Abstract
Fluorescent resonance energy transfer probes targeting the 16S rRNA gene were constructed for a sensitive and specific real-time PCR for identification and differentiation of Legionella pneumophila from other Legionella spp. For identification of non-L. pneumophila spp. by direct amplicon sequencing, two conventional PCR assays targeting the mip gene were established.
There are currently 50 species (http://www.dsmz.de/bactnom/bactname.htm) comprising about 70 distinct serogroups in the genus Legionella. Legionella pneumophila serogroup 1 accounts for the majority of infections of humans, but association with human disease has been reported for >20 of the species in the genus Legionella (4). It is, however, likely that most legionellae can cause human disease under appropriate conditions due to their capability for cellular invasion and intracellular growth (3, 4).
Diagnostic delay may result in increased mortality for patients with legionellosis (6). Culture is considered the “gold standard” for detection of legionellae, but due to the slow-growing and fastidious nature of legionellae, other strategies to ensure a rapid diagnosis of legionellosis have become imperative. Several PCR assays targeting Legionella sp. and L. pneumophila genes have been reported, including assays targeting the 16S rRNA gene (2, 5, 8, 9, 11, 12), the 5S rRNA gene (5), the 23S-5S spacer region (7), and the macrophage infectivity potentiator gene mip (1, 5, 10, 13). The aim of this study was to develop molecular tools enabling (i) rapid detection of Legionella spp. in clinical and environmental specimens, (ii) fast differentiation between L. pneumophila and other Legionella spp., and (iii) suitable amplicons for species identification by DNA sequencing.
The following Legionella reference strains were used: L. birminghamensis serogroup 1 (CCUG 31233 A), L. bozemanae serogroup 2 (CCUG 16416), L. cincinnatiensis serogroup 1 (CCUG 31230 A), L. dumoffii (CCUG 47789), L. hackeliae serogroup 1 (CCUG 31232 A), L. lansingensis serogroup 1 (CCUG 31227), L. longbeachae serogroup 1 (CCUG 28612), L. longbeachae serogroup 2 (CCUG 46623), L. micdadei serogroup 1 (CCUG 31229 A), three strains of L. pneumophila serogroup 1 (CCUG 33058, CCUG 13395, and CCUG 9568), L. pneumophila serogroup 2 (CCUG 13396), L. pneumophila serogroup 6 (CCUG 13400), L. pneumophila serogroup 14 (CCUG 44898), L. sainthelensi (CCUG 29672), L. tucsonensis (CCUG 31119), and L. wadsworthii (CCUG 16415). Legionellae were grown at 37°C on buffered charcoal-yeast extract (BCYE) agar for 48 to 72 h in humidified air. One colony was emulsified in 0.2 ml sterile water and heated for 15 min at 95°C. Bacterial DNA was purified using a DNeasy tissue kit (QIAGEN, Hilden, Germany).
A real-time PCR assay with hybridization probes targeting the bacterial multicopy 16S rRNA gene of Legionella spp. was established, using a primer pair described previously (8). By aligning published Legionella 16S rRNA gene sequences, the probes for fluorescent resonance energy transfer (FRET) technology were constructed to be 100% homologous to L. pneumophila and to have various numbers of mismatches to other Legionella spp. (Table 1). The PCR mixture consisted of 2 μl 10× LightCycler FastStart DNA Master Hybridization Probes mix (Roche Diagnostics, Basel, Switzerland), 3 mM MgCl2, 0.5 μM each primer, 0.2 μM each hybridization probe, 0.2 U of uracil-N-glycosylase (MedProbe, Oslo, Norway), and 2 μl of template DNA in a final volume of 20 μl. The PCR was monitored on a LightCycler device (Roche Diagnostics), starting with an initial denaturation step for 10 min at 95°C to activate the Taq DNA polymerase and proceeding with 50 cycles of amplification (5 s at 95°C, 15 s at 58°C, and 15 s at 72°C), followed by a melting curve analysis (40°C to 85°C with a heating rate of 0.1°C/s). A second PCR assay aiming at identification of Legionella species by DNA sequencing targeted the mip gene of Legionella spp. Based on alignment of mip sequences in the GenBank database, two primer sets were selected (Table 1). The PCR mixture consisted of 200 ng of each primer, 50 μM each deoxynucleoside triphosphate, 1× PCR buffer (Applied Biosystems, Foster City, CA), 1.5 mM MgCl2 (Applied Biosystems), 0.25 U of AmpliTaqGold DNA polymerase (Applied Biosystems), 2 μl template DNA, and sterile water to a final volume of 50 μl. Amplifying conditions were as follows: an initial denaturation step for 15 min at 94°C to activate the Taq polymerase; 35 cycles at 94°C for 1 min, 55°C for 1.5 min, and 72°C for 2 min; and finally a prolonged extension step for 7 min at 72°C. The amplicons were visualized by a Bioanalyzer 2100 (Agilent Technologies, Palo Alto, CA) or gel electrophoresis. The amplicons were purified with a QIAquick PCR purification kit (QIAGEN) and sequenced on a CEQ 8800 genetic analysis system (Beckman Coulter, Fullerton, CA), using a CEQ DTCS Quick Start kit (Beckman Coulter), 10 to 50 fmol purified PCR product, and 3.2 pmol primer. Primers used in the sequencing reaction were identical with the PCR primers. Sequence analysis was performed by using the Sequencher program (Gene Codes Corporation, Ann Arbor, MI). For all analyses, data obtained with the forward and reverse primers were combined and aligned manually. The consensus sequence was compared with sequences in the GenBank database for identification using NCBI BLAST (http://www.ncbi.nlm.nih.gov/BLAST/) or with sequences in the Legionella mip gene sequence database provided by the European Working Group for Legionella Infections (http://www.ewgli.org/).
TABLE 1.
Oligonucleotide primers and hybridization probes used in this study
| Oligonucleotide | Sequence (5′-3′)a | Gene | GenBank accession no. of reference sequence | Position | Reference |
|---|---|---|---|---|---|
| Leg primer 1 | AGGGTTGATAGGTTAAGAGC | 16S rRNA | M59157 | 451-470 | 8 |
| Leg primer 2 | CCAACAGCTAGTTGACATCG | 16S rRNA | M59157 | 836-817 | 8 |
| Leg probe 1 | GAGTCAACCAGTATTATCTGACCGTCCC-[FL] | 16S rRNA | M59157 | 653-626 | This study |
| Leg probe 2 | [Red 640]GGTTAAGCCCAGGAATTTCACAGATAACTTAATCA-[Ph] | 16S rRNA | M59157 | 624-590 | This study |
| mip FI | GGTCGCTGCAGCTGYCATRR | mip | S62141 | 700-719 | This study |
| mip RI | GCATTAATTGYARWGCTTCAGT | mip | S62141 | 1280-1259 | This study |
| mip FII | GGGGATTSTTTATGAAGATGA | mip | U91607 [S62141] | 467-487 [675-695]c | This study |
| mip RII | ACCAGCAGGCATTAATTGTAA | mip | U91607 [S62141] | 1050-1030 [1288-1268]b,c | This study |
[FL], fluorescein; [Red 640], LightCycler-Red 640-N-hydroxy-succinimide ester; [Ph], 3′-phosphate.
Four mismatches in this region compared to the mip RII primer.
For primer set FII/RII, the corresponding positions in S62141 are given in brackets.
The real-time PCR detected all 6 L. pneumophila strains and 11 of the 12 non-L. pneumophila reference strains. The sensitivity was estimated to be 1.4 fg per PCR of L. pneumophila DNA template, corresponding to less than 1 genome equivalent (data not shown). To investigate the specificity of the 16S real-time PCR, DNAs extracted from a number of commonly encountered microorganisms were analyzed. The species investigated included Staphylococcus epidermidis, Staphylococcus aureus, Enterococcus faecalis, Streptococcus agalactiae, Streptococcus pyogenes, Streptococcus pneumoniae, Neisseria meningitidis, Mycobacterium tuberculosis, Escherichia coli, Propionibacterium acnes, Bordetella pertussis, Chlamydia pneumoniae, Mycoplasma pneumoniae, Listeria monocytogenes, and Moraxella catarrhalis. Neither during amplification nor during the melting curve analysis did these specimens produce any FRET signal (data not shown).
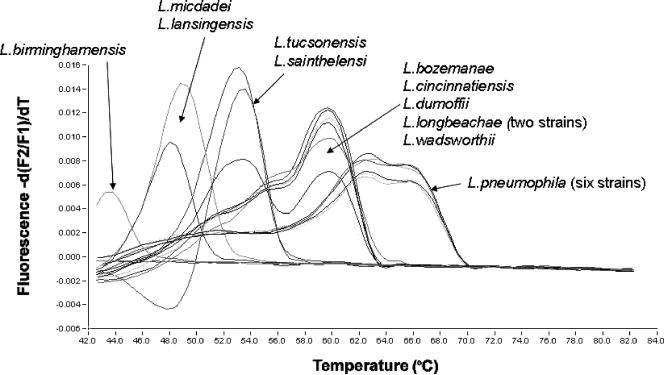
By melting curve analysis, L. pneumophila could easily be differentiated from the other Legionella species (Fig. 1). A hunchbacked melting curve was observed for all L. pneumophila strains and also for L. cincinnatiensis. The explanation for this phenomenon probably is related to the probe design, i.e., the melting temperatures (Tm) for the hybridization probes are approximately equal, and thus neither will work like an anchor probe. Nevertheless, it is highly feasible to distinguish L. pneumophila from the other legionellae by its melting curve. For the strains mentioned, the appearance of the melting curve will vary somewhat from batch to batch of the probe. For Tm determination, emphasis is placed on the peak with the higher Tm of the two. As expected, the deviation of the melting point observed was approximately proportional to the number of mismatches in the probe region compared to L. pneumophila (Table 2). The minimum and maximum observed differences in Tm between L. pneumophila and the non-L. pneumophila strains were 6°C and 22°C, corresponding to 4 and 11 polymorphic sites in the probe region, respectively. One of the reference strains that gave no FRET signal during either amplification or melting curve analysis (i.e., L. hackeliae) was amplified by the primers when analyzed by gel electrophoresis. Here the number of mismatches in the probe region was 15. By comparing the various 16S rRNA genes of non-L. pneumophila species in the GenBank database with the hybridization probe region, approximately 70% have fewer than 12 polymorphic sites in the probe region and are thus likely to be detected by the 16S real-time assay described here. In addition, none of these strains have fewer than 4 polymorphic sites in the probe region (i.e., the number of polymorphic sites is sufficient to differentiate them from L. pneumophila by melting curve analysis). The high melting temperatures of the hybridization probes allow multiple polymorphic sites in the probe region. The melting temperature for the probes when hybridized to L. pneumophila was shown to be 66°C. The L. pneumophila-specific hybridization probes presented by Reischl et al. (11) are shorter and thus have a lower melting temperature (61°C). Our strategy, by using somewhat longer hybridization probes, allows for multiple mismatches along the entire probe sequence and thus enables the simultaneous detection and differentiation of L. pneumophila and a number of non-L. pneumophila spp. Since the hybridization probe assay will not detect every Legionella species, it is possible on negative samples to do a post-PCR analysis of the 386-bp PCR product generated by the primers. For that purpose we are using a capillary electrophoresis system on a disposable chip (Bioanalyzer; Agilent Technologies) using 1 μl from the LightCycler capillary tube, allowing results within 30 min. For the past 3 years, this real-time PCR assay has been successfully implemented in our routine laboratory with several positive findings in cases of legionellosis.
FIG. 1.
Melting curve analysis with L. pneumophila-specific hybridization probes.
TABLE 2.
Polymorphism in the probe region among the various Legionella reference strains employed in this study in comparison with results of melting curve analysis
| Species | CCUG accession no. | Nucleotide sequence in the probe regiona | Melting temp (°C) | No. of nt mismatches in the probe region |
|---|---|---|---|---|
| L. pneumophila | 33058 | TGATTAAGTTATCTGTGAAATTCCTGGGCTTAACCTGGGACGGTCAGATAATACTGGTTGACTC | 66 | Perfect match |
| L. pneumophila | 44898 | **************************************************************** | 66 | Perfect match |
| L. pneumophila | 13400 | **************************************************************** | 66 | Perfect match |
| L. pneumophila | 9568 | **************************************************************** | 66 | Perfect match |
| L. pneumophila | 13396 | **************************************************************** | 66 | Perfect match |
| L. pneumophila | 13395 | **************************************************************** | 66 | Perfect match |
| L. bozemanae | 16416 | *********************C*****************CA********G************** | 60 | 4 |
| L. wadsworthii | 16415 | *********************C**C**************CA*********************** | 60 | 4 |
| L. cincinnatiensis | 31230A | *********************C*****************CA********G************** | 60 | 4 |
| L. dumoffii | 47789 | *********************C*****************CA********G************** | 60 | 4 |
| L. longbeachae | 46623 | *********************C*****************CA********G************** | 60 | 4 |
| L. longbeachae | 28612 | *********************C*****************CA********G************** | 60 | 4 |
| L. tucsonensis | 31119 | ***A*****************C*****************CA********G******T******* | 53 | 6 |
| L. sainthelensi | 29672 | *A*******************C*****************CA********G*********A**** | 53 | 6 |
| L. micdadei | 31229A | *TTA**************************C*****A**GT**********G****TGA****T | 49 | 12 |
| L. lansingensis | 31227 | *T*A*******G******************C********G*******C***G****T*A****T | 48 | 10 |
| L. birminghamensis | 31233A | *T*A*******A*********C*****************CA******T***G****T*A****A | 44 | 11 |
| L. hackeliae | 31232A | *T*A*******GT********C******************ATTG**CC***G****T*A****G | No data | 15 |
Asterisks indicate identity with L. pneumophila CCUG 33058.
To identify non-L. pneumophila species of legionellae, direct sequencing of the amplicon produced by the 16S rRNA gene primers can be performed (2). We found similarities in the 16S rRNA gene amplicon among the 11 non-L. pneumophila species ranging from 93.3 to 99.7% (data not shown). Based on comparison with sequences in the GenBank database, only L. dumoffi, L. birminghamensis, L. longbeachae, L. bozemanae, L. micdadei, and L. wadsworthii gave the expected result for the BLAST search (i.e., for 5 of the 12 non-L. pneumophila reference strains, the best hit in the GenBank database was different from the reference species).
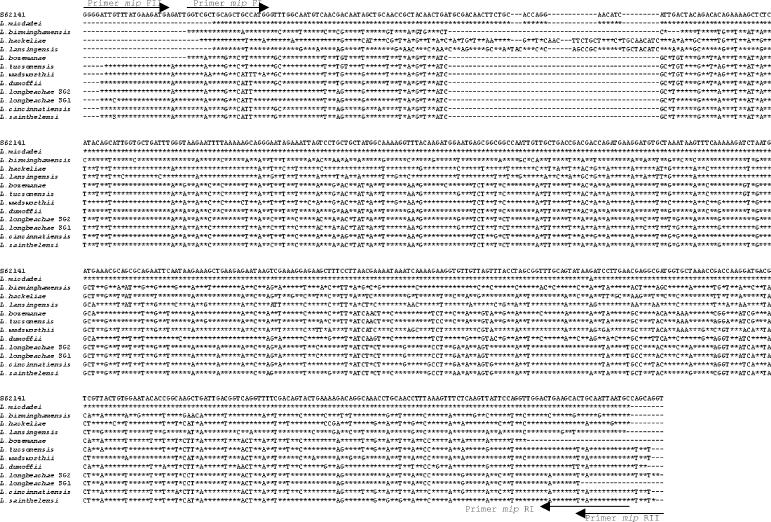
The mip gene has been reported to discriminate better among Legionella species, and identification through data comparison is available over the Internet (http://www.ewgli.org/). Ratcliff et al. (10) have previously described mip primers for this purpose, but these primers were to a very large extent degenerated. Based on alignment of mip sequences available in the GenBank database, we constructed two PCRs for detection and sequencing of non-L. pneumophila spp. Only 8 of 45 non-L. pneumophila mip sequences are not included in the assumed potential for identification by using these two primer sets. For these eight species, the mip sequences are not available for either or both of the primer binding sites, and thus the full potential for the modified primers cannot be assessed. Our main reason for constructing new primers is to avoid using too many degenerated base sites when the same primer sets are used for both PCR and sequencing. The conventional PCR assays targeting the mip gene detected all of the 12 non-L. pneumophila reference strains. By direct sequencing (Fig. 2), all of the non-L. pneumophila reference strains could be identified with high accuracy based on the GenBank data and the Legionella mip gene sequence database (http://www.ewgli.org/). These observations indicate that the mip gene sequence discriminates more reliably between Legionella species than does the 386-bp 16S rRNA gene sequence.
FIG. 2.
Alignment of the mip PCR products of the non-L. pneumophila reference strains. Asterisks indicate identity with the L. micdadei strain (GenBank accession no. S62141). A dash indicates a gap or no base analyzed.
In conclusion, we have established a sensitive and specific real-time PCR assay capable of identifying L. pneumophila and simultaneously differentiating L. pneumophila from other, non-L. pneumophila species by melting curve analysis. Additionally, we present a procedure for identification of non-L. pneumophila spp. based on mip sequencing.
REFERENCES
- 1.Bumbaugh, A., E. A. McGraw, K. L. Page, R. K. Selander, and T. S. Whittam. 2002. Sequence polymorphism of dotA and mip alleles mediating invasion and intracellular replication of Legionella pneumophila. Curr. Microbiol. 44:314-322. [DOI] [PubMed] [Google Scholar]
- 2.Cloud, J. L., K. C. Carroll, P. Pixton, M. Erali, and D. R. Hillyard. 2000. Detection of Legionella species in respiratory specimens using PCR with sequencing confirmation. J. Clin. Microbiol. 38:1709-1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fields, B. S. 1996. The molecular ecology of legionellae. Trends Microbiol. 4:286-290. [DOI] [PubMed] [Google Scholar]
- 4.Fields, B. S., R. F. Benson, and R. E. Besser. 2002. Legionella and Legionnaires' disease: 25 years of investigation. Clin. Microbiol. Rev. 15:506-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hayden, R. T., J. R. Uhl, X. Qian, M. K. Hopkins, M. C. Aubry, A. H. Limper, R. V. Lloyd, and F. R. Cockerill. 2001. Direct detection of Legionella species from bronchoalveolar lavage and open lung biopsy specimens: comparison of LightCycler PCR, in situ hybridization, direct fluorescence antigen detection, and culture. J. Clin. Microbiol. 39:2618-2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heath, C. H., D. I. Grove, and D. F. Looke. 1996. Delay in appropriate therapy of Legionella pneumonia associated with increased mortality. Eur. J. Clin. Microbiol. Infect. Dis. 15:286-290. [DOI] [PubMed] [Google Scholar]
- 7.Herpers, B., B. M. de Jongh, K. van der Zwaluw, and E. J. van Hannen. 2003. Real-time PCR assay targets the 23S-5S spacer for direct detection and differentiation of Legionella spp. and Legionella pneumophila. J. Clin. Microbiol. 41:4815-4816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jonas, D., A. Rosenbaum, S. Weyrich, and B. Sucharit. 1995. Enzyme-linked immunoassay for detection of PCR-amplified DNA of legionellae in bronchoalveolar fluid. J. Clin. Microbiol. 33:1247-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rantakokko-Jalava, K., and J. Jalava. 2001. Development of conventional and real-time PCR assay for detection of Legionella DNA in respiratory specimens. J. Clin. Microbiol. 39:2904-2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ratcliff, R. M., J. A. Lanser, P. A. Manning, and M. W. Heuzenroeder. 1998. Sequence-based classification scheme for the genus Legionella targeting the mip gene. J. Clin. Microbiol. 36:1560-1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reischl, U., H.-J. Linde, N. Lehn, O. Landt, K. Barrat, and N. Wellinghausen. 2002. Direct detection and differentiation of Legionella spp. and Legionella pneumophila in clinical specimens by dual-color real-time PCR and melting curve analysis. J. Clin. Microbiol. 40:3814-3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wellinghausen, N., C. Frost, and R. Marre. 2001. Detection of legionellae in hospital water samples by quantitative real-time LightCycler PCR. Appl. Environ. Microbiol. 67:3985-3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilson, D. A., B. Yen-Lieberman, U. Reischl, S. M. Gordon, and G. W. Procop. 2003. Detection of Legionella pneumophila by real-time PCR for the mip gene. J. Clin. Microbiol. 41:3327-3330. [DOI] [PMC free article] [PubMed] [Google Scholar]