 | 5.53 | 3.99 | 0.81 | 4.14 | <0.001 |
| RYC2056-A | 110 | 78.1 | <1.70 | 5.96 | 3.50 | 1.27 | 3.53 | |
| BTHPH-H | 73 | 98.6 | <1.70 | 5.86 | 4.19 | 0.68 | 4.17 | <0.001 |
| BTHPH-A | 71 | 7.0 | <1.70 | 3.08 | 1.76 | 0.25 | <1.70 | |
Labels of tracers are shown in the list of variables in Table 2. H, samples of human origin; A, samples of animal origin.
Value of P from Student's t test.
TABLE 7.
Concentrations of sterols in human samples and animal samples
| Tracera | No. of samples | Concn of sterols (μg/g)
|
P valueb | ||||
|---|---|---|---|---|---|---|---|
| Minimum | Maximum | Mean | SD | Median | |||
| COP-H | 92 | 5.0 | 6,476 | 413.79 | 1,096.44 | 60.10 | 0.054 |
| COP-A | 85 | 0.1 | 4,080 | 162.92 | 492.07 | 30.00 | |
| ETHYLCO-H | 92 | 0.1 | 2,985 | 117.92 | 344.19 | 15.50 | <0.001 |
| ETHYLCO-A | 85 | 0.1 | 9,360 | 1,305.36 | 2,302.33 | 154.60 | |
| EPICOP-H | 92 | 0.1 | 2,035 | 85.63 | 247.15 | 7.40 | 0.001 |
| EPICOP-A | 85 | 0.1 | 6,390 | 494.87 | 1,206.28 | 19.80 | |
| CHOL-H | 92 | 0.1 | 2,884 | 215.69 | 407.55 | 32.20 | 0.009 |
| CHOL-A | 85 | 0.1 | 6,600 | 529.46 | 1,058.37 | 114.70 | |
Labels of tracers are shown in the list of variables in Table 2. H, samples of human origin; A, samples of animal origin.
Value of P from Student's t test.
In order to improve the chances of obtaining good predictive models, 12 new variables were derived by combining some of the 20 single variables (as sums or ratios). Samples containing incomplete or outlying parameter values likely to be attributable to technical error were discarded. Consequently, a data matrix with 38 variables and 103 samples (called here “observations”) was obtained (Table 2). Techniques for selection of variables (32) (notably, the Relief algorithm [31]) were used in conjunction with several statistical or machine learning methods (45) in a series of experiments. The objective of these experiments was to find the most prominent subset of variables that yielded the highest discriminatory power with the lowest number of variables. Using this subset, predictive models for accurate microbial source tracking could be obtained.
The methods chosen were the k nearest-neighbor technique (with Euclidean distance), the linear and quadratic Bayesian classifiers (two discriminant analysis methods) (14), and the support vector machine (11). The development of predictive models was carried out using 81 of the 103 observations (hereafter referred to as the “training set”) and using cross-validation, as explained below. The remaining 22 observations (the “test set”) were withheld for an independent and unbiased assessment of the feasibility of the predictive models. These holdout observations presented unequivocally distinct values according to their origins (11 from waters polluted by human fecal sources and 11 from waters polluted by nonhuman fecal sources). These analyses were performed using the software package WEKA (60).
RESULTS
Directly quantifiable microorganisms.
Table 3 summarizes the descriptive statistics relating to the numbers of bacterial and bacteriophage tracers studied. With the exception of clostridia and total Bifidobacterium species, the numbers of each tracer in human samples differed significantly (Student's t test) from the numbers in animal samples (P < 0.01). However, two groups of source indicators can be distinguished. The first group includes fecal coliforms, enterococci, clostridia, somatic coliphages, and total bifidobacteria. These were detected in almost all samples (other than a single sample in the case of total bifidobacteria) of both human and animal origin. They were more abundant in the animal samples than in the human samples, but this seems to be due to the higher fecal load of these samples, since relative densities were similar in both groups of samples. In addition, no appreciable differences were observed between geographical areas with respect to this group of microorganisms, as supported by the low standard deviations. Conversely, the other group (which included F-specific RNA phages, phages infecting B. fragilis RYC2056, phages infecting B. thetaiotaomicron GA17, and sorbitol-fermenting Bifidobacterium species) showed a different pattern. Tracers in this second group either were not detected in animal samples or were present at significantly lower levels (P < 0.001) than in human samples. However, each tracer in this group was detected in some animal samples. Phages infecting B. thetaiotaomicron and sorbitol-fermenting bifidobacteria showed the greatest variation (P < 0.01) between animal and human samples. Additionally, both parameters showed a minor number of overlapping results when we tried to establish reference values; that is, a few animal samples gave higher values compared with the lowest values for a few human samples. Again, no appreciable differences between geographical areas were observed with regard to this second group of microorganisms (as demonstrated by the low standard deviations shown in Table 3). However, in all samples from the United Kingdom, more than 90% of the plaques of phages infecting B. thetaiotaomicron were very turbid. These plaques were barely visible to the untrained eye and needed to be counted by a more experienced operator. The results reported herein include results from the turbid plaques detected by the more experienced operator. In the other geographical areas, the great majority of plaques reported for phages infecting B. thetaiotaomicron were clear and were easily detected by less experienced operators.
Distribution of genotypes of F-specific RNA phages.
The descriptive statistics for the distribution of genotypes of F-specific RNA bacteriophages are shown in Table 4. As described elsewhere previously (52), the method used here (53) gave a percentage of plaques (ranging from 0 to 10% in different samples) that hybridized with the probes of two different genotypes. These plaques were assigned to the genotype that showed the stronger hybridization signal. Percentages of genotypes were also calculated by deleting the counts of the plaques hybridizing with two probes. The final calculations of percentages of genotypes with both approaches were similar (data not shown). The relative distributions of genotypes I, II, and IV in human samples and animal samples were significantly different. Genotypes I and IV were significantly more abundant in animal samples, and genotype II was significantly more abundant in human samples (P < 0.001). The percentages of genotype III in human and animal samples did not differ significantly, although the average percentage in human samples was slightly higher than that in animal samples. The major differences between human and animal samples were shown by genotype II. However, the percentage of genotype II in some animal samples (6%) was the highest among genotypes I to IV, and in 5% of human samples, it was the lowest among all the genotypes. The rule that genotypes II and III were higher in humans and I and IV were higher in animals complied in all human samples but failed in 35% of the animal samples.
TABLE 4.
Percentages of the four genotypes of F-specific RNA phages in human and animal samples
| Tracera | No. of samples | % Positive samples | % of tracer in samples
|
P valueb | ||||
|---|---|---|---|---|---|---|---|---|
| Minimum | Maximum | Mean | SD | Median | ||||
| FRNAPH I-H | 103 | 89.3 | 0 | 22 | 5.02 | 5.16 | 4.00 | <0.001 |
| FRNAPH I-A | 82 | 95.1 | 0 | 98 | 35.19 | 30.90 | 28.30 | |
| FRNAPH II-H | 103 | 99.0 | 0 | 100 | 49.56 | 28.35 | 46.90 | <0.001 |
| FRNAPH II-A | 82 | 65.8 | 0 | 69 | 7.61 | 11.78 | 3.85 | |
| FRNAPH III-H | 103 | 96.1 | 0 | 100 | 40.23 | 26.28 | 38.00 | 0.072 |
| FRNAPH III-A | 82 | 87.8 | 0 | 98 | 32.23 | 33.87 | 16.00 | |
| FRNAPH IV-H | 103 | 65.0 | 0 | 34 | 5.18 | 6.68 | 2.10 | <0.001 |
| FRNAPH IV-A | 82 | 90.2 | 0 | 99 | 24.94 | 27.82 | 14.80 | |
Labels of tracers are shown in the list of variables in Table 2. H, samples of human origin; A, samples of animal origin.
Value of P from Student's t test.
Phenotyping of fecal coliforms and enterococci.
The descriptive statistics of the different diversity indices and percentages of species, which were calculated from the phenotyping of fecal coliform bacteria and enterococci, are shown in Table 5. Diversity indices for fecal coliforms and enterococci (DiC and DiE, respectively) were higher in human samples than in animal samples. CNFC, ECP, FMFS, and HiR also differed significantly in the human and animal samples (P < 0.01). E. faecalis and E. faecium dominated enterococcal populations in human samples, but E. hirae dominated in animal samples. The samples of municipal, hospital, and military camp wastewaters also showed a lower ECP than did samples of animal wastewaters and slurries. No differences were observed between geographical areas with regard to diversity indices and the calculated percentages of different populations related to human or animal samples. However, in spite of the significant differences among human and animal samples, it is very difficult to establish differentiating reference percentages of CNFC, ECP, FMFS, and HiR, and there are many overlaps. For example, the percentage of human samples in which FMFS is lower than the average is 38%, and the percentage of animal samples in which FMFS is higher than the average from the human samples is 24%. The trend is similar for HiR, ECP, and CNFC. Therefore it is very difficult to use any of these data alone to differentiate fecal sources.
TABLE 5.
Levels of Simpson's diversity index and percentages of different microorganisms in human and animal samples
| Tracera | No. of samples | Value of tracer in samplesc
|
P valueb | ||||
|---|---|---|---|---|---|---|---|
| Minimum | Maximum | Mean | SD | Median | |||
| DiE-H | 103 | 0.16 | 1 | 0.88 | 0.14 | 0.94 | 0.001 |
| DiE-A | 110 | 0.09 | 1 | 0.80 | 0.21 | 0.89 | |
| FMFS-H | 103 | 13.00 | 100 | 72.54 | 18.34 | 75.00 | <0.001 |
| FMFS-A | 109 | 0.00 | 100 | 50.16 | 29.34 | 54.00 | |
| HiR-H | 103 | 0.00 | 54 | 12.30 | 11.88 | 12.00 | <0.001 |
| HiR-A | 109 | 0.00 | 96 | 24.55 | 27.61 | 13.00 | |
| DiC-H | 105 | 0.38 | 1 | 0.92 | 0.10 | 0.96 | 0.001 |
| DiC-A | 110 | 0.38 | 1 | 0.87 | 0.11 | 0.91 | |
| ECP-H | 105 | 8.00 | 100 | 78.73 | 20.97 | 87.00 | <0.001 |
| ECP-A | 105 | 32.00 | 100 | 96.52 | 9.20 | 100 | |
| CNFC-H | 104 | 8.00 | 100 | 72.97 | 21.84 | 75.00 | <0.001 |
| CNFC-A | 109 | 29.00 | 100 | 92.81 | 12.10 | 100.00 | |
Labels of tracers are shown in the list of variables in Table 2. H, samples of human origin; A, samples of animal origin.
Value of P from Student's t test.
Values for FMFS, HiR, ECP, and CNFC are percentages; all other values are diversity indexes expressed as rates of 1.
Bifidobacterium.
The descriptive statistics for the data relating to the ratio between the number of sorbitol-fermenting bacteria (SFBIF) and the number of total bifidobacteria (SFBIF/TBIF) are shown in Table 6. The difference is significant (P < 0.001). It is difficult to establish a value for the differentiation of origins. Thus, taking the SFBIF/TBIF log values to be ≥0.5 for human samples and ≤0.5 for animal samples, there is still a 5% failure rate. However, this may be considered the best value for this variable to differentiate sources attending to the percentage of correct sample classification achieved. The selection of other values as reference criteria resulted in more failures.
TABLE 6.
Ratios between the values in log10 units of sorbitol-fermenting bifidobacteria and those of total bifidobacteria in human and animal samples
| Tracera | No. of samples | Value
|
P valueb | ||||
|---|---|---|---|---|---|---|---|
| Minimum | Maximum | Mean | SD | Median | |||
| SFBIF/TBIF-H | 54 | 0.7 | 1.0 | 0.91 | 0.05 | 0.92 | <0.001 |
| SFBIF/TBIF-A | 56 | 0.0 | 1.0 | 0.08 | 0.20 | 0.01 | |
Labels of tracers are shown in the list of variables in Table 2. H, samples of human origin; A, samples of animal origin.
Value of P from Student's t test.
With regard to the detection of Bifidobacterium dentium and Bifidobacterium adolescentis, there were significant percentages of negatives in human samples (45% and 6.3%, respectively) and positives in animal samples (9.5% and 24.5%, respectively). Consequently, it is also difficult to use these tracers alone to identify fecal sources.
Fecal sterols.
The descriptive statistics for fecal sterol concentrations are shown in Table 7. The concentrations of 24-ethylcoprostanol, epicoprostanol, and cholestanol showed significant differences between human and animal samples, whereas coprostanol did not (P > 0.01), although it gave a P value of 0.054. Concentrations of 24-ethylcoprostanol varied the greatest between human and animal samples. Although in all cases, this was the fecal sterol (among those analyzed) that differed the most, the high number of overlaps prevented the establishment of a reference concentration for differentiation being established. The concentration of 24-ethylcoprostanol being greater than the concentration of coprostanol in animal samples, and vice versa in human samples, seems to be the more discriminant criterion among the data reported here. The percentage of incorrectly classified samples based on this criterion was 6.5%.
Correlation, regression, and discriminant analyses.
High linear correlation was found between the derived variable SOMCPH/BTHPH (ratio of the number of somatic coliphages [SOMCPH] to the number of isolates of B. thetaiotaomicron GA17 [BTHPH]) and the class variable (r = 0.886) and also between the derived variable FC/BTHPH (ratio of the number of fecal coliforms [FC] to the number of isolates of B. thetaiotaomicron GA17) and the class variable (r = 0.847). The correlation between these two derived variables was very high (r = 0.912). Best-subset regression performed with the 26 initial variables indicated that subsets with as few as seven variables (number of fecal enterococci [FE], percentage of genotype II of F-specific RNA bacteriophages [FRNAPH II], FRNAPH IV, concentration of epicoprostanol or coprostanol [EPICOP], FMFS, ECP, and detection of the presence or absence of Bifidobacterium adolescentis [BA]) gave an explanatory power (85.1%) almost equal to that provided by using all the single variables (85.7%). This fact points to high redundancy in terms of the available variables, and a level of redundancy in the data that is too high may reduce performance (38). However, subsets of the variables obtained may be taken as a first indication of relevance. Two-group discriminant analyses (for human or nonhuman fecal samples) using all 26 measured microbiological and chemical parameters provided a correct classification in 100% of the cases. The performance of all the microbial and chemical indicators allowed a predictive classification of cases by discriminant analysis. The question remains whether the same performance can be achieved using a lower number of variables, which would decrease the number of parameters measured, reduce costs, and provide simpler models that are easier to analyze from a microbiological point of view. Specifically, we were interested in finding the smallest subset of variables that was able to provide a correct classification in 100% of the cases. This was a difficult undertaking that could not be addressed by “generate-and-test” methods and is one of the main reasons why we expanded the toolbox to consider other statistical or machine learning methods. Before doing this, the same discriminant analyses were performed using only the subsets of variables that were considered meaningful. For instance, when only the bacterial indicators analyzed in this study were used (Table 3), 75% of nonhuman samples were correctly classified as nonhuman (25% of nonhuman samples were classified as human samples), and 3.7% of samples of human origin were misclassified as nonhuman samples (96.3% of human samples were classified as human samples). Classification using only the four genotypes of F-specific RNA phages allowed 98% of nonhuman and 85% of human samples to be classified correctly (with false-positive and false-negative rates being 0.02 and 0.14, respectively). The fecal sterols studied did not show better correct classifications, since only 38% of nonhuman samples were correctly classified, although 98% of human samples were correctly classified. Similar levels of correct classification were found for the phenotypic analysis of fecal coliforms and enterococcal populations: 79% of nonhuman and 89% of human samples were correctly determined (with false-positive and false-negative rates being 0.17 and 0.13, respectively). Finally, the classification functions developed using the results from the enumeration of the various bacteriophages (somatic coliphages, F-specific RNA phages, bacteriophages infecting B. fragilis RYC2056, and bacteriophages infecting B. thetaiotaomicron GA17) provided a correct classification (human versus nonhuman samples) in all cases.
Machine learning methods.
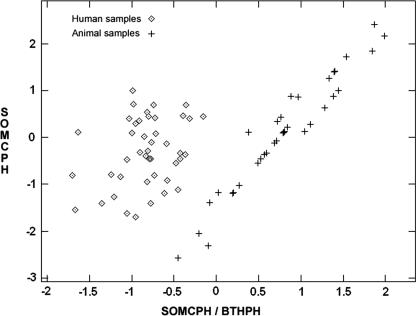
The Relief algorithm provided a list of individual variables arranged according to their discriminatory power. The top three variables in this list were SOMCPH/BTHPH, FC/BTHPH, and FRNAPH II. The next group was a group formed by the variables FMFS, FRNAPH II + FRNAPH III (sum of the percentages of genotypes II and III of F-specific bacteriophages), and FRNAPH I + FRNAPH IV. This outcome was used to build an optimal solution by using the four methods indicated above (Euclidean one-nearest-neighbor technique, linear Bayesian classifier, quadratic Bayesian classifier, and support vector machine). The main finding was that a set of just two variables, (SOMCPH/BTHPH and SOMCPH) provided a training set with 100% correct classification for all the inductive learning methods. The two variables FC/BTHPH and FC also provided excellent results using the four methods (100%, 98.8%, 98.8%, and 100% correct classification rates, respectively). The observations can be displayed in a two-dimensional scatter plot with no loss of information (Fig. 1). It is clear that observations of samples of human and samples of nonhuman origin are neatly separated. With this information, a linear separation is feasible. These two pairs of variables also gave 100% correct classification for the 22 samples in the withheld test set. Also noteworthy is the fact that there were no apparent differences between the various geographical sites. In other words, there were no subclusters. A secondary finding was that other subsets of three or more variables also showed good discriminating ability (for example, FRNAPH I, FRNAPH II, ECP, BA, and SFBIF). However, these variables gave some incorrect classifications and lower identification rates overall (between 85and 95%).
FIG. 1.
Distribution of training observations according to the variables SOMCPH/BTHPH and SOMCPH. Values are standardized to zero mean and unit standard deviation.
DISCUSSION
Accurately determining the occurrence and level of tracers in source feces may not be feasible because of the need to study many samples. Therefore, in order to meet our objectives, we studied only heavily contaminated waters such as municipal and hospital wastewaters, wastewater from abattoirs, and slurries emanating from at least 10 individual farm animals. We are aware that this approach results in the detection of only human or animal population tracers rather than tracers of individuals. However, the water quality problems that may be addressed through source tracking studies are more likely to arise from population-based contamination, and therefore, tracing of population-based contamination is more applicable to “real-world” scenarios than tracing of fecal contamination from individuals. Also, the effects of die-off (whether during wastewater treatment processes or in the natural environment) and dilution, as well as the effects of differing physical and chemical characteristics of the target water, will influence the detection of tracers in different water bodies. These are issues that are best addressed after determining which tracer or tracers best discriminate at the source level. As has been described above, source tracking is a complex task, and progress towards defining suitable tracers and methods will be achieved only by tackling relevant questions in a logical and linear sequence.
Results reported herein provide interesting information on the various conventional fecal indicators tested because of the broad spectrum of wastewaters and geographical areas tested. Fecal coliforms, enterococci, clostridia, total bifidobacteria, somatic coliphages, F-specific RNA phages, and phages infecting strain RYC2056 of B. fragilis had similar relative densities in municipal or human-derived wastewaters in the different geographical areas studied. No significant differences were observed between samples of human origin (hospital, military camp, and municipal wastewaters), regardless of the size of human communities (which ranged from a population of hundreds for hospital samples to 1.5 million for municipal wastewater samples). Consequently, wastewater samples from communities of around 100 inhabitants were shown to be representative. Also, for these indicators, geographical differences between animal samples were not evident.
With regard to the potential of the various microbial and chemical parameters studied as tracers of source pollution, there are a number of observations worthy of discussion. No differences were observed in the ratios between the values of fecal coliforms (taken as the reference value of fecal load) and those of bifidobacteria, enterococci, clostridia, and somatic coliphages in human and animal samples. Conversely, F-specific RNA phages and phages infecting B. fragilis RYC2056 showed differences, since the ratios of their numbers to numbers of fecal coliforms were clearly lower in animal samples, although the differences are not sufficient to allow source differentiation. Among the culture-based microbiological methods tested, which are independent of the characterization of the isolates, enumeration of phages infecting B. thetaiotaomicron GA17 and the ratio between numbers of total bifidobacteria and the numbers of sorbitol-fermenting bifidobacteria discriminated the most samples according to origin. No differentiated clusters were observed in the sets of values of all these nondiscriminant and discriminant indicators. Therefore, it can be concluded that the numbers of all of them are comparable in the various geographical areas studied. The only geographical difference detected was in the characteristics of the plaques of the phages detected by strain B. thetaiotaomicron GA17 in the United Kingdom. Most of these plaques were turbid and required a well-trained operator to count the phages accurately. This fact complicates the use of this method in this location. However, a recent investigation has shown that obtaining a geographically useful Bacteroides host with a performance similar to that of strain GA17 is feasible (49).
Genotypic methods (F-specific RNA genotypes and molecular detection of Bifidobacterium dentium and Bifidobacterium adolescentis) allowed differentiation but with a percentage of failures. As described previously, genotypes II and III predominated in human samples, and genotypes I and IV predominated in animal samples (12, 47, 52, 53). However, in this work, there was an unexpectedly high proportion of animal samples (33%) with high percentages of genotype III, similar to the ones in samples of human origin. There was also a small proportion of samples that gave misleading values, showing inverted percentages to those expected. Additionally, the percentage ranges of each of the genotypes or combinations of genotypes found in the different kinds of samples make the establishment of a threshold for this method difficult.
Conversely, the percentages of both Bifidobacterium dentium and Bifidobacterium adolescentis by molecular detection differed significantly in samples of human and nonhuman origin, being more common in human samples than in animal samples. Both species have been specifically associated with human intestinal microbiota. However, both species were not detected by multiplex PCR (7) in some human samples, and positive results were also observed for some animal samples. Both species were detected in water polluted by feces of human origin and not of animal origin. The detection method needs to be improved in order to detect these species at the lower densities commonly found in human samples to validate negative results in human samples. Additionally, an explanation for their presence in a percentage of animal samples should be sought as well. Although Bifidobacterium adolescentis has been described as a species that is related to humans exclusively (44), it was reported to have been found in samples from poultry (7).
Although some variables derived from phenotypic parameters are more related to nonhuman sources (percentage of E. hirae among the enterococci and percentages of E. coli Phene-Plate profiles or non-cellobiose-fermenting fecal coliforms among the total fecal coliforms) and others are more related to human fecal sources (percentage of E. faecium plus E. faecalis), these variables alone could fail to provide a correct identification of fecal source in some cases. On the other hand, phenotyping with the Phene-Plate system has previously been proven to be useful to identify specific animal species as contamination sources in surface water in Australia (1).
The relationships between the β-sterols coprostanol and 24-ethylcoprostanol were different in human and nonhuman samples, as reported elsewhere previously (37), but there was a percentage of failures that prevented the effective application of these chemical indicators as fecal source discriminators.
Two-group discriminant analysis showed that using the entire set of microbial and chemical indicators measured in this study enabled the fecal source in wastewater or slurries to be ascertained. However, testing of over 20 tracers is not feasible for routine analyses because of the high cost, the time required, and the need for staff trained in a wide variety of analytical fields (which is beyond the reach of many laboratories). Furthermore, discriminant analysis carried out using different subsets of the parameters showed some promising results. A subset of parameters consisting of the enumeration of the four bacteriophage groups was able to successfully distinguish the source of fecal pollution in the wastewaters and slurries analyzed. It was also observed that only bacteriophages infecting the host strain B. thetaiotaomicron GA17 showed a high specificity to human samples. Enumeration of all these four groups of bacteriophages provided information that complemented the enumeration of bacteriophages infecting strain GA17, achieving 100% correct classification. Conversely, the variable subgroup consisting of the enumeration of different bacterial groups, genotypes of F-specific bacteriophages, fecal sterols, or bacterial phenotypes alone did not determine the fecal source with a 100% correct classification. Again, individual variables within these subgroups (for example, sorbitol-fermenting bifidobacteria) showed great differences between water samples with human fecal contamination and those with nonhuman fecal contamination. Consequently, other combinations of the most promising tracers should be considered in order to determine the lowest number of variables needed to maintain the highest possible discrimination rate of fecal source. Note that some combinations need not include the enumeration of bacteriophages infecting B. thetaiotaomicron strain GA17, which demonstrated geographical differences with regard to the clarity of plaques. We were thus especially interested in finding combinations specifically including or excluding this tracer. However, to resolve the problem of geographical variation, new host strains of Bacteroides, either B. thetaiotaomicron or other species, should be obtained for each specific geographical site in order to facilitate the enumeration of this group of phages (49). Furthermore, the variations in results of discriminant analyses of the parameter sets coincide with previous studies by other authors who have studied the assessment of statistical methods using library-dependent tests for microbial source tracking (48, 51, 58). Those authors also reported a high degree of variability in the correct classification rates among statistical methods and observed that no commonly used statistical technique emerges as superior. Our results are in general agreement with this observation; but additionally, our results suggest that some combinations of library-independent methods that showed a consistently high degree of discriminatory power could determine the origin of fecal pollution. Consequently, the use of alternative statistical methods that could determine the optimal combination of discriminant parameters and thus facilitate the development of predictive models is the logical next stage in the development of data analysis for fecal source tracking.
To this end, several statistical and machine learning methods were applied to the 38 single and derived microbial and chemical variables. The obtained predictive models provided 100% correct classification in the distinction of wastewaters of human origin and those of nonhuman origin. No differences were found between the various European geographical locations in this prediction model. A predictive model using the pair of variables SOMCPH/BTHPH and SOMCPH emerged as the optimal model and allowed the successful classification of fecal source in all cases (in both training and test sets). It was noted that the variable SOMCPH/BTHPH accounts for the greatest part of the classification. In light of this observation, the variable BTHPH alone might suffice. This could be achieved by determining a reference level for BTHPH that differentiates human samples (values above this reference) from nonhuman samples (values below this reference). This reference level could be obtained by taking the middle value between the two closest known observations (in the training set), one of which is human and the other of which is animal. This simpler rule actually achieved 100% correct classification. However, this approach may be unstable because of the closeness of the values to the reference value, especially when factors such as fecal aging or dilution in waters modify the concentrations of the parameters (named variables in statistical or machine learning analyses). A wider margin of separation is necessary in order to obtain a more robust and stable discrimination. This is accomplished by using the set of two variables SOMCPH/BTHPH and SOMCPH, in which case the margin is greater.
Alternative predictive models that do not use the variable BTHPH were also found. However, these models showed lower rates of correct classification and required more parameters and thus entailed higher costs and resource requirements. Furthermore, their percentages of correct classification were more dependent on the statistical method used. For instance, the pair of variables SOMCPH and FRNAPH II showed a 96% correct classification using a quadratic Bayesian classifier, and sets of three (FRNAPH I, FRNAPH II, and ECP) or four (e.g., SOMCPH, FRNAPH II, BA, and SFBIF) variables were needed to provide 100% correct classification when using the Euclidean one-nearest-neighbor classifier.
In conclusion, none of the tested microbial and chemical parameters were alone able to determine the source of fecal pollution in wastewaters and slurries of known human or nonhuman origin, and therefore, a suite of parameters was required. However, we demonstrated that there are a number of potentially good tracers showing high discriminatory capabilities, and hence, there is a need for alternative numerical approaches to the data analysis. The concentration of phages infecting certain strains of Bacteroides is the parameter showing the greater discriminatory power. Host strains to detect and enumerate phages of Bacteroides seem to be geographically dependent, but a method for the isolation of geographically specific host strains for the enumeration of phages infecting Bacteroides has recently been published (49). Other tracers such as FRNAPH I, FRNAPH II, ECP, BA, and SFBIF also showed a good discriminatory ability when groups of three or more variables were used. Combinations of variables based on a discriminating tracer and a universal fecal indicator seem to offer the best solutions. The universal and nondiscriminant fecal indicator provides information on the fecal load of the sample at the time it is taken. The discriminant indicator (tracer) contributes to the identification of source. If both indicators have similar persistence in the environment, their combined use could be the best way of defining predictive models suitable for any environmental water sample. Such combinations may also offer advantages when samples different from the ones tested here are analyzed (such as diluted, aged, and mixed samples). Finally, the use of different statistical or machine learning methods in conjunction with algorithms for variable selection was shown to be a feasible numerical analysis for the development of predictive models for microbial source tracking in waters. The experimental approach used in this study aimed to provide a preliminary model suitable for wastewaters and slurries, which are considered the most important starting points for fecal pollution of surface waters. Any subset of methods selected for predictive models must be effective at this level of fecal pollution. Otherwise, there is no sense in applying it to surface waters or other kinds of waters with lower values for the indicators and parameters involved. Following our experimental approach, the next stage in the development of predictive models should consider additional factors such as dilution, specific types of animal sources, persistence of microbial tracers, and complex mixtures from different sources. All these factors will progressively add complexity to the models and bring them closer to “real-world” scenarios so as to provide effective and practical solutions for fecal pollution problems.
Supplementary Material
Acknowledgments
This study was supported by European Union project EVK1-2000-22080 and Spanish Government research project CGL2004-04702-C02-01/02.
We thank the Scientific-Technical Services at the University of Barcelona for their technical support in the analysis of fecal sterols.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Ahmed, W., R. Neller, and M. Katouli. 2005. Host species-specific metabolic fingerprint database for enterococci and Escherichia coli and its application to identify sources of fecal contamination in surface waters. Appl. Environ. Microbiol. 71:4461-4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atlas, R. M. 1984. Use of microbial diversity measurements to assess environmental stress, p. 540-545. In M. J. Klug and C. A. Reddy (ed.), Current perspectives in microbial ecology. American Society for Microbiology, Washington, D.C.
- 3.Beelwilder, J., R. Niewenhuizen, A. H. Havelaar, and J. van Duin. 1996. An oligonucleotide hybridization assay for the identification and enumeration of F-specific RNA phages in surface waters. J. Appl. Bacteriol. 80:179-186. [DOI] [PubMed] [Google Scholar]
- 4.Bianchi, M. A. G., and A. J. M. Bianchi. 1982. Statistical sampling of bacterial strains and its use in bacterial diversity measurement. Microb. Ecol. 8:61-69. [DOI] [PubMed] [Google Scholar]
- 5.Blanch, A. R., J. L. Caplin, A. Iversen, I. Kühn, A. Manero, H. D. Taylor, and X. Vilanova. 2003. Comparison of enterococcal populations related to urban and hospital wastewater in various climatic and geographic European regions. J. Appl. Microbiol. 94:994-1002. [DOI] [PubMed] [Google Scholar]
- 6.Blanch, A. R., L. Belanche-Muñoz, X. Bonjoch, J. Ebdon, C. Gantzer, F. Lucena, J. Ottoson, C. Kourtis, A. Iversen, I. Kühn, L. Moce, M. Muniesa, J. Schwartzbrod, S. Skraber, G. Papageorgiou, H. D. Taylor, J. Wallis, and J. Jofre. 2004. Tracking the origin of faecal pollution in surface water. An ongoing project within the European Union Research Programme. J. Water Health 2:249-260. [PubMed] [Google Scholar]
- 7.Bonjoch, X., E. Ballesté, and A. R. Blanch. 2004. Multiplex PCR with 16S rRNA gene-targeted primers of Bifidobacterium spp. to identify sources of fecal pollution. Appl. Environ. Microbiol. 70:3171-3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonjoch, X., E. Ballesté, and A. R. Blanch. 2005. Enumeration of bifidobacterial populations with selective media to determine the source of waterborne faecal pollution. Water Res. 39:1621-1627. [DOI] [PubMed] [Google Scholar]
- 9.Brion, G. M., T. R. Neelakantan, and S. Lingireddy. 2002. A neural-network-based classification scheme for sorting sources and ages of fecal contamination in water. Water Res. 36:3765-3774. [DOI] [PubMed] [Google Scholar]
- 10.Carson, C. A., B. L. Shear, M. R. Ellersieck, and J. D. Schnell. 2003. Comparison of ribotyping and repetitive extragenic palindromic-PCR for identification of fecal Escherichia coli from humans and animals. Appl. Environ. Microbiol. 69:1836-1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christianini, N., and J. Shawe-Taylor. 2000. An introduction to support vector machines, p. 189. Cambridge University Press, Cambridge, United Kingdom.
- 12.Cole, D., S. C. Long, and M. D. Sobsey. 2003. Evaluation of F+ RNA and DNA coliphages as source-specific indicators of fecal contamination in surface waters. Appl. Environ. Microbiol. 69:6507-6514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dombeck, P. E., L. K. Johson, S. J. Zimmerley, and M. J. Sadowsky. 2000. Use of repetitive DNA sequences and the PCR to differentiate Escherichia coli isolates from human and animal sources. Appl. Environ. Microbiol. 66:2572-2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duda, R. O., P. E. Hart, and L. Stork. 2001. Pattern classification, p. 654. John Wiley & Sons, Inc., New York, N.Y.
- 15.Field, K. G., A. E. Bernhard, and T. J. Brodeur. 2003. Molecular approaches to microbiological monitoring: fecal source detection. Environ. Monit. Assess. 81:313-326. [PubMed] [Google Scholar]
- 16.Hall, B. G., and W. Faunce. 1987. Functional genes for cellobiose utilization in natural isolates of Escherichia coli. J. Bacteriol. 169:2713-2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harwood, V. J., J. Whitlock, and V. Withington. 2000. Classification of antibiotic resistance patterns of indicator bacteria by discriminant analysis: use in predicting the source of fecal contamination in subtropical waters. Appl. Environ. Microbiol. 66:3698-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hertz, J., A. Krogh, and R. G. Palmer. 1991. Introduction to the theory of neural computation, p. 327. Addison-Wesley, Redwood City, Calif.
- 19.Hunter, P. R., and M. A. Gaston. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.International Standardisation Organisation. 1980. Water quality. Sampling. Part 1. Guidance on the design of sampling programmes. ISO 5667-1. International Standardisation Organisation, Geneva, Switzerland.
- 21.International Standardisation Organisation. 1982. Water quality. Sampling. Part 2. Guidance on sampling techniques. ISO 5667-2. International Standardisation Organisation, Geneva, Switzerland.
- 22.International Standardisation Organisation. 1983. Microbiology—general guidance for the preparation of dilutions for microbiological examination. ISO 6887. International Standardisation Organisation, Geneva, Switzerland.
- 23.International Standardisation Organisation. 1984. Detection and enumeration of fecal streptococci in water. Part 2. Method by membrane filtration. ISO 7899/1. International Standardisation Organisation, Geneva, Switzerland.
- 24.International Standardisation Organisation. 1985. Water quality. Sampling. Part 3. Guidance on the preservation and handling of samples. ISO 5667-3. International Standardisation Organisation, Geneva, Switzerland.
- 25.International Standardisation Organisation. 1986. Detection and enumeration of the spores of sulphite-reducing anaerobes. Clostridia. Part 1. Method by enrichment in a liquid medium. ISO 6461-1. International Standardisation Organisation, Geneva, Switzerland.
- 26.International Standardisation Organisation. 1992. Quantities and units. Part 0. General principles. ISO 31-0. International Standardisation Organisation, Geneva, Switzerland.
- 27.International Standardisation Organisation. 1995. Detection and enumeration of bacteriophages. Part 1. Enumeration of F-specific RNA bacteriophages. ISO 10705-1. International Standardisation Organisation, Geneva, Switzerland.
- 28.International Standardisation Organisation. 2000. Detection and enumeration of E. coli and coliform bacteria. Part 1. Membrane filtration method. ISO 9308-1. International Standardisation Organisation, Geneva, Switzerland.
- 29.International Standardisation Organisation. 2000. Water quality. Detection and enumeration of bacteriophages. Part 2. Enumeration of somatic coliphages. ISO 10705-2. International Standardisation Organisation, Geneva, Switzerland.
- 30.International Standardisation Organisation. 2001. Water quality. Detection and enumeration of bacteriophages. Part 2. Enumeration of bacteriophages infecting Bacteroides fragilis. ISO 10705-4. International Standardisation Organisation, Geneva, Switzerland.
- 31.Kira, K., and L. A. Rendell. 1992. The feature selection problem: traditional methods and a new algorithm, p. 129-134. In Proceedings of the Ninth National Conference on Artificial Intelligence. AAAI Press, Menlo Park, Calif.
- 32.Kohavi, R., and G. H. John. 1997. Wrappers for feature subset selection, Artif. Intell. 97:273-324. [Google Scholar]
- 33.Kühn, I. 1985. Biochemical fingerprinting of Escherichia coli: a simple method for epidemiological investigations. J. Microbiol. Methods 3:159-170. [Google Scholar]
- 34.Kühn, I., A. Iversen, L. G. Burman, B. Olsson-Liljequist, A. Franklin, M. Finn, F. Aarestrup, A. M. Seyfarth, A. R. Blanch, X. Vilanova, H. Taylor, J. Caplin, M. A. Moreno, L. Dominguez, I. A. Herrero, and R. Mollby. 2003. Comparison of enterococcal populations in animals, humans, and the environment—a European study. Int. J. Food Microbiol. 88:133-145. [DOI] [PubMed] [Google Scholar]
- 35.Kühn, I., G. Allestam, M. Engdahl, and T. A. Stenstrom. 1997. Biochemical fingerprinting of coliform bacterial populations—comparisons between polluted river water and factory effluents. Water Sci. Technol. 35:343-350. [Google Scholar]
- 36.Kühn, I., G. Allestam, T. A. Stenstrom, and R. Möllby. 1991. Biochemical fingerprinting of water coliform bacteria, a new method for measuring phenotypic diversity and for comparing different bacterial populations. Appl. Environ. Microbiol. 57:3171-3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leeming, R., A. Ball, N. Ashbolt, and P. D. Nichols. 1996. Using faecal sterols from humans and animals to distinguish faecal pollution in receiving waters. Water Res. 30:2893-2900. [Google Scholar]
- 38.Liu, H., and H. Motoda. 1998. Feature selection for knowledge discovery and data mining, p. 214. Springer-Verlag, New York, N.Y.
- 39.Lucena, F., X. Mendez, A. Moron, E. Calderon, C. Campos, A. Guerrero, M. Cardenas, C. Gantzer, L. Shwartzbrood, S. Skraber, and J. Jofre. 2003. Occurrence and densities of bacteriophages proposed as indicators and bacterial indicators in river waters from Europe and South America. J. Appl. Microbiol. 94:808-815. [DOI] [PubMed] [Google Scholar]
- 40.Malakoff, D. 2002. Microbiologists on the trail of polluting bacteria. Science 295:2352-2353. [DOI] [PubMed] [Google Scholar]
- 41.Manero, A., and A. R. Blanch. 1999. Identification of Enterococcus spp. with a biochemical key. Appl. Environ. Microbiol. 65:4425-4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mara, D. D., and J. I. Oragui. 1983. Sorbitol-fermenting bifidobacteria as specific indicators of human faecal pollution. J. Appl. Bacteriol. 55:349-357. [DOI] [PubMed] [Google Scholar]
- 43.Martellini, A., P. Payment, and R. Villemur. 2005. Use of eukaryotic mitochondrial DNA to differentiate human, bovine, porcine and ovine sources in fecally contaminated surface water. Water Res. 39:541-548. [DOI] [PubMed] [Google Scholar]
- 44.Matsuki, T., K. Watanabe, R. Tanaka, M. Fukuda, and H. Oyaizu. 1999. Distribution of bifidobacterial species in human intestinal microflora examined with 16S rRNA-gene-targeted species-specific primers. Appl. Environ. Microbiol. 65:4506-4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mitchell, M. 1997. Machine learning, p. 414. McGraw-Hill Higher Education, New York, N.Y.
- 46.Myoda, S. P., C. A. Carson, J. J. Fuhrmann, B. K. Hahm, P. G. Hartel, H. Yampara-Lquise, L. Johnson, R. L. Kuntz, C. H. Nakatsu, M. J. Sadowsky, and M. Samadpour. 2003. Comparison of genotypic-based microbial source tracking methods requiring a host origin database. J. Water Health 1:167-180. [PubMed] [Google Scholar]
- 47.Noble, R., S. Allen, A. Blackwood, W. Chu, S. Jiang, G. Lovelace, M. Sobsey, J. Stewart, and D. Wait. 2003. Use of viral pathogens and indicators to differentiate between human and non-human fecal contamination in a microbial source tracking comparison study. J. Water Health 1:195-207. [PubMed] [Google Scholar]
- 48.Parveen, S., K. M. Portier, K. Robinson, L. Edmiston, and M. L. Tamplin. 1999. Discriminant analysis of ribotype profiles of Escherichia coli for differentiating human and nonhuman sources of fecal pollution. Appl. Environ. Microbiol. 65:3142-3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Payán, A., J. Ebdon, H. Taylor, C. Gantzer, J. Ottoson, G. T. Papageorgiou, A. R. Blanch, F. Lucena, J. Jofre, and M. Muniesa. 2005. Method for isolation of Bacteroides bacteriophage host strains suitable for tracking sources of fecal pollution in water. Appl. Environ. Microbiol. 71:5659-5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Puig, A., N. Queralt, J. Jofre, and R. Araujo. 1999. Diversity of Bacteroides fragilis strains in their capacity to recover phages from human and animal wastes and from fecally polluted wastewater. Appl. Environ. Microbiol. 65:1772-1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ritter, K. J., E. Carruthers, C. A. Carson, R. D. Ellender, V. J. Harwood, K. Kingsley, C. Nakatsu, M. Sadowsky, B. Shear, B. West, J. E. Whitlock, B. A. Wiggins, and J. D. Wilbur. 2003. Assessment of statistical methods used in library-based approaches to microbial source tracking. J. Water Health 1:209-223. [PubMed] [Google Scholar]
- 52.Schaper, M., and J. Jofre. 2000. Comparison of methods for detecting genotypes of F-specific RNA bacteriophages and fingerprinting the origin of faecal pollution in water samples. J. Virol. Methods 89:1-10. [DOI] [PubMed] [Google Scholar]
- 53.Schaper, M., J. Jofre, M. Uys, and W. O. K. Grabow. 2002. Distribution of genotypes of F-specific RNA bacteriophages in human and non-human sources of faecal pollution in South Africa and Spain. J. Appl. Microbiol. 92:3605-3613. [DOI] [PubMed] [Google Scholar]
- 54.Scott, T. M., J. B. Rose, T. M. Jenkins, S. R. Farrah, and J. Lukasik. 2002. Microbial source tracking: current methodology and future directions. Appl. Environ. Microbiol. 68:5796-5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Simpson, J. M., J. W. Santo Domingo, and D. J. Reasoner. 2002. Microbial source tracking: state of the science. Environ. Sci. Technol. 36:5279-5288. [DOI] [PubMed] [Google Scholar]
- 56.Stewart, J. R., R. D. Ellender, J. A. Gooch, S. Jiang, S. P. Myoda, and S. B. Weisberg. 2003. Recommendations for microbial source tracking: lessons from a methods comparison study. J. Water Health 1:225-231. [PubMed] [Google Scholar]
- 57.Whitlock, J. E., D. T. Jones, and V. J. Harwood. 2002. Identification of the sources of fecal coliforms in an urban watershed using antibiotic resistance analysis. Water Res. 36:4273-4282. [DOI] [PubMed] [Google Scholar]
- 58.Wiggins, B. A. 1996. Discriminant analysis of antibiotic resistance patterns in fecal streptococci, a method to differentiate human and animal sources of fecal pollution in natural waters. Appl. Environ. Microbiol. 62:3997-4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wiggins, B. A., P. W. Cash, W. S. Creamer, S. E. Dart, P. P. Garcia, T. M. Gerecke, J. Han, B. L. Henry, K. B. Hoover, E. L. Johnson, K. C. Jones, J. G. McCarthy, J. A. McDonough, S. A. Mercer, M. J. Noto, H. Park, M. S. Phillips, S. M. Purner, B. M. Smith, E. N. Stevens, and A. K. Varner. 2003. Use of antibiotic resistance analysis for representativeness testing of multiwatershed libraries. Appl. Environ. Microbiol. 69:3399-3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Witten, I. H., and E. Frank. 2005. Data mining: practical machine learning tools and techniques, 2nd ed, p. 369. Morgan Kaufmann, San Francisco, Calif.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.