Abstract
Streptococcus mutans, the major pathogen responsible for dental caries in humans, is a biofilm-forming bacterium. In the present study, 17 different pulsed-field gel electrophoresis patterns of genomic DNA were identified in S. mutans organisms isolated clinically from whole saliva. The S. mutans isolates showed different abilities to form biofilms on polystyrene surfaces in semidefined minimal medium cultures. Following cultivation in a flow cell system in tryptic soy broth with 0.25% sucrose and staining using a BacLight LIVE/DEAD system, two strains, designated FSC-3 and FSC-4, showed the greatest and least, respectively, levels of biofilm formation when examined with confocal laser scanning microscopy. Further, image analyses of spatial distribution and architecture were performed to quantify the merged green (live cells) and red (dead cells) light. The light intensity of the FSC-3 biofilm was greater than that of the FSC-4 biofilm in the bottom area but not in the top area. S. mutans whole-genome array results showed that approximately 3.8% of the genes were differentially expressed in the two strains, of which approximately 2.2%, including bacitracin transport ATP-binding protein gene glrA and a BLpL-like putative immunity protein gene, were activated in FSC-3. In addition, about 1.6% of the genes, including those associated with phosphotransferase system genes, were repressed. Analyses of the glrA-deficient strains and reverse transcription-PCR confirmed the role of the gene in biofilm formation. Differential assessment of biofilm-associated genes in clinical strains may provide useful information for understanding the morphological development of streptococcal biofilm, as well as for colonization of S. mutans.
Streptococcus mutans, a biofilm-forming bacterium considered to be the primary etiological agent of human dental caries (16), possesses a variety of abilities to colonize tooth surfaces. S. mutans, under certain conditions, is numerically significant in cariogenic biofilms and forms biofilms with other organisms in the oral cavity (6) following colonization and the eruption of primary teeth (3). Further, epidemiological surveys have confirmed that greater numbers of S. mutans organisms in children are associated with a higher incidence of decayed, missing, and filled (dmf) teeth (13, 25).
Recently, considerable research with a variety of microscopy techniques has focused on the structure of intact visible biofilm. In previous studies, the spatial arrangement of microorganisms in dental biofilm samples showed voids outlined by layers of vital bacteria, which themselves were packed in layers of dead materials (2, 18). Although the structure of biofilm has been reported, few studies have quantified the differences in structure, although studies of the cellular functions that are modified during the cellular transition from the planktonic to the biofilm state are in progress, with some findings recently reported (23).
Isolation and characterization of the genes involved with biofilm formation may contribute to elucidating how S. mutans responds to environmental signals in the oral cavity. Previous studies have indicated the roles of sucrose and glucosyltransferases in S. mutans biofilm formation (48). Others have implicated several genes that are associated with genetic competence (24), including com genes (25, 50), which play other regulatory roles, together with one or more other genes, including ccpA and brpA (lytR) (43) and luxS (29). Those genes have also been shown to function with putative two-component response regulators (4), which are involved in biofilm formation. DNA microarrays have been used to monitor global gene expression profiles in response to different stimuli (44), such as heat shock and other stresses (17, 45), quorum sensing (10, 35), anaerobic metabolism (49), sporulation (11), and biofilm formation (35, 44). For example, a recent study of Pseudomonas aeruginosa by using a DNA microarray technique showed that only 1% of the genes were differentially expressed in biofilms compared to those expressed in suspension cells (44). In contrast, a proteomics approach used to study P. aeruginosa-associated biofilms showed a greater number of changes than with transcription profiling (42). Although there have been studies on biofilm formation by a variety of bacteria by using DNA microarrays, little is known about that associated with S. mutans. In addition, many reports have compared planktonic cells with biofilm cells by using laboratory strains (36, 37, 44); however, there is scant information regarding differential gene expression in biofilm formation in clinical strains. Clinical strains can survive in severe conditions involving various antibacterial agents, such as seen in the oral cavity, which leads to the expression of several genes involved with biofilm formation. In contrast, laboratory strains may not require those genes while being cultured under mild and planktonic conditions and may lose their expression.
In the present study, we attempted to quantify the intensity of biofilm formation by S. mutans clinical strains by measuring biofilm volume with confocal laser scanning microscopy (CLSM). Further, we analyzed how gene expression differs between various isolated S. mutans strains that showed either high or low levels of ability to form biofilm, and we compared the results with those regarding biofilm formation ability by using DNA microarray and reverse transcription-PCR (RT-PCR) methods.
MATERIALS AND METHODS
Subjects.
One hundred seventy-four pairs of mothers and children (3 years old) from the city of Yokohama in Japan participated in this study. The aim and details of the experiments were explained, and consent was obtained from all subjects prior to obtaining the samples. The study was conducted according to the ethical guidelines of Asahikawa Medical College and the Helsinki Declaration. Dental examinations were conducted under artificial white light by trained dentists. According to WHO criteria (47), scores for dmf teeth were recorded, along with findings of dental caries.
Human saliva collection.
Whole saliva samples were collected on cotton swabs from the subjects, after stimulation by biting paraffin gum for 5 min, and then placed in transport fluid (0.4% agar, 0.15% thioglycolate/phosphate-buffered saline [PBS]) and sent to Bio Medical Laboratory (BML) (Tokyo, Japan) for measurement of mutans streptococci (mS) and total streptococci (tS). Other saliva samples from three healthy human subjects (28 to 42 years old) were also collected after stimulation by biting paraffin gum and placed into ice-chilled sterile bottles over a period of 5 min. The samples were then clarified by centrifugation at 10,000 × g for 10 min, filter sterilized, and used immediately for biofilm assays, utilizing a flow cell system and a 96-well plate.
Sampling and bacteriological methods.
At BML, each saliva sample were poured onto mitis salivarius agar (MTS) (Nippon Becton Dickinson Co. Ltd., Tokyo, Japan) or modified MTS containing 0.2 U/ml of bacitracin (MMSB) (38), using an Eddy Jet spiral plating system (IUL, S.A., Barcelona, Spain), and incubated at 37°C under anaerobic conditions for 48 h. MTS and MMSB were used to count tS and mS, respectively. tS and mS colonies were identified by their characteristic appearance, and the mS ratio was calculated as the number of mS colonies/number of tS colonies × 100. Further, colonies were collected at random and tested using a PCR method with the GTF primers listed in Table 1, as previously reported (32). DNA from the bacteria was extracted using a DNeasy tissue kit (QIAGEN GmbH, Hilden, Germany) according to the manufacturer's instructions. The PCR assays were designed to discriminate between S. mutans and Streptococcus sobrinus by targeting the genes encoding the glucan-synthesizing enzyme (gtfB and gtfI). S. mutans isolates were propagated overnight in brain heart infusion (BHI) (Difco Laboratories, Detroit, MI) broth and frozen at −80°C, after the addition of glycerol to 25%, for subsequent use in genomic DNA preparations that were analyzed by pulsed-field gel electrophoresis (PFGE).
TABLE 1.
Oligonucleotide sequences of PCR primers
| Primer | Directiona | Sequenceb | Target gene product | Product size (bp) |
|---|---|---|---|---|
| PS0941-a | Fw (EcoRI) | 5′-CCGAATTCACTTGTCCAAACGATATGGA-3′ | Bacitracin transport ATP-binding | 1,047 |
| Rev (PstI) | 5′-CCCTGCAGTGTGGAAGCTGGATACCAA-3′ | protein GlrA | ||
| PS0941-b | Fw | 5′-GCGATCAAAGAATTTCGGC-3′ | 434 | |
| Rev | 5′-TTCTAAATCTTGGCGTTTG-3′ | |||
| PS1731 | Fw | 5′-AGACTTTTAACAGCGCAGC-3′ | BLpL-like putative immunity protein | 338 |
| Rev | 5′-CCCCATAATAAAAGAAGAGAG-3′ | |||
| PS0092 | Fw | 5′-CTCATAATCATATGGCATCAG-3′ | Putative PTS system IIA component | 386 |
| Rev | 5′-CTCATCCCCCTCATCATTT-3′ | |||
| PS1365 | Fw | 5′-GATGAAGAGGAAAGCGTATT-3′ | Conserved hypothetical protein S1365 | 340 |
| Rev | 5′-GCCTCCCGAAATGGTTTTA-3′ | |||
| LARNA5 | Fw | 5′-GTTGTCCGGATTTATTGGG-3′ | Control primer | 248 |
| LARNA6 | Rev | 5′-GGGTATCTAATCCTGTTCGC-3′ | ||
| GTFB-F | Fw | 5′-ACTACACTTTCGGGTGGCTTGG-3′ | Glucosyltransferase GtfB | 517 |
| GTFB-R | Rev | 5′-CAGTATAAGCGCCAGTTTCATC-3′ | ||
| GTFB-FIN | Fw | 5′-AAAGCAGATTCTAATGAATCGA-3′ | Glucosyltransferase GtfB (inner primer) | 469 |
| GTFB-RIN | Rev | 5′-AATGTAAAATTTTGCCATCAGC-3′ | ||
| GTFI-F | Fw | 5′-GATAACTACCTGACAGCTGACT-3′ | Glucosyltransferase GtfI | 712 |
| GTFI-R | Rev | 5′-AAGCTGCCTTAAGGTAATCACT-3′ | ||
| GTFI-FIN | Fw | 5′-TGGTATCGTCCAAAATCAATCC-3′ | Glucosyltransferase GtfI (inner primer) | 664 |
| GTFI-RIN | Rev | 5′-AGATTTGCAGTTGGTCAGCATC-3 |
Fw, forward; Rev, reverse.
Restriction site sequences are underlined.
PFGE.
PFGE analysis of mutans streptococcal DNA was performed as described elsewhere (19, 20), with minor modifications. Frozen stock cultures were poured onto BHI agar plates and incubated at 37°C under anaerobic conditions (BBL GasPak Systems; Becton Dickinson and Company, Franklin Lakes, NJ). Colonies on the BHI agar plates were scraped off using a sterilized spoon and resuspended in 200 μl of distilled water (DW) (optical density of 0.2). An equal amount of 1% low-melting-point agarose (Bio-Rad Laboratories, Richmond, CA) was then prepared in DW, mixed gently, melted in a microwave oven until liquid, and added to each cell suspension, after which the mixture was transferred to Bio-Rad plug molds (two wells per sample) with a pipette. Two plugs were prepared for each isolate and refrigerated on ice for 20 min.
For restriction endonuclease digestion, one half of a plug slice from each isolate was suspended overnight in a mixed solution (3 μl NotI or SmaI, 10 μl H or T buffer, 10 μl 0.1% bovine serum albumin, and up to 100 μl DW) at 37°C. The plug slices were then loaded and run on a 1% pulsed field certified agarose (Bio-Rad) gel in 45 mM Tris-borate-1 mM EDTA buffer. PFGE was performed using a CHEF-DRII apparatus (Bio-Rad Laboratories) in 0.5× 45 mM Tris-borate-1 mM EDTA buffer at 10 to 12°C. The switch times used for directional change of the electrical fields for NotI and SmaI were 40 seconds for 20 h at 180 V and 1 to 30 seconds for 20 h at 200 V, respectively. A lambda liner DNA ladder (Bio-Rad) ranging from 48.5 to 970 kbp was used as the size standard. Following PFGE, the gels were stained with ethidium bromide (0.2 μg/ml) and photographed under UV transillumination.
Biofilm formation assay.
Biofilm formation by all strains was assayed using a method described previously (24, 26), with some modifications. To evaluate the biofilms formed by the isolated S. mutans strains, 40 μl of cell suspension (8 × 106 CFU) and 160 μl of semidefined minimal medium (SDM) (chosen a minimal medium) (24) or tryptic soy broth without dextrose (Difco) supplemented with 0.25% sucrose (TSB) (chosen a rich medium) were mixed in the wells of 96-well (flat-bottom) microtiter plates (Sumitomo Bakelite, Tokyo, Japan). Before addition of the cell suspension, the wells were coated with whole saliva in TSB culture but were not coated in SDM, as biofilms are unable to form in saliva-coated wells in the presence of SDM. After the plates were incubated at 37°C for 16 h under anaerobic conditions, the liquid medium was removed and the wells were rinsed a second time with sterilized DW. The plates were then air dried and stained with 0.25% safranin for 15 min. After being stained, the plates were rinsed with DW to remove excess dye and then air dried. Each biofilm was examined without dissolving with solvent, using an enzyme-linked immunosorbent assay microplate reader (Multiskan Bichromatic; Laboratory Japan, Tokyo, Japan), as the biofilm was applied uniformly onto the bottoms of the wells in the 96-well plates. Quantification of the stained biofilm was performed by measuring A492 absorbance.
Flow chamber experiments.
Biofilm samples were cultivated at 37°C in three-channel flow cells (46) with individual channel dimensions of 1 by 4 by 40 mm and supplied with a flow of TSB, as biofilms under flow show better formation when cultured in TSB than in SDM. The flow system (Stovall Flowcell; Stovall Life Science Inc., Greensboro, NC) was assembled and prepared as described by Christensen et al. (8). The substratum consisted of a microscope glass coverslip, and the flow cells were covered with filter-sterilized human saliva samples and left for 30 min. After the saliva was carried away, the cells were inoculated with an overnight culture of each strain diluted to 1 × 106 to 10 × 106 CFU/ml. Following inoculation, the medium flow was stopped for 1 h. Next, the flow was started again and the medium was pumped through the flow cells at a constant rate of 3 ml/hour for 20 h using a peristaltic pump (Ismatec; IDEX Corp., Glattbrugg-Zürich, Switzerland).
CLSM.
Noninvasive confocal imaging of fully hydrated biofilms was performed using a Fluoview CLSM (Olympus, Tokyo, Japan) fitted with a water immersion dipping objective lens (100×) and a Kr-Ar laser. The specimens were stained for 30 min with BacLight LIVE/DEAD bacterial viability kit solution (4 ml of DW containing 6 μl each of components A and B) (Molecular Probes, Leiden, The Netherlands). The biofilm structure was analyzed using a series of horizontal (xy) optodigital sections, each 5.0 μm thick, with the intervening gaps between the horizontal sections ranging from 0 to 50 mm over the entire height (z axis) of the biofilm. In addition, we analyzed the vertical (xz) sections, which were recorded from the center of each biofilm, to determine the architecture. Each biofilm was scanned at five randomly selected positions away from the disk edge. The digital images were processed using Fluoview software version 2.0 (Olympus).
Image analysis.
The intensity profiles of both viable (488-nm) and nonviable (568-nm) channels for the image stacks were calculated using Olympus Fluoview software. This application is able to save images in several formats and also allows numerical analysis of images, such as average pixel counts, and then saves the data as an Excel file for later analysis (28). The data were normalized against the maximum image intensity for the total intensities of both channels. Briefly, the software divided the optional area (141.145 μm by 141.145 μm) into a grid of 64 by 64 masses in xy planes and presented light intensity as a value of 0 to 4,095 for each mass (see Fig. 3C). The rate of light intensity per highest intensity value (4,095) was calculated in each mass. If the average of intensity rate or area (μm2) was more than 25%, the intensity rate was recalculated as the volume of biofilm formation of green light (live cells) and red light (dead cells).
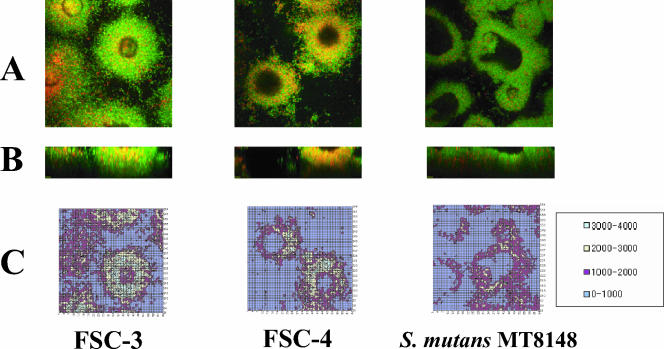
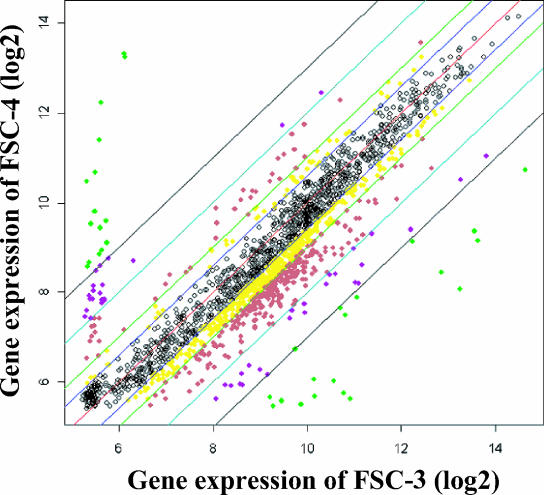
FIG. 3.
(A) CLSM images showing xy planes (141.145 μm by 141.145 μm) of the biofilm bottom area. Viable cells are colored green, and nonviable cells are colored red. (B) CLSM images showing xz planes (141.145 μm by 50.0 μm) recorded from the center of a single biofilm shown in panel A. (C) Two-dimensional contour map generated from the CLSM image shown in panel A. As a control, S. mutans MT8148 organisms were analyzed. Light intensities were divided into four color groups as shown in the box. The data are representative of three independent experiments.
Cell harvest and total RNA isolation.
To analyze the expression of mRNA in the S. mutans biofilms, biofilm cells were harvested from two sources: the microscope glass coverslips after removal from the flow cell chambers and the culture plates. As in the production of biofilm on six-well culture plates (Corning Incorporated, Corning, NY), 400 μl of cell suspension (8 × 106 CFU) and 1,600 μl of SDM were mixed and cultivated in the wells at 37°C for 16 h under anaerobic conditions. The biofilm cells were then scraped with a sterilized scraper after being washed three times with sterilized PBS. The planktonic cells were also cultivated in BHI. All cells were also harvested at 500 × g and 4°C and washed with sterilized PBS three times. Total RNA was isolated from the biofilm and planktonic cells by using a modified protocol with an RNeasy minikit (QIAGEN), which included an on-column DNase digestion with RNase-free DNase I (QIAGEN). Briefly, the harvested cells were suspended in PBS and sonicated, and then the samples were centrifuged at 5,000 × g at 4°C for 5 min. Next, the precipitate was resuspended in 100 μl of Tris-EDTA buffer containing 3 mg/ml lysozyme and 40 U of mutanolysin, followed by incubation at 37°C for 1 h, according to a modification of the manufacturer's instructions reported previously (51). Total RNA from extractions of flow cell biofilm samples performed four times was pooled and mixed to provide a sufficient quantity of RNA for analysis in the microarray. For the DNA microarray and RT-PCR analyses, 50 μg of total RNA from the flow cell biofilms and 1 mg of total RNA from biofilms or planktonic cells cultured in six-well plates were used.
DNA microarrays.
DNA microarray analyses were performed using a NimbleGen system (NimbleGen Systems, Inc., Madison, WI). These arrays can detect 1,960 of the 1,963 S. mutans UA159 open reading frames (ORFs) (1). An array with three sets of S. mutans ORFs was used to precisely detect the relative gene expressions of the FSC-3 and -4 strains. Data analysis and determination of absent and present cells were performed using raw fluorescence intensity values with Microarray Suite version 5.0 (Affymetrix, Santa Clara, CA). Gene expressions of mRNA were normalized to frequency values by using the robust multichip average (5). The gene functions were obtained from the National Center for Biotechnology Information database (http://www.ncbi.nlm.nih.gov/).
RT-PCR.
For RT-PCR, cDNA templates were created from 1 μg of RNA by using the SuperScript first-strand synthesis system (Invitrogen Corp., Carlsbad, CA) according to the recommended procedure, with the primers shown in Table 1. To check for DNA contamination, purified total RNA without reverse transcriptase served as a negative control. The resulting cDNA and negative control were amplified using a TaKaRa PCR amplification kit (Takara Bio Inc). We assayed the FSC-3 and -4 strains by using cDNA synthesized from RNA extracted from two independent biofilms grown in SDM and TSB cultures, as well as from planktonic cells grown in BHI in triplicate for each gene. The results were normalized using the conserved primers LARNA5 and LARNA5, which were selected on the basis of a comparison between the available 16S rRNA sequences of lactobacilli and gram-positive bacteria, including oral streptococci (27), as internal controls. To analyze the gene expression quantitatively in RT-PCR, PCR was performed at various cycles (24 to 50). The gene expression between FSC-3 and FSC-4 was assessed at the acceptable PCR cycle to each primer for PCR production before stationary phase.
Construction of glrA mutant.
An ORF for the glrA gene region was identified in the S. mutans UA159 database at the University of Oklahoma Advanced Center for Genomic Technology (http://www.genome.ou.edu/smutans.html). A mutant of the glrA gene was created using a double-crossover homologous recombination by insertion of an erythromycin resistance determinant into the gene. The plasmid used for inactivation of the glrA gene was prepared as follows. The PCR fragment of the glrA region containing the BamHI site within the gene was amplified with the forward primer for glrA (EcoRI) and the reverse primer for glrA (PstI) (Table 1, PS0941-a), using chromosomal DNA from S. mutans FSC-3 as the template. The amplified fragment was then inserted into multiple cloning sites of pUC19 (Invitrogen) following EcoRI-PstI digestion. The resultant plasmid was digested with BamHI, after which the erythromycin cassette isolated from pResEmMCS10 was inserted (34). The plasmid was linearized with SphI, and S. mutans FSC-3 was transformed with the resultant linear plasmid. Confirmation that plasmid insertion caused gene disruption was by either Southern blotting or PCR (data not shown).
Statistical analysis.
Comparisons of biofilm formation levels between FSC-3 and -4 and between FSC-3 and FSC-3D glrA were performed using a Mann-Whitney U test. Differences of 0.05 were considered statistically significant.
RESULTS
Isolation of S. mutans from human subjects.
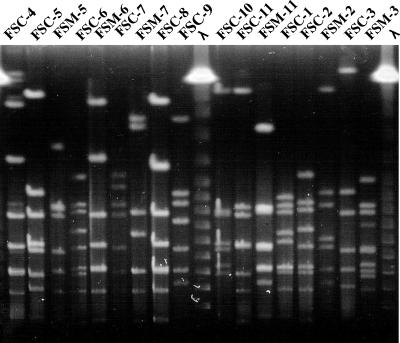
S. mutans was isolated from both mother and child in 19 (10.9%) of 174 mother-child pairs, of which 11 (57.9%) paired samples were identified by PCR and cultivated for use in the following experiments. Some of the S. mutans organisms from the other eight pairs died during transit to our laboratory from BML. A single colony was randomly taken from one MMSB plate per subject in our laboratory. The PFGE pattern of DNA digested with NotI or SmaI from the S. mutans sample from a mother was identical to that for the S. mutans sample from her child for 5 (45.5%) of the 11 mother-child pairs. Following digestion with NotI from S. mutans, 17 different patterns of genomic DNA were found, including the 5 fragment patterns of genomic DNA from children that were identical to those from their mothers (FSC-1, FSC-4, FSC-8, FSC-9, and FSC-10) (Fig. 1). Thus, various S. mutans genetic types were isolated from our human subjects.
FIG. 1.
PFGE patterns of genomic DNA from S. mutans isolates. At least 17 different PFGE patterns were found in the paired samples from mothers and children. NotI was used for digestion. λ, lambda DNA ladder molecular size standards; FSC, strain from child; FSM, strain from mother; 1 to 11, subject numbers. FSC and FSM strains with the same number are child-mother pairs. The data shown are representative of three independent experiments.
Evaluation of biofilm formation by isolated S. mutans.
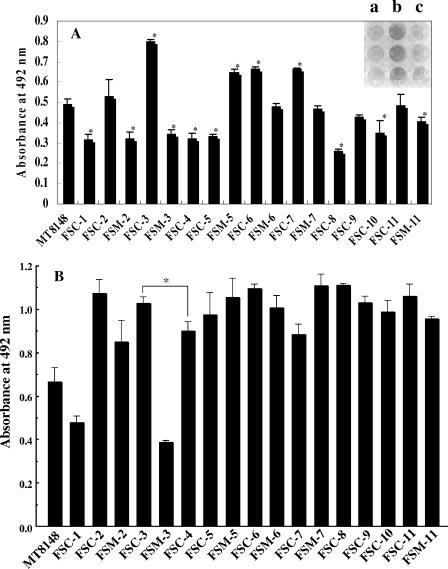
Seventeen different types of S. mutans on polystyrene surfaces in SDM were characterized in an in vitro biofilm formation assay (Fig. 2A). In SDM, strains FSC-3, FSM-5, FSC-6, and FSC-7 showed greater levels of biofilm formation, while FSC-1, FSM-2, FSM-3, FSC-4, FSC-5, FSC-8, and FSC-10 showed lower levels of biofilm formation, than a laboratory strain (S. mutans MT8148) (Fig. 2A). In the TSB cultures, all strains except FSC-1 and FSM-3 showed greater levels of biofilm formation than S. mutans MT8148 (Fig. 2B), while S. mutans MT8148 showed a level of biofilm formation similar to that of the other laboratory strains, S. mutans MT6229 and OMZ175, in both SDM and TSB cultures (data not shown). The growth rates in planktonic cell culture were comparable for all clinical strains and laboratory strains (data not shown). These results indicated multiple and medium nutrition-dependent variations in the abilities of S. mutans with different PFGE patterns to form biofilms on polystyrene surfaces. The 11 strains from the FSC child group in SDM cultures could be equivalently divided into two groups: those with high biofilm formation (greater than that of MT8148) (FSC-2, -3, -6, -7, and -11) and those with low biofilm formation (lower than that of MT8148) (FSC-1, -4, -5, -8, -9, and -10). Comparisons between the high and low biofilm formation groups were performed for incidence of dmf teeth, mS number, and mS ratio in saliva. The incidence of dmf teeth, mS number, and mS ratio were each greater in the high biofilm formation group (3.0 ± 4.7, 17.2 × 103 ± 27.8 × 103 CFU/ml, and 3.1% ± 5.5%, respectively) than in the low biofilm formation group (1.2 ± 2.2, 9.6 × 103 ± 18.1 × 103, and 0.8% ± 0.9%, respectively), whereas there were no significant differences in clinical status. Thereafter, the FSC-3 and FSC-4 strains were selected and used as typical strains showing greater and lower, respectively, biofilm formation in the following experiments using the flow cell system, while S. mutans MT8148 served as a control.
FIG. 2.
Biofilm formation by 17 S. mutans genotypes. Graphs show quantification of biofilms formed after 16 h of culture in SDM (A) and TSB (B). The upper right photograph shows typical biofilms grown on polystyrene microtiter plates (a, FSM-2; b, FSC-3; c, FSM-3). The results are expressed as the means ± standard deviations from three independent assays. MT8148, S. mutans MT8148. Asterisks denote significantly different (P < 0.05) relative levels of biofilm formation (A, other clinical strains versus MT8148; B, FSC-3 versus FSC-4).
Image analysis of FSC-3 and FSC-4 biofilms.
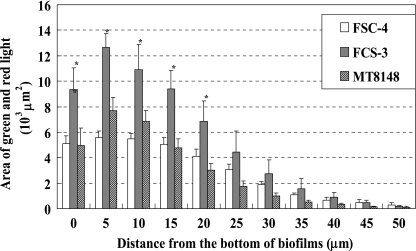
The biofilm formations of FSC-3 and FSC-4 in the flow cell system were observable in the condition using TSB but not in the condition using SDM. Therefore, TSB was chosen as a better experimental medium to compare biofilm formation between FSC-3 and FSC-4 in the flow cell system. Optical sections 5 μm apart on two axes were analyzed in CLSM images of the FSC-3 and FSC-4 biofilms. Eleven sections (0 to 50.0 μm) and image analysis of the bottom area (0 μm) are shown in Fig. 3. FSC-4 and S. mutans MT8148 biofilms had voids similar to those described by Auschill et al. (2). In contrast, FSC-3 showed a greater level of biofilm formation than the others and did not show a void (Fig. 3A and B). In the xy planes, we analyzed the average light area and light intensity to determine the quality of biofilm formation and found that the light area and intensity decreased from the bottom to the top in all of the samples. Although there was a clear difference between the volumes of biofilm formation by FSC-3 and FSC-4 in the bottom portions of the biofilms (approximately 1.8-fold), there was no difference in the top areas (Fig. 4). Further, the relative tendency of light intensity was similar to that of light area for all three strains (data not shown).
FIG. 4.
Quantification of merged light areas of green and red in xy planes of CLSM images. Each biofilm was scanned at five randomly selected positions. Data are representative of three independent experiments. The results are expressed as the means ± standard deviations of triplicate assays. Asterisks denote significantly different relative levels of biofilm formation (P < 0.05; versus MT8148).
Genes differentially expressed in two clinical strains.
Microarray analysis was performed to compare gene expression in the FSC-3 and FSC-4 biofilms. The array contained 1,960 of the 1,963 S. mutans UA159 ORFs. Of the total number of predicted ORFs, 63% are considered to have a putative function, 21% have homologs with different species (though their function is unknown), and 16% are unique to S. mutans (1). A few of these genes showed differential expression in FSC-3 biofilms compared with FSC-4 biofilms (Fig. 5). In total, 74 genes (3.8%) showed differential expression, 43 (2.2%) were activated (genes for bacteriocin secretion protein, bacitracin transport ATP-binding protein GlrA, and BLpL-like putative immunity protein, as well as others), and 31 (1.6%) were repressed (genes for phosphoenolpyruvate:sugar phosphotransferase system [PTS] IIA, B, C, and D and the Na+-driven multidrug efflux pump, as well as others) (genes exhibiting a four- to eightfold difference are shown in Table 2, and those exhibiting a greater-than-eightfold difference are shown in Table 3). Further, 30 genes (12 of those showing a four- to eightfold difference and 18 of those showing a greater-than-eightfold difference) selected randomly were examined by RT-PCR to confirm the gene expression shown by the microarray results. The gene expressions in the RT-PCR results showed tendencies of difference between FSC-3 and FSC-4 that were similar to those in the microarray results (data not shown).
FIG. 5.
Scatter plot showing intensities of the spots on an S. mutans microarray. Each gene expression was normalized using the robust multichip average, estimates of which were based on a robust average of log2 [B(PM)], where B(PM) represented background corrected PM (perfect match) intensities. Black dots, genes that showed less than 1.5-fold regulation; yellow dots, 1.5- to 2.0-fold; red dots, 2.0- to 4.0-fold; violet dots, 4.0- to 8.0-fold; green dots, more than 8.0-fold.
TABLE 2.
Genes differentially expressed (four- to eightfold difference) in two clinical strains
|
S. mutans ORF
|
Fold changea | |
|---|---|---|
| Product | No. | |
| Transmembrane protein Tmp5 | S1870 | 4.1 |
| Dipeptidyl aminopeptidase/ acylaminoacyl-peptidase-related protein | S0669 | 4.2 |
| Hemolysin III | S0852 | 4.2 |
| Murein hydrolase export regulator | S0522 | 4.3 |
| SUN protein | S0435 | 4.6 |
| UDP-glucose 4-epimerase | S1869 | 4.7 |
| DNA-damage-inducible protein J, putative | S1611 | 5.0 |
| Citrate transporter | S0921 | 5.3 |
| PTS system, cellobiose-specific IIC component | S1445 | 5.3 |
| Sterol biosynthesis methyltransferase related (Pyrococcus abyssi) | S0338 | 5.5 |
| PTS system, lactose-specific IIA component | S1447 | 6.1 |
| Putative ABC transporter, ATP-binding protain | S1704 | 6.5 |
| Acetylornithine aminotransferase | S0341 | 6.5 |
| BlpU (S. pneumoniae) | S1725 | 6.7 |
| Bactoprenol glucosyl transferase | S1871 | 7.1 |
| Transport protein ComB | S1720 | −4.0 |
| Putative transposase, ISSmu1 | S1713 | −4.5 |
| Transcriptional regulator, MutR family | S1367 | −4.6 |
| Dihydrolipamide acetyltransferase component of pyruvate dehydrogenase complex | S0115 | −4.7 |
| Putative site-specific DNA-methyltransferase restriction-modification protein | S0039 | −4.8 |
| Phage-related protein | S1273 | −5.3 |
| Putative transposase, ISSmu1 | S1279 | −5.5 |
| PTS system, IIC component | S0090 | −5.6 |
| Putative transposase, ISSmu1 | S0395 | −5.9 |
| Sucrose operon repressor | S0094 | −7.9 |
| Hypothetical proteins | S0043 | −7.8 |
| S0149 | 6.7 | |
| S0150 | 7.0 | |
| S0229 | 5.9 | |
| S0670 | 4.6 | |
| S1362 | 4.6 | |
| S1712 | −7.8 | |
| Conserved hypothetical proteins | S040 | −4.5 |
| S0174 | 5.4 | |
| S0354 | 7.6 | |
| S0657 | 5.3 | |
| S0743 | −4.4 | |
| S1446 | 7.9 | |
Fold change calculated as the log2 of gene expression in FSC-3 versus FSC-4. Positive values represent activation; negative values represent repression.
TABLE 3.
Genes differentially expressed (more-than-eightfold difference) in two clinical strains
|
S. mutans ORF
|
Fold changea | |
|---|---|---|
| Product | No. | |
| Putative bacteriocin secretion protein | S1724 | 8.6 |
| Bacitracin transport ATP-binding protein GlrA | S0941 | 9.0 |
| Contains similarity to (p)ppGpp synthase (GTP pyrophosphokinase) | S1099 | 13.9 |
| Phosphoglycerol transferase | S0754 | 14.5 |
| Glucosyltransferase (side chain biosynthesis) | S0755 | 16.9 |
| Putative immunity protein, BLpL-like | S1731 | 21.2 |
| Glycosyltransferase involved in cell wall biogenesis | S0757 | 22.4 |
| L-type calcium channel, alpha 1 subunit | S1729 | 22.4 |
| Transcriptional regulator | S0344 | 23.4 |
| Large-conductance mechanosensitive protein | S0742 | 32.5 |
| Bactoprenol glucosyltransferase | S0756 | 39.2 |
| Na+-driven multidrug efflux pump | S1364 | −9.4 |
| Putative PTS system, IID component | S0091 | −9.5 |
| PTS system, IIB component | S0089 | −9.8 |
| Alpha-glucosidase | S0093 | −9.9 |
| Heme biosynthsis protein | S1366 | −17.3 |
| Putative PTS system, IIA component | S0092 | −20.2 |
| BlpJ (S. pneumoniae) | S1715 | −97.0 |
| BlpU (S. pneumoniae) | S1710 | −137.5 |
| BlpU (S. pneumoniae) | S1716 | −147.8 |
| Hypothetical proteins | S0029 | −14.3 |
| S1098 | 14.5 | |
| S1437 | 20.7 | |
| S1610 | 11.6 | |
| S1705 | 10.0 | |
| S1711 | −14.5 | |
| S1721 | −56.3 | |
| S1722 | −10.2 | |
| S1727 | 18.2 | |
| S1728 | 14.9 | |
| S1730 | 35.8 | |
| Conserved hypothetical proteins | S0042 | −10.2 |
| S0048 | −35.6 | |
| S0049 | −36.2 | |
| S0261 | 8.4 | |
| S1365 | −19.8 | |
Fold change calculated as the log2 of gene expression in FSC-3 versus FSC-4. Positive values represent activation; negative values represent repression.
RT-PCR analysis of biofilm and planktonic cells.
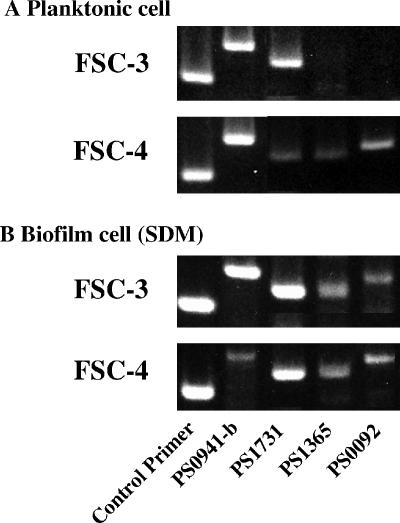
The genes for GlrA, the BLpL-like protein, PTS IIA, and hypothetical protein S1365 were randomly selected from the protein groups with positive and negative fold changes and the conserved hypothetical protein group (Table 3), respectively, as target genes to confirm their contribution to biofilm formation in the experimental stage, with two paired primers (PS0941-b, PS1731, PS1365, and PS0092) designated for these genes (Table 1). Amplifications with PS1731 and with PS1365 and PS0092 resulted in both higher and lower expressions of FSC-3 than of FSC-4 in planktonic cell (Fig. 6). Further, amplifications with PS1731, PS1365, and PS0092 showed a significant and similar expression of mRNA in the biofilm cells from both the FSC-3 and FSC-4 strains (Fig. 6). Amplification with PS0941-b showed a higher expression of FSC-3 compared to FSC-4 in biofilm cells and a similar expression of FSC-3 and FSC-4 in planktonic cells (Fig. 6). No PCR products were obtained from total RNA preparations that had not been initially reverse transcribed (data not shown).
FIG. 6.
RT-PCR analyses of biofilm and planktonic cells from S. mutans clinical strains. For FSC-3 and -4, we used the primers PS0941-b, PS1731, PS1365, and PS0092 to amplify the target genes described in Table 1. A control primer was used to normalize the expression of the test genes. Total bacterial RNA was isolated from biofilm and planktonic samples, and RT-PCR was performed as described in Materials and Methods. The data shown are representative of two independent assays.
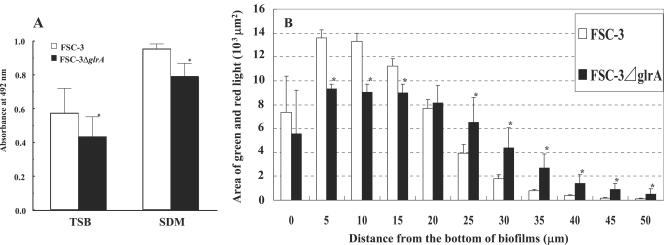
Biofilm formation by glrA deficient mutant.
The glrA was enhanced ninefold or greater in FSC-3 biofilms compared to the FSC-4 biofilms in the DNA microarray results (Table 3). Further, a higher expression of glrA was also shown in FSC-3 biofilms compared to FSC-4 biofilms in our RT-PCR analysis (Fig. 6). These results suggest that glrA interacts with this stage of biofilm bottom formation. To confirm whether glrA contributes to the increased biofilm volume of FSC-3 in comparison with FSC-4, a glrA-deficient mutant of FSC-3 was constructed and compared with FSC-3 (wild type), using a 96-well microplate and flow cell biofilm formation (Fig. 7). FSC-3 ΔglrA showed significant differences in biofilm formation in comparison with FSC-3 in both TSB and SDM in 96-well microplate cultures (Fig. 7A). The growth rates were comparable in the deficient and wild-type strains (data not shown). In the flow cell system, horizontal biofilm formation volumes at 5, 10, and 15 μm from the bottom of the biofilm were significantly lower, while those at 25, 30, 35, 40, 45, and 50 μm from the bottom were significantly higher, in FSC-3 ΔglrA compared to FSC-3 (Fig. 7B).
FIG. 7.
Biofilm formation by FSC-3 ΔglrA and FSC-3. (A) Quantification of biofilm after 16 h of culture in TSB and SDM in 96-well microtiter plates. The results are expressed as the means ± standard deviations from three independent assays. (B) Quantification of biofilm after 20 h of culture in TSB using the flow cell system and fluorescence area in xy planes of CLSM images. Each biofilm was scanned at five randomly selected positions. The results are expressed as the means ± standard deviations from three independent assays. Asterisks denote significantly different relative levels of biofilm formation (P < 0.05; versus FSC-3).
DISCUSSION
In spite of recent increased interest in biofilm research, little information is available on the physiology of the organisms that comprise surface-associated communities. Biofilm cells growing on surfaces exhibit properties distinct from those of planktonic cells, such as increased resistance to biocides and antimicrobial agents. In the present study, we investigated these mechanisms by using morphological and genetic analyses of clinical isolates of S. mutans as a means to analyze their biofilms. Because the clinical isolated strains might have been transmitted from mother to child and had survived in the severe conditions of the oral cavity, they were considered superior for testing compared to laboratory strains and were thought to express important genes for biofilm formation. DNA microarray and RT-PCR analyses were used to obtain gene expression profiles for the biofilms. We attempted to observe the biofilm samples in a state where the medium was flowing, with fresh nutrition continuously supplied. We utilized a flow cell system (8, 46) that provided a suitable in vitro method of growing reproducible biofilms for modeling dental plaque.
Regardless of the state of the medium, i.e., with (TSB) or without (SDM) added sucrose, FSC-3 showed a greater ability to form a biofilm than FSC-4 in both the 96-well microplates cultivated with SDM and flow cell systems cultivated with TSB (Fig. 2 to 4). However, the amount of FSC-4 biofilm increased and approached that of FSC-3 cultivated with TSB in the 96-well microtiter plate (Fig. 2). It was suspected that glucan-dependent biofilm formations were essential in FSC-3 and FSC-4 in the plate culture with rich medium supplemented with sucrose. The relative difference in biofilm formation between FSC-3 and FSC-4 in the 96-well microtiter plates was not similar to that in the flow cell system with TSB. Therefore, it was considered that biofilm formation and development in the nonflow 96-well microplate system were different from those in the flow cell system, even when the medium nutrition components were identical and included sucrose. Biofilm formation by FSC-3 was higher than that by FSC-4 in SDM, which did not contain sucrose, in a 96-well microtiter plate. Therefore, it was considered that genes not related to sucrose contribute to S. mutans biofilm formation, as well as genes, such as gtf, related to sucrose.
In regard to clinical status, the high biofilm formation group showed a possible relationship, with increases in incidence of dmf teeth, mS number, and mS ratio in saliva in comparison with the low biofilm formation group. Therefore, the differences in volume between the FSC-3 and FSC-4 biofilms indicated that distinctive strains of S. mutans might have been transmitted to and involved in pathogenic activity during the development of oral cavities in the 3-year-old children. A number of studies that used a serotyping method have shown that the initial acquisition of mutans streptococci by a child comes from the mother during infancy (14, 15), as have studies that used bacteriocin typing (14, 41) and genotyping (14, 22).
The PTS is the primary means of sugar transport in oral streptococci, especially under carbohydrate-limited conditions, and it plays important roles in global control of gene expression (33, 39). A previous study of S. mutans strains demonstrated that glucose PTS activity was markedly decreased in cells grown at pH 5.5 compared to those grown at neutral pH and suggested that repression occurs at the level of EII synthesis (40). On the other hand, in a study of Streptococcus parasanguinis, the addition of glucose to different types of media enhanced biofilm formation (12), whereas enriched media inhibited biofilm formation by Streptococcus gordonii (26) and S. mutans (50) in other studies.
Our results showed that further induction of the PTS system gene components (IIA, B, C, and D) was not correlated with increased biofilm formation in the bottom area in a comparison between the FSC-3 and -4 strains in culture medium that included sucrose (Fig. 4; Tables 2 and 3). However, RT-PCR analysis demonstrated that the PTS IIA gene was expressed in biofilm cells from both FSC-3 and -4 strains cultured in SDM, which was similar to that for FSC-3 and FSC-4 biofilm cells cultured in the nonflow system (Fig. 6). In contrast, the gene was expressed in planktonic cells of FSC-4 but not those of FSC-3 in BHI culture. Thus, PTS activity may depend on the various states of biofilm formation in in vitro nonflow and flow systems.
S. mutans is known to be cariogenic and resistant to bacitracin, and a number of mechanisms of bacitracin resistance have been reported for various bacteria (7, 31, 37). In the bacitracin-producing organism Bacillus licheniformis, resistance is encoded by the glrABC genes, which encode a putative heterodimeric ATP-binding cassette (ABC) transporter that has been proposed to mediate the active efflux of bacitracin (30). Homologs of this transporter have been identified in Bacillus subtilis (31) and S. mutans (9, 37). In the present study, glrA was enhanced ninefold or greater in FSC-3 biofilms compared to the FSC-4 biofilms in the DNA microarray analysis (Table 3). Further, a higher expression of glrA was also shown in FSC-3 biofilms compared to FSC-4 biofilms in our RT-PCR analysis (Fig. 6). To confirm the contribution of glrA to biofilm formation, FSC-3 ΔglrA was constructed and its biofilm formation was compared with that of FSC-3. FSC-3 ΔglrA demonstrated a lower level of biofilm formation at the bottom area than FSC-3, whereas the mutant, surprisingly, showed a higher level of biofilm formation at the top (Fig. 7). These findings indicate that glrA has a relationship to the morphology of S. mutans biofilm.
In the present study, we found that several regulatory genes of the Blp protein and the transport protein ComB were particularly repressed in FSC-3, which showed a high ability to form biofilm (Tables 2 and 3). Those results demonstrate that these genes do not have an association with the stage of greater biofilm formation by S. mutans that occurs at the bottom. In addition, a recent study investigated the role of the S. mutans relA gene, which codes for guanosine tetraphosphate and guanosine pentaphosphate [(p)ppGpp] synthetase/hydrolase, in both biofilm formation and acid tolerance. It was also found that the expression of the luxS gene was increased by as much as fivefold in the relA mutant, suggesting a link between AI-2 quorum sensing and the stringent response (21). Our findings revealed that genes similar to the (p)ppGpp synthase gene were up-regulated in FSC-3.
Several genes involved in biofilm formation have been identified in a variety of organisms (24, 50). In the present study, we also found several genes that were associated with the ability of biofilm morphological formation in experiments using different clinical strains. However, little is known regarding the molecular interactions that are involved with transducing signals between the genes that trigger biofilm development. Therefore, definitive conclusions regarding how these signals are sensed, interacted with, and transduced in biofilm-forming bacteria, as well as definitions of the molecular mechanisms utilized to initiate the development of biofilm, require further investigation using genes from knockout and complement strains of S. mutans. Nevertheless, the present results provide useful information for understanding and assessing streptococcal biofilm, as well as determination of the potential risk for mother-child transmission and dental caries.
Acknowledgments
We thank Tsutomu Sato and Fusao Nishikawara for their technical support.
This work was supported in part by grants-in-aid for development of scientific research from the Ministry of Education, Science, and Culture of Japan (15390571) and from the Ministry of Health, Labor and Welfare (H16-Medical Services-014).
REFERENCES
- 1.Ajdic, D., W. M. McShan, R. E. McLaughlin, G. Savic, J. Chang, M. B. Carson, C. Primeaux, R. Tian, S. Kenton, H. Jia, S. Lin, Y. Qian, S. Li, H. Zhu, F. Najar, H. Lai, J. White, B. A. Roe, and J. J. Ferretti. 2002. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc. Natl. Acad. Sci. USA 99:14434-14439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Auschill, T. M., N. B. Arweiler, L. Netuschil, M. Brecx, E. Reich, and A. Sculean. 2001. Spatial distribution of vital and dead microorganisms in dental biofilms. Arch. Oral Biol. 46:471-476. [DOI] [PubMed] [Google Scholar]
- 3.Berkowitz, R. J., and H. V. Jordan. 1975. Similarity of bacteriocins of Streptococcus mutans from mother and infant. Arch. Oral Biol. 20:725-730. [DOI] [PubMed] [Google Scholar]
- 4.Bhagwat, S. P., J. Nary, and R. A. Burne. 2001. Effects of mutating putative two-component systems on biofilm formation by Streptococcus mutans UA159. FEMS Microbiol. Lett. 205:225-230. [DOI] [PubMed] [Google Scholar]
- 5.Bolstad, B., R. Irizarry, M. Astrand, and T. Speed. 2003. A comparison of normalization methods for high density oligonucleotide array data based on bias and variance. Bioinfomatics 19:185-193. [DOI] [PubMed] [Google Scholar]
- 6.Burne, R. A. 1998. Oral streptococci: products of their environment. J. Dent. Res. 77:445-452. [DOI] [PubMed] [Google Scholar]
- 7.Cain, B. D., P. J. Norton, W. Eubanks, H. S. Nick, and C. M. Allen. 1993. Amplification of the bacA gene confers bacitracin resistance to Escherichia coli. J. Bacteriol. 175:3784-3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christensen, B. B., C. Sternberg, J. B. Andersen, R. J. Palmer, A. T. Nielsen, M. Givskov, and S. Molin. 1999. Molecular tools for study of biofilm physiology. Methods Enzymol. 310:20-42. [DOI] [PubMed] [Google Scholar]
- 9.Cvitkovitch, D. G., J. A. Gutierrez, J. Behari, P. J. Youngman, J. E. Wetz, P. J. Crowley, J. D. Hillman, L. J. Brady, and A. S. Bleiweis. 2000. Tn917-lac mutagenesis of Streptococcus mutans to identify environmentally regulated genes. FEMS Microbiol. Lett. 182:149-154. [DOI] [PubMed] [Google Scholar]
- 10.DeLisa, M. P., C.-F. Wu, L. Wang, J. J. Valdes, and W. E. Bentley. 2001. DNA microarray-based identification of genes controlled by autoinducer 2-stimulated quorum sensing in Escherichia coli. J. Bacteriol. 183:5239-5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fawcett, P., P. Eichenberger, R. Losick, and P. Youngman. 2000. The transcriptional profile of early to middle sporulation in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 97:8063-8068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Froeliger, E. H., and P. Fives-Taylor. 2001. Streptococcus parasanguis fimbria-associated adhesin Fap1 is required for biofilm formation. Infect. Immun. 69:2512-2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Granath, L., P. Cleaton-Jones, L. P. Fatti, and E. S. Grossma. 1993. Prevalence of dental caries in 4- to 5-year-old children partly explained by presence of salivary mutans streptococci. J. Clin. Microbiol. 31:66-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gronroos, L., M. Saarela, J. Matto, U. Tanner-Salo, A. Vuorela, and S. Alaluusua. 1998. Mutacin production by Streptococcus mutans may promote transmission of bacteria from mother and child. Infect. Immun. 66:2595-2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hamada, S., N. Masuda, and S. Kotani. 1980. Isolation and serotyping of Streptococcus mutans from teeth and feces of children. J. Clin. Microbiol. 11:314-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hamada, S., and H. D. Slade. 1980. Biology, immunology, and cariogenicity of Streptococcus mutans. Microbiol. Rev. 44:331-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Helmann, J. D., M. F. W. Wu, P. A. Kobel, F.-J. Gamo, M. Wilson, M. M. Morshedi, M. Navre, and C. Paddon. 2001. Global transcriptional response of Bacillus subtilis to heat shock. J. Bacteriol. 183:7318-7328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hope, C. K., D. Clements, and M. Wilson. 2002. Determining the spatial distribution of viable and nonviable bacteria in hydrated microcosm dental plaques by viability profiling. J. Appl. Microbiol. 93:448-455. [DOI] [PubMed] [Google Scholar]
- 19.Jordan, C., and D. J. LeBlanc. 2002. Influences of orthodontic appliances on oral populations of mutans streptococci. Oral Microbiol. Immunol. 17:65-71. [DOI] [PubMed] [Google Scholar]
- 20.Kiska, D. L., and F. L. Macrina. 1994. Genetic analysis of fructan-hyperproducing strains of Streptococcus mutans. Infect. Immun. 62:2679-2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lemos, J. A., T. A. Brown, Jr., and R. A. Burne. 2004. Effects of RelA on key virulence properties of planktonic and biofilm populations of Streptococcus mutans. Infect. Immun. 72:1431-1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, Y., and P. W. Caufield. 1995. The fidelity of initial acquisition of mutans streptococci by infants from their mothers. J. Dent. Res. 74:681-685. [DOI] [PubMed] [Google Scholar]
- 23.Li, Y. H., N. Tang, P. C. Lau, J. H. Lee, R. P. Ellen, and D. G. Cvitkovitch. 2001. Natural gemetic transformation of Streptococcus mutans growing in biofilms. J. Bacteriol. 183:897-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li, Y. H., N. Tang, M. B. Aspiras, P. C. Lau, J. H. Lee, R. P. Ellen, and D. G. Cvitkovitch. 2002. A quorum-sensing signaling system essential for genetic competence in Streptococcus mutans is involved in biofilm formation. J. Bacteriol. 184:2699-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loesche, W. J. 1986. Role of Streptococcus mutans in human dental decay. Microbiol. Rev. 50:353-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loo, C. Y., D. A. Corliss, and N. Ganeshkumar. 2000. Streptococcus gordonii biofilm formation: identification of genes that code for biofilm phenotypes. J. Bacteriol. 182:1374-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lucchini, F., V. Kmet, C. Cesena, L. Coppi, V. Bottazzi, and L. Morelli. 1998. Specific detection of a probiotic Lactobacillus strain in faecal samples by using multiplex PCR. FEMS Microbiol. Lett. 158:273-278. [DOI] [PubMed] [Google Scholar]
- 28.Majewska, A., G. Yiu, and R. Yuste. 2000. A custom-made two-photon microscope and deconvolution system. Pflugers Arch. 441:398-408. [DOI] [PubMed] [Google Scholar]
- 29.Merritt, J., F. Qi, S. D. Goodman, M. H. Anderson, and W. Shi. 2003. Mutation of luxS affects biofilm formation in Streptococcus mutans. Infect. Immun. 71:1972-1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neumuller, A. M., D. Konz, and M. A. Marahiel. 2001. The two-component regulatory system BacRS is associated with bacitracin ‘self-resistance’ of Bacillus licheniformis ATCC 10716. Eur. J. Biochem. 268:3180-3189. [DOI] [PubMed] [Google Scholar]
- 31.Ohki, R., Giyanto, K. Tateno, W. Masuyama, S. Moriya, K. Kobayashi, and N. Ogasawara. 2003. The BceRS two-component regulatory system induces expression of the bacitracin transporter, BceAB, in Bacillus subtilis. Mol. Microbiol. 49:1135-1144. [DOI] [PubMed] [Google Scholar]
- 32.Oho, T., Y. Yamshita, Y. Shimazaki, M. Kushiyama, and T. Koga. 2000. Simple and rapid detection of Streptococcus mutans and Streptococcus sobrinus in human saliva by polymerase chain reaction. Oral Microbiol. Immunol. 15:258-262. [DOI] [PubMed] [Google Scholar]
- 33.Postma, P. W., J. W. Lengeler, and G. R. Jacobson. 1993. Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol. Rev. 57:543-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shiroza, T., and H. K. Kuramitsu. 1993. Construction of a model secretion system for oral streptococci. Infect. Immun. 61:3745-3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sperandio, V., A. G. Torres, J. A. Giron, and J. B. Kaper. 2001. Quorum sensing is a global regulatory mechanism in enterohemorrhagic Escherichia coli O157:H7. J. Bacteriol. 183:5187-5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stanley, N. R., R. A. Britton, A. D. Grossman, and B. A. Lazazzera. 2003. Identification of catabolite repression as a physiological regulator of biofilm formation by Bacillus subtilis by use of DNA microarrays. J. Bacteriol. 185:1951-1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsuda, H., Y. Yamashita, Y. Shibata, Y. Nakano, and T. Koga. 2002. Genes involved in bacitracin resistance in Streptococcus mutans. Antimicrob. Agents Chemother. 46:3756-3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsuha, Y., N. Hanada, T. Asano, T. Abei, S. Yamaguchi, M. A. Salam, R. Nakao, H. Takeuchi, N. Kurosaki, and H. Senpuku. 2004. Role of peptide antigen for induction of inhibitory antibodies to Streptococcus mutans in human oral cavity. Clin. Exp. Immunol. 137:393-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vadeboncoeur, C., and M. Pelletier. 1997. The phosphoenolpyruvate:carbohydrate phosphotransferase system of oral streptococci and its role in the control of sugar metabolism. FEMS Microbial. Rev. 19:187-207. [DOI] [PubMed] [Google Scholar]
- 40.Vadeboncoeur, C., S. St. Martin, D. Brochu, and I. R. Hamilton. 1991. Effect of growth rate and pH on intracellular levels and activities of the components of the phosphoenolpyruvate:sugar phosphotransferase system in Streptococcus mutans Ingbritt. Infect. Immun. 59:900-906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Loveren, C., J. F. Buijs, and J. M. ten Cate. 2000. Similarity of bacteriocin activity profiles of mutans streptococci within the family when the children acquire the strains after the age of 5. Caries Res. 34:481-485. [DOI] [PubMed] [Google Scholar]
- 42.Vilain, S., P. Cosette, M. Hubert, C. Lange, C. A. Junter, and T. Jouenne. 2004. Comparative proteomic analysis of planktonic and immobilized Pseudomonas aeruginosa cells: a multivariate statistical approach. Anal. Biochem. 329:120-130. [DOI] [PubMed] [Google Scholar]
- 43.Wen, Z. T., and R. A. Burne. 2002. Functional genomics approach to identifying genes required for biofilm development by Streptococcus mutans. Appl. Environ. Microbiol. 68:1196-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Whiteley, M., M. G. Bangera, R. E. Bumgarner, M. R. Parsek, G. M. Teitzel, S. Lory, and E. P. Greenberg. 2001. Gene expression in Pseudomonas aeruginosa biofilms. Nature 413:860-864. [DOI] [PubMed] [Google Scholar]
- 45.Wilson, M., J. DeRisi, H.-H. Kristensen, P. Imboden, S. Rane, P. O. Brown, and G. K. Schoolnik. 1999. Exploring drug-induced alterations in gene expression in Mycobacterium tuberculosis by microarray hybridization. Proc. Natl. Acad. Sci. USA 96:12833-12838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wolfaardt, G. M., J. R. Lawrence, R. D. Robarts, S. J. Caldwell, and D. E. Caldwell. 1994. Mullticelluar organization in a degradative biofilm community. Appl. Environ. Microbiol. 60:434-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.World Health Organization. 1986. Oral health surveys. Basic methods. World Health Organization, Geneva, Switzerland.
- 48.Yamashita, Y., W. H. Bowen, R. A. Burne, and H. K. Kuramitsu. 1993. Role of the Streptococcus mutans gtf genes in caries induction in the specific-pathogen-free rat model. Infect. Immun. 61:3811-3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ye, R. W., W. Tao, L. Bedzyk, T. Young, M. Chen, and L. Li. 2000. Global gene expression profiles of Bacillus subtilis grown under anaerobic conditions. J. Bacteriol. 182:4458-4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yoshida, A., and H. K. Kuramitsu. 2002. Multiple Streptococcus mutans genes are involved in biofilm formation. Appl. Environ. Microbiol. 68:6283-6291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yoshida, A., N. Suzuki, Y. Nakano, M. Kawada, T. Oho, and T. Koga. 2003. Development of a 5′ nuclease-based real-time PCR assay for quantitative detection of cariogenic dental pathogens Streptococcus mutans and Streptococcus sobrinus. J. Clin. Microbiol. 41:4438-4441. [DOI] [PMC free article] [PubMed] [Google Scholar]