Abstract
Hydrothermal venting and the formation of carbonate chimneys in the Lost City hydrothermal field (LCHF) are driven predominantly by serpentinization reactions and cooling of mantle rocks, resulting in a highly reducing, high-pH environment with abundant dissolved hydrogen and methane. Phylogenetic and terminal restriction fragment length polymorphism analyses of 16S rRNA genes in fluids and carbonate material from this site indicate the presence of organisms similar to sulfur-oxidizing, sulfate-reducing, and methane-oxidizing Bacteria as well as methanogenic and anaerobic methane-oxidizing Archaea. The presence of these metabolic groups indicates that microbial cycling of sulfur and methane may be the dominant biogeochemical processes active within this ultramafic rock-hosted environment. 16S rRNA gene sequences grouping within the Methylobacter and Thiomicrospira clades were recovered from a chemically diverse suite of carbonate chimney and fluid samples. In contrast, 16S rRNA genes corresponding to the Lost City Methanosarcinales phylotype were found exclusively in high-temperature chimneys, while a phylotype of anaerobic methanotrophic Archaea (ANME-1) was restricted to lower-temperature, less vigorously venting sites. A hyperthermophilic habitat beneath the LCHF may be reflected by 16S rRNA gene sequences belonging to Thermococcales and uncultured Crenarchaeota identified in vent fluids. The finding of a diverse microbial ecosystem supported by the interaction of high-temperature, high-pH fluids resulting from serpentinization reactions in the subsurface provides insight into the biogeochemistry of what may be a pervasive process in ultramafic subseafloor environments.
The Lost City hydrothermal field (LCHF) is located near the summit of the Atlantis Massif at a water depth of ∼750 m (37, 38). Long-lived faulting and extensive uplift at the massif have resulted in the exposure of magnesium-rich, variably altered ultramafic rocks with lesser gabbroic material that is 1.5 to 2 million years of age. Fluid circulation within the massif is driven by serpentinization reactions and the cooling of the underlying mantle rocks. These reactions result in a combination of extreme conditions never before seen in the marine environment, which include venting of high-pH (from pH 9 to 11), 40 to 91°C hydrothermal fluids with high concentrations of dissolved hydrogen (H2), methane (CH4), and other low-molecular-weight hydrocarbons (38). Mixing of the warm, high-pH fluids with seawater results in carbonate precipitation and growth of chimneys, which tower up to 60 m above the surrounding seafloor (38). Carbon-14 radioisotopic dating indicates that hydrothermal activity has been ongoing for at least 30,000 years (17). A large percentage of exposed seafloor on and near slow- and ultraslow-spreading ridges is likely to contain ultramafic rocks similar to those that host the LCHF (4, 12, 14). Therefore, this system offers a unique opportunity to study an ultramafic-rock-hosted submarine ecosystem that may be both widespread and stable over thousands of years.
The concentration of H2 in LCHF fluids varies widely by vent site, with values of 1 to 15 mM (38, 60). This variability may be due in part to biological hydrogen oxidation, as indicated by measurements of the deuterium content of dissolved H2 in LCHF fluids (60). In contrast to black smoker environments, the high-pH LCHF fluids contain CO2 concentrations that are small to below detection limits (38). Methane, by contrast, is present at a relatively constant concentration of 1 to 2 mM throughout the LCHF (38, 60). Both H2 and CH4 are products of serpentinization (7, 36, 44), so the highly variable H2 concentrations and consistent values of CH4 are intriguing. The present study examines the possibility that biological communities are acting as a sink for H2 and possibly both a source of and sink for CH4. The CH4 concentrations of the fluids are similar to those of many environments that support anaerobic methane oxidation (AMO) (55, 75, 76). Therefore, the possibility that AMO exists at the LCHF is specifically addressed.
Previous reports have shown that the most actively venting chimneys at the LCHF are dominated by a single phylotype of Methanosarcinales (Lost City Methanosarcinales [LCMS]) (65). In areas bathed in >80°C hydrothermal fluid, these organisms form thick biofilms, and they comprise nearly 100% of the archaeal community. Recently, methyl coenzyme M reductase (mcrA) gene sequences corresponding to both LCMS and ANME-1, a group of anaerobic methane-oxidizing Archaea, have also been recovered from LCHF carbonate chimneys (38). ANME-1 has been identified in many environments, including CH4 seeps in anoxic marine sediments, CH4 hydrates, carbonate reefs in the Black Sea, and mud volcanoes (3, 8, 46, 55, 72, 75). Genomic evidence indicates that anaerobic methane-oxidizing Archaea harbor nearly all of the genes necessary for methanogenesis, including mcrA (21, 45), so it is unclear whether LCMS and ANME-1 are sources of or sinks for CH4 within the Lost City system. The limited number of LCHF samples studied to date has not allowed firm conclusions regarding the distribution of LCMS and ANME-1 with respect to temperature. The temperatures of most other sites with high rates of AMO are not significantly different from those of background seawater (earning them the name “cold seeps”). However, Schouten et al. (64) identified lipids that are indicative of methanotrophic Archaea throughout a sediment core from hydrothermally influenced sediments of the Guaymas Basin reaching 95°C. Those authors claimed evidence for active microbes involved in AMO at temperatures of >30°C. Kallmeyer and Boetius (34) measured low rates of AMO at up to 85°C at the same site. Therefore, determining whether the ANME-1 phylotype discovered at the LCHF lives at high temperatures and whether the LCMS biofilms known to exist at high temperatures are methanogens or methanotrophs has biogeochemical and ecological importance.
In all known environments where AMO occurs, anaerobic methanotrophic Archaea co-occur with sulfate-reducing Bacteria (SRB), commonly in tightly coupled consortia (8), although not always in direct physical contact (56). Incubation experiments have shown that this consortium represents a syntrophic metabolic relationship between the methane-oxidizing Archaea and SRB (19, 24, 52, 53). The role of Bacteria in CH4 cycling at the LCHF, however, has not been thoroughly explored in previous studies, and it is not known whether SRB are present (38, 65). To further characterize the microbial communities of the LCHF, we have examined the phylogenetic diversity of both archaeal and bacterial 16S rRNA genes in geochemically distinct carbonate and fluid samples. Utilizing terminal restriction fragment length polymorphism (T-RFLP) analyses, the distribution of specific phylogenetic groups of Archaea and Bacteria was determined. The distribution of microbial groups within the LCHF shows important biogeochemical linkages between serpentinization reactions and microbial metabolism.
MATERIALS AND METHODS
Sample collection.
Carbonate chimney samples were collected from the LCHF with DSV Alvin (dives 3651 and 3862 to 3881) during cruises AT03-6 and AT07-34 aboard the R/V Atlantis in December 2000 and April to May 2003 (38). Shipboard, subsamples of chimney material were frozen immediately at −80°C and remained frozen until onshore analysis. Filtered fluid samples were obtained using a hydrothermal fluid and particle sampler (10). Approximately 1 liter of fluid was pumped through a Sterivex-GP (0.22-μm) filter while the pump rate and temperature of the fluid were measured. Once on deck, the filters were placed into 50-ml sterile Falcon tubes (BD Sciences Labware) and stored at −20°C until DNA extraction. Fluid samples were also preserved in a 2% formalin solution and stored at 4°C. Cell enumeration of DAPI (4′,6′-diamidino-2-phenylindole)-stained cells (59) was performed using a Nikon Eclipse E600 POL epifluorescence microscope.
Sample descriptions.
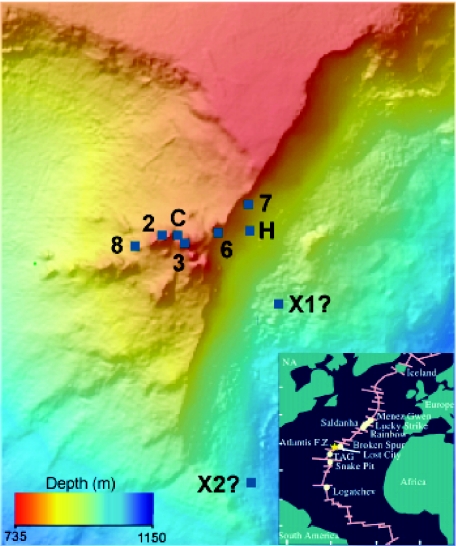
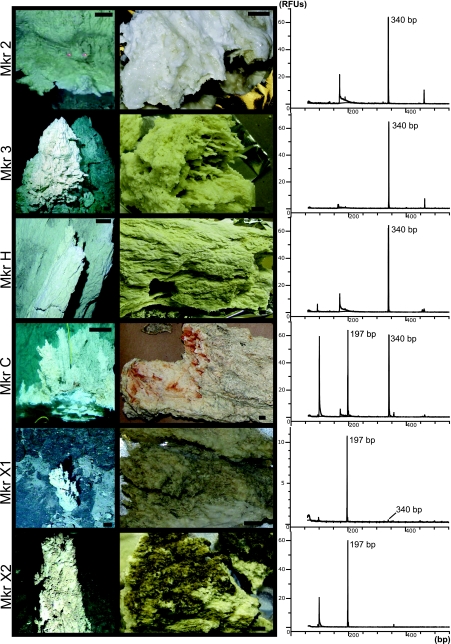
Samples were collected from carbonate structures with a range of venting temperatures and chemistries (Table 1), representing actively venting, hot chimneys (markers 2, 3, C, H, and 7) and less-active, cooler chimneys (markers X1 and X2). Active chimneys tended to cluster around the Poseidon complex (Fig. 1), while carbonate deposits at markers X1 and X2 were found at the periphery of the field. The chimney at marker 3 represents one of four pinnacles at the summit of the Poseidon complex, and the fluid sample from this chimney had the highest temperature (81°C) of any samples in this study and approached the maximum temperature of any vent at the LCHF (91°C). Two carbonate samples from near marker 2 were collected from the interior of a very large flange after the cooler, outer portions of the flange were cleared away by DSV Alvin. The carbonate material collected from markers 2 and 3 appeared to be the youngest samples in this study; i.e., the carbonate was very friable and not well lithified. These samples also had the largest quantity of a mucilaginous biofilm-like material, deposited throughout the carbonate matrix. Marker C is at a large flange composed of aragonite, calcite, and brucite [Mg(OH)2]. The top of the 70°C venting flange is comprised of well-lithified carbonate bathed in seawater. Markers H, 7, and 8 are located on somewhat cooler (46 to 53°C) chimneys and are located at the perimeter of the Poseidon complex on the east side of the field. Carbonate material from marker H appeared to be similar to the young carbonate deposits collected at markers 2 and 3. Marker 6 represents an actively venting chimney (70°C) located between the summit of Poseidon and markers H and 7. The lithified chimneys exhibiting little or no venting near markers X1 and X2 were discovered during exploratory DSV Alvin dives and are located several hundred meters downslope from the Poseidon complex. The marker X2 chimney was very highly lithified, having a dark gray and brownish color, while the chimney at marker X1 appeared to have recently precipitated carbonate in its interior. Sample characteristics are summarized in Tables 1 and 2, and Fig. 2 contains photographs of chimneys from which carbonate samples were analyzed.
TABLE 1.
Description of carbonate samples used for DNA extraction and phylogenetic analysisa
| Marker | Carbonate sample(s) | Fluid sample | Description | Temp (°C) | Mg level (range) (mM/kg) | H2 level (mmol/kg) | CH4 level (mmol/kg) | No. of cells/g carbonate | No. of cells/ml fluid |
|---|---|---|---|---|---|---|---|---|---|
| 2 | 3864-1524, 3864-1537, LC1149 | None | Venting flange, young looking | 62 | 2-39 | 3.77 | 1.28 | 1.4 × 109 | 4.7 × 104 |
| 3 | 3881-1408, LC1022 | FS242 | Peak of chimney complex | 81 | 3-39 | 13.26 | 1.55 | 2.0 × 108 | 3.9 × 105 |
| H | 3881-1228 | FS243 | Large chimney with flange | 46 | 28-46 | 2.21 | 1.44 | 3.6 × 108 | 1.4 × 105 |
| C | 3869-1404, 3869-1443, 3869-1446 | FS221 | Venting flange, more lithified | 70 | 12-28 | 14.38 | 1.98 | 1.2 × 109 | 1.4 × 105 |
| X1 | 3876-1133 | None | Lithified carbonate chimney in serpentinite | ND | ND | ND | ND | 1.6 × 108 | ND |
| X2 | 3880-1557 | None | Lithified carbonate chimney in serpentinite | ND | ND | ND | ND | 3.4 × 106 | ND |
| 6 | None | FS222 | Venting chimney | 68 | 45 | 14.19 | 1.61 | ND | 2.0 × 105 |
| 7 | None | FS197 | Venting chimney | 51 | 35-52 | 5.54 | 1.31 | ND | 2.8 × 105 |
| 8 | None | FS209 | Venting chimney | 53 | 7-53 | 4.20 | 1.84 | ND | 1.7 × 105 |
| None | LC1231 | None | Inactive chimney | 7 | ND | ND | ND | 1.2 × 107 | ND |
Coregistered hydrothermal fluids and carbonate deposits were sampled at markers 3, C, and H. Markers 2, X1, and X2 were sampled for carbonate only, and markers 6, 7, and 8 were sampled for fluid only. Mg concentrations are shown as a measure of the seawater component of the sample. ND, not determined.
FIG. 1.
Sample locations in the Lost City hydrothermal field analyzed in this study. Markers 2, 3, 6, 8, and C are within the central portion of the Poseidon complex of carbonate chimneys. Markers X1 and X2 are isolated chimneys exhibiting greatly reduced or no apparent hydrothermal activity.
TABLE 2.
Archaeal and bacterial phylotypes recovered in 16S rRNA gene clone libraries and OTUs resolved by T-RFLP analysis of carbonate chimney and fluid samplesa
| Sample | Archaeal phylotype (no. of clones) | Archaeal OTUs (no. identified) (HaeIII-MspI-BstUI) | Bacterial phylotype (no. of clones) | Bacterial OTUs (no. identified) (HaeIII-MspI-BstUI) |
|---|---|---|---|---|
| 3864-1524 | ND | ND | 7 (27) | 12 (4)-15 (5)-15 (4) |
| 3864-1537 | ND | 5 (1)-7 (2)-4 (1) | 15 (46) | 12 (8)-16 (11)-15 (6) |
| 3881-1408 | 1 (29) | 5 (1)-6 (2)-4 (2) | 37 (55) | 13 (6)-ND-13 (7) |
| FS242 | ND | 10 (4)-9 (1)-13 (2) | ND | 20 (14)-24 (13)-19 (9) |
| 3881-1228 | 2 (3) | 7 (2)-4 (3)-4 (3) | 24 (32) | ND |
| FS243 | 16 (57) | 9 (3)-8 (1)-11 (3) | ND | 24 (10)-22 (11)-23 (10) |
| 3869-1404 | 1 (3) | 4 (1)-3 (1)-4 (1) | 3 (3) | 23 (12)-26 (10)-18 (7) |
| 3869-1443 | ND | 7 (2)-7 (3)-5 (4) | 8 (9) | 27 (14)-37 (15)-20 (9) |
| 3869-1446 | ND | 10 (3)-7 (4)-9 (2) | 40 (72) | ND |
| FS221 | ND | ND | ND | 8 (6)-9 (4)-7 (2) |
| 3876-1133 | 1 (2) | 4 (2)-4 (2)-4 (2) | 30 (54) | 23 (12)-ND-22 (6) |
| 3880-1557 | 1 (2) | 2 (1)-3 (2)-4 (1) | ND | 21 (10)-24 (7)-20 (8) |
| FS222 | ND | 9 (2)-8 (1)-11 (1) | ND | 15 (7)-24 (13)-17 (5) |
| FS197 | 2 (3) | 12 (3)-12 (2)-15 (2) | ND | 24 (9)-16 (3)-17 (8) |
| FS209 | 12 (50) | ND | ND | 11 (4)-7 (4)-11 (5) |
Archaeal and bacterial phylotypes are defined as 97% DNA sequence similarity. The total number of clones in the library and the number of OTUs that were identified as corresponding to a terminal restriction fragment of a clone in Table 3 are shown in parentheses. Samples for which a clone library or T-RFLP analysis was not attained are indicated by “ND.”
FIG. 2.
T-RFLP peaks generated by BstUI digestion of archaeal 16S rRNA genes amplified from carbonate chimney samples. Photographs of the chimneys at each marker are shown (first column), as are the samples of each chimney used in the analysis (second column). The 340-bp LCMS fragment is present in the active chimneys at markers 2, 3, H, and C. The 196-bp ANME-1 fragment is also detected at marker C as well as a weakly venting chimney at marker X1 and an extinct chimney at marker X2. A very short peak corresponding to LCMS is also barely noticeable in the marker X1 profile. The carbonate material recovered from the active chimneys (at markers 2, 3, and H) was whiter and softer than the gray, lithified minerals of the extinct chimneys from markers X1 and X2. A milky, biofilm-like substance pervaded the sample from marker 2, as can been seen in the photo. The sample from the marker C flange exhibited both types of mineralogies; in the photo, bright white brucite deposits are evident in the actively venting portion underneath a gray, more-lithified zone that is bathed in background-temperature seawater (see the text for details). RFUs, relative fluorescent units.
Clone library construction and sequencing.
DNA was extracted from carbonate chimney material as previously described (6, 66), with the following modifications: 80 μl of 5 M NaCl and 80 μl of 10% cetyltrimethylammonium bromide in 0.7 M NaCl were added to 1 g of carbonate chimney sample and incubated at 50°C for 30 min after proteinase K treatment. To prevent shearing of genomic DNA, samples were not subjected to freeze-thaw treatment. Crude DNA extracts were purified with QiaQuick columns (QIAGEN) according to the manufacturer's instructions for PCR purification. Sterivex filters were extracted and purified as described previously (27). Amplification of the 16S rRNA gene via PCR was performed using the universal primers ARC-21F (5′-TTC CGG TTG ATC CYG CCG GA-3′) and ARC-958R (5′-YCC GGC GTT GAM TCC AAT T-3′) for Archaea and BAC-8F (5′-AGR GTT TGA TCC TGG CTC AG-3′) and BAC-1492R (5′-CGG CTA CCT TGT TAC GAC TT-3′) for Bacteria. The PCR-amplified DNA was reconditioned using the protocol described previously by Thompson et al. (74) and cloned using the TOPO-TA cloning kit (Invitrogen) according to the manufacturer's instructions. Nearly full-length sequences of cloned inserts were obtained using a MegaBACE 1000 apparatus (Molecular Dynamics) at the Marine Molecular Biotechnology Laboratory, University of Washington (UW), according to the methods described previously by Huber et al. (27) using the sequencing primers described previously by Huber et al. (29).
T-RFLP analysis.
T-RFLP analyses of carbonate chimney and fluid samples were performed using universal primers for archaeal and bacterial 16S rRNA genes. T-RFLP primer sequences for the 16S rRNA gene are as follows (51): ARC21F (5′-6-carboxyfluorescein-TTC [dP]GG TTG ATC C[dP]G CC[dK] GA-3′), ARC922R (5′-[dP]CCG GCG TTG A[dN]T CCA ATT-3′), BAC68F (5′-6-carboxyfluorescein-T[dN]A [dN]AC ATG CAA GTC G[dK][dK] CG-3′), and BAC1492R (5′-[dK]G[dP] TAC CTT GTT ACG ACT T-3′), where [dP] indicates the pyrimidine analog P, [dK] indicates the purine analog K, and [dN] is an equal mixture of the two analogs (Glen Research, Sterling, VA).
Aliquots of approximately 5 μl of purified PCR products (QiaQuick columns; QIAGEN) were digested overnight with 0.25 U of the restriction endonucleases HaeIII, MspI, and BstuI for 16S rRNA genes. Digests were immediately ethanol precipitated according to the manufacturer's instructions (Amersham Pharmacia Biotech Inc.) and analyzed using a MegaBACE 1000 apparatus (Molecular Dynamics). Electrophoretic profiles were visualized with DAx software (Van Mierlo Software Consultancy, The Netherlands) at the UW Marine Molecular Biotechnology Laboratory.
Predicted terminal restriction fragments (TRFs) of clones were obtained using Sequencher (version 4.1.2; Gene Codes Corp.). Actual TRF profiles of environmental samples and some clones were compared to the predicted TRFs. The presence or absence of phylogenetic groups (Table 2 and see Fig. 6) was determined by the positive identification of a predicted TRF in the actual TRF profile of an environmental sample. To score a phylogenetic group as “present,” two criteria were required: (i) the predicted TRF size matched the actual TRF size within ±2 bp, and (ii) at least two of the three restriction enzyme digests yielded a positive identification of a predicted TRF of a clone belonging to that group. Clones that had fewer than two predicted TRFs within the 50- to 550-bp size window were not included in the analysis. The counting of operational taxonomic units (OTUs) can yield various results depending on the threshold criteria and variation due to PCR errors (15, 41, 49, 62). For this study, an OTU in a TRF profile was defined as one bin of peaks within ±2 bp having a magnitude of relative fluorescent units greater than that of the signal bleed-through from the standard DNA marker channel. These criteria were validated by replicate PCRs and replicate digests of environmental samples as well as cloned DNA.
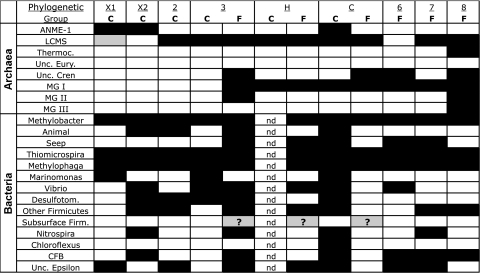
FIG. 6.
Presence or absence of phylogenetic groups in LCHF carbonate chimney and fluid samples. Colored and empty boxes indicate the presence and absence, respectively, of specific phylogenetic groups identified in Fig. 3 to 5. The gray box indicates the presence of a peak corresponding to LCMS of very low relative peak height, and the question marks indicate that TRFs corresponding to the “subsurface” clade were identified in the samples but did not match the sample profile within ±2 bp (see the text for details). “C” and “F” in the top row indicate carbonate chimney or fluid samples, respectively. “nd,” no data are available. Abbreviations: ANME-1, anaerobic methanotrophic Archaea; LCMS, Lost City Methanosarcinales; Thermoc., Thermococcus; Unc. Eury., uncultured Euryarchaeota; Unc. Cren, uncultured Crenarchaeota; MG I, MG II, and MG III, marine groups I, II, and III, respectively; CFB, Cytophaga-Flavobacterium-Bacteroides. Desulfotom., Desulfotomaculum. Unc. Epsilon, uncultured epsilon-proteobacteria.
Phylogenetic analysis.
Archaeal and bacterial 16S rRNA gene nucleotide sequences were aligned using ClustalX, version 1.81 (73). PCR primers and hypervariable regions were omitted from the alignments. Clones were checked for chimeric sequences using the CHIMERA_CHECK program of RDP II and the Bellerophon server (30). The Phylip version 3.62 software package (obtained from J. Felsenstein, UW) was used to construct maximum-likelihood (DNAML) trees. Confidence estimates for tree topology were calculated by bootstrap analysis (SEQBOOT) using 100 replicates. Trees were drawn with TreeViewPPC (version 1.6.6) and edited with Adobe Illustrator (version 10.0.3).
Nucleotide sequence accession numbers.
GenBank accession numbers for the clones reported here are listed in Table 3 and include accession numbers DQ228555 to DQ228583, DQ270590 to DQ270662, and DQ272585.
TABLE 3.
Archaeal and bacterial 16S rRNA gene clones generated in this study, in addition to previously published LCMS and marine group I clonesaa
| Domain | Clone | GenBank accession no. | Phylogenetic group | Fragment size (bp)
|
||
|---|---|---|---|---|---|---|
| HaeIII | MspI | BstuI | ||||
| Archaea | LC1022A-1 | AY299516 | LCMS | 219 | 130 | 340 |
| LC1149A-56 | AY299515 | LCMS | 219* | 130* | 340* | |
| LC1133A-9 | AY760632 | ANME-1 | 236* | 96* | 196* | |
| LC1133A-17 | DQ270605 | ANME-1 | 235 | 96 | 197 | |
| FS197A-23 | DQ270592 | Thermococcales | 154* | 78 | 73 | |
| FS197A-30 | DQ270593 | Thermococcales | 154 | 78 | 73 | |
| FS209A-41 | DQ270596 | Thermococcales | 154 | 78 | 73 | |
| FS243A-6 | DQ270598 | Unc. Eury. | 22 | 81 | 93 | |
| FS243A-60 | DQ270602 | Unc. Cren. | 152 | 124 | 331 | |
| FS209A-10 | DQ270595 | Unc. Cren. | 165 | 68 | 82 | |
| Bacteria | LC1228B-35 | DQ270650 | Actinobacteria | 187 | 426 | 184 |
| LC1022B-39 | DQ228573 | Actinobacteria | 191 | 105 | 472 | |
| LC1022B-1 | DQ228574 | Actinobacteria | 309 | 455 | 96 | |
| LC1022B-27 | DQ228572 | Actinobacteria | ND | ND | ND | |
| LC1133B-64 | DQ270644 | Alpha-proteobacteria | 188 | 399 | 58 | |
| LC1133B-90 | DQ270648 | Alpha-proteobacteria | 188 | 400 | 58 | |
| LC1133B-8 | DQ270640 | Alpha-proteobacteria | 188 | 400 | 58 | |
| LC1133B-74 | DQ270646 | Alpha-proteobacteria | 183 | 429 | 72 | |
| LC1133B-45 | DQ270642 | Alpha-proteobacteria | 163 | 410 | 167 | |
| LC1231B-170 | DQ228567 | Alpha-proteobacteria | 188 | 400 | 58 | |
| LC1022B-48 | DQ228566 | Alpha-proteobacteria | 188 | 399 | 58 | |
| LC1133B-23 | DQ270615 | Animal | 152 | 288 | 26 | |
| LC1228B-46 | DQ270625 | Animal | 151 | 286 | 26 | |
| LC1133B-24 | DQ270616 | Animal | 155 | 107 | 358 | |
| LC1524B-50 | DQ270636 | Beta-proteobacteria | 165 | 395 | 354 | |
| LC1537B-16 | DQ270638 | Beta-proteobacteria | 165 | 440 | 354 | |
| LC1022B-30 | DQ228559 | Beta-proteobacteria | 165 | 455 | 354 | |
| LC1537B-77 | DQ272585 | Chloroflexus | 181 | 174 | 361 | |
| LC1231B-173 | DQ228575 | Chloroflexus | 179 | 256 | 202 | |
| LC1408B-77 | DQ270634 | CFB | 361 | 442 | 65 | |
| LC1446B-1 | DQ270658 | CFB | 362 | 161 | 68 | |
| LC1524B-89 | DQ270637 | CFB | 376 | 458 | 351 | |
| LC1228B-138 | DQ270654 | CFB | 243 | 50 | 70 | |
| LC1133B-32 | DQ270641 | CFB | 368 | 50 | 70 | |
| LC1022B-31 | DQ228583 | CFB | 235 | 441 | 57 | |
| LC1228B-98 | DQ270651 | Desulfotomaculum | 270 | 131* | 77* | |
| LC1537B-22 | DQ270639 | Desulfotomaculum | 225 | 130 | 26 | |
| LC1404B-6 | DQ270655 | Epsilon-proteobacteria | 769 | 431 | 355 | |
| LC1133B-68 | DQ270645 | Epsilon-proteobacteria | 767 | 431 | 355 | |
| LC1228B-116 | DQ270653 | Epsilon-proteobacteria | 767 | 431 | 355 | |
| LC1133B-58 | DQ270643 | Epsilon-proteobacteria | 246 | 431 | 455 | |
| LC1408B-26 | DQ270633 | Epsilon-proteobacteria | 855 | 431 | 355 | |
| LC1149B-14 | DQ228557 | Epsilon-proteobacteria | 270 | 127 | 340 | |
| LC1149B-70 | DQ228579 | Epsilon-proteobacteria | 1,133 | 162 | 349 | |
| LC1149B-104 | DQ228555 | Epsilon-proteobacteria | 1,419 | 125 | 335 | |
| LC1231B-176 | DQ228582 | Epsilon-proteobacteria | ND | 436 | 360 | |
| LC1149B-9 | DQ228581 | Epsilon-proteobacteria | ND | ND | 353 | |
| LC1149B-130 | DQ228580 | Epsilon-proteobacteria | ND | 162 | 349 | |
| LC1149B-115 | DQ228556 | Epsilon-proteobacteria | ND | 124 | 264 | |
| LC1408B-88 | DQ270635 | Firmicutes | 267* | 435^ | 76* | |
| LC1022B-45 | DQ228570 | Firmicutes | 270 | 515 | 196 | |
| FS209A-1 | DQ270594 | Unc. Cren. | 192 | 124 | 332 | |
| FS197A-14 | DQ270591 | Marine group I | 214* | 284* | 460 | |
| FS243A-39 | DQ270601 | Marine group I | 214 | 284 | 460 | |
| FS243A-7 | DQ270590 | Marine group I | 214* | 285* | 461^ | |
| FS243A-90 | DQ270604 | Marine group I | 214 | 284 | 460 | |
| LC1231A-51 | AY505046 | Marine group I | 214 | 284 | 460 | |
| FS243A-20 | DQ270599 | Marine group II | 208 | 191 | 260 | |
| FS243A-33 | DQ270600 | Marine group II | 208 | 191 | 59 | |
| FS243A-89 | DQ270603 | Marine group II | 315 | 191 | 130 | |
| FS243A-3 | DQ270597 | Marine group III | 216 | 75 | 131 | |
| LC1022B-35 | DQ228568 | Firmicutes | 236 | 28 | 196 | |
| LC1149B-139 | DQ228571 | Firmicutes | 268 | 434 | 76 | |
| LC1022B-12 | DQ228569 | Firmicutes | 269 | 114 | 195 | |
| LC1022B-45 | DQ228570 | Firmicutes | 270 | 515 | 196 | |
| LC1537B-86 | DQ270611 | Marinomonas | 212 | 449 | 69 | |
| LC1228B-136 | DQ270627 | Methylobacter | 365 | 451 | 350 | |
| LC1133B-145 | DQ270623 | Methylobacter | 367 | 453 | 477 | |
| LC1133B-37 | DQ270617 | Methylobacter | 163 | 452 | 168 | |
| LC1446B-16 | DQ270628 | Methylobacter | 289 | 459 | 176 | |
| LC1133B-99 | DQ270621 | Methylobacter | 230 | 465 | 70 | |
| LC1446B-37 | DQ270629 | Methylobacter | 279 | 449 | 70 | |
| LC1133B-5 | DQ270612 | Methylobacter | 282 | 453 | 168 | |
| LC1133B-61 | DQ270619 | Methylobacter | 214 | 451 | 70 | |
| LC1133B-86 | DQ270620 | Methylobacter | 161 | 451 | 70 | |
| LC1231B-184 | DQ228565 | Methylobacter | 215 | 452 | 70 | |
| LC1408B-19 | DQ270606 | Methylophaga | 182 | 451 | 352 | |
| LC1446B-88 | DQ270631 | Methylophaga | 282 | 453 | 352 | |
| LC1231B-11 | DQ228564 | Methylophaga | 208 | 426 | 26 | |
| LC1443B-4 | DQ270657 | Nitrospira | 357* | 408* | 520^ | |
| LC1228B-113 | DQ270652 | Nitrospira | 355 | 406 | 518 | |
| LC1404B-28 | DQ270656 | Nitrospira | 357* | 408* | 520^ | |
| LC1446B-146 | DQ270662 | Planctomycetes | 247 | 88 | 74 | |
| LC1446B-77 | DQ270660 | Planctomycetes | 217 | 150 | 84 | |
| LC1231B-20 | DQ228576 | Planctomycetes | 173 | 103 | 20 | |
| LC1231B-166 | DQ228578 | Planctomycetes | 220 | 438 | 217 | |
| LC1231B-5 | DQ228577 | Planctomycetes | 245 | 134 | 112 | |
| LC1228B-91 | DQ270626 | Seep/hydrate | 152 | 455 | 26 | |
| LC1133B-18 | DQ270614 | Seep/hydrate | 216 | 104 | 72 | |
| LC1446B-28 | DQ270659 | Subsurface firm | 297 | 157 | 40 | |
| LC1133B-75 | DQ270647 | Subsurface firm | 297 | 157 | 40 | |
| LC1133B-108 | DQ270649 | Subsurface firm | 171 | 151 | 211 | |
| LC1446B-39 | DQ270630 | Thiomicrospira | 85 | 355 | 89 | |
| LC1537B-12 | DQ270608 | Thiomicrospira | 184 | 455 | 72 | |
| LC1408B-58 | DQ270607 | Thiomicrospira | 220 | 457 | 481 | |
| LC1537B-49 | DQ270609 | Thiomicrospira | 184 | 454 | 170 | |
| LC1133B-148 | DQ270624 | Thiomicrospira | 186 | 432 | 74 | |
| LC1133B-127 | DQ270622 | Thiomicrospira | 185 | 455 | 73 | |
| LC1149B-99 | DQ228561 | Thiomicrospira | 184 | 455 | 72 | |
| LC1022B-40 | DQ228560 | Thiomicrospira | 297 | 255 | 86 | |
| LC1537B-81 | DQ270610 | Vibrio | 174 | 464 | 181 | |
See reference 65. The predicted terminal restriction fragment size for each restriction enzyme is generated by identifying the first restriction site for each enzyme 3′ to the fluorescently tagged forward primer. Fragment sizes were experimentally verified within ±2 bp (*) or ±4 bp (^). The absence of a restriction site present in the portion of the clone that was sequenced is indicated by “ND.” Unc. Eury., uncultured Euryarchaeota; Unc. Cren, uncultured Crenarchaeota; CFB, Cytophaga-Flavobacterium-Bacteroides.
RESULTS
Archaeal 16S rRNA.
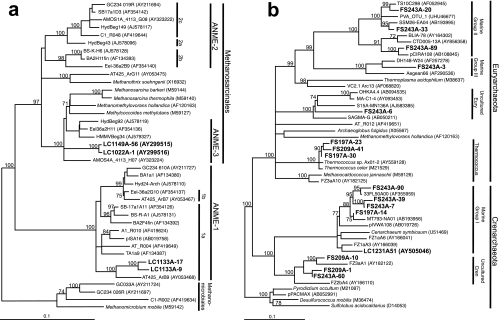
As reported previously (38, 65), samples of carbonate chimneys from the LCHF exhibited remarkably low archaeal diversity (Table 2). Phylogenetic analyses of 16S rRNA genes revealed a single phylotype each of Lost City Methanosarcinales (LCMS) and ANME-1 (clones LC1022A-1, LC1149A-56, LC1133A-9, LC1133A-17 in Fig. 3a and Table 3). The latter clones are most similar to the ANME-1a subgroup, which includes clones collected from a variety of CH4 seeps, hydrothermal sediments, and Black Sea carbonate reefs (23, 39, 40, 48, 55, 71, 72). LCMS has 93% similarity to a cluster of sequences that are derived from CH4 seep sediments and a mud volcano in the Barents Sea (20, 39, 47, 55).
FIG. 3.
Maximum-likelihood phylogenetic trees depicting the relationships of archaeal 16S rRNA gene sequences from LCHF carbonate chimneys to other ANME-related sequences. LC1022A-1 and LC1149A-56 (65) were cloned from carbonate mineral samples from active chimneys (a). LC1133A-9 and LC1133A-17 were cloned from sample X1, a chimney with very little venting fluid. Fluid samples (b) represent a mixture of hydrothermal fluid and ambient seawater (see Table 1 and the text for details). Methanomethylovorans hollandica is included in b as a representative of the Methanosarcinales. Lost City hydrothermal field environmental clones are in boldface type. Phylogenetic groups used in the T-RFLP analysis are labeled. Trees are unrooted, and scale bars represent 1 nucleotide change per 10 bases. GenBank accession numbers are in parentheses.
The entire suite of samples (Table 1) was assayed for TRFs corresponding to the 16S rRNA gene clones (Table 3). Actively venting, high-temperature chimneys such as those at markers 2, 3, and H were dominated by LCMS, while less-active carbonate veins hosted in serpentinite (markers X1 and X2) contained ANME-1 but not LCMS. A TRF of very low relative peak height corresponding to LCMS, however, was identified in the marker X1 sample. Peaks with a low relative height corresponding to LCMS were also identified in all fluid samples, likely due to the entrainment of portions of the densely populated LCMS biofilms into venting fluid. T-RFLP profiles for all three samples collected from marker C, which appears to consist of both young and old carbonate minerals (see Discussion), displayed peaks for both LCMS and ANME-1 TRFs (Fig. 2). Digests with alternative restriction enzymes (HaeIII and MspI) also yielded additional peaks but none with relative peak heights comparable to those of the peaks corresponding to LCMS or ANME-1. Although T-RFLP analysis is not a quantitative measurement and is susceptible to PCR biases, the order of magnitude difference in peak heights between the LCMS and ANME-1 peaks and those of all other peaks is consistent with the high percentage of LCMS previously found in phylogenetic and fluorescence in situ hybridization studies at this site (38, 65).
A somewhat wider diversity of archaeal 16S rRNA gene sequences was recovered from hydrothermal fluid samples (Fig. 3b). Many of the clones exhibited significant similarity to a variety of uncultured organisms belonging to the marine groups I, II, and III of the Archaea; these clones were not identified in any chimney samples by T-RFLP analysis, indicating that they are probably derived from the ambient seawater. Clones FS209A-1, FS243A-60, and FS209A-10, however, clustered with a novel group of uncharacterized Crenarchaeota that were identified within the walls of a high-temperature sulfide chimney (66). In addition, three sequences belonging to the Thermococcales (clones FS197A-23, FS197A-30, and FS209A-41) were identified in three fluid samples (from markers H, 7, and 8). These clones have extremely high sequence similarity (>99%) to an isolate from the Axial Volcano (28).
Bacterial 16S rRNA genes.
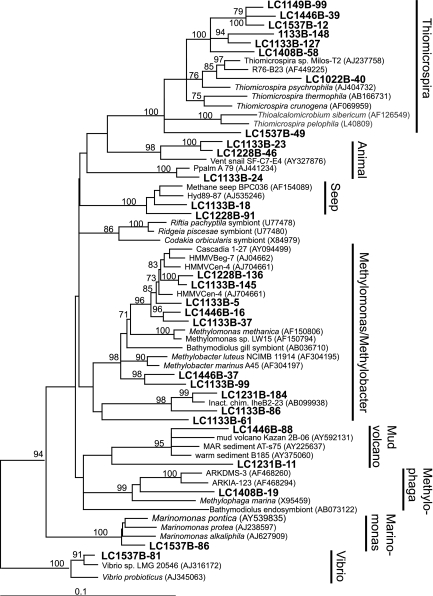
A high diversity of bacterial 16S rRNA gene sequences was recovered from LCHF carbonate chimney samples spanning many phyla (Fig. 4 and 5). The most diverse group was the gamma-proteobacteria (Fig. 4), which contained clones belonging to several methylotrophic genera including Methylobacter, Methylomonas, and Methylophaga. A number of clones formed a new clade within the genus Thiomicrospira (94% similar to Thiomicrospira sp. strain Milos-T2), a group of sulfur-oxidizing organisms commonly associated with hydrothermal vents. TRFs corresponding to clones from the Methylobacter/Methylomonas, Methylophaga, and Thiomicrospira clades (Table 3) were found in all carbonate chimney and fluid samples analyzed in this study (Table 1 and Fig. 5). The two clones representing the CH4 seep/hydrate group (clones LC1133B-18 and LC1228B-91) (Table 3 and Fig. 4) were present in four of the six fluid samples analyzed but in only one chimney sample.
FIG. 4.
Maximum-likelihood phylogenetic tree of gamma-proteobacterial 16S rRNA genes cloned from LCHF carbonate chimneys. Lost City hydrothermal field environmental clones are in boldface type. Labeled phylogenetic groups correspond to the groups used in the T-RFLP analysis (Fig. 6). Trees are unrooted, and scale bars represent 1 nucleotide change per 10 bases. GenBank accession numbers are in parentheses.
FIG. 5.
Maximum-likelihood phylogenetic tree of bacterial 16S rRNA genes cloned from LCHF carbonate chimneys similar to (a) Firmicutes and (b) epsilon-proteobacteria and their relatives. Lost City hydrothermal field environmental clones are in boldface type. Labeled phylogenetic groups correspond to the groups used in T-RFLP analysis (Fig. 6). Trees are unrooted, and scale bars represent 1 nucleotide change per 10 bases. GenBank accession numbers are in parentheses.
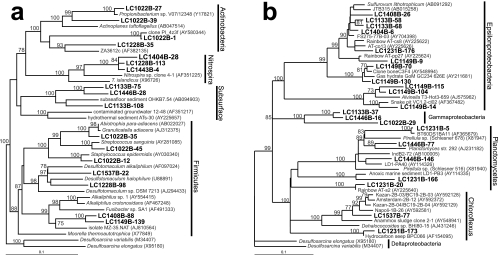
There was an unusually large diversity of Firmicutes in the carbonate chimneys (Fig. 5). Clones LC1537B-22 and LC1228B-98 have 88 to 92% sequence similarity to Desulfotomaculum halophilus and Desulfotomaculum alkaliphilus; many members of the Desulfotomaculum genus are known to be sulfate reducers (31) and are found in high-temperature, high-pH environments (61). In contrast to other AMO environments, there is a notable lack of delta-proteobacterial sequences in our Lost City clone libraries. The LCHF Desulfotomaculum clones, the only candidate sulfate-reducing Bacteria identified, were not detected in the ANME-1-dominated sample from marker X1 (Fig. 6), casting doubt on its ability to be the syntrophic partner in anaerobic methane oxidation.
Clones LC1133B-75, LC1446B-28, and LC1133B-108 consistently grouped apart from other sequences from Firmicutes spp. with both neighbor-joining (data not shown) and maximum-likelihood phylogenetic trees (Fig. 5), even though the three previously published sequences with which they group (88 to 89% sequence similarity among all six clones) have been described as low-GC-content, gram-positive Bacteria (43). Since all three previously published clones have been recovered from sediment or subsurface sources (43), these clones are tentatively identified as a “subsurface” clade in Fig. 3b. TRFs corresponding to clones from the “subsurface” clade were positively identified in three fluid samples (markers 3, H, and C) using the restriction enzyme HaeIII, but the predicted TRFs of these clones for MspI and BstuI either were outside the 50- to 500-bp analysis window of this study or did not match any environmental TRFs within 2 bp. Therefore, the uncertain presence of the “subsurface” clade in these fluid samples is indicated by the question marks in Fig. 6.
A number of clones group within the epsilon-proteobacteria (Fig. 5) and were identified in samples of carbonate chimneys and fluids of various temperatures and chemical characteristics (Fig. 5). The only cultured isolate with significant similarity to these clones is Sulfurovum lithotrophicum (94% sequence similarity to clones LC1133B-58 and LC1133B-68), a sulfur-oxidizing chemolithoautroph from hydrothermal sediments in a black smoker environment (33). Clone LC1537B-77 groups with environmental clones from three different mud volcanoes in the eastern Mediterranean (90 to 92% sequence similarity) (Fig. 5) (S. K. Heijs et al., unpublished data).
DISCUSSION
Archaea associated with active and inactive carbonate structures.
Phylogenetic and T-RFLP analyses of the LCHF show that the diversity of archaeal 16S rRNA genes is dominated by LCMS in sites of active venting and by ANME-1 in sites that are only weakly venting or inactive (Fig. 2 and 6). A temperature-controlled zonation of ANME communities at the Lost City is consistent with in vitro studies (53), which found that the temperature optimum for AMO was the biggest physiological difference between ANME-1 and ANME-2 communities.
The only exceptions to the segregation of LCMS and ANME-1 are data from the marker C samples, where both phylotypes were present. The underside of the flange at marker C hosts a shallow pool of 70°C hydrothermal fluid. Analyses of material from the flange interior showed that it is porous and comprised of carbonate minerals and brucite. Since brucite dissolves in seawater, it is likely that the porous interior of the flange was dominated by hydrothermal fluid. In contrast, the outer portion of the flange is well lithified, gray, and bathed in seawater (Fig. 2). Carbonate minerals sampled for T-RFLP analyses likely contained material from both the exterior and interior of the flange. Based on known environmental conditions at other sampling sites (markers 2, 3, X1, and X2), we infer that LCMS sequences were derived from the interior and that ANME-1 sequences were derived from the exterior of the flange at marker C.
The presence of LCMS in warm, actively venting chimneys and the presence ANME-1 in cold, inactive or very weakly venting structures suggest that ecological succession occurs in the microbial community as carbonate chimneys cool down, become less active, and are progressively bathed in seawater. Succession is also consistent with the increased number of bacterial OTUs (Table 2) identified in markers C, X1, and X2 compared to those at presumably younger chimneys. Preliminary age dating of a few carbonate samples, coupled with modeling, shows that hydrothermal activity at the Lost City may be remarkably stable and long lived, in contrast to the transient environments that typify most black smoker environments (35). Carbon isotopic analyses showed that venting has been ongoing for at least 30,000 years, with localized sites active for at least 50 to 300 years, and modeling results indicate that venting may last for 100,000 to up to 1 million years (17).
An important, unresolved question regarding LCMS and ANME-1 organisms at the LCHF is whether they are methanogens or methanotrophs. Knittel et al. (39) designated a cluster of sequences containing LCMS as “ANME-3,” which could indicate that these organisms are also involved in AMO. However, this seems unlikely at the LCHF, because the very high H2 concentrations (1 to 15 mM) should strongly favor methanogenesis. In addition, archaeal lipids collected from carbonate chimney samples at this site have a much higher δ13C level (−8.5‰ to 4.8‰ versus Pee Dee Belemnite) (A. S. Bradley, J. M. Hayes, and R. E. Summons, Eos Trans. AGU Fall Meet. Suppl. 85[47], abstr. B33B-0263, 2004) than would be predicted for organisms involved in AMO, whose lipids can have δ13C values of −85‰ or lower (38, 57). The abundant H2 and enriched 13C content of lipids must be reconciled, however, with the lack of dissolved inorganic carbon (DIC) available to putative methanogens living in an environment dominated by serpentinization-derived fluids. Unlike other H2- and CH4-rich subsurface environments where the δ13C values of CH4 are significantly more depleted than those of the source carbon and thus indicate a strong biological source (50, 70), the δ13C content of CH4 (−8.8‰ to −13.6‰) at the LCHF is similar to that of fluid DIC (−2‰ to −8‰), chimney carbonate minerals (+13‰ to −7‰), and chimney total organic carbon (−3.1‰ to −18.1‰) (38). The absence of 13C-depleted CH4 and archaeal lipids could be a result of carbon limitation due to high pH and an apparent absence of magmatic CO2 effectively eliminating available DIC. Because DIC is limiting and because CH4 has multiple possible sources (methanogens and serpentinization-associated reactions) and sinks (anaerobic and aerobic methanotrophy), stable isotope ratio measurements in this system are difficult to interpret. Mixing models of abiotic and biologically derived CH4 (42) may allow a more intricate understanding of the carbon budget of the LCHF. Nevertheless, it is interesting that the most 13C-depleted organic carbon from the LCHF is derived from archaeal lipids recovered from a sample at marker X2, which is dominated by ANME-1 (A. Bradley, personal communication). Therefore, while the stable isotope data are enigmatic, it is broadly consistent with a temperature-associated zonation of methanogenesis and AMO.
Archaea in hydrothermal fluid samples.
The microbial populations present in the fluid samples reflect the widespread mixing of seawater with serpentinization-derived fluids that occurs at the LCHF: 16S rRNA gene sequences likely originating from seawater (marine groups I, II, and III) as well as likely hydrothermal activity-associated organisms (novel Crenarchaeota and Thermococcus species) were recovered (Fig. 3). Three Crenarchaeota clones (Fig. 3b) have high sequence similarity (90 to 94%) to sequences collected from within the walls of a 300°C sulfide chimney from a hydrothermal vent on the Juan de Fuca Ridge (66). All LCHF clones similar to high-temperature Crenarchaeota species were recovered only from hydrothermal fluid samples (Fig. 6); none were detected in carbonate chimney samples. These relationships may reflect the entrainment of Crenarchaeota into upflowing fluids within large, open channels that did not come into direct contact with the carbonate chimneys. The presence of Thermococcales sequences has been used as a bioindicator of hot subseafloor habitats (25, 26, 28, 69). Their presence in moderate-temperature fluids (∼50°C) within the LCHF may reflect the presence of a hot subseafloor microbial habitat in the shallow mantle rocks beneath the field. This is consistent with modeling calculations that indicate temperatures of 150 to 250°C at depth (2).
Sulfur-cycling Bacteria.
Although LCHF fluids contain dissolved H2 concentrations that are several orders of magnitude higher than those present in other “hydrogen-based” ecosystems (11, 67), none of the H2-oxidizing Bacteria (Aquificales and delta-proteobacteria) found at those sites were identified in this study. Instead, the LCHF bacterial community appears to be dominated by aerobic or microaerophilic organisms that use reduced sulfur species as electron donors. All chimney and fluid samples analyzed in this study contained 16S rRNA gene sequences belonging to Thiomicrospira species (Fig. 6), known as chemolithoautotrophic sulfur and sulfide oxidizers in hydrothermal vent environments (9, 63). Thiomicrospira-like organisms from microbial mats have been shown to mediate the sequential oxidation of sulfide to sulfur and sulfate (54), which could obscure the chemical and isotopic signature of any sulfate-reducing Bacteria in the system. The epsilon-proteobacteria identified in this study (Fig. 5) are also putative sulfur-oxidizing chemolithoautotrophs, evidenced by their strong sequence similarity to Sulfurovum lithotrophicum. Building an integrated view of the LCHF ecosystem will require resolving the relationship, in terms of carbon and energy flow, between these ubiquitous sulfur-cycling Bacteria and the microbial mats of CH4-cycling Archaea within the chimneys.
In all known instances of AMO reported to date, Archaea partner with SRB to form an electron-shuttling couple that makes this process more metabolically feasible (8, 24, 57, 72, 75). At the LCHF, sequences belonging to Desulfotomaculum were recovered, but no clones exhibited any significant sequence similarity to organisms previously reported to be associated with AMO. Some species of Desulfotomaculum are known to be alkaliphilic (58) and have the dsrAB gene encoding dissimilatory sulfite reductase, the enzyme necessary for sulfate reduction (75, 77). The LCHF Desulfotomaculum-like clones (LC1537B-22 and LC1228B-98), however, were identified in high-temperature chimneys lacking ANME-1 and were not identified at marker X1, where ANME-1 dominates. Attempts to amplify the dsrAB gene from carbonate chimney samples have been unsuccessful (unpublished observations). However, the mcrA gene, encoding methyl coenzyme M reductase, which is present in all known methanogens and anaerobic methanotrophs, has been amplified and sequenced from the same samples (38). These phylogenetic data, therefore, are inconsistent with Desulfotomaculum playing a role in AMO. Although sulfate reducers belonging to the delta-proteobacteria have been found in all AMO-associated environments, microscopic observations indicate that a close association between sulfate reducers and methanotrophic Archaea, especially the ANME-1 group, is not necessary (56). Therefore, it is possible that sulfate reducers within the LCHF, if they exist, are only loosely associated with ANME-1.
On the basis of previous studies (5, 32, 43), clones LC1133B-75, LC1446B-28, and LC1133B-108 were expected to group among the other Firmicutes sequences identified in this study. However, the three clones repeatedly formed their own distinct clade, despite attempts to produce one monophyletic clade of Firmicutes by editing multiple sequence alignments, jumbling the order of sequence addition to the alignment, adding and subtracting outgroups, and using both neighbor-joining and maximum-likelihood treeing algorithms. The T-RFLP profiles from this study are ambiguous regarding the distribution of these three clones, but the tentative identification of their TRFs in three different fluid samples but in no chimney samples suggests that they are more prevalent in hydrothermal fluids than in carbonate chimneys. This possibility, coupled with the fact that previously described clones with high sequence similarity to these clones were recovered from sediments and subsurface sources, may indicate that these Firmicutes-like organisms represent another member of the subsurface community at the LCHF.
Biogeochemical cycling.
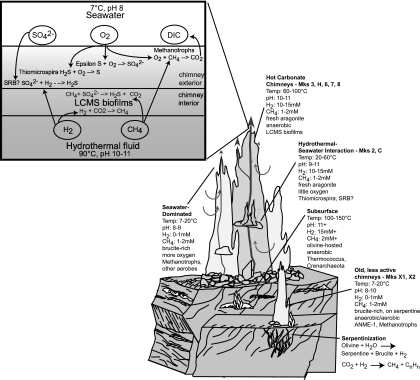
Clones with high sequence similarity to methanotrophic Bacteria were identified in six carbonate chimney samples and six hydrothermal fluid samples included in the T-RFLP survey (Fig. 6). The co-occurrence of CH4-oxidizing Bacteria with obligately anaerobic methanogens and anaerobic methane-oxidizing Archaea raises questions about microscale redox and temperature gradients present in this system. The highly porous chimney structures are likely to favor anaerobic niches within areas dominated by hydrothermal fluids but that are in close physical proximity to the seawater-influenced exterior of the chimney. Indeed, Schrenk et al. (65) have previously described distinct microbial communities inhabiting the inner and outer portions of LCHF carbonate chimneys. Figure 7 depicts a biogeochemical model of carbon and sulfur cycling in the carbonate chimneys. The system is characterized by the interaction of H2- and CH4-rich hydrothermal fluid with oxygenated seawater. Biofilms of LCMS are restricted to high-temperature anoxic zones within the carbonate matrix that are dominated by high-pH fluids, which lack measurable CO2. The biofilms may promote the maintenance of microscale anoxic niches by trapping dissolved H2 and CH4 for use as metabolic substrates and excluding dissolved O2 (68). Where highly reducing hydrothermal fluid meets seawater laden with abundant electron acceptors, a diverse bacterial community flourishes. As carbonate chimneys age, cool down, and are progressively bathed in fluids less enriched with H2, ANME-1 displaces LCMS as the dominant archaeal population, while the aerobic methanotrophic and sulfur-oxidizing bacterial community remains.
FIG. 7.
Model of biogeochemical zonation at the Lost City hydrothermal field. Exothermic reactions in the serpentinization reaction zone produce H2, methane, and other hydrocarbons to drive the ecosystem. Hyperthermophilic Thermococcales and Crenarchaeota inhabit the subsurface where both temperature and pH could be very high. LCMS biofilms, consuming the abundant H2 and/or methane present in hydrothermal fluids, coat the surface of fresh aragonite precipitated at actively venting carbonate chimneys. Electron acceptors are introduced via seawater circulating through the highly porous chimney structures, allowing aerobic sulfide oxidizers and methylotrophs to inhabit the exterior portions of the chimneys. As chimneys age and cool down, they vent less H2-dominated hydrothermal fluid but retain enough methane to support anaerobic methanotrophic Archaea (ANME-1), which replace LCMS as the dominant phylotype.
Many of the putative CH4- and sulfur-cycling organisms identified in this study have close phylogenetic relationships to organisms found at CH4 seeps and mud volcanoes. Although overlain by sediment, many mud volcanoes are also influenced by serpentinization reactions in the subsurface and are significant sources of CH4 (13). In addition to the CH4- and sulfur-cycling organisms discussed above, other clones in carbonate chimney and fluid samples represent uncultured lineages with unknown physiologies. Clone LC1537B-77, for example, shares significant sequence similarity with clones recovered from three different mud volcanoes in the eastern Mediterranean (78). Clones LC1231B-173, LC1149B-70, LC1149B-130, LC1446B-88, and LC1231B-11 also have high sequence similarity to clones recovered from CH4 seeps and mud volcanoes, and all represent uncultured lineages. Recently, a low-diversity community of anaerobic methanotrophic Archaea and diverse bacterial assemblages in terrestrial mud volcanoes were discovered in the Carpathian Mountains (1). There was also a report of an H2- and CH4-rich deep borehole dominated by methanogens and sulfate-reducing Desulfotomaculum (50). Thus, it appears that CH4 produced by abiotic processes has selected for similar microbial communities in geologically diverse sites. The LCHF represents the first hydrothermal chimney environment where such an assemblage of CH4-cycling organisms has been discovered.
With abundant dissolved H2 as an energy source and high concentrations of CH4 as energy and carbon sources, a geochemically complex ecosystem that includes methanogens, aerobic and anaerobic methanotrophs, sulfate reducers, and sulfur oxidizers is thriving at the LCHF. Further biochemical and genetic investigations are necessary to determine whether LCMS is a methanogen or methanotroph and to resolve the issues of very limited DIC concentrations at the LCHF and the ambiguous evidence for syntrophic SRB associated with LCMS biofilms. Considering the widespread global distribution of ultramafic subsurface environments (16) and the possibility of their existence on other planetary bodies such as Mars (18, 22), exploration of serpentinite-hosted ecosystems has the potential to yield profound discoveries with implications for understanding the linkages between abiotic water-rock reactions and microbial evolution.
Acknowledgments
We express our gratitude and appreciation to the crews of the R/V Atlantis and DSV Alvin and the scientific party of the 2003 Lost City Expedition. We also thank Sheryl Bolton, Craig Moyer, and Minhui Lin for technical assistance and Kristin Ludwig for helpful discussions.
This research was supported by the National Science Foundation (OCE0137206), the Washington Sea Grant (NA76RG0119), and the NASA Astrobiology Institute through the Carnegie Geophysical Institute.
REFERENCES
- 1.Alain, K., T. Holler, F. Musat, M. Elvert, T. Treude, and M. Kruger. 2005. Microbiological investigation of methane- and hydrocarbon-discharging mud volcanoes in the Carpathian Mountains, Romania. Environ. Microbiol. 8:574-590. [DOI] [PubMed] [Google Scholar]
- 2.Allen, D. E., and W. E. Seyfried. 2004. Serpentinization and heat generation: constraints from Lost City and rainbow hydrothermal systems. Geochim. Cosmochim. Acta 68:1347-1354. [Google Scholar]
- 3.Aloisi, G., I. Bouloubassi, S. K. Heijs, R. D. Pancost, C. Pierre, J. S. S. Damste, J. C. Gottschal, L. J. Forney, and J. M. Rouchy. 2002. CH4-consuming microorganisms and the formation of carbonate crusts at cold seeps. Earth Planet. Sci. Lett. 203:195-203. [Google Scholar]
- 4.Bach, W., N. R. Banerjee, H. J. B. Dick, and E. T. Baker. 2002. Discovery of ancient and active hydrothermal systems along the ultra-slow spreading Southwest Indian Ridge 10 degrees-16 degrees E. Geochem. Geophys. Geosyst. [Online.] doi: 10.1029/2001GC000279. [DOI]
- 5.Bakermans, C., and E. L. Madsen. 2002. Diversity of 16S rDNA and naphthalene dioxygenase genes from coal-tar-waste-contaminated aquifer waters. Microb. Ecol. 44:95-106. [DOI] [PubMed] [Google Scholar]
- 6.Barns, S. M., R. E. Fundyga, M. W. Jeffries, and N. R. Pace. 1994. Remarkable archaeal diversity detected in a Yellowstone-National-Park hot-spring environment. Proc. Natl. Acad. Sci. USA 91:1609-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berndt, M. E., D. E. Allen, and W. E. Seyfried. 1996. Reduction of CO2 during serpentinization of olivine at 300 degrees C and 500 bar. Geology 24:351-354. [Google Scholar]
- 8.Boetius, A., K. Ravenschlag, C. J. Schubert, D. Rickert, F. Widdel, A. Gieseke, R. Amann, B. B. Jorgensen, U. Witte, and O. Pfannkuche. 2000. A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature 407:623-626. [DOI] [PubMed] [Google Scholar]
- 9.Brinkhoff, T., and G. Muyzer. 1997. Increased species diversity and extended habitat range of sulfur-oxidizing Thiomicrospira spp. Appl. Environ. Microbiol. 63:3789-3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Butterfield, D. A., K. K. Roe, M. D. Lilley, J. A. Huber, J. A. Baross, R. W. Embley, and G. J. Massoth. 2004. Mixing, reaction, and microbial activity in the sub-seafloor revealed by temporal and spatial variation in diffuse flow vents at Axial Volcano. Am. Geophys. Union Monogr. 144:269-289. [Google Scholar]
- 11.Chapelle, F. H., K. O'Neill, P. M. Bradley, B. A. Methe, S. A. Ciufo, L. L. Knobel, and D. R. Lovley. 2002. A hydrogen-based subsurface microbial community dominated by methanogens. Nature 415:312-315. [DOI] [PubMed] [Google Scholar]
- 12.Dick, H. J. B., J. Lin, and H. Schouten. 2003. An ultraslow-spreading class of ocean ridge. Nature 426:405-412. [DOI] [PubMed] [Google Scholar]
- 13.Dimitrov, L. I. 2002. Mud volcanoes—the most important pathway for degassing deeply buried sediments. Earth Sci. Rev. 59:49-76. [Google Scholar]
- 14.Edmonds, H. N., P. J. Michael, E. T. Baker, D. P. Connelly, J. E. Snow, C. H. Langmuir, H. J. B. Dick, R. Muhe, C. R. German, and D. W. Graham. 2003. Discovery of abundant hydrothermal venting on the ultraslow-spreading Gakkel ridge in the Arctic. Nature 421:252-256. [DOI] [PubMed] [Google Scholar]
- 15.Egert, M., and M. W. Friedrich. 2003. Formation of pseudo-terminal restriction fragments, a PCR-related bias affecting terminal restriction fragment length polymorphism analysis of microbial community structure. Appl. Environ. Microbiol. 69:2555-2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fruh-Green, G. L., J. A. D. Connolly, A. Plas, D. S. Kelley, and B. Grobety. 2004. Serpentinization of oceanic peridotites: implications for geochemical cycles and biological activity. Am. Geophys. Union Monogr. 144:119-136. [Google Scholar]
- 17.Fruh-Green, G. L., D. S. Kelley, S. M. Bernasconi, J. A. Karson, K. A. Ludwig, D. A. Butterfield, C. Boschi, and G. Proskurowski. 2003. 30,000 years of hydrothermal activity at the Lost City vent field. Science 301:495-498. [DOI] [PubMed] [Google Scholar]
- 18.Gellert, R., R. Rieder, R. C. Anderson, J. Bruckner, B. C. Clark, G. Dreibus, T. Economou, G. Klingelhofer, G. W. Lugmair, D. W. Ming, S. W. Squyres, C. d'Uston, H. Wanke, A. Yen, and J. Zipfel. 2004. Chemistry of rocks and soils in Gusev crater from the alpha particle X-ray spectrometer. Science 305:829-832. [DOI] [PubMed] [Google Scholar]
- 19.Girguis, P. R., A. E. Cozen, and E. F. DeLong. 2005. Growth and population dynamics of anaerobic methane-oxidizing archaea and sulfate-reducing bacteria in a continuous-flow bioreactor. Appl. Environ. Microbiol. 71:3725-3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Girguis, P. R., V. J. Orphan, S. J. Hallam, and E. F. DeLong. 2003. Growth and methane oxidation rates of anaerobic methanotrophic archaea in a continuous-flow bioreactor. Appl. Environ. Microbiol. 69:5472-5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hallam, S. J., N. Putnam, C. M. Preston, J. C. Detter, D. Rokhsar, P. M. Richardson, and E. F. DeLong. 2004. Reverse methanogenesis: testing the hypothesis with environmental genomics. Science 305:1457-1462. [DOI] [PubMed] [Google Scholar]
- 22.Hamilton, V. E., and P. R. Christensen. 2005. Evidence for extensive, olivine-rich bedrock on Mars. Geology 33:433-436. [Google Scholar]
- 23.Hinrichs, K. U., J. M. Hayes, S. P. Sylva, P. G. Brewer, and E. F. DeLong. 1999. Methane-consuming archaebacteria in marine sediments. Nature 398:802-805. [DOI] [PubMed] [Google Scholar]
- 24.Hoehler, T. M., M. J. Alperin, D. B. Albert, and C. S. Martens. 1994. Field and laboratory studies of methane oxidation in an anoxic marine sediment—evidence for a methanogen-sulfate reducer consortium. Global Biogeochem. Cycles 8:451-463. [Google Scholar]
- 25.Holden, J. F., M. Summit, and J. A. Baross. 1998. Thermophilic and hyperthermophilic microorganisms in 3-30°C hydrothermal fluids following a deep-sea volcanic eruption. FEMS Microbiol. Ecol. 25:33-41. [Google Scholar]
- 26.Holland, M. E., J. A. Baross, and J. F. Holden. 2004. Illuminating subseafloor ecosystems using microbial tracers. Am. Geophys. Union Monogr. 144:291-303. [Google Scholar]
- 27.Huber, J. A., D. A. Butterfield, and J. A. Baross. 2002. Temporal changes in archaeal diversity and chemistry in a mid-ocean ridge subseafloor habitat. Appl. Environ. Microbiol. 68:1585-1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huber, J. A., D. A. Butterfield, and J. A. Baross. Diversity and distribution of subseafloor Thermococcales populations in diffuse hydrothermal vents at an active deep-sea volcano in the northeast Pacific Ocean. J. Geophys. Res. Biogeosci., in press.
- 29.Huber, J. A., H. P. Johnson, D. A. Butterfield, and J. A. Baross. 2006. Microbial life in ridge flank crustal fluids. Environ. Microbiol. 8:88-99. [DOI] [PubMed] [Google Scholar]
- 30.Huber, T., G. Faulkner, and P. Hugenholtz. 2004. Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20:2317-2319. [DOI] [PubMed] [Google Scholar]
- 31.Imachi, H., Y. Sekiguchi, Y. Kamagata, A. Loy, Y.-L. Qiu, P. Hugenholtz, N. Kimura, M. Wagner, A. Ohashi, and H. Harada. 2006. Non-sulfate-reducing, syntrophic bacteria affiliated with Desulfotomaculum cluster I are widely distributed in methanogenic environments. Appl. Environ. Microbiol. 72:2080-2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Inagaki, F., M. Suzuki, K. Takai, H. Oida, T. Sakamoto, K. Aoki, K. H. Nealson, and K. Horikoshi. 2003. Microbial communities associated with geological horizons in coastal subseafloor sediments from the Sea of Okhotsk. Appl. Environ. Microbiol. 69:7224-7235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Inagaki, F., K. Takai, K. H. Nealson, and K. Horikoshi. 2004. Sulfurovum lithotrophicum gen. nov., sp. nov., a novel sulfur-oxidizing chemolithoautotroph within the ɛ-Proteobacteria isolated from Okinawa Trough hydrothermal sediments. Int. J. Syst. Evol. Microbiol. 54:1477-1482. [DOI] [PubMed] [Google Scholar]
- 34.Kallmeyer, J., and A. Boetius. 2004. Effects of temperature and pressure on sulfate reduction and anaerobic oxidation of methane in hydrothermal sediments of Guaymas Basin. Appl. Environ. Microbiol. 70:1231-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kelley, D. S., J. A. Baross, and J. R. Delaney. 2002. Volcanoes, fluids, and life at mid-ocean ridge spreading centers. Annu. Rev. Earth Planet. Sci. 30:385-491. [Google Scholar]
- 36.Kelley, D. S., and G. L. Fruh-Green. 1999. Abiogenic methane in deep-seated mid-ocean ridge environments: insights from stable isotope analyses. J. Geophys. Res. Solid Earth 104:10439-10460. [Google Scholar]
- 37.Kelley, D. S., J. A. Karson, D. K. Blackman, G. L. Fruh-Green, D. A. Butterfield, M. D. Lilley, E. J. Olson, M. O. Schrenk, K. K. Roe, G. T. Lebon, P. Rivizzigno, et al. 2001. An off-axis hydrothermal vent field near the Mid-Atlantic Ridge at 30° N. Nature 412:145-149. [DOI] [PubMed] [Google Scholar]
- 38.Kelley, D. S., J. A. Karson, G. L. Fruh-Green, D. R. Yoerger, T. M. Shank, D. A. Butterfield, J. M. Hayes, M. O. Schrenk, E. J. Olson, G. Proskurowski, M. Jakuba, A. Bradley, B. Larson, K. Ludwig, D. Glickson, K. Buckman, A. S. Bradley, W. J. Brazelton, K. Roe, M. J. Elend, A. Delacour, S. M. Bernasconi, M. D. Lilley, J. A. Baross, R. T. Summons, and S. P. Sylva. 2005. A serpentinite-hosted ecosystem: the Lost City hydrothermal field. Science 307:1428-1434. [DOI] [PubMed] [Google Scholar]
- 39.Knittel, K., T. Losekann, A. Boetius, R. Kort, and R. Amann. 2005. Diversity and distribution of methanotrophic archaea at cold seeps. Appl. Environ. Microbiol. 71:467-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lanoil, B. D., R. Sassen, M. T. La Duc, S. T. Sweet, and K. H. Nealson. 2001. Bacteria and Archaea physically associated with Gulf of Mexico gas hydrates. Appl. Environ. Microbiol. 67:5143-5153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lehours, A. C., C. Bardot, A. Thenot, D. Debroas, and G. Fonty. 2005. Anaerobic microbial communities in Lake Pavin, a unique meromictic lake in France. Appl. Environ. Microbiol. 71:7389-7400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lollar, B. S., G. Lacrampe-Couloume, G. F. Slater, J. Ward, D. P. Moser, T. M. Gihring, L.-H. Lin, and T. C. Onstott. 2006. Unravelling abiogenic and biogenic sources of methane in the Earth's deep subsurface. Chem. Geol. 226:328-339. [Google Scholar]
- 43.Lopez-Garcia, P., S. Duperron, P. Philippot, J. Foriel, J. Susini, and D. Moreira. 2003. Bacterial diversity in hydrothermal sediment and epsilon proteobacterial dominance in experimental microcolonizers at the Mid-Atlantic Ridge. Environ. Microbiol. 5:961-976. [DOI] [PubMed] [Google Scholar]
- 44.McCollom, T. M., and J. S. Seewald. 2001. A reassessment of the potential for reduction of dissolved CO2 to hydrocarbons during serpentinization of olivine. Geochim. Cosmochim. Acta 65:3769-3778. [Google Scholar]
- 45.Meyerdierks, A., M. Kube, T. Lombardot, K. Knittel, M. Bauer, F. O. Glockner, R. Reinhardt, and R. Amann. 2005. Insights into the genomes of archaea mediating the anaerobic oxidation of methane. Environ. Microbiol. 7:1937-1951. [DOI] [PubMed] [Google Scholar]
- 46.Michaelis, W., R. Seifert, K. Nauhaus, T. Treude, V. Thiel, M. Blumenberg, K. Knittel, A. Gieseke, K. Peterknecht, T. Pape, A. Boetius, R. Amann, B. B. Jorgensen, F. Widdel, J. R. Peckmann, N. V. Pimenov, and M. B. Gulin. 2002. Microbial reefs in the Black Sea fueled by anaerobic oxidation of methane. Science 297:1013-1015. [DOI] [PubMed] [Google Scholar]
- 47.Milkov, A. V., P. R. Vogt, K. Crane, A. Y. Lein, R. Sassen, and G. A. Cherkashev. 2004. Geological, geochemical, and microbial processes at the hydrate-bearing Hakon Mosby mud volcano: a review. Chem. Geol. 205:347-366. [Google Scholar]
- 48.Mills, H. J., C. Hodges, K. Wilson, I. R. MacDonald, and P. A. Sobecky. 2003. Microbial diversity in sediments associated with surface-breaching gas hydrate mounds in the Gulf of Mexico. FEMS Microbiol. Ecol. 46:39-52. [DOI] [PubMed] [Google Scholar]
- 49.Moeseneder, M. M., C. Winter, J. M. Arrieta, and G. J. Herndl. 2001. Terminal-restriction fragment length polymorphism (T-RFLP) screening of a marine archaeal clone library to determine the different phylotypes. J. Microbiol. Methods 44:159-172. [DOI] [PubMed] [Google Scholar]
- 50.Moser, D. P., T. M. Gihring, F. J. Brockman, J. K. Fredrickson, D. L. Balkwill, M. E. Dollhopf, B. S. Lollar, L. M. Pratt, E. Boice, G. Southam, G. Wanger, B. J. Baker, S. M. Pfiffner, L.-H. Lin, and T. C. Onstott. 2005. Desulfotomaculum and Methanobacterium spp. dominate a 4- to 5-kilometer-deep fault. Appl. Environ. Microbiol. 71:8773-8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moyer, C. L., J. M. Tiedje, F. C. Dobbs, and D. M. Karl. 1998. Diversity of deep-sea hydrothermal vent Archaea from Loihi seamount, Hawaii. Deep-Sea Res. II 45:303-317. [Google Scholar]
- 52.Nauhaus, K., A. Boetius, M. Kruger, and F. Widdel. 2002. In vitro demonstration of anaerobic oxidation of methane coupled to sulphate reduction in sediment from a marine gas hydrate area. Environ. Microbiol. 4:296-305. [DOI] [PubMed] [Google Scholar]
- 53.Nauhaus, K., T. Treude, A. Boetius, and M. Kruger. 2005. Environmental regulation of the anaerobic oxidation of methane: a comparison of ANME-I and ANME-II communities. Environ. Microbiol. 7:98-106. [DOI] [PubMed] [Google Scholar]
- 54.Okabe, S., T. Ito, K. Sugita, and H. Satoh. 2005. Succession of internal sulfur cycles and sulfur-oxidizing bacterial communities in microaerophilic wastewater biofilms. Appl. Environ. Microbiol. 71:2520-2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Orphan, V. J., K. U. Hinrichs, W. Ussler III, C. K. Paull, L. T. Taylor, S. P. Sylva, J. M. Hayes, and E. F. DeLong. 2001. Comparative analysis of methane-oxidizing archaea and sulfate-reducing bacteria in anoxic marine sediments. Appl. Environ. Microbiol. 67:1922-1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Orphan, V. J., C. H. House, K. U. Hinrichs, K. D. McKeegan, and E. F. DeLong. 2002. Direct phylogenetic and isotopic evidence for multiple groups of Archaea involved in the anaerobic oxidation of methane. Geochim. Cosmochim. Acta 66:A571-A571. [Google Scholar]
- 57.Orphan, V. J., C. H. House, K. U. Hinrichs, K. D. McKeegan, and E. F. DeLong. 2001. Methane-consuming Archaea revealed by directly coupled isotopic and phylogenetic analysis. Science 293:484-487. [DOI] [PubMed] [Google Scholar]
- 58.Pikuta, E., A. Lysenko, N. Suzina, G. Osipov, B. Kuznetsov, T. Tourova, V. Akimenko, and K. Laurinavichius. 2000. Desulfotomaculum alkaliphilum sp. nov., a new alkaliphilic, moderately thermophilic, sulfate-reducing bacterium. Int. J. Syst. Evol. Microbiol. 50:25-33. [DOI] [PubMed] [Google Scholar]
- 59.Porter, K. G., and Y. S. Feig. 1980. The use of DAPI for identifying and counting aquatic microflora. Limnol. Oceanogr. 25:943-948. [Google Scholar]
- 60.Proskurowski, G., M. D. Lilley, D. S. Kelley, and E. J. Olson. 2006. Low temperature volatile production at the Lost City hydrothermal field, evidence from a hydrogen stable isotope geothermometer. Chem. Geol. 229:331-343. [Google Scholar]
- 61.Reysenbach, A.-L., D. Gotz, and D. Yernool. 2002. Microbial diversity of marine and terrestrial thermal springs, p. 345-421. In J. T. Staley and A.-L. Reysenbach (ed.), Biodiversity of microbial life. Wiley-Liss, Inc., New York, N.Y.
- 62.Rosch, C., and H. Bothe. 2005. Improved assessment of denitrifying, N2-fixing, and total-community bacteria by terminal restriction fragment length polymorphism analysis using multiple restriction enzymes. Appl. Environ. Microbiol. 71:2026-2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ruby, E. G., C. O. Wirsen, and H. W. Jannasch. 1981. Chemolithotrophic sulfur-oxidizing bacteria from the Galapagos Rift hydrothermal vents. Appl. Environ. Microbiol. 42:317-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schouten, S., S. G. Wakeham, E. C. Hopmans, and J. S. S. Damste. 2003. Biogeochemical evidence that thermophilic archaea mediate the anaerobic oxidation of methane. Appl. Environ. Microbiol. 69:1680-1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schrenk, M. O., D. S. Kelley, S. A. Bolton, and J. A. Baross. 2004. Low archaeal diversity linked to subseafloor geochemical processes at the Lost City hydrothermal field, Mid-Atlantic Ridge. Environ. Microbiol. 6:1086-1095. [DOI] [PubMed] [Google Scholar]
- 66.Schrenk, M. O., D. S. Kelley, J. R. Delaney, and J. A. Baross. 2003. Incidence and diversity of microorganisms within the walls of an active deep-sea sulfide chimney. Appl. Environ. Microbiol. 69:3580-3592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Spear, J. R., J. J. Walker, T. M. McCollom, and N. R. Pace. 2005. Hydrogen and bioenergetics in the Yellowstone geothermal ecosystem. Proc. Natl. Acad. Sci. USA 102:2555-2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stewart, P. S. 2003. Diffusion in biofilms. J. Bacteriol. 185:1485-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Summit, M., and J. A. Baross. 2001. A novel microbial habitat in the mid-ocean ridge subseafloor. Proc. Natl. Acad. Sci. USA 98:2158-2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Takai, K., T. Gamo, U. Tsunogai, N. Nakayama, H. Hirayama, K. H. Nealson, and K. Horikoshi. 2004. Geochemical and microbiological evidence for a hydrogen-based, hyperthermophilic subsurface lithoautotrophic microbial ecosystem (HyperSLiME) beneath an active deep-sea hydrothermal field. Extremophiles 8:269-282. [DOI] [PubMed] [Google Scholar]
- 71.Takai, K., and K. Horikoshi. 1999. Genetic diversity of Archaea in deep-sea hydrothermal vent environments. Genetics 152:1285-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Teske, A., K. U. Hinrichs, V. Edgcomb, A. D. Gomez, D. Kysela, S. P. Sylva, M. L. Sogin, and H. W. Jannasch. 2002. Microbial diversity of hydrothermal sediments in the Guaymas Basin: evidence for anaerobic methanotrophic communities. Appl. Environ. Microbiol. 68:1994-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Thompson, J. R., L. A. Marcelino, and M. F. Polz. 2002. Heteroduplexes in mixed-template amplifications: formation, consequence and elimination by ‘reconditioning PCR. ’ Nucleic Acids Res. 30:2083-2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Thomsen, T. R., K. Finster, and N. B. Ramsing. 2001. Biogeochemical and molecular signatures of anaerobic methane oxidation in a marine sediment. Appl. Environ. Microbiol. 67:1646-1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Torres, M. E., J. McManus, D. E. Hammond, M. A. de Angelis, K. U. Heeschen, S. L. Colbert, M. D. Tryon, K. M. Brown, and E. Suess. 2002. Fluid and chemical fluxes in and out of sediments hosting methane hydrate deposits on Hydrate Ridge, OR. I. Hydrological provinces. Earth Planet. Sci. Lett. 201:525-540. [Google Scholar]
- 77.Wagner, M., A. J. Roger, J. L. Flax, G. A. Brusseau, and D. A. Stahl. 1998. Phylogeny of dissimilatory sulfite reductases supports an early origin of sulfate respiration. J. Bacteriol. 180:2975-2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Werne, J. P., R. R. Haese, T. Zitter, G. Aloisi, L. Bouloubassi, S. Heijs, A. Fiala-Medioni, R. D. Pancost, J. S. S. Damste, G. de Lange, L. J. Forney, J. C. Gottschal, J. P. Foucher, J. Mascle, J. Woodside, et al. 2004. Life at cold seeps: a synthesis of biogeochemical and ecological data from Kazan mud volcano, eastern Mediterranean Sea. Chem. Geol. 205:367-390. [Google Scholar]