Abstract
Genes involved in magnetite biomineralization are clustered within the genomic magnetosome island of Magnetospirillum gryphiswaldense. Their transcriptional organization and regulation were studied by several approaches. Cotranscription of genes within the mamAB, mamDC, and mms clusters was demonstrated by reverse transcription-PCR (RT-PCR) of intergenic regions, indicating the presence of long polycistronic transcripts extending over more than 16 kb. The transcription start points of the mamAB, mamDC, and mms operons were mapped at 22 bp, 52 bp, and 58 bp upstream of the first genes of the operons, respectively. Identified −10 and −35 boxes of the PmamAB, PmamDC, and Pmms promoters showed high similarity to the canonical σ70 recognition sequence. The transcription of magnetosome genes was further studied in response to iron and oxygen. Transcripts of magnetosome genes were detected by RT-PCR both in magnetic cells grown microaerobically under iron-sufficient conditions and in nonmagnetic cells grown either aerobically or with iron limitation. The presence of transcripts was found to be independent of the growth phase. Further results from partial RNA microarrays targeting the putative magnetosome transcriptome of M. gryphiswaldense and real-time RT-PCR experiments indicated differences in expression levels depending on growth conditions. The expression of the mam and mms genes was down-regulated in nonmagnetic cells under iron limitation and, to a lesser extent, during aerobic growth compared to that in magnetite-forming cells grown microaerobically under iron-sufficient conditions.
Magnetic orientation in magnetotactic bacteria is based on the synthesis of magnetosomes, which consist of crystals of magnetite (Fe3O4) enclosed within intracytoplasmic vesicles of the magnetosome membrane (MM) (1, 29). The MM consists of a lipid bilayer, which provides spatial and physicochemical control over magnetite biomineralization and has a distinct biochemical composition. In the microaerophilic alphaproteobacterium Magnetospirillum gryphiswaldense, the MM is associated with a characteristic subset of magnetosome membrane proteins (MMPs), which are present in different quantities, with relative abundances between <1% and >15% (11). Classes of MMPs include those with presumed functions in the activation of magnetosomes, magnetosome-directed transport of iron, nucleation and control of crystal growth, and the assembly of magnetosome chains (16, 26, 28, 29). The targeting of MMPs to the MM is controlled by an unknown mechanism, but it can be assumed that stoichiometric synthesis of individual constituents is regulated for proper assembly of magnetosomes (29).
In M. gryphiswaldense, the MMPs are encoded within a hypervariable 130-kb genomic magnetosome island (MAI) (28, 35). In addition to all known magnetosome genes, the MAI contains further genes putatively involved in magnetosome biomineralization and is particularly rich in insertion elements. The mam (magnetosome membrane) and mms (magnetic particle membrane-specific) genes encode nearly all of the identified MMPs, along with several proteins of unknown function. They are located within <35 kb of the MAI and are organized within three gene clusters that are conserved among different magnetotactic bacteria (MTB) (12, 28). The mamAB cluster encompasses 17 colinear open reading frames (ORFs) extending for 16.4 kb of DNA. The 2.1-kb mamGFDC cluster is located 10 kb upstream of the mamAB cluster and comprises four ORFs. The 3.6-kb mms cluster is located 368 bp upstream of the mamGFDC cluster and contains five ORFs. The colinear organization and close spacing of genes within the three clusters suggest that they each might be transcribed as a polycistronic operon from a single promoter.
Magnetosome formation in M. gryphiswaldense and other MTB does not occur constitutively but is tightly regulated by growth conditions (2, 31). Magnetite biomineralization is induced by microaerobiosis, and there is a clear correlation between extracellular oxygen tension and magnetite content in M. gryphiswaldense (30). In oxystat experiments, magnetite biomineralization was induced only below a threshold value of 2 kPa O2, and the highest magnetosome numbers were found at 25 Pa O2, whereas higher oxygen levels entirely repressed magnetosome formation (15). Although it has been speculated that oxygen dependence is mediated at the level of differential regulation of magnetosome genes, the molecular mechanisms governing the expression of the magnetic phenotype are unknown (15). Beside the prevalence of microaerobic conditions, magnetosome biomineralization depends on the availability of iron. Synthesis of >80 magnetite crystals per cell is accompanied by intracellular iron accumulation of up to 4% of the cell dry weight, and iron uptake and magnetite biomineralization are saturated above an extracellular iron concentration of 50 μM (15, 31). Because of the extraordinarily large requirement for iron for magnetosome formation, its uptake and precipitation have to be genetically regulated via an unknown mechanism. Despite recent advances in the identification and characterization of genes controlling magnetosome biomineralization, their expression has not yet been addressed. Specifically, the transcriptional organization of genes identified within the MAI and information about their regulation in MTB are not available, and no promoter structures within the MAI have been identified so far.
In this study, we analyzed the transcriptional organization of the magnetosome gene clusters in Magnetospirillum gryphiswaldense and identified putative promoter structures which govern the transcription of long polycistronic operons of mam and mms genes. In addition, we examined the transcription levels of magnetosome genes in response to various iron and oxygen concentrations by partial differential RNA microarrays and quantitative real-time RT-PCR (qPCR). Hybridization experiments were performed with a three-color assay involving the isogenic mutant MSR-1B lacking all three magnetosome clusters, which provided an additional control for the specificities of hybridization signals.
MATERIALS AND METHODS
Bacterial strains.
Magnetospirillum gryphiswaldense strain MSR-1 (DSM 6361) (27, 32) and the nonmagnetic mutant strain MSR-1B, which contains a deletion of the MAI of 40.4 kb comprising the entire mam and mms clusters (28), were used in this study.
Oxystat cultivation.
Growth experiments were performed in a modified dual-vessel laboratory fermenter system (Biostat A Twin; B. Braun Biotech International, Melsungen, Germany) equipped for the automatic control of pH, temperature, and dissolved oxygen concentration (oxystat) as described previously (15). Soybean peptone was omitted from large-scale medium in order to create conditions of iron-limited growth because of the significant intrinsic iron content of peptone. Iron was added to the flask standard medium as ferric citrate, as indicated. RNAs for quantitative expression studies were isolated from cells cultivated under three different conditions, which were kept constant throughout the growth experiment: (i) microaerobic/iron sufficient (referred to as “standard growth”; 150 μM iron, 25 Pa O2), (ii) microaerobic/iron limited (iron-limited growth; ferric citrate was omitted from the medium, trace iron was <1 μM, 25 Pa O2), and (iii) high oxygen/iron sufficient (aerobic growth; 10,000 Pa O2, 150 μM iron). The media (10 liters) were inoculated with an initial cell density of approximately 1 × 108/ml, using 1-liter precultures that were grown under the same conditions.
Analytical methods.
Cell growth and magnetism were measured turbidimetrically at 565 nm. The average magnetic orientation of cell suspensions (magnetism) was assayed by an optical method as described previously (15, 33). Iron measurements were made with an atomic absorption spectrometer (model 3110; Perkin-Elmer, Überlingen, Germany) as described elsewhere (12, 15).
Isolation of total RNA from Magnetospirillum gryphiswaldense.
Among various tested protocols for RNA isolation, only the phenol-chloroform extraction method described by Oelmüller et al. (20) yielded satisfactory results with respect to low degradation and high yields of RNA. Briefly, cells of a 200-ml oxystat culture were harvested, washed in 1 ml of phosphate-buffered saline (150 mM NaCl, 10 mM sodium phosphate [pH 7]), and resuspended in 500 μl of ice-cold AE buffer (20 mM sodium acetate [pH 5.5], 1 mM EDTA). The solution was incubated with 1 ml of hot phenol-chloroform-isoamyl alcohol (25:24:1) and 10 μl of 25% (wt/vol) sodium dodecyl sulfate for 10 min at 60°C, cooled on ice, and centrifuged at 4°C. The aqueous phase was mixed with 62.5 μl of 2 M sodium acetate (pH 5.5) and 1 ml of phenol-chloroform-isoamyl alcohol for 5 min. The aqueous phase was again extracted with 1 ml of phenol-chloroform-isoamyl alcohol. After ethanol precipitation, the pellet was dissolved in 100 μl of 1× DNase buffer (10 mM Tris-HCl [pH 7.5], 2.5 mM MgCl2, 0.1 mM CaCl2) and incubated with 10 U of RNase-free DNase I (MBI Fermentas, St. Leon Roth, Germany) for 1 h at 37°C. The quality of the nucleic acid preparations was tested with an Agilent 2100 bioanalyzer (Agilent, Palo Alto, Calif.), and their quantity was measured by spectrophotometric measurement using an ND-1000 spectrophotometer (NanoDrop Technologies, Delaware).
cDNA synthesis for qualitative RT-PCR and qPCR.
The SuperScript III first-strand synthesis system for reverse transcription-PCR (RT-PCR) (Invitrogen, Karlsruhe, Germany) was used to synthesize cDNA in a 20-μl reaction mix with random hexamers applied according to the manufacturer's instructions.
Five micrograms of total RNA, 50 ng random hexamers, and a 0.5 mM concentration of each deoxynucleoside triphosphate were mixed first, heated to 70°C for 10 min, and placed on ice until the addition of the cDNA synthesis mix (20 mM Tris-HCl [pH 8.4], 50 mM KCl, 5 mM MgCl2, 10 mM dithiothreitol, 40 U RNase Out, 200 U SuperScript III RT). The reaction was incubated for 10 min at 25°C, followed by 50 min at 50°C, and was terminated by heat inactivation at 85°C for 5 min. Afterwards, cDNA was treated with 2 U RNase H for 20 min at 37°C and precipitated with ethanol. PCR was conducted as described elsewhere (28).
Primer extension analysis.
For all primers and probes used in this study (see Table S1 in the supplemental material), numbering of nucleotide positions and genetic nomenclature were done according to the sequence deposited under GenBank accession no. BX571797 (35).
Up to 50 μg total RNA, 2 μM antisense 6-carboxyfluorescein-labeled primer (MWG Biotech, Ebersberg, Germany) targeting the region 100 to 300 bp upstream of the start codon, and a 1 mM concentration of each deoxynucleoside triphosphate were mixed first, heated to 70°C for 10 min, and placed on ice until the addition of RT buffer (50 mM Tris-HCl [pH 8.3], 50 mM KCl, 5 mM MgCl2, 10 mM dithiothreitol) and 0.5 μg actinomycin. The reaction mix was incubated for 10 min at 37°C, and 200 U Revert Aid H Minus M murine leukemia virus reverse transcriptase (MBI Fermentas, St. Leon Roth, Germany) was added, followed by 60 min of incubation at 42°C and termination by heat inactivation at 70°C for 5 min. Afterwards, the cDNA was digested with 2 U RNase H for 20 min at 37°C. The extended product was ethanol precipitated and dissolved in 5 μl Tris-EDTA buffer. The product was analyzed with an ABI Prism 3100 genetic analyzer (Applied Biosystems, Foster City, Calif.) with the Genescan 500 ROX standard. The fragment analysis program had the following parameters: oven temperature, 60°C; prerun time, 180 s; run voltage, 15 kV; and run time, 1,800 s. The data were analyzed with GeneMapper software v.3.7 from Applied Biosystems.
Western blot analysis.
Crude extracts of various cultures were separated by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to a nitrocellulose membrane, and analyzed with an anti-MamC primary antibody (a gift of K. Grünberg, Bremen, Germany). Detection was conducted with a secondary antibody that was conjugated to alkaline phosphatase and could be detected with BCIP (5-bromo-4-chloro-3-indolylphosphate) tablets (Roche Biochemicals).
Sequence analysis.
Predictions of hairpin structures for putative transcriptional terminators were analyzed with RNAfold (19, 38). Binding sites of transcription factors were analyzed with DBTBS, release 3.4 (18).
DNA microarray analysis.
Isolated total RNAs of MSR-1 (standard growth, magnetic), MSR-1 (iron-limited growth), MSR-1 (aerobic growth), and MSR-1B (standard growth) were directly chemically labeled with the fluorescent dye Alexa 546, Alexa 647, or Alexa 488, using platinum-linked reporter technology purchased from Molecular Probes (Eugene, Oreg.). Labeling of 10 μg of total RNA was done according to the manufacturer's protocol, except that 10 and 2 μl of the labeling reagent for dyes Alexa 546/647 and Alexa 488, respectively, were added to the labeling buffer. The labeling efficiency and amount of labeled RNA were checked by spectrophotometric measurement using an ND-1000 spectrophotometer (NanoDrop Technologies).
Oligonucleotide probes of 65 to 70 nucleotides, targeting the mRNAs of nearly all genes of the magnetosome gene clusters of M. gryphiswaldense, were designed semiautomatically using the software package ARB (17) and the sequence deposited under GenBank accession no. BX571797 (35). The characteristics of the 22 probes evaluated in this study are listed in Table S2 in the supplemental material. DNA oligonucleotide probes were purchased from Biomers (Ulm, Germany) and spotted in four replicates onto GAPS II slides (Corning, Schiphol-Rijk, Netherlands), using a SpotArray 24 spotting device from Perkin-Elmer with Telechem Stealth pins (Sunnyvale, Calif.). The concentration of the unmodified oligonucleotides in Micro Spotting Solution Plus spotting buffer (Telechem) was 10 μM. Postprocessing of the slides was done according to the manufacturer's protocol, except that after rehydration, slides were dried for 10 s at 200°C on a 555 digital hotplate (VWR, Darmstadt, Germany). For covalent immobilization, both UV cross-linking, using a GS gene linker (Bio-Rad, Munich, Germany) at 245 nm and 350 mJ, and incubation at 80°C for 3 h were used. Slides were stored at room temperature in the dark. Spotted slides were blocked in a prehybridization solution containing 250 mM NaCl, 5 mM Tris-HCl at pH 8.0, 50% formamide, 0.5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.05% bovine serum albumin, and 1% blocking reagent from Roche (Mannheim, Germany) for at least 45 min at 46°C and air dried. For the parallel hybridization of three different target samples, a protocol was used as recently described by Peplies et al. (22), except that the hybridization buffer contained 50% formamide and 2 μg of labeled RNA of each of the transcriptomes was applied. Hybridization was conducted in an HS-400 hybridization station (Tecan, Crailsheim, Germany) at 50°C for at least 12 h. Afterwards, slides were washed in ULTRArray low-stringency wash buffer (Ambion, Huntington, United Kingdom) and dried under nitrogen. Slides were imaged at a resolution of 5 μm, using a ScanArray microarray scanner (Perkin-Elmer) with varied laser power and sensitivity levels of the photomultiplier tube for each slide. The image analysis software QuantArray 3.0 (Perkin-Elmer) was used for automatic spot detection and signal quantification. The raw data were then processed in terms of filtering, combination of probe replicates, and normalization by using the MicroArray Data Analysis (MADA) software tool (www.megx.net/mada). Spot replicates with poor homogeneity were removed from the data set according to the 50% outlier test of MADA. Non-background-corrected spot signal intensities were normalized to the mean global background of the corresponding dye. The signal of MSR-1B was used to determine the relative light units caused by nonspecific binding for every probe and was subtracted from hybridization signals (controlled-signal approach). Probes identified as nonspecific based on insignificant signal differences between MSR-1 and MSR-1B were marked as having “nonspecific hybridization” in Table 2 and omitted from subsequent analysis. Genes were considered up-regulated if the difference between signals for MSR-1 and MSR-1 grown in the presence of O2 or without Fe was significant. Significance was defined as a difference of more than the onefold signal standard deviation (t test; 15% false-positive results) in fluorescence emission after normalization of both channels.
TABLE 2.
Summary of regulatory characteristics of mam and mms genes analyzed by microarray profiling
| Gene | Cluster | Regulation
|
qPCR confirmation | |
|---|---|---|---|---|
| Iron-limited growth | Aerobic growth | |||
| mgI457 | mms | Down | Down | + |
| mgI458 | mms | Down | No regulation | |
| mgI460 | mms | Down | Down | + |
| mamG | mamGFDC | Down | No regulation | + |
| mamF | mamGFDC | Nonspecific hybridizationa | Nonspecific hybridizationa | + |
| mamD | mamGFDC | Down | Downa | + |
| mamH | mamAB | Down | Downa | |
| mamI | mamAB | Nonspecific hybridizationa | Nonspecific hybridizationa | |
| mamE | mamAB | Nonspecific hybridizationa | Nonspecific hybridizationa | |
| mamJ | mamAB | Downa | No regulation | |
| mamK | mamAB | Down | Down | |
| mamL | mamAB | Nonspecific hybridizationa | Nonspecific hybridizationa | |
| mamM | mamAB | Downa | Downa | + |
| mamN | mamAB | Nonspecific hybridizationa | Nonspecific hybridizationa | + |
| mamO | mamAB | Downa | No regulation | |
| mamP | mamAB | Nonspecific hybridizationa | Nonspecific hybridizationa | |
| mamA | mamAB | Downa | Downa | |
| mamQ | mamAB | Nonspecific hybridizationa | Nonspecific hybridizationa | |
| mamR | mamAB | Nonspecific hybridizationa | Nonspecific hybridizationa | |
| mamB | mamAB | Downa | No regulation | + |
| mamS | mamAB | Nonspecific hybridizationa | Nonspecific hybridizationa | |
| mamT | mamAB | Nonspecific hybridizationa | Nonspecific hybridizationa | |
Regulation was detected exclusively by use of the controlled-signal approach, which takes into account the signals generated by nonspecific binding of RNAs hybridized with the deletion mutant MSR-1B. “Nonspecific hybridization” indicates an insignificant signal difference between MSR-1 and MSR-1B.
qPCR.
The primers listed in Table S1 in the supplemental material were designed using MacVector 7.0 software and purchased from MWG Biotech (Ebersberg, Germany) and Carl Roth GmbH (Karlsruhe, Germany). PCRs were carried out in 96-well microtiter plate wells in a 25-μl reaction volume with Platinum SYBR green qPCR SuperMix UDG (Invitrogen, Karlsruhe, Germany) and 0.2 μmol of each primer (Table 1). An ABI Prism 7700 sequence detector (Perkin-Elmer-Applied Biosystems) was programmed for an initial step of 2 min at 50°C and 10 min at 95°C, followed by 40 thermal cycles of 15 s at 95°C and 1 min at 60°C. For each reaction, 10 ng of the individual RT reaction mix served as a template. Primer pairs complementary to an assortment of mam genes were used to amplify gene-specific products (see Table S1 in the supplemental material). The specificities of accumulated products were verified by melting-curve analysis. PCR efficiencies were determined as described previously by Pfaffl (see equation 3 in reference 23) and Ramakers et al. (24). Relative expression levels were determined by standard curves. For each tested RNA preparation, at least three independent real-time RT-PCR experiments were conducted. To determine relative transcript amounts, a standard curve for each gene was generated first. After adjusting the quantities to the size of the amplified PCR product, we revealed relative transcript amounts, as indicated.
TABLE 1.
Results of growth experiments with strains MSR-1 (wild type) and MSR-1B (nonmagnetic deletion mutant) under various conditions
| Strain (culture) | Extracellular iron concn (μM) | Oxygen partial pressure (Pa) | Final optical density at 565 nm | Intracellular iron content (μg/mg) | Magnetism (Cmag value) |
|---|---|---|---|---|---|
| MSR-1 (standard) | 150 | 25 | 0.99 | 13 | 1.54 |
| MSR-1B (standard) | 150 | 25 | 0.6 | 5 | 0 |
| MSR-1 (iron limited) | <1 | 25 | 0.46 | 4 | 0.28 |
| MSR-1 (aerobic) | 150 | 10,000 | 0.64 | 4 | 0 |
RESULTS
Transcriptional organization of the mamAB, mamDC, and mms gene clusters.
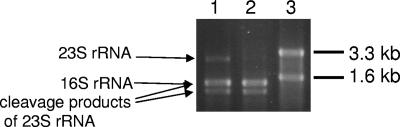
Among the various methods tested for isolation of RNA, only phenol-chloroform extraction yielded high-molecular-weight RNAs of sufficient quality from M. gryphiswaldense, as indicated by sharp bands of nondegraded rRNA species. We noticed that the intact 23S rRNA species running at 2.9 kb was not permanently detectable in our preparations, independent of the isolation procedure. Instead, additional smaller species migrating at about 1.3 and 1.6 kb in agarose gels were present, indicating that the 23S rRNA precursor was cleaved during rRNA maturation, similar to the case reported for numerous species of alpha- and other proteobacteria (8, 20). Interestingly, rRNA fragmentation was dependent on the growth phase: while the 23S rRNA precursor was detectable in growing cultures, this species was entirely degraded into cleavage products in late-exponential- and stationary-phase cells. Thus, RNA fragmentation may provide a means for monitoring the growth phase of an M. gryphiswaldense culture (Fig. 1).
FIG. 1.
Agarose gel showing different species of rRNA in preparations from M. gryphiswaldense (lane 1, exponential-phase culture; lane 2, stationary-phase culture) and E. coli (lane 3). A 23S rRNA precursor was cleaved into two species, of 1.6 kb and 1.3 kb (arrows). In stationary-phase cells, the 23S rRNA precursor was no longer detectable.
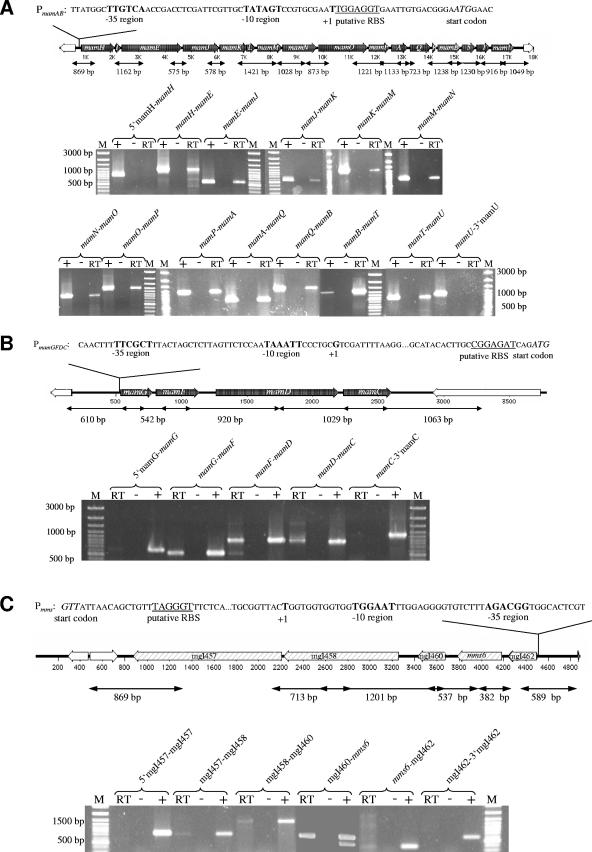
The operon-like, colinear organization of the mamAB, mamGFDC, and mms clusters suggested that they each might be cotranscribed within a single long message (Fig. 2). Sequence analysis did not reveal any characteristic rho-independent transcription terminator structures within the three clusters (data not shown). Initial attempts to directly identify transcripts from a variety of mam genes by Northern hybridization persistently resulted in a conspicuous smear, regardless of the RNA preparation methods and probes used. This might be due to common problems associated with the isolation of intact long transcripts in sufficient amounts (14). Therefore, we used RT-PCR to establish whether adjacent genes are cotranscribed, using primers amplifying the intergenic regions of the three clusters. cDNAs were synthesized from RNAs of magnetic cells, using specific primers situated at the 3′ end of each cluster (primer CW10_3R for the mamAB cluster, primer mamC_SSr for the mamDC cluster, and primer ORF000_SSf2 for the mms cluster). Transcripts were detected for all tested intergenic junctions but not for regions located 5′ and 3′ of the first and last genes of each of the clusters. Amplicons obtained from the cDNAs had the same sizes as the amplicons obtained from genomic DNA. Negative control experiments, which revealed the complete absence of DNA in the RNA samples, were performed by omitting the reverse transcriptase enzyme during RT reactions (Fig. 2A to C). These results suggested that at least one long, polycistronic transcript exists for any of the three clusters.
FIG. 2.
Transcriptional analysis by RT-PCR of the mamAB (A), mamGFDC (B), and mms (C) operons. Primers used in RT-PCR experiments are indicated by vertical marks. The expected sizes of PCR products are indicated below the arrows. Agarose gel electrophoresis of PCR products is shown at the bottom of each panel. Lanes: RT, RT-PCR; −, negative control with reverse transcriptase enzyme omitted; +, positive control with genomic DNA as the template; M, DNA size marker. Sequences of identified promoters (PmamAB, PmamGFDC, and Pmms) are indicated on top of each operon. The putative −10 and −35 regions are shown in bold, and the putative ribosome-binding site (RBS) is underlined. Start codons are shown in italics.
The mamAB, mamGFDC, and mms operons are each cotranscribed from single promoters.
The transcription start sites of the mamAB, mamDC, and mms operons were determined by primer extension analysis using RNAs isolated from magnetic cells grown under standard (microaerobic, iron-sufficient) conditions. The results of promoter mapping are shown in Fig. 2. As suspected from the RT-PCR results, promoters were identified upstream of the first genes of all three operons. Promoters Pmms, PmamDC, and PmamAB are located 58 nucleotides, 51 nucleotides, and 22 bp upstream of the translation start sites of mgI462, mamG, and mamH, respectively. The putative −10 and −35 regions display different degrees of similarity with the conserved consensus sequences of the vegetative sigma70 promoter (5′ TTGACA…TATAAT 3′) of Escherichia coli (Fig. 2).
In addition to the region located upstream of the initial mamH gene, regions preceding other genes of the mamAB operon were screened for the presence of promoters approximately every second gene, including mamE, mamJ, mamK, mamM, mamO, mamA, mamB, mamT, and mamU. However, we failed to identify internal transcriptional starting points in repeated attempts.
Magnetosome genes are transcribed under various growth conditions.
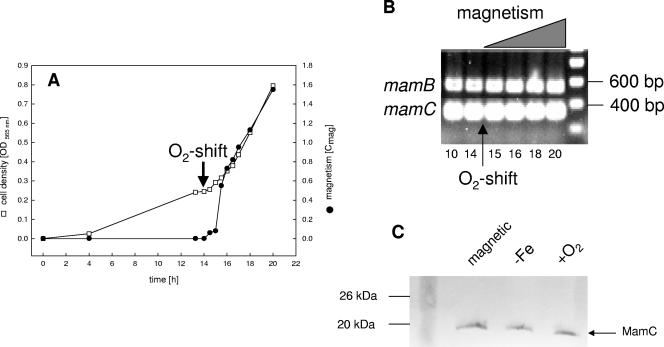
Transcription studies were carried out with cells grown at various defined oxygen concentrations in the fermenter. In an induction experiment, the culture was initially incubated at 3 kPa oxygen in iron-sufficient medium (aerobic growth). Under these conditions, cells grew at comparable rates to those for microaerobiosis, but they did not produce magnetite. After 14 h of exponential growth, the pO2 was shifted from 3 kPa to 300 Pa. Magnetite production became detectable 1.5 h after the shift, as revealed by a quantitative light-scattering assay (Cmag, 0.55), and the magnetism of the culture gradually increased until stationary phase (Cmag, 1.55). RT-PCR with RNAs from cells sampled at different time points revealed no difference in the presence of transcripts for mamB and mamC (Fig. 3B) as well as for mamA, -D, -E, -F, -G, -H, and -M (not shown), indicating that mam genes were expressed throughout growth. Further experiments, performed on cells grown at various constant oxygen concentrations (25, 50, 200, and 600 Pa O2) as well as at different iron concentrations (<1 μM to 150 μM), revealed the presence of transcripts for all magnetosome genes tested, independent of the formation of magnetite (data not shown). To verify at the protein level whether MMPs are in fact translated from the detected transcripts under conditions repressing magnetite formation, cell extracts from different growth conditions were analyzed with an antibody against MamC, which represents the most abundant MMP. Western blot analysis revealed that the MamC protein was present under all tested conditions in magnetic as well as nonmagnetic cells (Fig. 3C).
FIG. 3.
(A) Growth and magnetism of M. gryphiswaldense during oxystat cultivation. After 14 h, the pO2 was shifted from 3,000 Pa to 300 Pa (arrow). (B) RT-PCR with gene-specific primers and RNAs collected at different time points during the oxygen shift experiment. As an example, results for mamB and mamC are shown. Experiments with all other tested mam genes yielded identical results. (C) Western blot analysis of MamC expression in magnetic and nonmagnetic cells. Cultures were grown under standard, iron-limited (−Fe), and aerobic (+O2) conditions.
Expression profiling of magnetosome genes by RNA microarrays.
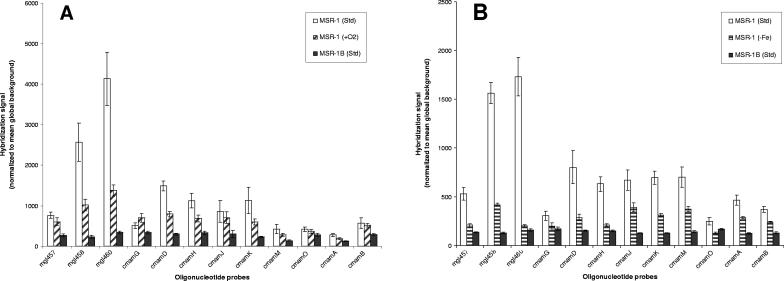
Since the experiments described above suggested either constitutive expression or a more subtle regulation of magnetosome genes, data from microarray experiments conducted with cells grown under different conditions were analyzed. RNA microarrays were designed to study the responses in gene expression to iron and oxygen, targeting most mam and mms genes of M. gryphiswaldense. Comparative microarray analyses of two different MSR-1 cultures in each case were performed with directly labeled RNAs extracted from cells grown in an oxystat-controlled fermenter under the conditions shown in Table 1. RNAs from the nonmagnetic mutant MSR-1B, labeled with a third fluorescent dye, were added to all hybridizations to serve as an internal negative control, since all targeted mam and mms genes are absent from this strain (28, 35). Interestingly, RNAs isolated from MSR-1B displayed weak signals for all probes applied and in each experiment performed, even with increased hybridization stringency (data not shown). However, based on the differences in signal intensities and taking into account the background hybridization signals of the MSR-1B mutant, the following genes can be considered down-regulated compared to those in magnetic cells grown under standard conditions: for iron-limiting conditions, mgI457, mgI458, mgI460, mamG, mamD, mamH, mamJ, mamK, mamM, mamA, mamO, and mamB; and for cells grown under aerobic conditions, mgI458, mgI460, mamD, mamH, mamK, mamM, and mamA (Table 2; Fig. 4). In summary, 12 and 7 of the 22 selected probes resulted in significantly stronger hybridization signals for the magnetic, wild-type MSR-1 strain than those in cells grown under iron-limited and oxygen-supplemented conditions, respectively. Based on the controlled-signal approach, the investigated genes can be separated into a set of 6 genes with no indication of regulation and 10 genes where the capture probes failed, as indicated by nonspecific hybridization in the MSR-1B control strain (Table 2).
FIG. 4.
Expression levels of selected genes, as determined by microarray experiments, showing the probes which yielded specific hybridization results. Error bars represent standard deviations for at least five spotted replicates. (A) Aerobic growth conditions (MSR-1 plus O2); (B) iron-limited (MSR-1 without Fe) growth conditions.
Expression profiling of selected magnetosome genes by qPCR.
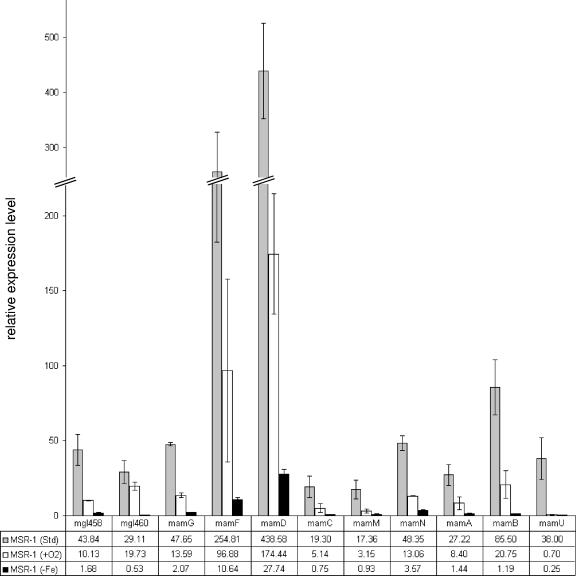
In order to verify the data obtained by microarray analysis, relative transcript amounts for 11 selected mam and mms genes (mgI458, mgI460, mamG, mamF, mamD, mamC, mamM, mamN, mamA, mamB, and mamU) were determined by qPCR (Table 1; Fig. 5). The selection was based on results of microarray analysis and on expression patterns predicted by operonal organization. As expected, no transcripts of any mam or mms genes were observed in the MSR-1B deletion strain. Cells grown under aerobic conditions showed down-regulation of the investigated mam and mms genes compared to the case under magnetite-forming conditions. For example, mamU and mamM were down-regulated about 55-fold and 6-fold, respectively, and displayed larger regulatory differences than other mam/mms genes that showed 2.5- to 4.3-fold down-regulation. In cells grown under iron-limited conditions, the expression of all investigated mam and mms genes was down-regulated. The main differences in regulation were found for mamU (137-fold) and mamB (55-fold). The regulation differences for the other genes of the mamAB and mamDC clusters were between 19- and 23-fold, with the exception of mamN (12-fold regulation).
FIG. 5.
Expression levels of selected genes under standard, iron-limited, and aerobic conditions, as determined with qPCR. Error bars represent data from three independent qRT-PCR experiments. The data given in the table are mean values.
DISCUSSION
We have shown that the previously identified mam and mms gene clusters are cotranscribed as polycistronic operons from single promoters. For the mamAB cluster, we found evidence for the existence of a long transcript extending >16 kb and comprising 17 genes. Although there are well-documented examples of large transcriptional units spanning operons as large as 35 kb (4, 6, 25), transcription in large operons is frequently governed from multiple promoters, such as that in the 10-kb gal-lac operon in Streptococcus salivarius (36), and multiple promoters have even been found in smaller operons, such as the ilv operon of Corynebacterium glutamicum, spanning only 4 kb (21). Although no internal promoters could be identified by primer extension analysis, we cannot entirely rule out that there are additional promoters located within the mamAB cluster that escaped detection by this method. Various transcript levels of individual genes from the same transcriptional unit, such as those observed for the mamGFDC operon, on the other hand, might be explained by differential internal stabilities within a polycistronic mRNA (3, 13).
The transcription start sites PmamAB, PmamDC, and Pmms are the first putative promoter sequences identified in M. gryphiswaldense and, to our knowledge, in a magnetotactic bacterium.
Sequence comparison revealed that the −10 regions show similarity to the E. coli σ70 −10 consensus sequence, whereas the −35 regions are more divergent. Moreover, the three identified promoter sequences show only weak conservation between each other and with promoters in other alphaproteobacteria, such as Rhizobium and Rhodobacter, controlled by σ70. Promoter regions in the closely related strains Magnetospirillum magnetotacticum MS-1 and Magnetococcus sp. strain MC-1 have not been identified experimentally so far. However, the genetic organization of magnetosome genes in these organisms seems conserved as far as revealed by available genome sequence data (12; http://genome.jgi=psf.org/mic_home.html). Interestingly, inspection of the sequence upstream of the mamH gene of M. magnetotacticum revealed a nearly identical promoter sequence, PmamAB, with highly conserved −10 and −35 regions but slight differences in the region between the two boxes (not shown), whereas Pmms and PmamDC seem to be poorly conserved in other MTB. Although magnetosome formation is tightly controlled by the extracellular iron and oxygen concentrations (15), transcripts of analyzed magnetosome genes were present under all tested conditions, and translation of the most abundant magnetosome protein, MamC, was also confirmed in nonmagnetic cells grown under these conditions. However, the transcription levels of several genes varied in response to iron and oxygen, as indicated by microarray and qPCR data. While the patterns of regulation detected by the two different methods were identical for most genes, regulation of mamB and mamG expression was only detectable by qRT-PCR. This difference might be explained by the targeting of different intragenic regions by PCR primers and capture probes, as pointed out by Etienne et al. (7). Regulated magnetosome genes showed maximum expression under magnetite-forming conditions, which resemble those encountered by MTB in their natural environment within the sediment (absence or low concentrations of oxygen and availability of micromolar amounts of iron) (9). While magnetite formation resumes with no delay when iron-starved cells are shifted to iron-sufficient conditions (15), we observed a lag of at least 1.5 h in magnetite formation in our growth experiments after oxygen induction, suggesting that protein synthesis is likely required after shifting from aerobiosis towards microaerobiosis. The tight repression of magnetite biomineralization at high oxygen levels is not consistent with the relaxed regulation pattern observed for magnetosome genes. Therefore, we have to postulate that the expression of the magnetic phenotype is not mainly controlled by differential transcription of the analyzed magnetosome genes but that other regulatory circuits are likely to exist to mediate repression or induction of magnetite biomineralization. Common mechanisms of regulation in response to oxygen are the ArcA (aerobic respiration control) and Fnr (fumarate and nitrate reduction) regulons (34), and a common regulator in response to iron is the Fur (ferric uptake regulator) regulon (10, 37). Although homologs of these regulatory proteins are present in the genome of M. gryphiswaldense, it is uncertain if they are involved in the regulation of the magnetic phenotype. The observed differences in expression determined by qPCR were within comparable ranges for most tested genes. However, some genes, such as the mamU gene, exhibited a larger amplitude of regulation, and differences in transcription levels of several genes encoding MMPs were not fully consistent with the observed abundances of gene products in a previous analysis of the magnetosome subproteome (11). A possible explanation could be different stabilities of mRNAs, other mechanisms of posttranscriptional regulation, or different labeling efficiencies with the fluorescent dye.
The three-color labeling method used in this study for microarray analysis in combination with the isogenic deletion mutant MSR-1B permitted increased sensitivity and specificity, which allowed us to identify additional differentially expressed genes. For example, the regulation of four genes by oxygen and six genes by iron would have escaped identification by the conventional “twofold-cutoff” method (Table 2) (5). Since the regulation was confirmed by qPCR experiments, the additional data provided by the deletion mutant MSR-1B gave valuable indications about the level of nonspecific target binding on microarrays. Currently, application of the “controlled-signal approach” is limited to the availability of appropriate isogenic mutants. However, it seems possible to apply these results as a starting point for a more general attempt to estimate nonspecific hybridization in microarrays in the future.
In conclusion, the results presented in this study reveal that gene clusters encoding magnetosome proteins are transcribed as operons. In addition, first insights into the promoter structures of magnetotactic bacteria have been obtained. Furthermore, the first results of microarray experiments will set the stage for a global analysis in order to explore the upcoming genome sequence of M. gryphiswaldense.
Supplementary Material
Acknowledgments
We thank Sylke Wohlrab, Ekaterina Schmidt, and Cornelia Stumpf for excellent technical assistance.
This study was supported by the BMBF BioFuture Program and the Max Planck Society.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Bazylinski, D. A., and R. B. Frankel. 2004. Magnetosome formation in prokaryotes. Nat. Rev. Microbiol. 2:217-230. [DOI] [PubMed] [Google Scholar]
- 2.Blakemore, R. P., K. A. Short, D. A. Bazylinski, C. Rosenblatt, and R. B. Frankel. 1985. Microaerobic conditions are required for magnetite formation within Aquaspirillum magnetotacticum. Geomicrobiol. J. 4:53-71. [Google Scholar]
- 3.Carrier, T. A., and J. D. Keasling. 1997. Controlling messenger RNA stability in bacteria: strategies for engineering gene expression. Biotechnol. Prog. 13:699-708. [DOI] [PubMed] [Google Scholar]
- 4.Dal, S., G. Trautwein, and U. Gerischer. 2005. Transcriptional organization of genes for protocatechuate and quinate degradation from Acinetobacter sp. strain ADP1. Appl. Environ. Microbiol. 71:1025-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeRisi, J. L., V. R. Iyer, and P. O. Brown. 1997. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278:680-686. [DOI] [PubMed] [Google Scholar]
- 6.Enguita, F. J., J. J. Coque, P. Liras, and J. F. Martin. 1998. The nine genes of the Nocardia lactamdurans cephamycin cluster are transcribed into large mRNAs from three promoters, two of them located in a bidirectional promoter region. J. Bacteriol. 180:5489-5494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Etienne, W., M. H. Meyer, J. Peppers, and R. A. Meyer, Jr. 2004. Comparison of mRNA gene expression by RT-PCR and DNA microarray. BioTechniques 36:618-626. [DOI] [PubMed] [Google Scholar]
- 8.Evguenieva-Hackenberg, E., and G. Klug. 2000. RNase III processing of intervening sequences found in helix 9 of 23S rRNA in the alpha subclass of Proteobacteria. J. Bacteriol. 182:4719-4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flies, C. B., H. M. Jonkers, D. de Beer, K. Bosselmann, M. E. Böttcher, and D. Schüler. 2005. Diversity and vertical distribution of magnetotactic bacteria along chemical gradients in freshwater microcosms. FEMS Microbiol. Ecol. 52:185-195. [DOI] [PubMed] [Google Scholar]
- 10.Grifantini, R., S. Sebastian, E. Frigimelica, M. Draghi, E. Bartolini, A. Muzzi, R. Rappuoli, G. Grandi, and C. A. Genco. 2003. Identification of iron-activated and -repressed Fur-dependent genes by transcriptome analysis of Neisseria meningitidis group B. Proc. Natl. Acad. Sci. USA 100:9542-9547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grünberg, K., E. C. Müller, A. Otto, R. Reszka, D. Linder, M. Kube, R. Reinhardt, and D. Schüler. 2004. Biochemical and proteomic analysis of the magnetosome membrane in Magnetospirillum gryphiswaldense. Appl. Environ. Microbiol. 70:1040-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grünberg, K., C. Wawer, B. M. Tebo, and D. Schüler. 2001. A large gene cluster encoding several magnetosome proteins is conserved in different species of magnetotactic bacteria. Appl. Environ. Microbiol. 67:4573-4582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grunberg-Manago, M. 1999. Messenger RNA stability and its role in control of gene expression in bacteria and phages. Annu. Rev. Genet. 33:193-227. [DOI] [PubMed] [Google Scholar]
- 14.Gupta, A. 1999. RT-PCR: characterization of long multi-gene operons and multiple transcript gene clusters in bacteria. BioTechniques 27:966-972. [DOI] [PubMed] [Google Scholar]
- 15.Heyen, U., and D. Schüler. 2003. Growth and magnetosome formation by microaerophilic Magnetospirillum strains in an oxygen-controlled fermentor. Appl. Microbiol. Biotechnol. 61:536-544. [DOI] [PubMed] [Google Scholar]
- 16.Komeili, A., H. Vali, T. J. Beveridge, and D. K. Newman. 2004. Magnetosome vesicles are present before magnetite formation, and MamA is required for their activation. Proc. Natl. Acad. Sci. USA 101:3839-3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Makita, Y., M. Nakao, N. Ogasawara, and K. Nakai. 2004. DBTBS: database of transcriptional regulation in Bacillus subtilis and its contribution to comparative genomics. Nucleic Acids Res. 32:D75-D77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mathews, D. H., J. Sabina, M. Zuker, and D. H. Turner. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288:911-940. [DOI] [PubMed] [Google Scholar]
- 20.Oelmüller, U., N. Krüger, A. Steinbüchel, and C. G. Friedrich. 1990. Isolation of prokaryotic RNA and detection of specific mRNA with biotinylated probes. J. Microbiol. Methods 11:73-84. [Google Scholar]
- 21.Pátek, M., J. Nesvera, A. Guyonvarch, O. Reyes, and G. Leblon. 2003. Promoters of Corynebacterium glutamicum. J. Biotechnol. 104:311-323. [DOI] [PubMed] [Google Scholar]
- 22.Peplies, J., F. O. Glöckner, and R. Amann. 2003. Optimization strategies for DNA microarray-based detection of bacteria with 16S rRNA-targeting oligonucleotide probes. Appl. Environ. Microbiol. 69:1397-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramakers, C., J. M. Ruijter, R. H. Deprez, and A. F. Moorman. 2003. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 339:62-66. [DOI] [PubMed] [Google Scholar]
- 25.Reeves, A. R., R. S. English, J. S. Lampel, D. A. Post, and T. J. Vanden Boom. 1999. Transcriptional organization of the erythromycin biosynthetic gene cluster of Saccharopolyspora erythraea. J. Bacteriol. 181:7098-7106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scheffel, A., M. Gruska, D. Faivre, A. Linaroudis, P. L. Graumann, J. M. Plitzko, and D. Schüler. 2006. An acidic protein aligns magnetosomes along a filamentous structure. Nature 440:110-114. [DOI] [PubMed] [Google Scholar]
- 27.Schleifer, K., D. Schüler, S. Spring, M. Weizenegger, R. Amann, W. Ludwig, and M. Köhler. 1991. The genus Magnetospirillum gen. nov., description of Magnetospirillum gryphiswaldense sp. nov. and transfer of Aquaspirillum magnetotacticum to Magnetospirillum magnetotacticum comb. nov. Syst. Appl. Microbiol. 14:379-385. [Google Scholar]
- 28.Schübbe, S., M. Kube, A. Scheffel, C. Wawer, U. Heyen, A. Meyerdierks, M. H. Madkour, F. Mayer, R. Reinhardt, and D. Schüler. 2003. Characterization of a spontaneous nonmagnetic mutant of Magnetospirillum gryphiswaldense reveals a large deletion comprising a putative magnetosome island. J. Bacteriol. 185:5779-5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schüler, D. 2004. Molecular analysis of a subcellular compartment: the magnetosome membrane in Magnetospirillum gryphiswaldense. Arch. Microbiol. 181:1-7. [DOI] [PubMed] [Google Scholar]
- 30.Schüler, D., and E. Baeuerlein. 1998. Dynamics of iron uptake and Fe3O4 biomineralization during aerobic and microaerobic growth of Magnetospirillum gryphiswaldense. J. Bacteriol. 180:159-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schüler, D., and E. Baeuerlein. 1996. Iron-limited growth and kinetics of iron uptake in Magnetospirillum gryphiswaldense. Arch. Microbiol. 166:301-307. [DOI] [PubMed] [Google Scholar]
- 32.Schüler, D., and M. Köhler. 1992. The isolation of a new magnetic spirillum. Zentbl. Mikrobiol. 147:150-151. [Google Scholar]
- 33.Schüler, D., R. Uhl, and E. Baeuerlein. 1995. A simple light-scattering method to assay magnetism in Magnetospirillum gryphiswaldense. FEMS Microbiol. Lett. 132:139-145. [Google Scholar]
- 34.Shalel-Levanon, S., K. Y. San, and G. N. Bennett. 2005. Effect of ArcA and FNR on the expression of genes related to the oxygen regulation and the glycolysis pathway in Escherichia coli under microaerobic growth conditions. Biotechnol. Bioeng. 92:147-159. [DOI] [PubMed] [Google Scholar]
- 35.Ullrich, S., M. Kube, S. Schübbe, R. Reinhardt, and D. Schüler. 2005. A hypervariable 130-kilobase genomic region of Magnetospirillum gryphiswaldense comprises a magnetosome island, which undergoes frequent rearrangements during stationary growth. J. Bacteriol. 185:7176-7184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vaillancourt, K., S. Moineau, M. Frenette, C. Lessard, and C. Vadeboncoeur. 2002. Galactose and lactose genes from the galactose-positive bacterium Streptococcus salivarius and the phylogenetically related galactose-negative bacterium Streptococcus thermophilus: organization, sequence, transcription, and activity of the gal gene products. J. Bacteriol. 184:785-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wan, X. F., N. C. Verberkmoes, L. A. McCue, D. Stanek, H. Connelly, L. J. Hauser, L. Wu, X. Liu, T. Yan, A. Leaphart, R. L. Hettich, J. Zhou, and D. K. Thompson. 2004. Transcriptomic and proteomic characterization of the Fur modulon in the metal-reducing bacterium Shewanella oneidensis. J. Bacteriol. 186:8385-8400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.