Abstract
Biosynthesis of the commercial carotenoids canthaxanthin and astaxanthin requires β-carotene ketolase. The functional importance of the conserved amino acid residues of this enzyme from Paracoccus sp. strain N81106 (formerly classified as Agrobacterium aurantiacum) was analyzed by alanine-scanning mutagenesis. Mutations in the three highly conserved histidine motifs involved in iron coordination abolished its ability to catalyze the formation of ketocarotenoids. This supports the hypothesis that the CrtW ketolase belongs to the family of iron-dependent integral membrane proteins. Most of the mutations generated at other highly conserved residues resulted in partial activity. All partially active mutants showed a higher amount of adonixanthin accumulation than did the wild type when expressed in Escherichia coli cells harboring the zeaxanthin biosynthetic gene cluster. Some of the partially active mutants also produced a significant amount of echinenone when expressed in cells producing β-carotene. In fact, expression of a mutant carrying D117A resulted in the accumulation of echinenone as the predominant carotenoid. These observations indicate that partial inactivation of the CrtW ketolase can often lead to the production of monoketolated intermediates. In order to improve the conversion rate of astaxanthin catalyzed by the CrtW ketolase, a color screening system was developed. Three randomly generated mutants, carrying L175M, M99V, and M99I, were identified to have improved activity. These mutants are potentially useful in pathway engineering for the production of astaxanthin.
Carotenoids are a class of diverse natural pigments produced from plants and microorganisms. Their physiological roles include tolerance against excess light and UV radiation, light harvesting, species-specific pigmentation, and protection against oxidation of polyunsaturated fatty acids (13, 31). Carotenoids are commercially used as food colorants in the aquaculture and poultry industries (3, 10, 11). They are also widely used as antioxidants in the nutraceutical industry. Currently, a majority of the commercial carotenoids, especially astaxanthin, are synthesized via a chemical route. The natural form of astaxanthin can be produced from the red yeast Xanthophyllomyces dendrorhous (12) and from the freshwater alga Haematococcus pluvialis (10). Genetic engineering of noncarotenogenic organisms for the production of existing carotenoids has also been explored (1, 14, 19, 23, 28). In addition, efforts have been made to diversify carotenoid biosynthetic pathways by directed evolution (32, 33).
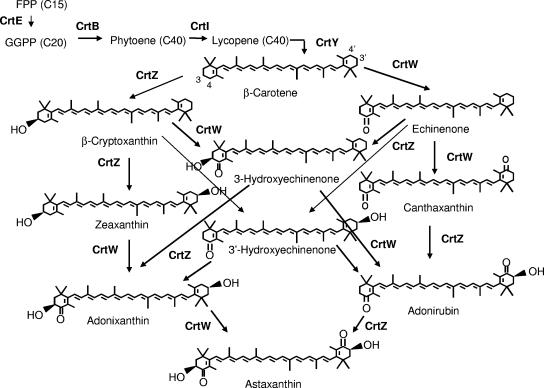
The biosynthetic route of carotenoids is derived from the isoprenoid pathway (3). Various genes involved in the biosynthesis of carotenoids have been identified and characterized (17, 18). Expression of four carotenogenic genes (crtE, crtB, crtI, and crtY) can lead to the biosynthesis of a β-carotene that contains two β-ionone rings in Escherichia coli and other microorganisms. The addition of two keto groups into the 4,4′ positions on the β-ionone rings is catalyzed by the carotenoid 4,4′-ketolase, which is encoded by crtW (16) or crtO (30). Coexpression of the crtW or crtO gene along with the crtEYIB cluster leads to the biosynthesis of canthaxanthin. Further addition of two hydroxyl groups into the 3,3′ positions leads to the biosynthesis of astaxanthin. This hydroxylation reaction is catalyzed by the carotenoid 3,3′-hydroxylase, encoded by crtZ or crtR (15). The hydroxylase can introduce hydroxyl groups into the 3,3′ positions on the β-ionone ring regardless of whether there are keto groups at the 4 or 4′ position (8). Likewise, the oxygenase can introduce keto groups at the 4,4′ positions regardless of the prior hydroxylation at the 3 or 3′ position. As a result, there are quite a few intermediates produced when a combination of crtW and crtZ genes is expressed for the biosynthesis of astaxanthin (Fig. 1). It has been found that the CrtW ketolase from Paracoccus sp. strain N81106 (formerly classified as Agrobacterium aurantiacum) has a strong substrate preference for carotenoids with nonhydroxylated β-ionone rings (9). Expression of this gene along with crtZ leads to the accumulation of adonixanthin and other intermediates. On the other hand, expression of the crtW gene from Brevundimonas sp. strain SD212 does not result in the accumulation of adonixanthin (5). This result suggests that the activity of the CrtW ketolases varies depending on the source.
FIG. 1.
Carotenoid biosynthetic pathway.
Based on amino acid sequences, CrtW ketolases have similarities to other oxygen-dependent and iron-containing integral membrane enzymes. Essentially, very little is known regarding the structure and function of this group of enzymes. In this study, we used alanine-scanning mutagenesis to investigate the conserved amino acid residues of CrtW ketolases for their functional roles in the conversion of β-carotene to canthaxanthin and astaxanthin in E. coli cells. Furthermore, we developed a color screening system that enabled us to identify random mutations that improved the activity of CrtW toward the biosynthesis of astaxanthin.
MATERIALS AND METHODS
Strains and plasmids.
Bacterial strains and plasmids used in this study are listed in Table 1. E. coli Top 10 cells were used for the study except as otherwise indicated. For routine maintenance, the E. coli strains harboring various plasmids with PBAD were grown without l-arabinose to avoid instability of the cloned genes.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| E. coli Top 10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara-leu)7697 galU | Invitrogen |
| Pantoea agglomerans DC404 | Source of the crtE-idi-crtYIB gene cluster | 25 |
| Plasmids | ||
| pSU18 | Cmr; derived from low-copy-number plasmid pACYC | 2 |
| pSUCMW | Derived from pSU18; Cmr; contains crtW under the control of Cm promoter | This study |
| pSUKMBC | Derived from pSU18; Kmr, Cmr; contains β-carotene biosynthetic crtE-idi-crtYIB cluster under the control of Km promoter | This study |
| pSUKMZX | Derived from pSU18; Kmr, Cmr; contains zeaxanthin cluster crtZE-idi-crtYIB under the control of Km promoter | This study |
| pBAD/His | Apr; araBAD promoter (PBAD); derived from pBR322 | Invitrogen |
| pBADW | Apr; contains ketolase gene crtW under the control of PBAD in pBAD | This study |
| pBADBC | Apr; contains β-carotene biosynthetic cluster crtE-idi-crtYIB under the control of PBAD in pBAD | This study |
| pBADZX | Apr; contains zeaxanthin biosynthetic cluster crtZE-idi-crtYIB under the control of PBAD in pBAD | This study |
For the alanine-scanning experiment, a synthetic codon-optimized crtW gene from Paracoccus sp. strain N81106 (formerly classified as Agrobacterium aurantiacum) was cloned into the vector pBAC/His under the control of PBAD. The coding region was amplified as a BsrGI and HindIII fragment with the primer set GCATTGTACAATGAGCGCCCATGCCCTGCCGAAAGCCGA (forward) and GATCAAGCTTTCACGCGGTGTCGCCTTTGGTGCGGGT (reverse). PCR was carried out with a high-fidelity PCR polymerase (AccuPrime Pfx SuperMix; Invitrogen). The pBAD/His vector was modified to contain the BsrGI site with the primer set ATTCGAAGCTTGGCTGTTTTGGCGGA (forward) and GCATTGTACATTCCTCCTGTTAGCCCAAAAAACGGGTATGGA (reverse). After digestion with BsrGI and HindIII, the amplified crtW coding region was cloned into the modified pBAD/His vector. Positive clones were identified by PCR amplification with the same primer set and were further confirmed by DNA sequencing. The resulting construct was designated pBADW.
Assembly of zeaxanthin biosynthetic gene clusters.
Biosynthesis of zeaxanthin in E. coli requires the expression of the crtZ gene and the β-carotene biosynthetic cluster crtEYIB. To combine these genes into an operon, a ligation-independent cloning (LIC) method based on PTO (α-phosphorothioate) nucleotide-modified primers was used. In this procedure, multiple α-thiol linkages are introduced in the primers to stop the digestion of the T7 exonuclease at a specific site from the 5′ end of the DNA strand. This leads to the generation of complementary region for annealing. This LIC method does not rely on the presence of restriction sites, and DNA fragments can be assembled in any given order. The annealed structure can be directly transformed into competent E. coli cells without ligation. For assembly of the zeaxanthin gene cluster, the four DNA fragments containing crtZ, crtE-idi, crtYIB, and vector were amplified and a 14-nucleotide complementary region was used for annealing. The cloning vector was based on pSU18, containing a kanamycin (Km) resistance marker (2). The vector was amplified with the primer set TGTTACAACCAATTAXAXCXCXAATTC (reverse) and CGCGCTAGAATTCCAXCXCXAXTCATACACTAAATCAGTAAG (forward). The “X” in the primers indicates PTO modification. The crtZ gene from Paracoccus sp. strain N81106 was amplified with primer set AATTGGTTGTAACAGXAXAXTXTCCCTAGGTCTAGAAAGGAGGAATAAACCATGACCA (forward) and CTCGACTAGAATTGTXGXTXAXCATTAGGTGCGTTCTTGGGCTTCGGCA (reverse). The crtE and idi genes from the environmental isolate Pantoea agglomerans DC404 (25) were obtained with PCR primers CAATTCTAGTCGAGAXCXGXCXCGGGTACCAACCATGACAAGACCCTTTGAAACACATCC (forward) and ATCAGATCCCATTTTXTXTXCXATACCGCTCCCCGGTATAA (reverse). Finally, the fragment containing the crtYIB genes from DC404 was amplified with primers AAATGGGATCTGATTXCXTXGXGTCGGCG (forward) and GGAATTCTAGCGCGGXGXCXGXCTGCCAG (reverse). After PCR amplification, the samples were treated with DpnI and DNA fragments were purified from the agarose gel. In the standard LIC reaction of 20 μl in a PCR tube, 30 ng of each DNA fragment was mixed and treated with 10 units of T7 exonuclease (New England Biolabs, Beverly, MA) for 5 min at 30°C in NT buffer. NT buffer consisted of NEB buffer 4 supplemented with Tris-HCl (pH 8.0) and MgCl2 at final concentrations of 50 mM and 10 mM, respectively. The LIC reaction was carried out in a thermocycler with the following conditions: 5 min at 30°C for enzyme digestion and 3 min at 72°C for denaturing of the single-stranded ends, followed by one cycle of annealing (65°C, 1 min; 60°C, 1 min; 55°C, 1 min; 50°C, 1 min; 45°C, 1 min; 40°C, 1 min; 37°C, 10 min; 22°C, 10 min). An aliquot (2 μl) was used for transformation with chemically competent Top 10 cells. The transformants were grown on LB plates supplemented with Km (50 μg/ml). After overnight growth, yellow colonies were picked and plasmid DNA was isolated. DNA sequencing was used to confirm the proper assembly of the gene cluster. The resulting construct was designated pSUKMZX (Table 1). To construct the plasmid for β-carotene biosynthesis, the crtZ gene was deleted by digesting the pSUKMZX with restriction enzymes BsrGI and XbaI. The ends were repaired with the End-It DNA End-Repair Kit (Epicenter, Madison, WI). The fragment was then ligated with the Fast-Link DNA Ligation and Screening Kit from the same company. The resulting construct was designated pSUKMBC.
To express the zeaxanthin gene cluster under the control of PBAD, the crtZE-idi-crtYIB cluster was isolated as an EcoRI fragment from pSUKMZX and cloned into the corresponding site in the pBAD/His vector. This led to the construction of pBADZX. Cloning of the β-carotene biosynthetic cluster crtE-idi-crtYIB without the crtZ gene generated pBADBC.
Alanine-scanning mutagenesis for the crtW gene.
Alanine-scanning mutagenesis was used to replace individual conserved amino acids with alanine to study the importance of amino acid side chains for the function of the CrtW ketolase. The method of making the specific change was oligonucleotide directed and based on the QuikChange II Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA). The DNA template for PCR was pBADW. The PCR conditions were essentially the same as described in the manual except that an extension time of 5.5 min was used. After DpnI treatment, the reaction mixture was transformed into Top 10 cells. DNA sequencing was used to identify the desired mutations, which were transformed into two types of cells for in vivo functional analysis. To analyze their activity toward the production of astaxanthin, cells harboring pSUKMZX, containing the crtZ gene, were used. To analyze their activity toward the production of canthaxanthin, mutations were expressed in cells harboring pSUKMBC for β-carotene production.
Development of a color screening system for crtW mutants with improved selectivity for astaxanthin.
Our screening system was based on the color appearance of the E. coli cells harboring the two plasmids containing the crtW gene and the biosynthetic gene cluster crtZE-idi-crtYIB. The plasmid pSU18 was used for cloning and expressing of the crtW gene. The vector backbone containing the chloramphenicol (Cm) resistance marker was amplified with the primers TAACGATGCAAAACGCATCCTGCCACCATCATACACTAAATCAGTAAG (forward) and TTTCAGCGACAACTCCTGCATTGGCATACGAGCCGGAAGCATAAAGTG (reverse). The crtW gene was obtained with the primers TCATCCGGAATTCACTAGTAAGGAGG (forward) and ATCAATTGGAGCTCGTTTATTCCTCCTTTCTAGATCACG (reverse). An EcoRI site was incorporated in the forward primer, while MfeI and SacI sites were introduced in the reverse primer. After amplification, both the vector and the crtW DNA fragments were purified, ligated, and transformed into competent E. coli Top 10 cells containing pBADZX. The construct with the crtW gene in the same orientation as the Cm marker was designated pSUCMW (Table 1).
Mutagenesis of the crtW gene by error-prone PCR.
In vitro mutagenesis of the crtW gene was carried out with the GeneMorph II Random Mutagenesis kit (Stratagene, La Jolla, CA). PCR was carried out with plasmid DNA of pSUCMW as the template and the same set of primers as used for the cloning of the crtW gene. The reaction conditions were designed to achieve a frequency of 0 to 4 mutations per kb of DNA, according to the instructions. The PCR products were gel purified and digested with EcoRI and SacI. The cloning vector was prepared by digesting the pSUCMW construct with the same restriction enzymes. After ligation, the reaction mixture was transformed into E. coli Top 10 cells harboring the pBADZX plasmid via electroporation. The E. coli culture was spread on large LB agar plates (9- by 9-in. low-profile square bioassay dishes; Corning, Corning, NY). The LB agar medium was supplemented with 100 μg/liter ampicillin, 12.3 μg/liter Cm, and 0.002% l-arabinose. Spreading of the transformation sample was aided by glass beads. The plates were incubated at 37°C overnight, followed by another overnight incubation at room temperature. Those colonies having a bright orange color were chosen for further analysis by DNA sequencing. The amount of carotenoid products in these cells was analyzed by high-pressure liquid chromatography (HPLC).
HPLC analysis of carotenoid intermediates.
In order to investigate the carotenoid intermediate profiles, strains containing the wild-type as well as the mutated crtW were grown in 25 ml LB solution supplemented with appropriate antibiotics and 0.002% l-arabinose. After 8 h of growth, the samples were collected in a Corning 50-ml disposable polypropylene tube and centrifuged at 6,600 × g for 10 min. The pellet was frozen at −80°C, or pigments were extracted immediately.
To extract the pigments, the cell pastes were first resuspended by vortexing with a small amount of liquid remaining in the pellets. Then, 0.1-mm glass beads were added, followed by 2 ml of ethanol. The mixture was vortexed until the entire pellet dissolved. Next, 3 ml of dichloromethane was added, and the samples were vortexed again for about 2 minutes. After centrifugation at 6,600 × g with a table-top centrifuge for 10 min, the supernatant was transferred to a new 50-ml Corning polypropylene centrifuge tube and was dried under a stream of nitrogen. The residue was dissolved completely in 90 μl of chloroform followed by the addition of 1,910 μl of n-hexane (HPLC grade). Before injection into the HPLC, the samples were filtered with a 0.2-μm Gelman Teflon filter. The extract (20 μl) was analyzed by the HPLC system equipped with a model 168 diode array detector (Beckman, Fullerton, CA). A 250- by 4.6-mm Brownlee Spheri-5 Silica 5-μm normal-phase HPLC column from Perkin Elmer (Norwalk, CT) was used. The mobile phase consisted of 14% acetone and 86% n-hexane. The flow rate was 1.5 ml per min.
Spectral data for individual carotenoids.
All carotenoids identified by HPLC were compared with standards purchased from CaroteNature GmbH (Lupsingen, Switzerland). They were further confirmed with spectral data obtained by scanning and mass spectrometry. For astaxanthin: retention time (RT) 9.5 min, λmax 473 nm; MS, m/z 597 [M+H]+. For adonixanthin: RT 12.5 min, λmax 462 nm; MS, m/z 583 [M+H]+. For adonirubin: RT 6.6 min, λmax 467 nm; MS, m/z 581 [M+H]+. For zeaxanthin, RT 15.4 min, λmax 451, 478 nm; MS, m/z 569 [M+H]+. For 3-hydroxyechinenone, RT 6.8 min, λmax 458 nm; MS, m/z 567 [M+H]+. For lycopene, RT 2.0 min, λmax 445, 472, 503 nm; m/z 537 [M+H]+. For canthaxanthin, RT 4.7 min, λmax 466 nm; MS m/z 565 [M+H]+. For echinenone, RT 2.9 min, λmax 453 nm; m/z 551 [M+H]+. For β-carotene, RT 1.9 min, λmax 452, 478 nm; m/z 537 [M+H]+.
RESULTS
Topology model of the CrtW ketolase from Paracoccus sp. strain N81106.
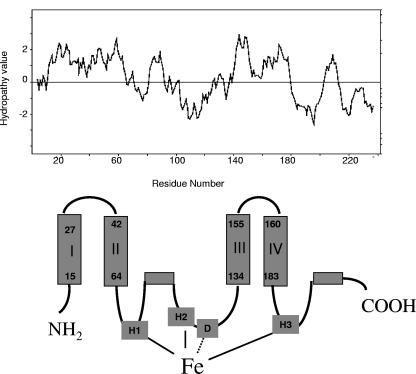
Based on amino acid sequences and catalytic characteristics, the membrane-bound β-carotene ketolases belong to a superfamily of integral membrane proteins that catalyze oxidative reactions. This group of proteins share the putative iron-binding His motifs (6, 24, 27) and have transmembrane domains. Figure 2 shows the similarities in amino acid sequences for a group of CrtW ketolases. All of them share the His-rich motifs HX(3 or 4)H, HX(2 or 3)HH, and HX(2 or 3)HH. The hydropathy and transmembrane prediction analyses (29) indicate that the CrtW enzyme from Paracoccus sp. strain N81106 also contains four transmembrane domains and two other hydrophobic regions. Here we propose a topology model for the CrtW ketolase that is very similar to those for fatty acid desaturases (Fig. 2 and 3). In this model, the first His motif involved in iron coordination comprises the H65 and H69 residues. This motif is located right after transmembrane domain II. The second His motif consists of H103, H106, and H107. The histidine residues in the third His motif are H218, H221, and H222. These residues in the His motifs are predicted to be located in the cytoplasmic phase rather than in membrane-spanning domains. The location of the iron active center near the membrane surface provides easy access to the electron donors, which are likely either soluble or peripheral membrane proteins. On the other hand, all of the carotenoid substrates are highly hydrophobic and likely partition into the lipid bilayer. As a result, the substrate binding site is likely located inside the membrane. In addition to the transmembrane domains, the CrtW ketolase appears to have two hydrophobic patches, one between domains II and III and one near the C terminus.
FIG. 2.
Similarity comparison of amino acid sequences of CrtW ketolases from Paracoccus sp. strain N81106 (16) (NCBI accession number BAE47465), Brevundimonas sp. strain SD212 (21) (GenBank accession number AB181388), and Alcaligenes sp. (16) (NCBI accession number BAA09596). The four transmembrane domains and three conserved His motifs involved in iron binding are indicated. Highly conserved amino acids are in bold and underlined..
FIG. 3.
Hydropathy analysis and topology model of the CrtW enzyme from Paracoccus sp. strain N81106 (formerly classified as Agrobacterium aurantiacum).
Functional analysis of histidine residues in the conserved His motifs.
To investigate the functional role of the His residues in the His motifs that are involved in iron coordination, site-directed mutagenesis was used to change them individually to Ala residues. The expression construct pBADW (Table 1) was used as the DNA template in the site-directed mutagenesis reaction. Positive clones were transformed into cells harboring either the zeaxanthin or the β-carotene expression construct for functional analysis (see Materials and Methods). If the enzyme remained functional after the His-to-Ala mutation, carotenoid products with a keto group(s) in the β-ionone rings should be detected. In the strain background expressing the zeaxanthin gene cluster crtZE-idi-crtYIB, the ketolated products included adonixanthin and astaxanthin (Fig. 1). In the strain background expressing the β-carotene biosynthetic cluster, crtE-idi-crtYIB, the products included echinenone and canthaxanthin. In the first His motif, a change to an alanine residue at H65 essentially abolished the CrtW ketolase activity, since no detectable products with ketolation were observed in either strain background (Tables 2 and 3). Surprisingly, the mutant with the His-to-Ala mutation at residue H69 retained partial activity. This result indicates that only the H65 residue is essential for enzymatic activity in the first conserved His motif. In the second conserved His motif, all three residues, H103, H106, and H107, appeared to be essential for activity, since strains expressing these mutations did not produce ketolated products. In the third cluster, the H221 and H222 residues were found to be essential, while partial activity was observed for the H218 residue. Next to the H218 residue, there is another histidine residue, H219, which is conserved in some of the CrtW enzymes (Fig. 2). The mutant carrying the His-to-Ala mutation at this position rendered the enzyme partially active in cells containing the zeaxanthin gene cluster. In cells expressing the β-carotene gene cluster, the activity was similar to that of the wild type based on the amount of canthaxanthin produced. It can be concluded that H219 may not play a significant role in iron coordination, but it can have an impact on substrate utilization. Results of site-directed mutagenesis of these three His motifs were consistent with the hypothesis that they are essential for enzymatic function due to their role in iron coordination. However, not all of the conserved His residues in these motifs are essential for this role.
TABLE 2.
Percentages of different carotenoid products in cells expressing a mutated crtW gene in the E. coli host harboring the zeaxanthin biosynthetic gene cluster crtZE-idi-crtYIBa
| Mutation | % ofb:
|
||
|---|---|---|---|
| Zeaxanthin | Adonixanthin | Astaxanthin | |
| H65A | 95.0 | 1.0 | — |
| H69A | 55.9 | 35.6 | 2.9 |
| H103A | 90.8 | — | — |
| H106A | 91.1 | — | — |
| H107A | 91.7 | — | — |
| H218A | 53.0 | 40.8 | 2.2 |
| H219A | 1.0 | 45.7 | 47.7 |
| H221A | 93.8 | — | — |
| H222A | 92.0 | — | — |
| H184A | 92.1 | 1.2 | 3.4 |
| D117A | 77.2 | 20.3 | — |
| F130A | 33.6 | 56.5 | 1.3 |
| Y134A | 25.6 | 63.1 | 1.6 |
| Y91A | 10.2 | 65.9 | 11.2 |
| F176A | 23.4 | 64.5 | 3.0 |
| F118A | 13.9 | 67.6 | 10.4 |
| P116A | 1.5 | 62.5 | 20.9 |
| W126A | 5.8 | 60.6 | 25.4 |
| W229A | 1.2 | 64.0 | 25.5 |
| L232A | 1.7 | 65.8 | 24.3 |
| W26A | 1.5 | 21.7 | 67.5 |
| N80A | — | 5.0 | 80.1 |
| G84A | 1.5 | 23.3 | 66.1 |
| W96A | 1.5 | 26.2 | 60.7 |
| D114A | 1.1 | 17.8 | 69.1 |
| D115A | — | 6.2 | 81.4 |
| P228A | 0.9 | 17.7 | 70.1 |
| Wild type | 1.6 | 16.1 | 72.0 |
The strain background contained the pSUKMZX construct (Table 1). The total carotenoid titer in this strain was 0.6 mg/g (dry cell weight). The carotenoid products were determined by HPLC with E. coli cells harvested after 8 h of growth. These products were also confirmed by spectral data as described in Materials and Methods.
—, trace or nondetectable amount.
TABLE 3.
Percentage of carotenoid products in cells expressing a mutated crtW gene in the E. coli host containing the β-carotene biosynthetic gene cluster crtE-idi-crtYIBa
| Mutation | % ofb:
|
||
|---|---|---|---|
| β-Carotene | Echinenone | Canthaxanthin | |
| H65A | 96.2 | 1.9 | — |
| H69A | 24.5 | 31.1 | 39.6 |
| H103A | 98.1 | — | — |
| H106A | 98.4 | — | — |
| H107A | 98.6 | — | — |
| H218A | 12.5 | 27.8 | 39.4 |
| H219A | 1.0 | 10.1 | 87.2 |
| H221A | 98.9 | — | — |
| H222A | 96.4 | — | — |
| H184A | 85.6 | 11.8 | — |
| D117A | 14.4 | 62.1 | 22.8 |
| F130A | 8.4 | 45.2 | 46.5 |
| Y134A | 3.0 | 18.7 | 72.4 |
| Y91A | 7.1 | 15.9 | 77.1 |
| F176A | 4.1 | 17.7 | 76.5 |
| F118A | 2.9 | 10.5 | 80.5 |
| P116A | 4.2 | 12.4 | 76.7 |
| W126A | 3.1 | — | 89.6 |
| W229A | 2.2 | — | 92.0 |
| L232A | 1.9 | — | 93.3 |
| W26A | 2.5 | 2.7 | 85.8 |
| N80A | 2.4 | 3.6 | 93.4 |
| G84A | 2.9 | 2.9 | 85.8 |
| W96A | 3.6 | 2.8 | 84.3 |
| D114A | 3.3 | 2.8 | 85.5 |
| D115A | 3.1 | 2.4 | 85.5 |
| P228A | 2.9 | — | 88.3 |
| Wild type | 3.9 | 2.5 | 91.3 |
The strain background contained the pSUKMBC construct (Table 1). The total carotenoid titer in this strain was 0.9 mg/g (dry cell weight). The carotenoid products were determined by HPLC with E. coli cells harvested after 8 h of growth. These products were also confirmed by spectral data as described in Materials and Methods.
—, trace or nondetectable amount.
It is worthwhile to note that all three partially active mutants, with mutations H65A, H218A, and H219A, accumulated adonixanthin as the dominant intermediate, producing very little astaxanthin (Table 2). A significant amount of echinenone was also observed when they were expressed in the host harboring only the β-carotene biosynthetic gene cluster (Table 3). Both adonixanthin and echinenone are monoketolated carotenoid products.
Functional characterization of other conserved amino acid residues outside the His clusters.
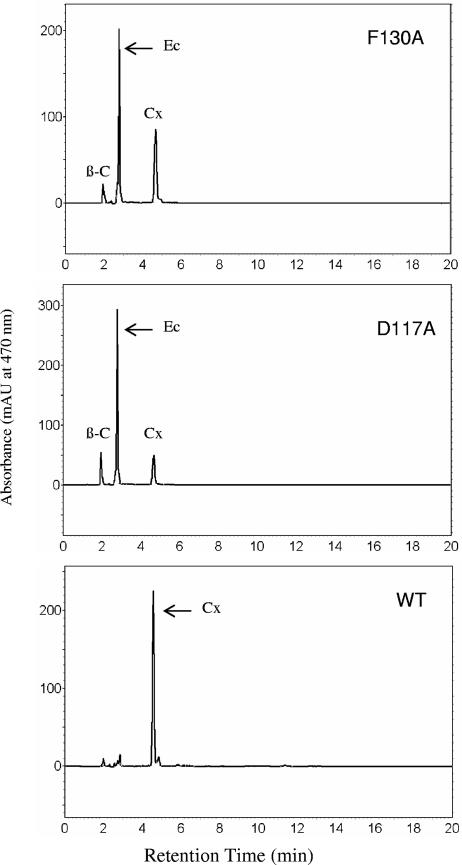
Besides the three His clusters that are putatively involved in iron coordination, the importance of highly conserved amino acid residues in the rest of the protein was investigated with the same alanine-scanning mutagenesis approach. The histidine residue at position 184 is not part of the His clusters. However, this residue appears to be essential, since expression of the crtW gene carrying the H184A mutation accumulated only zeaxanthin or β-carotene as the predominant product in both E. coli hosts (Tables 2 and 3). A change to alanine for the aspartate residue at position 117 severely impacted the function of the crtW gene. When the D117A mutation was expressed in the host containing the crtZE-idi-crtYIB cluster, the cells accumulated about 20% adonixanthin and very little astaxanthin (Table 2). A majority of the total carotenoid remained as zeaxanthin. Interestingly, when this mutation was expressed in the β-carotene-producing host, a majority of the intermediate was echinenone (Fig. 4). This result suggested that the mutant carrying the D117A change was active enough to convert a large portion of β-carotene in the cells but had difficulty using the monoketolated intermediate echinenone as a substrate. The D117 residue was part of the highly conserved DPDF(Y) motif located in a hydrophilic region with close proximity to His cluster II (Fig. 2 and 3). In addition to D117A, cells expressing the F130A mutation also produced a significant amount of echinenone when β-carotene was the substrate (Table 3). F130 is part of the WX(3)FX(3)Y motif found in most of the CrtW ketolases (Fig. 2).
FIG. 4.
HPLC analysis of carotenoid products in E. coli cells expressing wild-type (WT) CrtW and D117A and F130A mutants. The crtW genes were cloned in the pBAD vector under the control of PBAD. The reporting construct was pSUKMBC, containing the β-carotene biosynthetic cluster, crtE-idi-crtYIB. The E. coli cells were harvested after 8 h of growth. The carotenoids identified were β-carotene (β-C), echinenone (Ec), and canthaxanthin (Cx).
When the wild-type crtW gene was expressed in the host strain containing the zeaxanthin biosynthetic cluster, crtZE-idi-crtYIB, the E. coli cells produced about 70% astaxanthin after 8 h of incubation. A change to alanine for many other conserved amino acid residues in the protein resulted in partial activity. The E. coli cells expressing partially active mutants accumulated adonixanthin as the dominant intermediate and produced less than 30% astaxanthin (Table 2). This group of mutants included the Y134A, F176A, F118A, P116A, W126A, W229A, and L232A mutations. However, when they were expressed in the host harboring the β-carotene biosynthetic cluster crtE-idi-crtYIB, the final product, canthaxanthin, was the predominant pigment. This suggested that the impact of these mutations on the ability to catalyze β-carotene was less severe. As for the other mutants listed in Table 2 and 3, the product profiles in the two E. coli hosts were very similar to those containing the wild-type crtW gene. This indicated that not all of the conserved amino acids are important for function.
Development of a color screening system for random mutants that have improved efficiency for astaxanthin production.
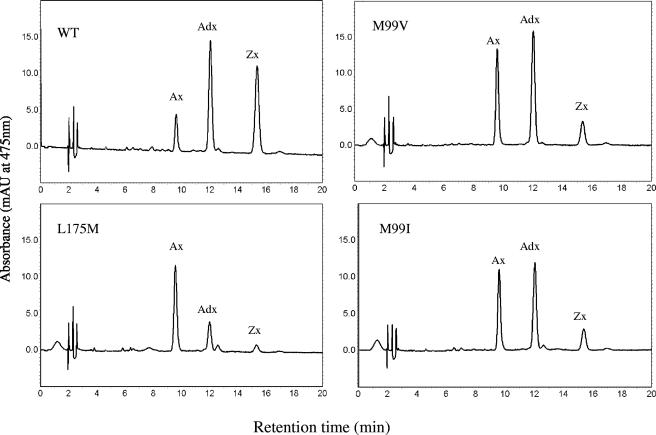
Adonixanthin is one of the common intermediates produced during biosynthesis of astaxanthin. In order to identify mutations that favor the completion of the reaction, an efficient screening system was necessary. We found that the relative balance in the expression levels of the crtW and crtZ genes and the overall flux dictated the intermediate profiles of carotenoids in the cell culture. When the two plasmids pSUCMW, containing the crtW gene, and pBADZX, with the zeaxanthin gene cluster, were combined in the E. coli cells and grown in the presence of 0.002% l-arabinose, the colonies appeared yellow for at least 24 h. HPLC analysis showed that this strain containing the wild-type crtW gene accumulated mainly the pigments adonixanthin and zeaxanthin (Fig. 5). As more astaxanthin was accumulated after a couple of days, the cells started to turn orange. A screening system based on the development of a bright orange color during the initial 48 h was employed to identify randomly generated mutants that may have improved selectivity for astaxanthin.
FIG. 5.
HPLC analysis of carotenoid products in E. coli cells expressing wild-type (WT) CrtW and M99I, M99V, and L175M mutants. E. coli cells contained the plasmid pSUBCMW and the reporting construct pBADZX (Table 1). Pigments were extracted from the cells after 8 h of growth. The carotenoids identified were astaxanthin (Ax), adonixanthin (Adx), and zeaxanthin (Zx).
Characterization of mutants with improved astaxanthin selectivity.
Libraries of CrtW mutants were generated by error-prone PCR. Positive clones identified by the color screening method were further analyzed by DNA sequencing. A mutant carrying one change, L175M, was isolated repeatedly. HPLC analysis showed that the strain expressing this mutant produced astaxanthin as the predominant intermediate (Fig. 5). Site saturation mutagenesis carried out at this position resulted in isolation of clones with the same leucine-to-methionine mutation, suggesting that substitution with other amino acid residues did not yield a better result. Screening of the crtW random libraries also led to the isolation of two additional mutants, M99I and M99V mutants. E. coli cells expressing these two mutants showed a moderate increase in astaxanthin accumulation and a moderate reduction in the level of zeaxanthin.
All three mutants were further characterized in the high-flux E. coli strain MG1655 PT5-dxs (34). In this strain, the carbon flux for carotenoid biosynthesis is increased due to the overexpression of isoprenoid genes. Both the reporter plasmid pBADZX and plasmid pSUCMW, containing the wild-type and mutated crtW, were introduced into the high-flux strain. Expression of the zeaxanthin gene cluster on pBADZX was induced by 0.002% l-arabinose. Analysis of the carotenoid profile in the strain with the wild-type crtW showed the presence of 53% astaxanthin and 33% adonixanthin. In the strain containing L175M, on the other hand, up to 78% of the total carotenoid was in the form of astaxanthin and the level of the adonixanthin intermediate was lowered to only 5%. An increase in astaxanthin production and a reduction in adonixanthin accumulation were also observed with M99I and M99V. This result indicated that a high conversion efficiency of adonixanthin to astaxanthin contributed to a lower intermediate level in E. coli strains expressing these three mutants.
To investigate the efficiency of β-carotene conversion for L175M, M99I, and M99V mutants, the constructs containing the wild-type and mutated crtW were introduced into the E. coli host harboring the plasmid pBADBC (Table 1). In the cells containing the wild-type crtW gene, up to 90% of the total carotenoid was canthaxanthin. A similar level of canthaxanthin production was found in cells expressing L175M, M99I, and M99V. This result suggested that the efficiency of conversion of β-carotene to canthaxanthin was not affected in these mutants.
DISCUSSION
Based on structural and catalytic similarities, the β-carotene ketolase encoded by crtW gene belongs to the superfamily of integral membrane oxidative enzymes along with fatty acid desaturases, alkane hydroxylase, xylene monooxygenase, sterol methyloxidase, and β-carotene hydroxylase (4, 6, 24, 26, 27). Although there is a lack of three-dimensional structural information for these membrane proteins, the easily noticeable features include the three histidine-rich motifs for iron coordination and hydrophobicity patterns. It has been observed that all eight conserved His residues in the His motifs for the rat Δ9 desaturase are essential for activity (27). It has also been observed that the corresponding conserved histidines are essential for enzymatic function of the AlkB ω hydroxylase from Pseudomonas oleovorans (26) and the β-carotene hydroxylase from pepper fruits (4). Our experiments, on the other hand, showed that six of the eight conserved histidine residues, H65, H103, H106, H107, H221, and H222, were essential for activity of the CrtW enzyme from Paracoccus sp. strain N81106 (Tables 2 and 3). Partial activity was found for H69A and H218A mutants. This suggests that only one histidine residue in the first motif is adequate to confer enzymatic function. The difference between our results and previous results obtained for other enzymes in this family could be due to the unique differences in structure and function of the CrtW ketolase.
In addition to the histidines, carboxylate ligands are present in the dinuclear ion centers that have been characterized so far (22). Mutation of the aspartate residue at position 181 to an alanine residue resulted in a loss of activity for the AlkB ω hydroxylase (26). The corresponding aspartate residue for the CrtW protein appears to be D117, located in the aspartate-rich region DDPDFD, which resides in a relatively hydrophilic region with close proximity to His cluster II (Fig. 2 and 3). Asp-117 is highly conserved among CrtW ketolases and partially conserved across the eukaryotic desaturases, hydroxylases, epoxygenases, acetylenases, and conjugases (26). Although the crtW gene carrying the D117A mutation did not result in a complete loss of activity in our experiment, the enzymatic function was severely impacted (Tables 2 and 3). This mutant showed a lack of activity toward both zeaxanthin and echinenone. Results from studying the D117A mutation raises the possibility that the carboxyl group from this aspartate residue is located in the di-iron center near or on the surface of the cytoplasmic membrane, playing a role in bridging the di-iron center or in the electron transfer reaction. The Asp-117 residue could also provide flexibility and additional negative charge to the site, thus stabilizing the higher oxidation states of the metal, as previously suggested (22). Since expression of the mutant carrying D117A led to accumulation of echinenone as the predominant product, it can be very useful for pathway engineering if this carotenoid is of commercial interest. It is worth noting that the asymmetrical catalysis of β-carotene to echinenone is a characteristic of the CrtO enzyme found in cyanobacteria (7, 20).
It has been proposed that the hydrophobic segment between the first two His clusters in the Bacillus subtilis acyl-lipid desaturase may correspond to a protein region that is involved in substrate interaction (6). In the fatty acid desaturase of Caenorhabditis elegans, the C-terminal domain and one of the hydrophobic segments determine the regioselectivity (24). In the CrtW proteins, there are at least two hydrophobic patches, one located between the first two His clusters and the other near the C terminus (Fig. 3). In addition, the possibility of a small hydrophobic region preceding transmembrane domain III in some CrtW ketolases exists. The hydrophobic patches within the CrtW protein could be a part of the enzyme active site, involved in the interaction with the hydrophobic carotenoid substrates.
Alanine-scanning mutagenesis with the CrtW ketolase showed that many conserved residues outside the His motifs were important for full enzyme function. A change to alanine residues in these locations rendered the enzyme partially active when expressed in the host harboring the zeaxanthin gene cluster, crtZE-idi-crtYIB. The common observation with these partially active mutants was the accumulation of large amounts of the precursor intermediate adonixanthin (Table 2). However, the CrtW ketolase with these mutations retained most, if not all, activity for canthaxanthin production from β-carotene (Table 3). The intermediate profiles indicate that these partially active CrtW mutants were capable of catalyzing nonhydroxylated substrate β-carotene and, to a certain degree, hydroxylated substrate zeaxanthin. On the other hand, the ability to catalyze adonixanthin, which is a hydroxylated and monoketolated intermediate, was more problematic. A number of mutants, such as those carrying H69A, H218A, F130A, and Y134A, also showed a significant accumulation of echinenone (Table 3). This intermediate is a nonhydroxylated but monoketolated carotenoid.
Production of intermediates can present a challenge for pathway engineering for the production of astaxanthin. In fact, there are eight possible intermediates produced by the actions of the CrtW ketolase and the CrtZ hydroxylase for astaxanthin biosynthesis (Fig. 1). As a result, adequate activities for both hydroxylase and ketolase are required to achieve a high selectivity for astaxanthin. Here we developed a rapid color screening system that allows the identification of CrtW mutants having improved activity for astaxanthin production. This system was based on changes in the color of E. coli colonies when an increased amount of astaxanthin was present. The key feature of the system was the combination of two expression constructs, pSUCMW and pBADZX. With this combination, activity of the CrtW enzyme was relatively low, since the cells harboring these two constructs produced a very small amount of astaxanthin while the predominant products were zeaxanthin and adonixanthin (Fig. 5). Using this screening system, we identified three improved mutants, those carrying L175M, M99I, and M99V. It is worthwhile noting that the Met-99 residue is located between the first hydrophobic patch and the second His cluster for iron coordination (Fig. 2 and 3). A change from a methionine residue to either valine or isoleucine at this position could have altered the electron transport or the substrate specificity of the enzyme that accounts for an improvement in astaxanthin selectivity. Leu-175 is located in transmembrane domain IV (Fig. 2). Due to a lack of detailed structural information for the CrtW ketolase and other membrane-bound oxidative enzymes in the family, it is difficult to understand the structural effect of the methionine substitution at this position. Nevertheless, the improvement in astaxanthin selectivity was more significant in E. coli cells expressing L175M than in those expressing M99I or M99V. Isolation of desirable mutants from this study demonstrates that protein engineering is another viable option for manipulating the carotenoid biosynthetic pathway for astaxanthin production.
Acknowledgments
We thank Qiong Cheng for providing plasmid pDCQ334, containing the carotenoid genes used in this study. We thank Dennis Arcilla and Tina Henry-Smith for help with HPLC analysis. We also thank Pamela Sharpe, Jean-Francois Tomb, and Rob Scott for insightful discussions.
Strong support for the carotenoid project by Ethel Jackson and Martin Odom is greatly appreciated.
REFERENCES
- 1.Alper, H., K. Miyaoku, and G. Stephanopoulos. 2005. Construction of lycopene-overproducing E. coli strains by combining systematic and combinatorial gene knockout targets. Nat. Biotechnol. 23:612-616. [DOI] [PubMed] [Google Scholar]
- 2.Bartolomé, B., Y. Jubete, E. Martinez, and F. de la Cruz. 1991. Construction and properties of a family of pACYC-derived cloning vectors compatible with pBR322 and its derivatives. Gene 102:75-78. [DOI] [PubMed] [Google Scholar]
- 3.Bhosale, P., and P. S. Bernstein. 2005. Microbial xanthophylls. Appl. Microbiol. Biotechnol. 68:445-455. [DOI] [PubMed] [Google Scholar]
- 4.Bouvier, F., Y. Keller, A. d'Harlingue, and B. Camara. 1998. Xanthophyll biosynthesis: molecular and functional characterization of carotenoid hydroxylases from pepper fruits (Capsicum annuum L.). Biochim. Biophys. Acta 1391:320-328. [DOI] [PubMed] [Google Scholar]
- 5.Choi, S. K., Y. Nishida, S. Matsuda, K. Adachi, H. Kasai, X. Peng, S. Komemushi, W. Miki, and N. Misawa. 2005. Characterization of β-carotene ketolases, CrtW, from marine bacteria by complementation analysis in Escherichia coli. Mar. Biotechnol. (New York) 7:515-522. [DOI] [PubMed] [Google Scholar]
- 6.Diaz, A. R., M. C. Mansilla, A. J. Vila, and D. de Mendoza. 2002. Membrane topology of the acyl-lipid desaturase from Bacillus subtilis. J. Biol. Chem. 277:48099-48106. [DOI] [PubMed] [Google Scholar]
- 7.Fernandez-Gonzalez, B., G. Sandmann, and A. Vioque. 1997. A new type of asymmetrically acting beta-carotene ketolase is required for the synthesis of echinenone in the cyanobacterium Synechocystis sp. PCC 6803. J. Biol. Chem. 272:9728-9733. [DOI] [PubMed] [Google Scholar]
- 8.Fraser, P. D., Y. Miura, and N. Misawa. 1997. In vitro characterization of astaxanthin biosynthetic enzymes. J. Biol. Chem. 272:6128-6135. [DOI] [PubMed] [Google Scholar]
- 9.Fraser, P. D., H. Shimada, and N. Misawa. 1998. Enzymic confirmation of reactions involved in routes to astaxanthin formation, elucidated using a direct substrate in vitro assay. Eur. J. Biochem. 252:229-236. [DOI] [PubMed] [Google Scholar]
- 10.Guerin, M., M. E. Huntley, and M. Olaizola. 2003. Haematococcus astaxanthin: applications for human health and nutrition. Trends Biotechnol. 21:210-216. [DOI] [PubMed] [Google Scholar]
- 11.Higuera-Ciapara, I., L. Felix-Valenzuela, and F. M. Goycoolea. 2006. Astaxanthin: a review of its chemistry and applications. Crit. Rev. Food Sci. Nutr. 46:185-196. [DOI] [PubMed] [Google Scholar]
- 12.Johnson, E. A. 2003. Phaffia rhodozyma: colorful odyssey. Int. Microbiol. 6:169-174. [DOI] [PubMed] [Google Scholar]
- 13.Krinsky, N. I. 1989. Antioxidant function of carotenoids. Free Radic. Biol. Med. 7:617-635. [DOI] [PubMed] [Google Scholar]
- 14.Lee, P. C., and C. Schmidt-Dannert. 2002. Metabolic engineering towards biotechnological production of carotenoids in microorganisms. Appl. Microbiol. Biotechnol. 60:1-11. [DOI] [PubMed] [Google Scholar]
- 15.Masamoto, K., N. Misawa, T. Kaneko, R. Kikuno, and H. Toh. 1998. Beta-carotene hydroxylase gene from the cyanobacterium Synechocystis sp. PCC6803. Plant Cell Physiol. 39:560-564. [DOI] [PubMed] [Google Scholar]
- 16.Misawa, N., S. Kajiwara, K. Kondo, A. Yokoyama, Y. Satomi, T. Saito, W. Miki, and T. Ohtani. 1995. Canthaxanthin biosynthesis by the conversion of methylene to keto groups in a hydrocarbon β-carotene by a single gene. Biochem. Biophys. Res. Commun. 209:867-876. [DOI] [PubMed] [Google Scholar]
- 17.Misawa, N., M. Nakagawa, K. Kobayashi, S. Yamano, Y. Izawa, K. Nakamura, and K. Harashima. 1990. Elucidation of the Erwinia uredovora carotenoid biosynthetic pathway by functional analysis of gene products expressed in Escherichia coli. J. Bacteriol. 172:6704-6712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Misawa, N., Y. Satomi, K. Kondo, A. Yokoyama, S. Kajiwara, T. Saito, T. Ohtani, and W. Miki. 1995. Structure and functional analysis of a marine bacterial carotenoid biosynthesis gene cluster and astaxanthin biosynthetic pathway proposed at the gene level. J. Bacteriol. 177:6575-6584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miura, Y., K. Kondo, T. Saito, H. Shimada, P. D. Fraser, and N. Misawa. 1998. Production of the carotenoids lycopene, β-carotene, and astaxanthin in the food yeast Candida utilis. Appl. Environ. Microbiol. 64:1226-1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mochimaru, M., H. Masukawa, and S. Takaichi. 2005. The cyanobacterium Anabaena sp. PCC 7120 has two distinct beta-carotene ketolases: CrtO for echinenone and CrtW for ketomyxol synthesis. FEBS Lett. 579:6111-6114. [DOI] [PubMed] [Google Scholar]
- 21.Nishida, Y., K. Adachi, H. Kasai, Y. Shizuri, K. Shindo, A. Sawabe, S. Komemushi, W. Miki, and N. Misawa. 2005. Elucidation of a carotenoid biosynthesis gene cluster encoding a novel enzyme, 2,2′-β-hydroxylase, from Brevundimonas sp. strain SD212 and combinatorial biosynthesis of new or rare xanthophylls. Appl. Environ. Microbiol. 71:4286-4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nordlund, P., and H. Eklund. 1995. Di-iron-carboxylate proteins. Curr. Opin. Struct. Biol. 5:758-766. [DOI] [PubMed] [Google Scholar]
- 23.Sandmann, G. 2001. Genetic manipulation of carotenoid biosynthesis: strategies, problems and achievements. Trends Plant Sci. 6:14-17. [DOI] [PubMed] [Google Scholar]
- 24.Sasata, R. J., D. W. Reed, M. C. Loewen, and P. S. Covello. 2004. Domain swapping localizes the structural determinants of regioselectivity in membrane-bound fatty acid desaturases of Caenorhabditis elegans. J. Biol. Chem. 279:39296-39302. [DOI] [PubMed] [Google Scholar]
- 25.Sedkova, N., L. Tao, P. E. Rouviere, and Q. Cheng. 2005. Diversity of carotenoid synthesis gene clusters from environmental Enterobacteriaceae strains. Appl. Environ. Microbiol. 71:8141-8146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shanklin, J., and E. Whittle. 2003. Evidence linking the Pseudomonas oleovorans alkane omega-hydroxylase, an integral membrane diiron enzyme, and the fatty acid desaturase family. FEBS Lett. 545:188-192. [DOI] [PubMed] [Google Scholar]
- 27.Shanklin, J., E. Whittle, and B. G. Fox. 1994. Eight histidine residues are catalytically essential in a membrane-associated iron enzyme, stearoyl-CoA desaturase, and are conserved in alkane hydroxylase and xylene monooxygenase. Biochemistry 33:12787-12794. [DOI] [PubMed] [Google Scholar]
- 28.Shimada, H., K. Kondo, P. D. Fraser, Y. Miura, T. Saito, and N. Misawa. 1998. Increased carotenoid production by the food yeast Candida utilis through metabolic engineering of the isoprenoid pathway. Appl. Environ. Microbiol. 64:2676-2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sonnhammer, E. L., G. von Heijne, and A. Krogh. 1998. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 6:175-182. [PubMed] [Google Scholar]
- 30.Tao, L., and Q. Cheng. 2004. Novel beta-carotene ketolases from non-photosynthetic bacteria for canthaxanthin synthesis. Mol. Genet. Genomics 272:530-537. [DOI] [PubMed] [Google Scholar]
- 31.Telfer, A. 2005. Too much light? How β-carotene protects the photosystem II reaction centre. Photochem. Photobiol. Sci. 4:950-956. [DOI] [PubMed] [Google Scholar]
- 32.Umeno, D., A. V. Tobias, and F. H. Arnold. 2005. Diversifying carotenoid biosynthetic pathways by directed evolution. Microbiol. Mol. Biol. 69:51-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang, C. W., and J. C. Liao. 2001. Alteration of product specificity of Rhodobacter sphaeroides phytoene desaturase by directed evolution. J. Biol. Chem. 276:41161-41164. [DOI] [PubMed] [Google Scholar]
- 34.Yuan, L. Z., P. E. Rouviere, R. A. Larossa, and W. Suh. 2006. Chromosomal promoter replacement of the isoprenoid pathway for enhancing carotenoid production in E. coli. Metab. Eng. 8:79-90. [DOI] [PubMed] [Google Scholar]