Abstract
Biofouling communities contribute significantly to aquatic ecosystem productivity and biogeochemical cycling. Our knowledge of the distribution, composition, and activities of these microbially dominated communities is limited compared to other components of estuarine ecosystems. This study investigated the temporal stability and change of the dominant phylogenetic groups of the domain Bacteria in estuarine biofilm communities. Glass slides were deployed monthly over 1 year for 7-day incubations during peak tidal periods in East Sabine Bay, Fla. Community profiling was achieved by using 16S rRNA genes and terminal restriction fragment length polymorphism (T-RFLP) of 16S rRNA genes in combination with ribotyping, cloning, and sequencing to evaluate diversity and to identify dominant microorganisms. Bacterial community profiles from biofilms grown near the benthos showed distinct periods of constancy within winter and summer sampling periods. Similar periods of stability were also seen in T-RFLP patterns from floating biofilms. Alternating dominance of phylogenetic groups between seasons appeared to be associated with seasonal changes in temperature, nutrient availability, and light. The community structure appeared to be stable during these periods despite changes in salinity and in dissolved oxygen.
Biofilms develop on all surfaces in aquatic environments and are defined as matrix-enclosed microbial populations adherent to each other and/or surfaces (1, 32). A substantial part of the microbial activity in nature is associated with surfaces (12). Surface association (biofouling) is an efficient means for bacteria to proliferate in both favorable and sometimes hostile environments. By adopting a sessile mode of life, microbes can achieve several advantages over their planktonic counterparts (38), including the ability to capture and concentrate nutrients from the water column in the existing exopolysaccharide matrix, cometabolic interactions with neighboring microorganisms (17), and resistance to harmful chemicals (2) and environmental stress.
Biofilm-associated microbes, because of their ubiquity, diverse metabolic capabilities, and high enzymatic activity, play a crucial role in biogeochemical cycling. Direct observations show that biofilm-associated organisms (photo- and heterotrophic) account for a major part of ecosystem processes, both numerically and metabolically (12). Biofilm communities in nature play a key role in the production and degradation of organic matter, the degradation of environmental pollutants, and the cycling of limiting nutrients.
Biofilm formation and persistence in estuarine environments is governed by a suite of complex physical, chemical, and biological processes (4, 44, 45). Many of these parameters can vary significantly over different time scales. For example, nutrient availability can vary over diel light cycles, daily tidal cycles, and with rainfall events and seasonal change. Patterns in particulate and dissolved nutrient input to estuarine systems may influence shifts in biofilm bacterial community structure as select guilds respond to their required conditions.
The growth of biofilms on new surfaces is affected by the range of ambient conditions during development. Initial stages of biofilm accumulation are dominated by physical adsorption processes and growth dynamics that are controlled by planktonic conditions (13). Biomass and polymer accumulation from growth of attached organisms increasingly buffers the community from external influences, as internal nutrient recycling and energy production increasingly influence the nascent community (8).
The specific aim of this investigation was to evaluate stability and change in the bacterial community structures of nascent bacterial biofouling communities over a seasonal cycle to assess the potential for use of biofilm information as an environmental indicator. Community stability, species richness, and dominant functional groups were tracked in response to temporal variations in abiotic environmental factors.
MATERIALS AND METHODS
Periphytometers.
Polyvinyl chloride (PVC) periphytometers (24) were modified for this study to hold artificial substrata for biofilm growth at the surface and near-benthos. Periphytometers (60 cm in length) with styrofoam floats held vertically positioned Plexiglas plates (250 cm2) ∼3 cm apart and parallel to each other. Plexiglas slide racks were attached to the PVC frames to hold vertically positioned microscope slides ∼3 cm apart. Benthic samplers (42) held vertical Plexiglas plates (∼382 cm2) ∼3 cm apart. Slide racks were also affixed to the benthic samplers. Benthic periphytometers were anchored by encasing PVC pipe frames in cement (Quikrete) to hold substrates approximately 15 cm above the benthos.
Sampling scheme.
Biofilms were grown on the western edge of a sea grass bed located in East Sabine Bay, Gulf Breeze, Fla. (30°20′19"N, 87°09′21"W). Benthic and floating periphytometers were deployed for 7-day incubations every month over a period of 12 or 14 months, respectively (benthic, November 2002 to December 2003; floating, January 2003 to December 2003). Spring tide periods (http://tbone.biol.sc.edu/tide/tideshow.cgi?site=Pensacola%2C+Florida) were chosen within each month. For each sampling event, three benthic samplers were deployed at a depth of ∼1.5 m within a 5-m radius and were accompanied by two floating periphytometers. Floating periphytometers were anchored at one end to allow racks to float parallel to surface currents in a 2-m radius. To avoid shading benthic of samplers, floating samplers were offset by 2 m. Benthic periphytometers were oriented east to west to minimize shading effects among parallel slides and plates. Biofilms were allowed to accumulate on artificial substrata for 7-day periods to maximize microbial growth while minimizing colonization by calcareous invertebrates (data not shown).
Physical water quality parameters during incubations (salinity, temperature, pH, and dissolved oxygen [DO]) were measured with a Hydrolab DataSonde (version 4a) programmed to record at 15-min intervals. Photosynthetically active radiation (PAR) was monitored at the benthos and surface by light probes (LICOR Co.). Light readings were averaged for 30-min intervals. Benthic values were expressed as percent transmission of surface values.
Dissolved and total nutrient concentrations (nitrate-nitrite, orthophosphate, and total Kjeldahl nitrogen [TKN]) were determined from samples taken intermittently throughout incubations. For dissolved nutrient analysis, particulate matter was removed from ambient seawater by filtration with Whatman 0.7-μm GFF filters. Filtrate was collected in 20-ml polypropylene scintillation vials and stored at −80°C until processing. Raw seawater (100 ml) for total nutrient analysis was preserved with 0.4 ml of concentrated sulfuric acid (H2SO4). Nutrient concentrations were determined at the Wetlands Research Laboratory facility (UWF, Pensacola, Fla. [certification no. E71176]) with a BRAN+LUEBB Auto-Analyzer in accordance with EPA standard operating procedures (300.00, 300.01, 300.03).
Immediately upon retrieval, biofilms were carefully scraped from randomly selected plates and slides and rinsed with 0.2-μm-filtered (Durapore) ambient seawater into sterile glass filtration columns. Biofilm material for analysis of community structure was filtered onto sterile 0.2-μm Durapore membrane filters. Biofilm material used for biomass estimation was filtered onto 0.7-μm glass fiber (Whatman GFF) filters. Filters were individually stored in 1.5-ml Eppendorf tubes at −80°C until further processing.
Estimation of biofilm biomass.
Biofouled Plexiglas plates were randomly selected and allowed to air dry intact overnight on racks for the determination of optical density. Dried biofilms from floating and benthic samplers were scanned at 300 dpi on a flat-bed scanner (Epson 636) in transparency mode with Adobe Photoshop software. Images were analyzed with NIH Image software (http://www.nih.gov/) to provide estimates of average pixel density and standard deviations of pixel density on a gray scale of 0 to 256.
Material harvested from benthic plates was also quantified by ash-free dry weight (AFDW) analysis according to EPA method 10300D. Filters were place in aluminum dishes and dried to a constant weight at 105°C (Baxter drying oven DX-61). Filters were cooled overnight in a desiccator and placed in prewashed, prefired, and tared crucibles. Samples were weighed (Sartorius R160p) and placed in an electric muffle furnace (Fisher Scientific) at 500°C for 1 h. Following furnace cool-down, filters were rewetted with reagent-grade H2O and dried to a constant weight at 105°C. Final weights were determined following cooling of crucibles in a desiccator for 12 h. Total organic content was estimated from differences in pre- and postignition filter weights.
Genomic DNA extraction.
Biofilm material from six random samples from each sampling period was removed from filters with 2% NaCl and combined in sterile 50-ml Falcon tubes. Material was transferred to sterile 1.5-ml microcentrifuge tubes and pelleted by centrifugation at 8,000 rpm for 7 min at 4°C. The supernatant was carefully removed, and the pellet was resuspended in 50 μl of lysis buffer (0.05 M NaCl, 50 mM Tris-HCl [pH 7.5], 50 mM EDTA, 0.3 M sucrose, 1.5% hexadecyltrimethylammonium bromide).
Material was sonicated with two 15-s bursts (Ultrasonics Microtip 415 attached to a model W225R sonicator, power level 3; Heat Systems Inc., Farmingdale, N.Y.). Tubes were chilled on ice for 2 to 3 min between bursts to avoid excessive warming of the samples. Samples were subjected to enzymatic lysis by adding 32 μl lysozyme (100 mg/ml), 3.5 μl pronase E (100 mg/ml), 3 μl mutanolysin (2,500 U/ml), 13 μl sodium dodecyl sulfate (SDS) (20%, wt/vol), and 2 μl RNase (100 mg/ml), followed by incubation for 1 h in a 37°C water bath. Next, 17.5 μl proteinase K (100 mg/ml) was added to the mixture and incubated for another 30 min at 37°C, followed by incubation at 55°C for 30 min. SDS and NaCl concentrations were then increased through addition of 70 μl SDS and 170 μl NaCl (5 M), followed by addition of 95 μl N-laurylsarcosine (10%, wt/vol). This mixture was incubated in a hot block for 30 min at 65°C with mixing by inverting tubes gently every 10 min. Following incubation, extraction mixtures were subjected to three freeze-thaw cycles (5 min each) with liquid nitrogen (−170°C) and a hot block (65°C). The resultant aqueous lysate was centrifuged (10 min at 13,000 rpm), and the supernatant was transferred to new microcentrifuge tubes.
Lysates of organic material were purified by adding equal volumes of phenol (water saturated, pH 8.0; phenol-Tris, 10 mM). This was followed by two additional extractions with phenol-chloroform-isoamyl alcohol (25:24:1). Remaining traces of phenol were removed by extraction with equal volumes of chloroform-isoamyl alcohol (24:1). Upon each addition of the organic solvents, mixtures were vortexed for 30 s, followed by centrifugation at 13,000 rpm for 5 min. DNA was precipitated by addition of 0.7 volumes of 100% isopropanol, followed by centrifugation at 13,000 rpm for 30 min. Residual salt was removed by rinsing DNA pellets with 1 ml of 70% ethanol followed by centrifugation for 5 min and removal of the ethanol. DNA pellets were air-dried under a sterile hood (Airclean 600) and resuspended in 50 μl of Tris-EDTA buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA) at 55°C for 1 h.
PCR amplification.
Amplification of 16S rRNA genes was accomplished by using the universal bacterial primers 27F (5′-CACCAGAGTTTGATCA/CTGGCTCAG-3′, corresponding to positions 8 to 27 of E. coli) and 1492R (5′-GGC/TTACCTTGTTACGACTT-3′, corresponding to positions 1510 to 1492 of E. coli). For terminal restriction fragment length polymorphism (T-RFLP) analysis, the forward primer was labeled at the 5′ end with 6-carboxyfluorescein (Sigma-Genosys). The 50-μl reaction mixture contained 15 to 30 ng template DNA (15 to 30 ng/μl), 5 μl of 10× reaction buffer (containing 2 mM MgCl), 2.5 mM deoxynucleoside triphosphates, 10 pmol of each primer, and 0.25 U of Extaq polymerase (Takara Mirus Bio, Madison, Wis.). The amplification conditions were as follows: initial denaturation (1.5 min at 94°C); 30 cycles of denaturing (30 sec at 94°C), annealing (1 min at 50°C), and extension (1.5 min at 72°C); and a final extension at 72°C for 7 min. No final extension period was used for amplification of DNA for directional cloning. Aliquots (5 μl) of PCR products were visualized after electrophoresis on a 1% agarose gel stained with ethidium bromide (0.5 μg/ml).
T-RFLP analysis.
PCR products used for T-RFLP analysis were purified by 0.8% agarose gel electrophoresis. Bands were excised with a sterile scalpel, and the DNA was extracted with a QIAprep gel extraction kit (QIAGEN, Valencia, Calif.) according to the manufacturer's instructions. PCR product (∼200 to 300 ng) was digested with the endonucleases CfoI, MspI, and RsaI (Roche Diagnostics) in separate reactions. Each digestion was performed at 37°C for 4 h in a thermal cycler (GeneAmp; Applied Biosystems, Foster City, Calif.) in a total volume of 40 μl containing 4 μl of 10× buffer L (provided with the enzyme) and 1.25 U of the corresponding enzyme. DNA was precipitated at −20°C for at least 1 h after the addition of 0.1 volumes of 3 M sodium acetate (pH 5.2) and 2.5 volumes of ethanol (95% p.a. grade). DNA was pelleted at 13,000 rpm at 4°C for 1 h. The DNA pellet was desalted with 70% ethanol, air-dried, and resuspended in 10 μl double-distilled H2O. DNA (e.g., 3 μl) was combined with 0.25 μl of DNA standard (Mapmarker 1000; Bioventures Inc., Murfreesboro, Tenn.) and denatured after addition of 2 volumes of formamide at 95°C for 5 min. Samples were directly chilled on ice to prevent renaturation. Fragments were separated on an automated DNA sequencer (ABI Prism 310 genetic analyzer; Applied Biosystems) in GeneScan mode (15 kV, 5-s injection, 60°C for 40 min) using a 36-cm capillary and Performance Optimized Polymer 4 (Applied Biosystems). Fluorescence intensities of major peaks of community profiles were adjusted by increasing or decreasing template volumes and/or injection times so that they fell in a range of between 6,500 and 7,500 fluorescence units to ensure comparability of profiles. The lengths of the 5′-terminal restriction fragments (T-RFs) and corresponding fluorescence intensities (peak areas) were calculated with DAx software (Van Mierlo Software). A threshold excluding values below 150 fluorescence units was used to separate real signals from background noise.
Amplified rRNA gene restriction analysis (ARDRA).
Amplification of DNA from January, April, and August 2003 samples was performed in triplicate and pooled. PCR products were purified on 0.8% agarose gels followed by phenol extraction, as previously described. Purified DNA products were directionally cloned into plasmids with the pENTR Directional TOPO cloning kit (Invitrogen, Carlsbad, Calif.) according to the manufacturer's instructions. Plasmids were transformed into ONE SHOT competent E. coli cells (Invitrogen) according to the manufacturer's directions. Cells containing inserts were screened by selective growth on LB medium agar plates containing kanamycin (50 μg/ml). Bacteria were grown overnight (∼16 h) at 37°C. Cell colonies were transferred onto duplicate plates with sterile toothpicks and grown for an additional 10 to 12 h at 37°C.
Cell mass from each plate colony (roughly 0.5 to 1.0 μl) was resuspended in a tube containing 40 μl of Tris-EDTA buffer. Lysates were prepared by using a hot block (100°C) and centrifugation for 2 min at 13,000 rpm. Supernatant (2 μl) was reamplified with primers M13F (5′-GTAAAACGACGGCCAG-3′) and M13R (5′-CACGAAACAGCTATGAC-3′), which flanked the insertion sites. PCR conditions were as follows: initial denaturation for 1.5 min at 94°C; 30 cycles of denaturation (30 s at 94°C), annealing (30 s at 55°C), and extension (1.5 min at 72°C); and a final extension step of 7 min at 72°C. PCR product (8 μl) was digested with CfoI, as previously described for T-RFLP analysis. Restriction fragments were separated on 3% (wt/vol) agarose gels in 1× TAE (20 mM acetic acid, 40 mM Tris, 1 mM EDTA). Bands were visualized under UV illumination, digitized, and analyzed with GelComparII software. PCR products with similar banding patterns were digested further with MspI and RsaI endonucleases in order to define operational taxonomic units (OTUs). Approximately 150 to 180 clones were evaluated for each month.
Phylogenetic analysis of clones and 16S rRNA gene sequencing.
Plasmid DNA was prepared from E. coli carrying plasmids with 16S rRNA gene inserts of common ribotypes (>1) with the QIAprep Spin Miniprep kit (QIAGEN, Valencia, Calif.) according to the manufacturer's instructions. Plasmids were sent to the Genomics Technology Support Facility (Michigan State University, Lansing, Mich.) for high-throughput sequencing with M13F primers. Unaligned sequences were entered into the basic local alignment search tool (BLAST) program of the National Center for Biotechnology information (NCBI) for phylogenetic categorization by the maximum-likelihood algorithm. 16S rRNA gene sequences were subjected to a ClustalW multiple alignment analysis with the MacVector 7.1.1 program (Accelrys). Sequence data were analyzed by the neighbor-joining method in the PHYLIP software package (version 3.5) (15). Bootstrapping analysis was performed with results from 1,000 replications.
Data analysis.
The number of unique T-RFs in each restriction was used as a surrogate measure of the richness of each community. Peak area as a percentage of total peak area was used as an estimate of relative abundance. Relative T-RF peak areas were binned according to fragment length (±1). Richness and abundance data were analyzed by ordination of T-RFLP patterns though correlation-based principal component (PC) analysis. PC values were presented graphically in three-dimensional space with Deltagraph 4.5 (Red Rock Software, Inc.). Axes were adjusted to account for the variance associated with each PC value.
The diversity of OTUs was estimated for each community comparison (January, April, and August sample months) through the use of various taxonomic distinctness measures modified to accept sequence data: a taxonomic diversity (Δ) index empirically related to the Shannon-Weaver index, a measure of taxonomic distinctness (Δ*) (46, 47), average taxonomic distinctness (Δ+) (10), total taxonomic distinctness (sΔ+), and the variation in taxonomic distinctness (Λ+) (11). These indexes, originally designed to integrate taxonomic differences between species within sample groups, were modified to incorporate genetic differences in bacterial clones. Therefore, we replaced the term “taxonomic” with “genetic” in order to avoid confusion.
Phylogenetic distances were determined for clone sequences (∼650 to 675 bp), based on relative similarities (0.1-bp substitutions per 100 nucleotides) of aligned sequences. Clone similarity distances obtained from distance matrix files were rounded off to the nearest whole number and entered in “sample files” by using Primer 5 software (Primer-E Ltd., Plymouth, United Kingdom), along with the corresponding number of common ribotypes (OTU abundance). In place of species in which hierarchical levels are assigned weights, similarity distances from 1 to 100 were entered. Weights were incrementally assigned to each distance by the Primer 5 program, with higher values placed on longer phylogenetic tree distances.
RESULTS
Physical and chemical characteristics of the biofilm and water column.
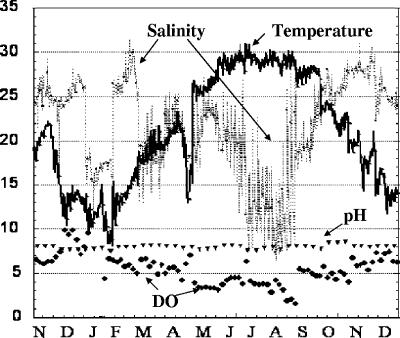
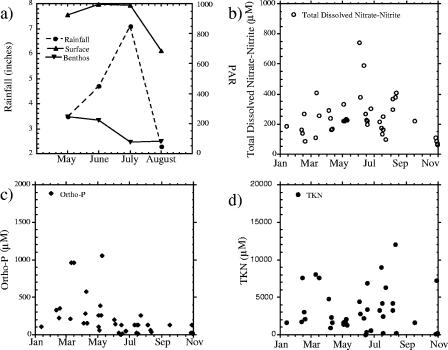
Seasonal change was documented by water quality and ambient light availability measurements. Dissolved oxygen followed a decreasing trend with increasing temperature (Fig. 1). Mild hypoxia (≤3 mg/liter) occurred in July, August, and September 2003. Average pH was stable throughout the year. The greatest variation was in temperature and salinity. The water temperature was at its lowest in January (8°C) and increased to a maximum of 31°C in July. Salinity ranged from 6.28 to 32.1 ppt throughout the 14-month sampling period (November 2002 to December 2003) without a clear seasonal trend. High fluctuations in salinity occurred during summer incubation periods (July, 7.54 to 22.83 ppt; August, 6.28 to 28.43 ppt). Average salinity estimates were lowest (∼12.48 ± 2.17 and 12.10 ± 4.35) in the summer months (Fig. 1) following rain events (Fig. 2a). These changes in salinity were coincident with water column production as the nitrogen content of particulate matter increased and decreased (Fig. 2d). Dissolved nitrate-nitrite concentrations were typically less than about 71.4 μM throughout the year and displayed no obvious seasonal trends (Fig. 2b). Dissolved orthophosphate was higher in the winter and decreased during the warmer months, presumably as a result of demand (Fig. 2c). TKN concentrations gradually increased throughout spring and summer, reaching maxima in August and reflecting increases in planktonic biomass (Fig. 2d).
FIG. 1.
Seasonal variations (November 2002 to December 2003) in ambient water parameters (salinity, temperature, DO, and pH) at the sample site.
FIG. 2.
(a) Average rainfall and PAR (at the surface and at depth) during summer monthly incubation periods (May to August 2003). Subplots for January to December 2003 show temporal distributions of total dissolved nitrate-nitrite (b), dissolved orthophosphate (c), and total particulate TKN (d).
Biomass estimates.
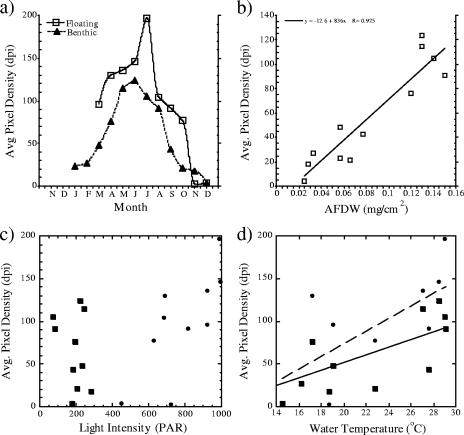
Biofilm biomass from benthic plates increased fivefold in the summer months relative to winter, with AFDW ranging from 0.033 mg/cm2 in January samples to 0.15 mg/cm2 in June. Optical densities of the benthic plates also increased from a mean of 23 dpi in January to 124 dpi in June (Fig. 3a). Optical densities of floating samples were on average 31% higher (12 to 72%) than those of benthic samples for the year, excluding the months of November and December 2003, when surface biomass estimates were lower than those of benthic samples. Biomass estimates by AFDW and optical density were highly correlated (Fig. 3b). Seasonal changes in light availability did not appear to be a limiting factor in the benthic incubations (Fig. 3c), and biofilm growth had a strong positive correlation with temperature (Fig. 3d).
FIG. 3.
Estimated biomass for floating and benthic biofilms harvested from periphytometers after 7-day incubation periods. (a) Representation of the seasonal trend in average pixel density. (b) Correlation of pixel density estimates and estimated AFDW of benthic samples. (c) Correlation analyses of biomass versus light: optical density of biofilms (squares, benthic; circles, floating) and ambient PAR. (d) Optical density estimates and average ambient water temperatures.
T-RFLP analysis.
Analysis of replicate benthic plate samples from August and November 2003 showed variation in the absence or presence of two to four T-RFs with low peak heights (<200 fluorescence units; data not shown); otherwise, the reproducibility between plates was reasonably high. The three restriction enzymes (CfoI, MspI, and RsaI) used in this study provided 5 to 71 T-RFs for analysis. The range of T-RFs from the floating samples was greater than that determined from benthic material.
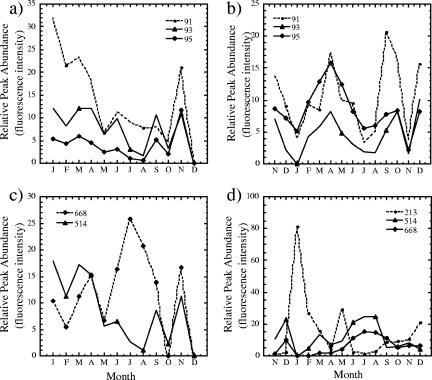
To assess similarity among bacterial communities, the 5′ T-RFLP patterns were used as “community fingerprints,” incorporating the presence or absence of T-RFs and their relative areas as analogs of species richness and evenness, respectively. The T-RFLP profiles of both benthic and floating samples showed dramatic seasonal differences. For example, the CfoI-derived T-RF at 213 bp was a major peak in January to March and in May benthic samples but occurred as a minor, albeit consistent, peak in June to September benthic samples and was again a major peak in October to December (Fig. 4c). Conversely T-RFs B and C (514 and 668 bp) peaked in magnitude during the summer months and were either absent or present as low peak areas in the remaining samples. Additionally, the peaks at 93, 95, and 98 bp were present throughout all sample months (excluding January), displayed some seasonal change, and fluctuated in contribution to the overall community structure (Fig. 4a).
FIG. 4.
Temporal representation of relative dominant peak intensities (93, 95, 98, 514, and 668 bp) from T-RFLP profiles of CfoI-digested PCR-amplified DNA from monthly benthic (a and c) and floating (b and d) samples.
Benthic and floating communities were relatively similar but displayed some striking differences. Phylogenetic groups yielding fragments at 93, 95, 98, 514, and 668 bp were consistently present yet variable in peak areas between floating and benthic samples over time (Fig. 4). There were observable differences between the floating and benthic sets of profiles. Certain T-RFs observed in the benthic CfoI-restricted profiles were not present in the T-RFLP fingerprints of the floating communities, such as the prominent 213-bp peak in the CfoI-restricted benthic samples. Several additional minor T-RFs appeared periodically and were restricted to benthic or floating habitats.
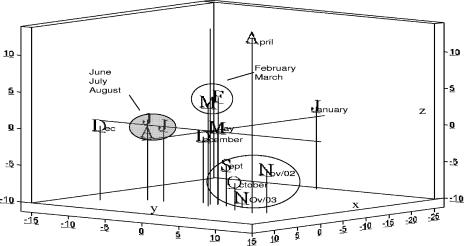
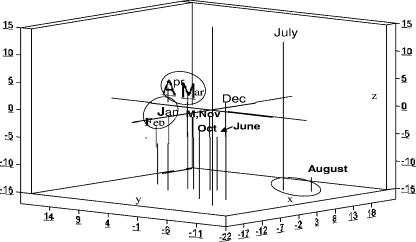
Substantial temporal variation in community structure was confirmed through graphic illustrations of PC ordinate scores of each monthly sample. Samples taken in winter (January to March) and summer (June to August) for both benthic (Fig. 5) and floating (Fig. 6) bacterial communities showed pronounced within-season consistencies in community patterns. Biofilms collected from floating samples showed less seasonal change than did the benthic biofilms. The similarity between benthic and floating communities was greatest during the summer incubation periods (data not shown).
FIG. 5.
Plot of the first three coordinate scores from PC analyses of T-RFLPs generated from benthic samples. The relative similarity of samples is characterized by proximity to each other. Graph axes (x, y, and z) were adjusted according to the percent variance accounted for by each PC.
FIG. 6.
Plot of the first three coordinate scores from PC analyses of T-RFLPs generated from floating samples. The relative similarity of samples is characterized by proximity to each other. Graph axes (x, y, and z) were adjusted according to the percent variance accounted for by each PC.
Analysis of 16S rRNA gene sequences and identification of T-RFs.
Higher-resolution analysis of the benthic community profiles was achieved by constructing 16S rRNA gene clone libraries from the January, April, and August benthic samples, representing the winter, transition, and summer periods as suggested by T-RF analysis. Clone libraries consisting of 187, 140, and 145 clones were created for January, April, and August benthic samples, respectively. For the January library, 136 out of 187 clones analyzed produced different RFLPs, defined as unique OTUs. For the April clone library, 115 of 140 were unique OTUs. For August, there were 89 OTUs out of 145 clones. The low numbers of duplicate clones indicated high genetic diversity.
Representative clones of each OTU were partially sequenced for phylogenetic identification. Table 1 summarizes clones representing ≥2% abundance of each clone library and having ≥90% similarity to species found in the GenBank database. Sequenced clones from the January clone library fell into four major bacterial groups: the γ, δ, and β subdivisions of the Proteobacteria (31, 13.6, and 10%, respectively, and the Bacteroides group (13.5%). OTUs representing 30 of the 187 clones evaluated were highly similar (>98%) to existing 16S rRNA gene sequences of Stenotrophomonas maltophilia. CfoI restriction sites of these clones generated a 213-bp peak, corresponding to the dominant peak in the January benthic community profile. In addition, all clone sequences representing the δ-Proteobacteria contained CfoI restriction sites, generating a T-RF located at 95 bp. The most frequently recovered sequences of this group were relatives of δ-proteobacterial sulfate-reducing bacteria, more specifically, the Olavius and Flexibacter spp. Nearly a third of the clones (∼32%) did not match existing sequences, though some were closely related to sequences representing chloroplast 16S rRNA genes, presumably from microalgae present in the biofilms.
TABLE 1.
Key OTUs (≥2% abundance) and their closest relatives (≥90% similarity) in the GenBank database
| OTU | Most closely related species (GenBank accession no.) | Similarity (%) | Abundance (%) |
|---|---|---|---|
| Jan-112 | Stenotrophomonas maltophilia (AB180661) | 98 | 16 |
| Jan-117 | Uncultered δ proteobacterium (AB031598) | 94 | 6 |
| Jan-185 | Alcanivorax sp. (AB053131) | 99 | 4 |
| Jan-68 | Acidovorax sp. (Y18617) | 99 | 4 |
| Jan-113 | Bacteriodetes bacterium (AY521223) | 94 | 3 |
| Jan-95 | Cytophaga sp. (AF427479) | 92 | 2 |
| Jan-56 | Flexibacter tractuosus (AB031598) | 93 | 2 |
| Jan-162 | Olavius algarvensis (AF328857) | 92 | 2 |
| Jan-118 | Robiginitalea biformata (AY424899) | 94 | 2 |
| Apr-116 | Stenotrophomonas maltophilia (AB180661) | 99 | 8 |
| Apr-148 | Unidentified γ proteobacterium (AB015576) | 94 | 4 |
| Apr-12 | Olavius algarvensis (AF328857) | 94 | 2 |
| Apr-46 | Flavobacteriaceae bacterium (AY177723) | 98 | 2 |
| Apr-66 | Synechococcus sp. (AF001480) | 98 | 2 |
| Apr-50 | Uncultured Holophaga sp. (AJ241004) | 90 | 2 |
| Apr-75 | Uncultured Verrucomicrobia (AJ401121) | 92 | 2 |
| Apr-210 | Sulfur-oxidizing bacterium (AF170422) | 92 | 2 |
| Apr-253 | Planctomycetales strain (AJ231181) | 90 | 2 |
| Aug-132 | Uncultured γ proteobacterium (AJ240987) | 92 | 30 |
| Aug-80 | Olleya marilimosa (AY586527) | 93 | 15 |
| Aug-58 | Uncultured CFB group bacterium (AJ441241) | 91 | 9 |
| Aug-180 | Synechococcus sp. (BX569694) | 93 | 4 |
| Aug-9 | Uncultured Planctomycetales (AJ290183) | 96 | 2 |
| Aug-187 | Legionella pneumophila (AF134574) | 95 | 2 |
In the April clone library, the highest percentage of clones identified were closely related (>97%) to S. maltophilia; however, the relative OTU abundance (8%) was substantially lower than in the January clone library. Other sequences were affiliated with the phyla Planctomycetales (13.6%), Verrucomicrobia (4.5%), and Cyanobacterium (4.5%).
Sequenced clones from the August libraries fell into three major bacterial lineages, two of which exhibited highly abundant OTUs (70%) that were assigned to the Bacteroides group (40%) and the γ subdivision of the Proteobacteria (30%). Overall, 47% of the clones sequenced were affiliated with the Proteobacteria. Clone sequences affiliated with the Cyanobacteria (4%) correlated with the CfoI-restricted 668-bp T-RF in the August benthic community profile. Only 2% of the clones were affiliated with the Planctomycetales. No clones were identified as being similar to S. maltophilia, in contrast to the other sampling periods. MspI and RsaI restriction sites for abundant OTUs in the January, April, and August clone libraries were found to correlate with dominant peaks in the corresponding T-RFLP profiles. No chimeric sequences were identified by the CHECK_CHIMERA program of Ribosomal Database Project II (29).
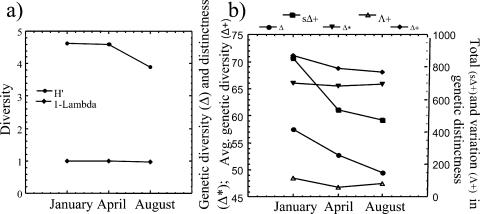
Diversity values generated from Simpson's index showed no change in diversity between the January, April, and August clone libraries. Shannon values showed minimal change between the January and April communities and a notable decrease in diversity between the April and August bacterial communities (Fig. 7a). Genetic diversity values showed a decrease in diversity from winter to summer (Fig. 7b). Average genetic (Δ+) and total genetic (sΔ+) distinctness values yielded similar but lower gradients of diversity with the onset of the summer season. There were moderate fluctuations in variation in genetic distinction, with the lowest value given for the April library (Fig. 7b). This is due to the even distribution (approximately two clones per OTU) of representative OTUs present as opposed to the January and August clone libraries, which had anywhere from 2 to 26 clones with identical restriction patterns. No trend was observed in the genetic distinctness values.
FIG. 7.
(a) Shannon (H′) and Simpson (D) diversity values based on OTUs from January, April, and August sample clone libraries. (b) Genetic diversity and distinctness values based on sequence similarities and respective OTU numbers: taxonomic diversity (Δ) index empirically related to the Shannon-Weaver index, taxonomic distinctness (Δ*), average taxonomic distinctness (Δ+), total taxonomic distinctness (sΔ+), and variation in taxonomic distinctness (Λ+).
DISCUSSION
Biofilm growth and community structure were responsive to seasonal changes in abiotic conditions and differences at the small vertical scale of 1.5-m depth between surface and benthic biofilms. Both T-RFLP and ARDRA analyses showed distinct seasonal variations in the benthic and floating biofilms that included periods of stasis and transition. Some elements of the community were found throughout the year yet followed seasonal trends. T-RFs at 514 and 668 bp represented this type, exhibiting increased abundance during summer months. Several CfoI-derived T-RFs (e.g., 93, 95, and 98 bp) were prominent in most of the benthic and floating community profiles and appeared to follow similar trends in abundance (Fig. 4a and b). Others, such as T-RFs at 95 and 98 bp (representative of the δ and γ subdivisions of the Proteobacteria) were found throughout the year but did not exhibit any definitive seasonal pattern, possibly due to the great physiological diversity among microorganisms in these groups (6, 16).
Moderate deviations in environmental parameters appeared to have little effect on major phylogenetic groups within these microbial communities. High stability and similarity were found in T-RFLP profiles from the winter and summer seasons. Despite marked changes in salinity, the summer benthic profiles exhibited striking consistency in phylogenetic composition and relative abundance. In this respect, biofilm community profiles appear to be less responsive to short-term changes in salinity and moderate DO fluctuations and appear to be responsive to growth temperatures and energy bioavailability.
Temporal and spatial variation in microbial communities has not been extensively investigated, especially under variable salinity, although the results of this analysis are not without precedent. Brummer et al. (6), in a comparative investigation of phylogenetic groups of bacteria present in river biofilms, revealed seasonal patterns of abundance of rarer phylogenetic groups. Specifically, they observed seasonal cycles of abundance in the Planctomycetes and Cytophaga-Flavobacterium groups, trends that were supported by culture and culture-independent studies. Wolsing and Prieme (48) revealed, by T-RFLP, distinct seasonal variation in community structures of denitrifying bacteria in soil. Seasonal (28, 40) and spatial (27) differences in the dominant members of soil microbial communities have also been demonstrated. Additional studies focusing on soil bacteria in salt marshes (21) have shown variations in both the size and structure of microbial communities over time. Others (35) have found little or no variation with increased temporal or spatial distance. However, some of these studies have focused on species richness and not on community structure per se, which also involves the proportional abundance of the community members.
It is possible that the seasonal fluctuations of dominant phylogenetic groups observed in this investigation were reflective of temperature-dependent biofilm growth stages. Biofilm formation as a successional process is influenced by both external and internal processes. This is supported by a recent investigation of freshwater biofilms grown on artificial substrata in which a substantial increase in community stability was shown after 7 days (3). Temperature limitations on growth may have forestalled autochthonous influences that could have changed biofilm microbial community structure. Manz et al. (30) showed clear variations in relative abundance of the α and β subdivisions of the Proteobacteria and bacteria affiliated with the Cytophaga-Flavobacterium group with respect to biofilm developmental time in lotic biofilms grown on polycarbonate slides. Taxonomic diversity shown by fluorescent in situ hybridization using rRNA-targeted probes demonstrated domination by the β-Proteobacteria during early biofilm stages, followed by rapid increases in and continual prevalence of the α-Proteobacteria and Cytophaga-Flavobacterium groups in biofilm maturity. Distinctive growth patterns for culturable bacteria on surfaces were also illustrated as a species- or strain-specific feature (23).
The dissimilarity between the T-RFLP profiles of the benthic and floating biofilms is intriguing considering the shallow depth of the water column at the site. Variation in light energy availability was apparent, and even though light was not a significant correlate for benthic biofilm biomass (Fig. 3c), reduction of light transmission was evident in the summer months due to water column particulates. Moderate hypoxia was recorded for the benthic biofilms in the summer months, but surface DO was not measured, so it is not known whether this variable changed between the benthos and surface. In addition, macro- and micrograzers may have been more available to the benthic biofilms from the surrounding sea grasses.
The use of ARDRA in combination with T-RFLP permitted the taxonomic assignment of bacteria to specific T-RFs. Construction of a 16S rRNA gene clone library from the January sample enabled the identification of the 95- and 213-bp T-RFs (Table 1). 16S rRNA genes from five distinct OTUs in the January clone library were most closely matched to Stenotrophomonas maltophilia. S. maltophilia, previously Pseudomonas maltophilia (20), has been reported to be an opportunistic psychrotrophic pathogen, ubiquitous in a wide variety of habitats and having optimal temperatures between 14 and 22°C (43). This agrees with recorded ambient water temperatures during the periods of highest abundance in benthic T-RFLP (213-bp peak) and ARDRA analyses and underscores adaptation to environmental conditions rather than truncated succession as a cause for temporal changes in the biofilm community structure.
Some sequences in the April and August clone libraries were affiliated with the Cyanobacteria (12%) and correlated with the 668-bp T-RF in the benthic community profiles. April and August ARDRA analyses marked an 8% (4.5 to 12%) increase in Cyanobacteria. In estuarine plankton, cyanobacteria become increasingly prominent in warm water months (34, 36). This was mirrored by T-RFLP analyses, with an estimated 13% (1.6 to 14.7%) increase in relative abundance of the 668-bp peak for the summer samples.
Estimates of diversity by using clone libraries and resultant OTUs as “species” have been generated with Shannon-Weaver and related indexes for comparing various aquatic and terrestrial microbial communities (19, 41). However, these approaches do not incorporate information on the degree of relatedness among community members. A community may have a large number of OTUs but also high genetic variability. Species diversity indices such as Shannon-Weaver will not distinguish between this and a community of equivalent richness but lower genetic relatedness.
A truer measure of biodiversity should be informative about how different the inhabitants of a community are from each other (18). This study employed a modified approach for the investigation of microbial biodiversity through implementation of the taxonomic distinctness concept (46, 47). Biodiversity indices originally developed for surveying benthic invertebrate communities were modified to incorporate DNA sequence information and genetic similarities with their relative frequencies to obtain estimates of genetic diversity. This modified approach encapsulates both major features of the Shannon-Weaver and related indices while also incorporating a value of relatedness between clonal sequences. Genetic diversity and total genetic distinctness values showed a decrease in biodiversity among clones from January through August. Also, a practical advantage of taxonomic diversity/distinctness indices is that they are less influenced by sample size disparities (9), allowing investigators to compare clone libraries of conflicting sizes.
Some, but not all, of the major T-RFs detected in the T-RFLP analysis were found in the clone libraries. Also, identified T-RFs were not necessarily reflective of dominant microbes, due to peaks representing a combination of many taxa or species or due to biases introduced through cloning and T-RFLP techniques. The ARDRA technique has been commonly used to assess community structure and to detect changes in communities (33, 39). However, ARDRA appears to have little efficacy in estimating diversity or for following specific ribotypes. In the present case, a single species (i.e., S. maltophilia) was represented by several restriction fragment patterns. In a multifaceted community with many dissimilar species, an ARDRA profile may become excessively complex and, consequently, may lose the phylogenetic information that is important in community analysis. Although T-RF peaks can incorporate multiple species and collapse community structure to relatively simplistic patterns, T-RFLP methodologies avoid some of the problems associated with ARDRA analysis and yield community profiles that are amenable to community structure analysis (26). Restriction fragment lengths can be determined from ribosomal databases, adding some phylogenetic information to T-RFLP profiles for comparison of bacterial communities.
Despite its utility as a surrogate measure of community structure, microbial diversity is underestimated by T-RFLP analyses. T-RFs may represent particular species but are more likely to represent known bacterial groups or even groups of unrelated organisms. In order to increase the resolution of the analytical method, three different restriction enzyme digests were used in this study. Exclusion of organisms also occurred in a few community profiles (September to December 2003 benthic; July to December 2003 floating), where a small number of peaks fell outside the range of the T-RFLP standards. In addition, it is possible that in some sequences the primer and restriction sites were located in close proximity, resulting in T-RFs that were too short to be detected. Moreover, preferential DNA extraction, well-known inherent PCR biases (25, 37), and differences in the 16S rRNA gene copy number between species (14) can distort relative abundances. This analysis is a relative and not absolute measure of community structure. The inclusiveness of the T-RFLP and ARDRA analyses for community structure analysis is also dependent upon the specificity of the primers used. The primers used (primers 27F and 1492R) are universal for amplifying eubacterial 16S rRNA genes and have been found to work well for evaluation of bacterial populations (5, 22, 33), although primer 27F has been known to contain mismatches to certain bacterial groups (i.e., Planctomycetales); therefore, a possibility exists that diversity was underestimated by the analysis (31). None of the recognized universal primers for 16S rRNA genes have been shown to amplify all sequences from the bacterial, eukaryal, and archaeal domains (7).
The results of this study indicate that microbial biofilm communities shift in composition and biomass with seasonal change but are stable within seasonal limits. Temperature and rainfall appeared to be the primary factors responsible for the transition from one seasonal community type to another in this subtropical estuary. The high stability and similarity of phylogenetic groups within seasonal profiles suggest that biofilm microorganisms are tolerant of fluctuations in salinity and suggest that biofilms as a physical habitat provide some buffering of minor fluctuations in ambient water quality parameters. This stability in the presence of normal estuarine variability in an area of relatively healthy sea grass habitat suggests that biofilm microbial communities may be useful in monitoring estuarine ecosystem conditions. Further investigations of the responsiveness of biofilm communities to estuarine environmental conditions will aid in utilizing the information content of these microbial communities as sentinels of deleterious environmental change.
Acknowledgments
This research was supported by a grant from the U.S. Environmental Protection Agency's Science To Achieve Results (STAR) Estuarine and Great Lakes (EaGLe) Coastal Initiative through funding to the CEER-GOM project, U.S. EPA agreement EPA/R-82945801.
REFERENCES
- 1.Allison, D. G., and P. Gilbert. 1992. Bacterial biofilms. Sci. Prog. 76:305-321. [Google Scholar]
- 2.Anwar, H., J. L. Strap., and J. W. Costerton. 1992. Establishment of aging biofilms: a possible mechanism of bacterial resistance to antimicrobial therapy. Antimicrob. Agents Chemother. 36:1347-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Araya, R., K. Tani, T. Takagi, N. Yamaguchi, and M. Nasu. 2003. Bacterial activity and community composition in stream water and biofilm from an urban river determined by fluorescent in situ hybridization and DGGE analysis. FEMS Microbiol. Ecol. 43:111-119. [DOI] [PubMed] [Google Scholar]
- 4.Boivin, J., and J. W. Costerton. 1991. Biofilms and biodeteriation, p. 53-62. In H. W, Rossmore (ed.), Biodeterioration and biodegradation. Elsevier Applied Science, London, England.
- 5.Bowman, J. P., S. A. McCammon, M. V. Brown, D. S. Nichols, and T. A. McMeekin. 1997. Diversity and association of psychrophilic bacteria in Antarctic sea ice. Appl. Environ. Microbiol. 63:3068-3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brummer, I. H. M., W. Fehr, and I. Wagner-Dobler. 2000. Biofilm community structure in polluted rivers: abundance of dominant phylogenetic groups over a complete annual cycle. Appl. Environ. Microbiol. 66:3078-3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brunk, C. F., A. Aghajani, and C. A. Brunk. 1996. A complete analysis of primer and probe hybridization potential with bacterial small sub-unit sequences. Appl. Environ. Microbiol. 12:872-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Busscher, H. J., and H. C. Van der Mei. 1995. Use of flow chamber devices and image analysis methods to study microbial adhesion. Methods Enzymol. 253:455-476. [DOI] [PubMed] [Google Scholar]
- 9.Clarke, K. R., and R. M. Warwick. 1998. A taxonomic distinctness index and its statistical properties. J. Appl. Ecol. 35:423-531. [Google Scholar]
- 10.Clarke, K. R., and R. M. Warwick. 1999. The taxonomic distinctness measure of biodiversity: weighting of step lengths between hierarchical levels. Mar. Ecol. Prog. Ser. 184:21-29. [Google Scholar]
- 11.Clarke, K. R., and R. M. Warwick. 2001. A further biodiversity index applicable to species lists: variation in taxonomic distinctness. Mar. Ecol. Prog. Ser. 216:265-278. [Google Scholar]
- 12.Costerton, J. W., Z. Lewandowski, D. E. Caldwell, D. R. Korber, and H. M. Lappin-Scott. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49:711-745. [DOI] [PubMed] [Google Scholar]
- 13.Davies, D. G., A. M. Chakrabarty, and G. G. Geesey. 1993. Exopolysaccharide production in biofilms: substratum activation of alginate gene expression by Pseudomonas aeruginosa. Appl. Environ. Microbiol. 59:1181-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farrelly, V., F. A. Rainey, and E. Stackebrandt. 1995. Effect of genome size and rrn gene copy number on PCR amplification of 16S rRNA genes from a mixture of bacterial species. Appl. Environ. Microbiol. 61:2798-2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Felsenstein, J. 1989. PHYLIP—Phylogenetic Inference Package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 16.Gupta, R. S. 2000. The phylogeny of proteobacteria: relationships to other eubacterial phyla and eukaryotes. FEMS Microbiol. Rev. 24:367-402. [DOI] [PubMed] [Google Scholar]
- 17.Hamilton, W. A. 1985. Sulfate-reducing bacteria and anaerobic corrosion. Annu. Rev. Microbiol. 39:195-217. [DOI] [PubMed] [Google Scholar]
- 18.Harper, J. L., and D. L. Hawksworth. 1994. Biodiversity: measurement and estimation. Phil. Trans. R. Soc. Lond. Ser. 345:5-12. [DOI] [PubMed] [Google Scholar]
- 19.Huber, J. A., D. A. Butterfield, and J. A. Baross. 2004. Bacterial diversity in a subseafloor habitat following a deep-sea volcanic eruption. FEMS Microbiol. Ecol. 43:393-409. [DOI] [PubMed] [Google Scholar]
- 20.Jackson, S. M., and E. B. G. Jones. 1991. Interactions within biofilms: the disruption of biofilm structure by protozoa. Kiel. Meeresforsch. Sonderh. 8:264-268. [Google Scholar]
- 21.Keith-Roach, M. J., N. D. Bryan, R. D. Bardgett, and F. R. Livens. 2002. Seasonal changes in the microbial community of a salt marsh, measured by phospholipid fatty acid analysis. Biogeochemistry 60:77-96. [Google Scholar]
- 22.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley and Sons, New York, N.Y.
- 23.Lawrence, J. R., P. J. Delaquis, D. R. Korber, and D. E. Caldwell. 1989. Behaviour of Pseudomonas fluorescens in the hydrodynamic boundary layer of surface microenvironments. Microb. Ecol. 14:1-14. [DOI] [PubMed] [Google Scholar]
- 24.Lewis, M. A., D. A. Weber, L. R. Goodman, R. S. Stanley, G. Craven, J. M. Patrick, R. L. Quarles, T. H. Roush, and J. M. Macauley. 2000. Periphyton and sediment bioassessment in North Florida Bay. Environ. Monit. Assess. 65:503-522. [Google Scholar]
- 25.Liesack, W., H. Weyland, and E. Stackebrandt. 1991. Potential risks of gene amplification by PCR as determined by 16S rDNA analysis of a mixed-culture of strict barophilic bacteria. Microb. Ecol. 21:191-198. [DOI] [PubMed] [Google Scholar]
- 26.Liu, W.-T., T. L. Marsh, H. Cheng, and L. J. Forney. 1997. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl. Environ. Microbiol. 63:4516-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ludemann, H., I. Arth, and W. Liesack. 2000. Spatial changes in the bacterial community structure along a vertical oxygen gradient in flooded paddy soil cores. Appl. Environ. Microbiol. 66:754-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lukow, T., P. F. Dunfield, and W. Liesack. 2000. Use of the T-RFLP technique to assess spatial and temporal changes in the bacterial community structure within an agricultural soil planted with transgenic and non-transgenic potato plants. FEMS Microbiol. Ecol. 32:241-247. [DOI] [PubMed] [Google Scholar]
- 29.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. Parker, P. R. Saxman, J. M. Stredwick, G. M. Garrity, B. Li, G. J. Olsen, S. Pramanik, T. M. Schmidt, and J. M. Tiedje. 2000. The RDP (Ribosomal Database Project) continues. Nucleic Acids Res. 28:173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Manz, W., K. Wendt-Potthoff, T. R. Neu, U. Szewyck, and J. R. Lawrence. 1999. Phylogenetic composition, spatial structure, and dynamics of lotic bacterial biofilms investigated by fluorescent in situ hybridization and confocal laser scanning microscopy. Microb. Ecol. 37:225-237. [DOI] [PubMed] [Google Scholar]
- 31.Marchesi, J. R., S. Takuichi, A. J. Weightman, T. A. Martin, J. C. Fry, S. J. Hiom, and G. M. Wade. 1998. Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNA. Appl. Environ. Microbiol. 64:795-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marshall, K. C. 1992. Biofilms: an overview of bacterial adhesion, activity, and control at surfaces. ASM News 58:202-207. [Google Scholar]
- 33.Michaud, L., F. Di Cello, M. Brilli, R. Fani, A. L. Giudice, and V. Bruni. 2004. Biodiversity of cultivable psychrotrophic marine bacteria from Terra Nova Bay (Ross Sea, Antarctica). FEMS Microbiol. Lett. 230:63-71. [DOI] [PubMed] [Google Scholar]
- 34.Millie, D. F., H. J. Carrick, P. H. Doering, and K. A. Steidinger. 2004. Intra-annual variability of water quality of phytoplankton in the North Fork of the St. Lucie River Estuary, Florida (USA): a quantitative assessment. Estuar. Coast. Shelf Sci. 61:137-149. [Google Scholar]
- 35.Mummey, D. L., and P. Stahl. 2003. Spatial and temporal variability of bacterial 16S rDNA-based T-RFLP patterns derived from soil of two Wyoming grassland ecosystems. FEMS Microbiol. Ecol. 46:113-120. [DOI] [PubMed] [Google Scholar]
- 36.Murrell, M. C., and E. M. Lores. 2004. Phytoplankton and zooplankton seasonal dynamics in a subtropical estuary: importance of cyanobacteria. J. Plankton Res. 26:371-382. [Google Scholar]
- 37.Schmalenberger, A., F. Schwieger, and C. C. Tebbe. 2001. Effect of primers hybridizing to different evolutionary conserved regions of the small-subunit rRNA gene in PCR-based microbial community analyses and genetic profiling. Appl. Environ. Microbiol. 67:3557-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shirtliff, M. E., J. T. Mader, and A. K. Camper. 2002. Molecular interactions in biofilms. Chem. Biol. 9:859-871. [DOI] [PubMed] [Google Scholar]
- 39.Smit, E., P. Leeflang, and K. Wernars. 1997. Detection of shifts in microbial community structure and diversity in soil caused by copper contamination using amplified ribosomal DNA restriction analysis. FEMS Microbiol. Ecol. 23:249-261. [Google Scholar]
- 40.Smit, E., P. Leeflang, S. Gommans, J. van der Broek, S. van Mil, and K. Warners. 2001. Diversity and seasonal fluctuations of the dominant members of the bacterial soil community in a wheat field as determined by cultivation and molecular methods. Appl. Environ. Microbiol. 67:2284-2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith, N. R., Z. Yu, and W. W. Mohn. 2003. Stability of the bacterial community in a pulp mill effluent treatment system during normal operation and a system shutdown. Water Res. 37:4873-4884. [DOI] [PubMed] [Google Scholar]
- 42.Snyder, R. A., M. A. Lewis, A. Nocker, and J. E. Lepo. 2005. Microbial biofilms as integrative sensors of environmental quality, p. 111-122. In S. A. Bortone (ed.), Estuarine indicators. CRC Press, Boca Raton, Fla.
- 43.Soares, A., B. Guieysse, O. Delgado, and B. Mattiasson. 2003. Aerobic biodegradation of nonylphenol by cold-adapted bacteria. Biotechnol. Lett. 25:731-738. [DOI] [PubMed] [Google Scholar]
- 44.van den Heuvael, J. C. 1992. Mass transfer in and around biofilms, p. 239-250. In L. F. Melo, T. R. Bott, M. Fletcher, and B. Capdeville (ed.), Biofilms: science and technology. Kluwer, Dordrecht, The Netherlands.
- 45.Viera, M. J., L. F. Melo., and M. M. Pinheiro. 1993. Biofilm formation: hydrodynamic effects on internal diffusion and structure. Biofouling 7:67-80. [Google Scholar]
- 46.Warwick, R. M., and K. R. Clarke. 1995. New “biodiversity” measures reveal a decrease in taxonomic distinctness with increasing stress. Mar. Ecol. Prog. Ser. 129:301-305. [Google Scholar]
- 47.Warwick, R. M., and K. R. Clarke. 1998. Taxonomic distinctness and environmental assessment. J. Appl. Ecol. 35:532-543. [Google Scholar]
- 48.Wolsing, M., and A. Prieme. 2004. Observation of high seasonal variation in community structure of denitrifying bacteria in arable soil receiving artificial fertilizer and cattle manure by determining T-RFLP of nir gene fragments. FEMS Microbiol. Ecol. 48:261-271. [DOI] [PubMed] [Google Scholar]