Abstract
Free-living amoebae in water are hosts to many bacterial species living in such an environment. Such an association enables bacteria to select virulence factors and survive in adverse conditions. Waterborne mycobacteria (WBM) are important sources of community- and hospital-acquired outbreaks of nontuberculosis mycobacterial infections. However, the interactions between WBM and free-living amoebae in water have been demonstrated for only few Mycobacterium spp. We investigated the ability of a number (n = 26) of Mycobacterium spp. to survive in the trophozoites and cysts of Acanthamoeba polyphaga. All the species tested entered the trophozoites of A. polyphaga and survived at this location over a period of 5 days. Moreover, all Mycobacterium spp. survived inside cysts for a period of 15 days. Intracellular Mycobacterium spp. within amoeba cysts survived when exposed to free chlorine (15 mg/liter) for 24 h. These data document the interactions between free-living amoebae and the majority of waterborne Mycobacterium spp. Further studies are required to examine the effects of various germicidal agents on the survival of WBM in an aquatic environment.
Mycobacteria are a large group of microorganisms that inhabit a diverse range of natural environments. Environmental mycobacteria are a frequent cause of opportunistic infection in human beings and livestock (23, 56, 76). There is growing recognition in recent years that water is an important vehicle of transmission of environmental mycobacteria. This is based on the fact that in the recent past contaminated water supply systems have been responsible for several hospital and community outbreaks of mycobacterial infections (16, 77, 78, 79, 82, 83). These included infections as diverse as life-threatening pneumonia in patients with artificial ventilation, cystic fibrosis (54), and chronic granulomatous disease (79); outbreaks of skin infection following liposuction (51); furunculosis after domestic footbaths (77, 83); mastitis after body piercing (73); and abscess formation in intravenous drug users (26). In one instance (79, 82), workers exposed to contaminated industrial effluents developed pneumonia due to an environmental mycobacterium.
In hospital therapy pools, waterborne mycobacteria (WBM) may represent about 33% of the microorganisms present in water and about 80% of those in air in the vicinity of such a contaminated water source (5). WBM include both rapidly and slowly growing Mycobacterium spp., depending on whether they require a week or less for the production of visible colonies in solid medium. Examples of WBM include among other species the Mycobacterium avium complex, Mycobacterium gordonae, Mycobacterium malmoense, Mycobacterium simiae, Mycobacterium marinum, Mycobacterium tusciae, and Mycobacterium lentiflavum, which have been found in natural fresh waters (72, 76). Mycobacterium mucogenicum, Mycobacterium kansasii, M. gordonae, Mycobacterium flavescens, Mycobacterium aurum, Mycobacterium fortuitum, Mycobacterium peregrinum, and Mycobacterium chelonae have been isolated from public potable water (17, 47, 76). In water, M. avium and Mycobacterium intracellulare can survive, even under low oxygen tension (13, 41). Furthermore, M. intracellulare can remain viable for a year in deionized sterile water (6). Some WBM such as the M. avium complex, Mycobacterium xenopi, Mycobacterium phlei, and M. chelonae can withstand extreme temperatures and contaminate ice machine water (61). M. fortuitum and M. chelonae can form biofilms on Silastic rubber disks even under running water (34).
Free-living amoebae including Acanthamoeba spp. are commonly found in natural aquatic systems, water supplies, and cooling systems (37, 50), usually feeding on bacteria (68). It was shown that amoebae host several intracellular pathogens including Legionella spp., Chlamydia spp., Parachlamydia spp., Listeria spp., Burkholderia spp., Campylobacter jejuni, Helicobacter pylori, Pasteurella multocida, Salmonella enterica, Francisella tularensis, and Simkania negevensis (10, 32, 38). The association, mentioned above, of Legionella pneumophila with Acanthamoeba spp. may select organisms better capable of surviving in the hostile environment of mammalian phagocytic cells (35). This is supported by the fact that Legionella spp. use the same set of genes to establish themselves in Acanthamoeba spp. and in mammalian cells (27, 62).
The interactions between WBM and free-living amoebae are poorly understood. A number of Mycobacterium species, including M. avium, M. marinum, M. simiae, M. phlei, Mycobacterium smegmatis, and M. fortuitum, live intracellularly in amoebae (42). M. avium was shown to grow in Acanthamoeba spp. (15, 66). Recently, we used an amoebal coculture system for the isolation of Mycobacterium massiliense from clinical specimens (1).
In our present work, we compared levels of intra-amoebal penetration and intracystic survival of 26 environmental Mycobacterium spp. We also examined the effects of chlorination on the intra-amoebal growth and survival of these Mycobacterium species.
MATERIALS AND METHODS
Mycobacterium strains.
The type strains used in this study are listed in Table 1. The identification of species was establishedusing 16S rRNA and rpoB gene sequences. The primers used were Fd1 (5′-AGAGTTTGATCATGGCTCAG-3′) and Rp2 (5′-ACGGCTACCTTGTTACGACTT-3′) for 16S rRNA gene (81) and Myco-F (5′-GGCAAGGTCACCCCGAAGGG-3′) and Myco-R (5′-AGCGGCTGCTGGGTGATCATC-3′) for the rpoB gene (2). The strains had been held in skim milk and kept at −20°C before they were used. When required, each strain was cultured in Middelbrook 7H9 liquid medium. For subculture, we used Middelbrook and Cohn 7H10 agar (Becton Dickinson, Le Pont de Claix, France). All subcultures were done at 30°C. The minimum durations of incubation were 5 and 7 days for rapidly growing mycobacteria (RGM) and slowly growing mycobacteria (SGM), respectively.
TABLE 1.
List of Mycobacterium sp. strains used in this study
| Species | Strain | Source | Survival after infection of A. polyphaga in:
|
Reference | |
|---|---|---|---|---|---|
| Trophozoites | Cysts | ||||
| M. abscessus | CIP 104536T | Abscess | + | + | 44 |
| M. chelonae | CIP 104535T | Tortoise, tubercle | + | + | 43 |
| M. immunogenum | CIP 106684T | Metal working fluid | + | + | 82 |
| M. mucogenicum | ATCC 49650T | Thyroglossal duct cyst | + | + | 64 |
| M. fortuitum | CIP 104534T | Abscess | + | + | 20 |
| M. peregrinum | CIP 105382T | Bronchial aspiration | + | + | 44 |
| M. smegmatis | ATCC 19420T | Genital secretions | + | + | 48 |
| M. goodii | ATCC 700504T | Heel | + | + | 14 |
| M. mageritense | CIP 104973T | Sputum | + | + | 22 |
| M. aurum | CIP 104465T | Soil | + | + | 74 |
| M. kansasii | CIP 104589T | Fatal infection | + | + | 36 |
| M. szulgai | CIP 104532T | —a | + | + | 49 |
| M. malmoense | CIP 105775T | Lung tissue | + | + | 60 |
| M. terrae | CIP 104321T | Sputum, gastric lavage | + | + | 80 |
| M. marinum | CIP 104528T | Saltwater fish | + | + | 8 |
| M. tusciae | CIP 106367T | Cervical lymph node | + | + | 71 |
| M. gordonae | CIP 104529T | Gastric lavage | + | + | 12 |
| M. intracellulare | CIP 104243T | — | + | + | 58 |
| M. avium subsp. avium | CIP 104244T | Diseased hen, liver | + | + | 69 |
| M. simiae | CIP 104531T | Indian monkey | + | + | 39 |
| M. bohemicum | CIP 105811T | Sputum | + | + | 57 |
| M. lentiflavum | CIP 105465T | Spondylodiscitis | + | + | 65 |
| M. massiliense | CIP 108297T | Bronchoalveolar fluid | + | + | 1 |
| M. septicum | ATCC 700731T | Catheter tip of centrally located Hickman | + | + | 59 |
| M. porcinum | CIP 105392T | Swine, lymph node | + | + | 75 |
| M. gastri | CIP 104530T | Gastric lavage fluid | + | + | 80 |
—, source unknown.
Amoebal coculture and light-microscopic detection of Acanthamoeba polyphaga infected with mycobacteria.
The strain (Linc-AP1) of A. polyphaga used was a donation by T. J. Rowbotham, Public Health Laboratory, Leeds, United Kingdom. It was grown at 28°C for 4 days in 150-cm3 culture flasks (Corning, New York) containing 30 ml of peptone-yeast extract-glucose (PYG) broth (29, 30, 46). Amoebal cells were harvested when their average concentration reached a level of 5 × 105 cells/ml of broth. The harvested cells were then centrifuged at 2,000 rpm for 10 min, and the pellet was suspended twice in 30 ml of Page's modified Neff's amoeba saline (PAS) (29, 46). One milliliter of this suspension was dropped into each well of a 12-well microplate (Corning, New York) and incubated at 33°C for 7 days. The microplate, prepared as described above, was used for culturing mycobacteria. Each well of the microplate was inoculated with mycobacterium cells (concentration, 105 mycobacteria/ml) suspended in phosphate-buffered saline. The final concentration of mycobacterium cells in the well was 104 mycobacteria/ml. As a control, 1 μl of the suspension of mycobacterium cells (concentration, 104 mycobacteria/ml of PAS) was dropped into each well of a 12-well control microplate. The microplate was centrifuged at 4,000 rpm for 30 min and incubated at 33°C under an atmosphere humidified with 5% CO2. After incubation for 48 h, the monolayer was washed three times with PAS, before being treated with amikacin (100 μg of amikacin for each milliliter of PAS) for 2 hours. This was done to kill any remaining extracellular or adherent mycobacteria (15, 67). Amikacin was removed from the monolayer by washing with PAS. After the washing, the monolayer was incubated in 1 ml of PAS for 3 days. In parallel, the supernatant obtained after washing was cultured in appropriate axenic medium. The microplate was examined daily to detect the presence of cytopathic effect. After gentle shaking and cytocentrifugation at 800 rpm for 10 min, mycobacteria were detected inside amoebal trophozoites as described by Gimenez (28), by Ziehl-Neelsen staining, and by Gram staining in amoebal cocultures that were 1 day, 3 days, and 5 days old, respectively. Intra-amoebal mycobacteria were released by lysing the monolayer with 1 ml of 0.5% sodium dodecyl sulfate, followed by two successive passages through a 27-gauge needle. The presence of viable mycobacteria was documented by detecting the presence of a colony of mycobacteria formed on Middlebrook 7H10 agar that had been inoculated with 200 μl of the cell lysate. All experiments were repeated three times.
Encystment.
For encystment, we used amoebal cocultures that were 48 hours old. Ten milliliters of amoebal coculture (concentration, 5 × 105 amoebal cells/ml of PAS) was taken in 25-cm3 culture flasks (Corning) and infected with 1 ml (concentration, 105 mycobacterium cells/ml of PAS) of mycobacterium suspension in PAS. The supernatant was discarded, and the amoebal monolayer was rinsed once with encystment buffer (0.1 M KCl, 0.02 M Tris, 8 mM MgSO4, 0.4 mM CaCl2, 1 mM NaHCO3) before being incubated (at 33°C for 3 days) in fresh encystment buffer (66). The cysts formed were centrifuged at 4,000 rpm for 30 min. Then, they were washed three times with PAS by centrifugation. In order to kill extracellular mycobacteria, the pellet was resuspended in sodium hypochlorite (Javel oxena, Portes-Les-Valences, France) solution (concentration, 15 mg of chlorine/ml of the solution) and held at 33°C for 24 h. The concentration of chlorine we used was well in accord with that recommended for decontamination of water reservoirs throughout France. The cysts were washed thrice with PYG medium. One half of the sample so washed was processed for electron microscopy and the other half incubated (at 33°C for 7 days) in PYG medium. Viable cysts became trophozoites and eventually lysed due to the large number of intracellular mycobacteria. The process of excystment was verified by light-microscopic examination of Ziehl-Neelsen smears and by observing viable mycobacteria grown in Middlebrook 7H10 agar medium. In parallel with electron microscopy studies, cysts were prepared and stored at room temperature for 15 days. All experiments were repeated three times.
Ultrastructural study.
To eliminate noningested mycobacteria, the amoebal monolayer inoculated with mycobacteria and amoebal cysts were washed twice with sterile phosphate-buffered saline. They were then incubated overnight in monophosphate buffer (pH 12) before being fixed (1 h at 4°C) with 1% osmium tetroxide. The fixed samples were dehydrated by successive washing with graded (from 25 to 100%) concentrations of acetone. Then, the samples were successively incubated (for 1 h) in a 50% (vol/vol) acetone-Epon suspension and Epon (overnight) before being embedded in Araldite (Fluka, St Quentin Fallavier, France). Ultrathin sections were cut from the blocks using an ultracut microtome (Reichert-Leica, Marseille, France) before being deposited on copper grids coated with Formvar (Sigma-Aldrich, Taufkirchen, Germany). The ultrathin sections so mounted were stained for 10 min with methanol-uranyl acetate and lead nitrate with sodium citrate solution (30). The grids were examined under a transmission electron microscope (Morgani 268D; Philips, Eindhoven, The Netherlands).
Stastitical analysis.
Repeated-measures analysis of variance was performed using the SAS v9.1.3 (SAS Institute Inc., Cary, NC) software to test whether the amoebal infection rates differed within and between mycobacterial types. Tests were two sided, and P values <0.05 were considered significant.
RESULTS
Survival of mycobacteria in A. polyphaga.
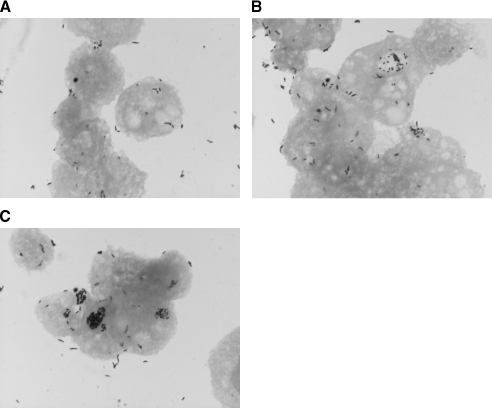
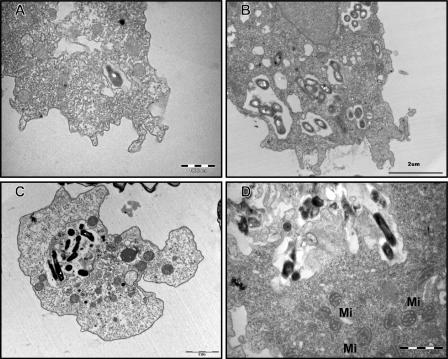
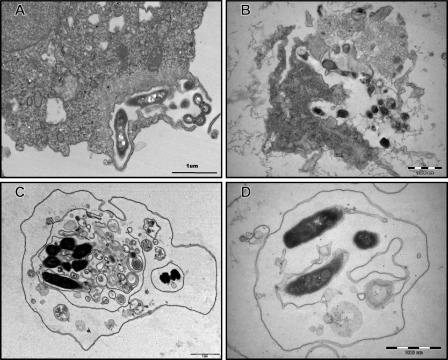
All the species of Mycobacterium tested survived in PAS. However, they did not grow or show residual growth in this nutrient-limiting medium after 5 days. Mycobacteria could be seen both unattached and in close association with amoebal cells (Fig. 1A, B, and C). Based on multigenic phylogeny analysis (3, 21), six mycobacterial species representative of the mycobacterial groups under study, i.e., M. chelonae, M. mucogenicum, M. septicum, M. kansasii, M. bohemicum, and M. szulgai, did not grow after exposure of extra-amoebal organisms to amikacin at 100 mg/ml for 2 h. As demonstrated by microscopic examination after Gimenez and Ziehl-Neelsen stainings (Fig. 1A, B, and C), all the species of Mycobacterium tested were able to enter A. polyphaga at 33°C. The percentage of infected amoeba increased significantly from 93% to 99% over 5 days (P = 0.0005) (Table 2). This percentage was not significantly different between RGM and SGM (P = 0.34). Species were able to survive in the intracellular milieu of the trophozoites of A. polyphaga for the duration of the experiment (5 days). This was confirmed by positive subcultures although quantification of subcultured mycobacteria was not attempted. Mycobacterial species could be easily observed in the amoebal cytoplasm even on day 1 postinfection. The majority of RGM caused extensive vacuolization of trophozoites after 48 h of coculture. In contrast, it took at least 96 h for SGM to induce the formation of visible vacuoles inside amoeba. The amoeba cell infected as described above showed one or more than one vacuole containing mycobacteria (Fig. 1A, B, and C). The presence of mycobacteria inside the vacuole was confirmed by transmission electron microscopy (Fig. 2A, B, C, and D). The vacuoles containing mycobacteria were surrounded by a large number of host mitochondria (Fig. 2D). However, we did not observe any ultrastructural change in the mitochondria of the host. A. polyphaga released free mycobacteria (Fig. 3A and B) or 2.5- to 6.5-μm vesicles containing mycobacteria (Fig. 3C and D).
FIG. 1.
A. polyphaga trophozoite infected with M. bohemicum and stained by Ziehl-Nielsen at (A) day 1, (B) day 3, and (C) day 5.
TABLE 2.
Percentages of A. polyphaga trophozoites infected with different species of nontuberculous mycobacteria over a 5-day coculturea
| Age of A. polyphaga culture (days) | Mean %b (±SD) of A. polyphaga trophozoites infected by:
|
|||
|---|---|---|---|---|
| M. mucogenicum | M. septicum | M. bohemicum | M. szulgai | |
| 1 | 93.0 ± 3.0 | 93.0 ± 4.6 | 96.0 ± 3.6 | 97.6 ± 3.2 |
| 3 | 97.3 ± 1.5 | 97.3 ± 2.1 | 99.0 ± 1.0 | 98.3 ± 1.5 |
| 5 | 96.7 ± 3.2 | 98.3 ± 1.5 | 96.7 ± 1.5 | 99.7 ± 0.6 |
The observed percentages were not significantly different.
Percentage of A. polyphaga cells with at least one intravacuolar mycobacterium organism.
FIG. 2.
Transmission electron-microscopic observation of M. mucogenicum (2-h coculture; A), M. massiliense (3-day coculture; B), M. septicum (5-day coculture; C), and M. terrae (5-day coculture; D) cocultivated with A. polyphaga trophozoites. The bacteria are seen inside an amoebal vacuole. Mi, mitochondria.
FIG. 3.
Transmission electron-microscopic observation of A. polyphaga relapsing free M. massiliense (A), relapsing M. terrae through lysis (B), and relapsing vesicles containing M. septicum (C and D).
Mycobacteria inside A. polyphaga cysts.
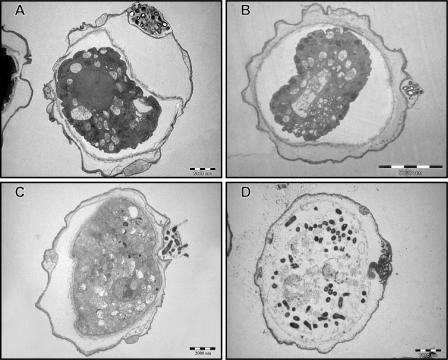
All the 26 Mycobacterium spp. tested survived inside A. polyphaga cysts for 15 days at room temperature. We examined the fate of mycobacteria inside A. polyphaga cysts by transmission electron microscopy. Figure 4A and B document one such cyst containing numerous M. chelonae and M. abscessus bacilli, respectively. They were visible in the spaces between the two walls (i.e., inner and outer walls) of the cyst. Some species (e.g., M. septicum) were observed to have been present on the inner side of the outer wall and in the cytoplasm of the cyst (Fig. 4C and D). Furthermore, M. septicum was present in dead cysts (data not shown). Less than 3% of infected cysts harbored bacilli in the cytoplasm, and <1% of infected cysts harbored bacilli in the outer wall.
FIG. 4.
Transmission electron-microscopic observation of A. polyphaga cysts containing M. chelonae (A) and M. abscessus (B) within a double cell wall and M. septicum within a double cell wall (C) and in the cytoplasm (D).
The effect of chlorination on the viability of mycobacterium was examined. The mycobacterium species exposed, as described above, to free chlorine (15 mg/liter) for 24 h did not grow in Middelbrook 7H10 agar medium. In contrast, the concentration of chlorine we used was not destructive to A. polyphaga cysts, irrespective of whether they had been infected or were noninfected. This is evident by the fact that well over 50% of the cysts that were exposed to free chlorine for 24 h excysted in PYG medium. The abilities of amoeba cysts, both infected with mycobacteria and noninfected, to revert to trophozoites under optimal growth conditions were examined by microscopy. Samples were obtained from these cultures and subcultured in Middlebrook 7H10 agar. The results were positive for mycobacteria. The morphological features of the bacterial colonies grown in Middlebrook 7H10 agar were consistent with those of acid-fast bacilli.
DISCUSSION
Twenty-six Mycobacterium spp. tested survived within A. polyphaga trophozoites for 5 days (Table 1). We observed that the addition of amikacin to the incubation buffer after 2 hours of coculture killed extra-amoebal organisms due to the bactericidal effect of this antibiotic, as previously described (15, 67). No difference was observed in the numbers of intracellular mycobacteria in the presence or in the absence of amikacin (15). Previous studies have suggested an association between free-living amoeba and various Mycobacterium spp. in aquatic environments (23, 32, 56, 76). Recently, such an association has been documented in a hospital water supply system (68). Our data show, for example, that free-living amoeba may be part of the reservoir of M. septicum. This newly described mycobacterium was isolated from human samples (4, 59) and further from biofilms in a drinking water distribution system (63) and was recently associated with a devastating outbreak in a fish colony (40).
Furthermore, these species survived within the cysts of A. polyphaga for 15 days. The possibility that the cysts of A. polyphaga might have been contaminated by mycobacteria from an external source was considered extremely unlikely. To avoid contamination, we carefully and repeatedly washed the amoebal monolayer. For each washing step, we used fresh encystment buffer. We also exposed the cysts to chlorination (15 mg/liter) for 24 h. We have herein demonstrated that this level and duration of exposure to free chlorine was germicidal to the Mycobacterium spp. tested. The cysts are double-walled, resilient entities which survive exposure to temperature between −20°C and +42°C (7). We found that M. chelonae and M. abscessus survive within the amoeba cyst. This fact could explain why these species, which cannot grow at 42°C as free organisms, were recovered in large numbers from a hot-water drinking water distribution system (25). Also, M. chelonae can withstand extreme temperatures and can contaminate ice machine water (61).
The cysts of A. polyphaga can withstand germicidal compounds commonly used for decontaminating bronchoscopes (31). This means that the cysts of A. polyphaga may remain viable in bronchoscopes even after they have been decontaminated with germicides. In fact, contaminated bronchoscopes have been the sources of infections due to some nontuberculosis mycobacterial species, including M. chelonae and M. kansasii, in immunocompromised patients (18, 55, 78, 84). Likewise, contamination of hospital equipment and medication was traced to the ubiquitous presence of these organisms in tap water (70, 78). Their resistance to commonly used disinfectants was responsible for pseudo-outbreaks of infections associated with surgical implants, health care-associated septicemia, and lung disease following bronchoscopy (9, 24, 45, 78). Our data also indicate that mycobacterium-infected cysts can withstand a chlorine concentration of 15 mg/liter whereas the trophozoites of A. polyphaga are sensitive to chlorine at a concentration of 1.25 mg/liter (19). Indeed, we recovered viable mycobacteria from the cysts of A. polyphaga that had been exposed to such a chlorine concentration. This is a source of concern. These data suggest that encystment of mycobacteria may be one of the many mechanisms used by these organisms to resist the germicidal effect of chlorine in water distribution systems. These data may also explain the differences in the mycobacterium populations observed in treated and untreated waters (72). In one pilot study, treatment of a water system with ozone or chlorine resulted in a dramatic shift in the bacterial population to the Actinomyces family, which includes mycobacterial species (53).
Previous studies have also documented the presence and growth of M. avium inside the trophozoites of Acanthamoeba castellanii (15). The growth in trophozoites enhanced the entry of this mycobacterium into amoebae, the intestinal epithelial cell line (HT-29), and macrophages (15). It also enhanced the abilities of M. avium to colonize the intestine and replicate in the livers and spleens of mice in a murine model of M. avium infection (15). Furthermore, it was found that mycobacteria cultured into amoebas were better able to survive within macrophages and that interaction of M. avium with environmental amoebae enhances virulence (15). Our findings are in agreement with the speculation that bacteria resistant to amoebae may be resistant to macrophages and vice versa and therefore that amoebae select pathogenic microorganisms (33). Compared to those living within macrophage-like cell lines (e.g., U937), M. avium living inside the trophozoites of A. castellanii were shown to be more resistant to rifabutin, azithromycin, and clarithromycin (52). Our data now extend previous data to M. intracellulare, a species closely related to M. avium. We have also demonstrated the survival of M. smegmatis inside the trophozoites of A. polyphaga. This contradicts the findings of others (15), while M. smegmatis was shown to have persisted inside HEp-2 epithelial cells (11).
In conclusion, 26 environmental Mycobacterium spp. survived within amoebal trophozoites and cysts for various lengths of time. This fact may have implications for the mode of transmission of these microorganisms. It has been shown previously that the association between amoeba and Legionella pneumophila in an aquatic environment enabled this bacterium to select virulence factors and survive in such a hostile environment (27). This may also be true for environmental Mycobacterium spp. The environmental Mycobacterium spp. living within the cysts of A. polyphaga can withstand the concentration of free chlorine normally used for the treatment of water in municipal water supply systems. We believe that testing encysted mycobacteria may be necessary to properly evaluate decontamination procedures for water and endoscopes.
Acknowledgments
This work was in part supported by the project Interreg IIIB of the Université Euro-Mediterranéenne.
We thank Lina Barrassi for technical assistance, Nicolas Aldrovandi and Bernard Campagna for electron microscopy, and Mohammad Khan for expert review of the manuscript.
REFERENCES
- 1.Adékambi, T., M. Reynaud-Gaubert, G. Greub, M. J. Gevaudan, B. La Scola, D. Raoult, and M. Drancourt. 2004. Amoebal coculture of “Mycobacterium massiliense” sp. nov. from the sputum of a patient with hemoptoic pneumonia. J. Clin. Microbiol. 42:5493-5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adékambi, T., P. Colson, and M. Drancourt. 2003. rpoB-based identification of nonpigmented and late-pigmenting rapidly growing mycobacteria. J. Clin. Microbiol. 41:5699-5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adékambi, T., and M. Drancourt. 2004. Dissection of phylogenic relationships among nineteen rapidly growing mycobacterium species by 16S rRNA, hsp65, sodA, recA, and rpoB gene sequencing. Int. J. Syst. Evol. Microbiol. 54:2095-2105. [DOI] [PubMed] [Google Scholar]
- 4.Adékambi, T., and M. Drancourt. 2006. Isolation of Mycobacterium septicum from the sputum of patient suffering hemoptoic pneumonia. Res. Microbiol. 157:466-470. [DOI] [PubMed] [Google Scholar]
- 5.Angenent, L. T., S. T. Kelley, A. St Amand, N. R. Pace, and M. T. Hernandez. 2005. Molecular identification of potential pathogens in water and air of a hospital therapy pool. Proc. Natl. Acad. Sci. USA 102:4860-4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Archuleta, R. J., P. Mullens, and T. P. Primm. 2002. The relationship of temperature to desiccation and starvation tolerance of the Mycobacterium avium complex. Arch. Microbiol. 178:311-314. [DOI] [PubMed] [Google Scholar]
- 7.Armstrong, M. 2000. The pathogenesis of human Acanthamoeba infection. Infect. Dis. Rev. 2:65-73. [Google Scholar]
- 8.Aronson, J. D. 1926. Spontaneous tuberculosis in salt water fish. J. Infect. Dis. 39:315-320. [Google Scholar]
- 9.Ashford, D. A., S. Kellerman, M. Yarkrus, S. Brin, R. C. Good, L. Fineli, W. R. Jarvis, and M. M. McNeil. 1997. Pseudo-outbreak of septicemia due to rapidly growing mycobacteria associated with extrinsic contamination of culture supplement. J. Clin. Microbiol. 35:2040-2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Axelsson-Olsson, D., J. Waldenstrom, T. Broman, B. Olsen, and M. Holmberg. 2005. Protozoan Acanthamoeba polyphaga as a potential reservoir for Campylobacter jejuni. Appl. Environ. Microbiol. 71:987-992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bermudez, L. E., K. Shelton, and L. S. Young. 1995. Comparison of the ability of Mycobacterium avium, M. smegmatis and M. tuberculosis to invade and replicate within HEp-2 epithelial cells. Tuber. Lung Dis. 76:240-247. [DOI] [PubMed] [Google Scholar]
- 12.Bojalil, L. F., J. Cerbon, and A. Trujillo. 1962. Adansonian classification of mycobacteria. J. Gen. Microbiol. 28:333-346. [DOI] [PubMed] [Google Scholar]
- 13.Brooks, R. W., K. L. George, B. C. Parker, J. O. Falkinham III, and H. Gruff. 1984. Recovery and survival of nontuberculous mycobacteria under various growth and decontamination conditions. Can. J. Microbiol. 30:1112-1117. [DOI] [PubMed] [Google Scholar]
- 14.Brown, B. A., B. Springer, V. A. Steingrube, R. W. Wilson, G. E. Pfyffer, M. J. Garcia, M. C. Menendez, B. Rodriguez-Salgado, K. C. Jost, Jr., S. H. Chiu, G. O. Onyi, E. C. Bottger, and R. J. Wallace, Jr. 1999. Mycobacterium wolinskyi sp. nov. and Mycobacterium goodii sp. nov., two new rapidly growing species related to Mycobacterium smegmatis and associated with human wound infections: a cooperative study from the International Working Group on Mycobacterial Taxonomy. Int. J. Syst. Bacteriol. 49:1493-1511. [DOI] [PubMed] [Google Scholar]
- 15.Cirillo, J. D., S. Falkow, L. S. Tompkins, and L. E. Bermudez. 1997. Interaction of Mycobacterium avium with environmental amoebae enhances virulence. Infect. Immun. 65:3759-3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Conger, N. G., R. J. O'Connell, V. L. Laurel, K. N. Olivier, E. A. Graviss, N. Williams-Bouyer, Y. Zhang, B. A. Brown-Elliott, and R. J. Wallace, Jr. 2004. Mycobacterium simae outbreak associated with a hospital water supply. Infect. Control. Hosp. Epidemiol. 25:1050-1055. [DOI] [PubMed] [Google Scholar]
- 17.Covert, T. C., M. R. Rodgers, A. L. Reyes, and G. N. Stelma, Jr. 1999. Occurrence of nontuberculous mycobacteria in environmental samples. Appl. Environ. Microbiol. 65:2492-2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cox, R., K. deBorja, and M. C. Bach. 1997. A pseudo-outbreak of Mycobacterium chelonae infections related to bronchoscopy. Infect. Control Hosp. Epidemiol. 18:136-137. [DOI] [PubMed] [Google Scholar]
- 19.Cursons, R. T., T. J. Brown, and E. A. Keys. 1980. Effect of disinfectants on pathogenic free-living amoebae in axenic conditions. Appl. Environ. Microbiol. 40:62-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Da Costa Cruz, J. 1938. “Mycobacterium fortuitum”, um novo bacilo acido-resistente patogenico para o homen. Acta Med. 1:297-301. [Google Scholar]
- 21.Devulder, G., M. Perouse de Montclos, and J. P. Flandrois. 2005. A multigene approach to phylogenetic analysis using the genus Mycobacterium as a model. Int. J. Syst. Evol. Microbiol. 55:293-302. [DOI] [PubMed] [Google Scholar]
- 22.Domenech, P., M. S. Jimenez, M. C. Menendez, T. J. Bull, S. Samper, A. Manrique, and M. J. Garcia. 1997. Mycobacterium mageritense sp. nov. Int. J. Syst. Bacteriol. 47:535-540. [DOI] [PubMed] [Google Scholar]
- 23.Falkinham, J. O., III. 2002. Nontuberculous mycobacteria in the environment. Clin. Chest Med. 23:529-551. [DOI] [PubMed] [Google Scholar]
- 24.Ferguson, D. D., K. Gershman, B. Jensen, M. J. Arduino, M. A. Yakrus, R. C. Cooksey, and A. Srinivasan. 2004. Mycobacterium goodii infections associated with surgical implants at Colorado hospital. Emerg. Infect. Dis. 10:1868-1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fischeder, R., R. Schulze-Röbbecke, and A. Weber. 1991. Occurrence of mycobacteria in drinking water samples. Zentbl. Hyg. Umweltmed. 192:154-158. [PubMed] [Google Scholar]
- 26.Galil, K., L. A. Miller, M. A. Yakrus, R. J. Wallace, Jr., D. G. Mosley, B. England, G. Huitt, M. M. McNeil, and B. A. Perkins. 1999. Abscesses due to Mycobacterium abscessus linked to injection of unapproved alternative medication. Emerg. Infect. Dis. 5:681-687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao, L. Y., O. S. Harb, and K. Y. Abu. 1997. Utilization of similar mechanisms by Legionella pneumophila to parasitize two evolutionarily distant host cells, mammalian macrophages and protozoa. Infect. Immun. 65:4738-4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gimenez, D. F. 1964. Staining rickettsiae in yolk-sac cultures. Stain Technol. 39:135-140. [DOI] [PubMed] [Google Scholar]
- 29.Greub, G., S. B. La, and D. Raoult. 2004. Amoebae-resisting bacteria isolated from human nasal swabs by amoebal coculture. Emerg. Infect. Dis. 10:470-477. [DOI] [PubMed] [Google Scholar]
- 30.Greub, G., and D. Raoult. 2002. Crescent bodies of Parachlamydia acanthamoeba and its life cycle within Acanthamoeba polyphaga: an electron micrograph study. Appl. Environ. Microbiol. 68:3076-3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Greub, G., and D. Raoult. 2003. Biocides currently used for bronchoscope decontamination are poorly effective against free-living amoebae. Infect. Control Hosp. Epidemiol. 24:784-786. [DOI] [PubMed] [Google Scholar]
- 32.Greub, G., and D. Raoult. 2004. Microorganisms resistant to free-living amoebae. Clin. Microbiol. Rev. 17:413-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Greub, G., J. L. Mege, and D. Raoult. 2003. Parachlamydia acanthamoebae enters and multiplies within human macrophages and induces their apoptosis. Infect. Immun. 71:5979-5985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hall-Stoodley, L., and H. Lappin-Scott. 1998. Biofilm formation by the rapidly growing mycobacterial species Mycobacterium fortuitum. FEMS Microbiol. Lett. 168:77-84. [DOI] [PubMed] [Google Scholar]
- 35.Harb, O. S., L. Y. Gao, and K. Y. Abu. 2000. From protozoa to mammalian cells: a new paradigm in the life cycle of intracellular bacterial pathogens. Environ. Microbiol. 2:251-265. [DOI] [PubMed] [Google Scholar]
- 36.Hauduroy, P. 1955. A new group of mycobacteria: the abnormal mycobacteria. Ann. Inst. Pasteur 89:673-675. [PubMed] [Google Scholar]
- 37.Hoffmann, R., and R. Michel. 2001. Distribution of free-living amoebae (FLA) during preparation and supply of drinking water. Int. J. Hyg. Environ. Health 203:215-219. [DOI] [PubMed] [Google Scholar]
- 38.Hundt, M. J., and C. G. Ruffolo. 2005. Interaction of Pasteurella multocida with free-living amoebae. Appl. Environ. Microbiol. 71:5458-5464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karassova, V., J. Weissfeiler, and E. Krasznay. 1965. Occurrence of atypical mycobacteria in Macacus rhesus. Acta Microbiol. Acad. Sci. Hung. 12:275-282. [PubMed] [Google Scholar]
- 40.Kent, M. L., C. M. Whipps, J. L. Matthews, D. Florio, V. Watral, J. K. Bishop-Stewart, M. Poort, and L. Bermudez. 2004. Mycobacteriosis in zebrafish (Danio rerio) research facilities. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 138:383-390. [DOI] [PubMed] [Google Scholar]
- 41.Kirschner, R. A., B. C. Parker, and J. O. Falkinham. 1999. Humic and fulvic acids stimulate the growth of Mycobacterium avium. FEMS Microbiol. Ecol. 30:327-332. [DOI] [PubMed] [Google Scholar]
- 42.Krishna-Prasad, B. N., and S. K. Gupta. 1978. Preliminary report on engulfment and retention of mycobacteria by trophozoites of axenically grow Acanthamoeba castellanii Douglas. Curr. Sci. 47:245-247. [Google Scholar]
- 43.Kubica, G. P., I. Baess, R. E. Gordon, P. A. Jenkins, J. B. Kwapinski, C. McDurmont, S. R. Pattyn, H. Saito, V. Silcox, J. L. Stanford, K. Takeya, and M. Tsukamura. 1972. A co-operative numerical analysis of rapidly growing mycobacteria. J. Gen. Microbiol. 73:55-70. [DOI] [PubMed] [Google Scholar]
- 44.Kusunoki, S., and T. Ezaki. 1992. Proposal of Mycobacterium peregrinum sp. nov., nom. rev., and elevation of Mycobacterium chelonae subsp. abscessus (Kubica et al.) to species status: Mycobacterium abscessus comb. nov. Int. J. Syst. Bacteriol. 42:240-245. [DOI] [PubMed] [Google Scholar]
- 45.Lai, K. K., B. A. Brown, J. A. Westerling, S. A. Fontecchio, Y. Zhang, and R. J. Wallace, Jr. 1998. Long-term laboratory contamination by Mycobacterium abscessus resulting in two pseudo-outbreaks: recognition with the use of random amplified polymorphic DNA (RAPD) polymerase chain reaction. Clin. Infect. Dis. 27:169-175. [DOI] [PubMed] [Google Scholar]
- 46.La Scola, B., L. Mezi, P. J. Weiller, and D. Raoult. 2001. Isolation of Legionella anisa using an amoebic coculture procedure. J. Clin. Microbiol. 39:365-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Le Dantec, C., J. P. Duguet, A. Montiel, N. Dumoutier, S. Dubrou, and V. Vincent. 2002. Occurrence of mycobacteria in water treatment lines and in water distribution systems. Appl. Environ. Microbiol. 68:5318-5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lehmann, K. B., and R. O. Neumann. 1931. Mycobacterium smegmatis, p. 755-757. In A. Hafner (ed.), Bacteriology, especially determinative bacteriology. G. E. Stechert and Co., New York, N.Y.
- 49.Marks, J., P. A. Jenkins, and M. Tsukamura. 1972. Mycobacterium szulgai—a new pathogen. Tubercle 53:210-214. [DOI] [PubMed] [Google Scholar]
- 50.Marshall, M. M., D. Naumovitz, Y. Ortega, and C. R. Sterling. 1997. Waterborne protozoan pathogens. Clin. Microbiol. Rev. 10:67-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meyers, H., B. A. Brown-Elliott, D. Moore, J. Curry, C. Truong, Y. Zhang, and R. J. Wallace, Jr. 2002. An outbreak of Mycobacterium chelonae infection following liposuction. Clin. Infect. Dis. 34:1500-1507. [DOI] [PubMed] [Google Scholar]
- 52.Miltner, E. C., and L. E. Bermudez. 2000. Mycobacterium avium grown in Acanthamoeba castellanii is protected from the effects of antimicrobials. Antimicrob. Agents Chemother. 44:1990-1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Norton, C. D., and M. W. LeChevallier. 2000. A pilot study of bacteriological population changes through potable water treatment and distribution. Appl. Environ. Microbiol. 66:268-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Olivier, K. N., D. J. Weber, J. H. Lee, A. Handler, G. Tudor, P. L. Molina, J. Tomashefski, and M. R. Knowles. 2003. Nontuberculous mycobacteria. II: nested-cohort study of impact on cystic fibrosis lung disease. Am. J. Respir. Crit. Care Med. 167:835-840. [DOI] [PubMed] [Google Scholar]
- 55.Parkash, U. 1993. Does the bronchoscope propagate infection? Chest 104:552-559. [DOI] [PubMed] [Google Scholar]
- 56.Primm T. P., C. A. Lucero, and J. O. Falkinham III. 2004. Health impacts of environmental mycobacteria. Clin. Microbiol. Rev. 17:98-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reischl, U., S. Emler, Z. Horak, J. Kaustova, R. M. Kroppenstedt, N. Lehn, and L. Naumann. 1998. Mycobacterium bohemicum sp. nov., a new slow-growing scotochromogenic mycobacterium. Int. J. Syst. Bacteriol. 48:1349-1355. [DOI] [PubMed] [Google Scholar]
- 58.Runyon, E. H. 1965. Typical mycobacteria: their classification. Am. Rev. Respir. Dis. 91:288-289. [DOI] [PubMed] [Google Scholar]
- 59.Schinsky, M. F., M. M. McNeil, A. M. Whitney, A. G. Steigerwalt, B. A. Lasker, M. M. Floyd, G. G. Hogg, D. J. Brenner, and J. M. Brown. 2000. Mycobacterium septicum sp. nov., a new rapidly growing species associated with catheter-related bacteraemia. Int. J. Syst. Evol. Microbiol. 50:575-581. [DOI] [PubMed] [Google Scholar]
- 60.Schroder, K. H. 1977. Atypical mycobacteria. Med. Klin. 72:1796-1802. [PubMed] [Google Scholar]
- 61.Schulze-Robbecke, R., and K. Buchholtz. 1992. Heat susceptibility of aquatic mycobacteria. Appl. Environ. Microbiol. 58:1869-1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Segal, G., and H. A. Shuman. 1999. Legionella pneumophila utilizes the same genes to multiply within Acanthamoeba castellanii and human macrophages. Infect. Immun. 67:2117-2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.September, S. M., V. S. Brozel, and S. N. Venter. 2004. Diversity of nontuberculoid Mycobacterium species in biofilms of urban and semiurban drinking water distribution systems. Appl. Environ. Microbiol. 70:7571-7573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Springer, B., E. C. Bottger, P. Kirschner, and R. J. Wallace, Jr. 1995. Phylogeny of the Mycobacterium chelonae-like organism based on partial sequencing of the 16S rRNA gene and proposal of Mycobacterium mucogenicum sp. nov. Int. J. Syst. Bacteriol. 45:262-267. [DOI] [PubMed] [Google Scholar]
- 65.Springer, B., W. K. Wu, T. Bodmer, G. Haase, G. E. Pfyffer, R. M. Kroppenstedt, K. H. Schroder, S. Emler, J. O. Kilburn, P. Kirschner, A. Telenti, M. B. Coyle, and E. C. Böttger. 1996. Isolation and characterization of a unique group of slowly growing mycobacteria: description of Mycobacterium lentiflavum sp. nov. J. Clin. Microbiol. 34:1100-1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Steinert, M., K. Birkness, E. White, B. Fields, and F. Quinn. 1998. Mycobacterium avium bacilli grow saprozoically in coculture with Acanthamoeba polyphaga and survive within cyst walls. Appl. Environ. Microbiol. 64:2256-2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Taylor, S. J., L. J. Ahonen, F. A. de Leij, and J. W. Dale. 2003. Infection of Acanthamoeba castellanii with Mycobacterium bovis and M. bovis BCG and survival of M. bovis within the amoebae. Appl. Environ. Microbiol. 69:4316-4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Thomas, V., K. Herrera-Rimann, D. S. Blanc, and G. Greub. 2006. Biodiversity of amoebae and amoeba-resisting bacteria in a hospital water network. Appl. Environ. Microbiol. 72:2428-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Thorel, M. F., M. Krichevsky, and V. V. Levy-Frebault. 1990. Numerical taxonomy of mycobactin-dependent mycobacteria, emended description of Mycobacterium avium, and description of Mycobacterium avium subsp. avium subsp. nov., Mycobacterium avium subsp. paratuberculosis subsp. nov., and Mycobacterium avium subsp. silvaticum subsp. nov. Int. J. Syst. Bacteriol. 40:254-260. [DOI] [PubMed] [Google Scholar]
- 70.Tiwari T. S., B. Ray, K. C. Jost, Jr., M. K. Rathod, Y. Zhang, B. A. Brown-Elliott, K. Hendricks, and R. J. Wallace, Jr. 2003. Forty years of disinfectant failure: outbreak of postinjection Mycobacterium abscessus infection caused by contamination of benzalkonium chloride. Clin. Infect. Dis. 36:954-962. [DOI] [PubMed] [Google Scholar]
- 71.Tortoli, E., R. M. Kroppenstedt, A. Bartoloni, G. Caroli, I. Jan, J. Pawlowski, and S. Emler. 1999. Mycobacterium tusciae sp. nov. Int. J. Syst. Bacteriol. 49:1839-1844. [DOI] [PubMed] [Google Scholar]
- 72.Torvinen, E., S. Suomalainen, M. J. Lehtola, I. T. Miettinen, O. Zacheus, L. Paulin, M. L. Katila, and P. J. Martikainen. 2004. Mycobacteria in water and loose deposits of drinking water distribution systems in Finland. Appl. Environ. Microbiol. 70:1973-1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Trupiano, J. K., B. A. Sebek, J. Goldfarb, L. R. Levy, G. S. Hall, and G. W. Procop. 2001. Mastitis due to Mycobacterium abscessus after body piercing. Clin. Infect. Dis. 33:131-134. [DOI] [PubMed] [Google Scholar]
- 74.Tsukamura, M., and S. Tsukamura. 1966. Mycobacterium aurum; a new species. Igaku To Seibutsugaku 72:270-273. [PubMed] [Google Scholar]
- 75.Tsukamura, M., H. Nemoto, and H. Yugi. 1983. Mycobacterium porcinum sp. nov., a porcine pathogen. Int. J. Syst. Bacteriol. 33:162-165. [Google Scholar]
- 76.Vaerewijck, M. J., G. Huys, J. C. Palomino, J. Swings, and F. Portaels. 2005. Mycobacteria in drinking water distribution systems: ecology and significance for human health. FEMS Microbiol. Rev. 29:911-934. [DOI] [PubMed] [Google Scholar]
- 77.Vugia, D. J., Y. Jang, C. Zizek, J. Ely, K. L. Winthrop, and E. Desmond. 2005. Mycobacteria in nail salon whirlpool footbaths, California. Emerg. Infect. Dis. 11:616-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wallace, R. J., Jr., B. A. Brown, and D. E. Griffith. 1998. Nosocomial outbreaks/pseudo-outbreaks caused by nontuberculous mycobacteria. Annu. Rev. Microbiol. 52:453-490. [DOI] [PubMed] [Google Scholar]
- 79.Wallace, R. J., Jr., Y. Zhang, R. W. Wilson, L. Mann, and H. Rossmoore. 2002. Presence of a single genotype of the newly described species Mycobacterium immunogenum in industrial metalworking fluids associated with hypersensitivity pneumonitis. Appl. Environ. Microbiol. 68:5580-5584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wayne, L. G. 1966. Mycobacterial classification. Am. Rev. Respir. Dis. 93:958-959. [DOI] [PubMed] [Google Scholar]
- 81.Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wilson, R. W., V. A. Steingrube, E. C. Bottger, B. Springer, B. A. Brown-Elliott, V. Vincent, K. C. Jost, Jr., Y. Zhang, M. J. Garcia, S. H. Chiu, G. O. Onyi, H. Rossmoore, D. R. Nash, and R. J. Wallace, Jr. 2001. Mycobacterium immunogenum sp. nov., a novel species related to Mycobacterium abscessus and associated with clinical disease, pseudo-outbreaks and contaminated metalworking fluids: an international cooperative study on mycobacterial taxonomy. Int. J. Syst. Evol. Microbiol. 51:1751-1764. [DOI] [PubMed] [Google Scholar]
- 83.Winthrop, K. L., M. Abrams, M. Yakrus, I. Schwartz, J. Ely, D. Gillies, and D. J. Vugia. 2002. An outbreak of mycobacterial furunculosis associated with footbaths at a nail salon. N. Engl. J. Med. 346:1366-1371. [DOI] [PubMed] [Google Scholar]
- 84.Zhang, Q., R. Kennon, M. A. Koza, K. Hulten, and J. E. Clarridge III. 2002. Pseudoepidemic due to a unique strain of Mycobacterium szulgai: genotypic, phenotypic, and epidemiological analysis. J. Clin. Microbiol. 40:1134-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]