Abstract
Bacteria growing in the human large intestine live in intimate association with the host and play an important role in host digestive processes, gut physiology, and metabolism. Fecal bacteria have been investigated extensively, but few studies have been done on biofilms that form on digestive wastes in the large bowel. The aims of this investigation were to investigate the composition and metabolic activities of bacterial communities that colonize the surfaces of food residues in fecal material, with respect to their role in the fermentation of complex carbohydrates. Fresh stools were obtained from 15 healthy donors, and food residues were separated by filtration. Adherent bacteria were removed by surfactant treatment for microbiological analysis and fermentation studies. Scanning electron microscopy and fluorescent in situ hybridization in conjunction with confocal laser scanning microscopy (CLSM) were used to visualize intact biofilms. Results showed that bacterial populations strongly adhering to particulate matter were phenotypically similar in composition to unattached communities, with bacteroides and bifidobacteria predominating. Biofilms comprised a mixture of living and dead bacteria, and CLSM showed that bifidobacteria in the biofilms occurred as isolated dispersed cells and in microcolonies near the interface with the substratum. Fermentation experiments with a variety of complex carbohydrates demonstrated that biofilm populations were more efficient in digesting polysaccharides, while nonadhering communities fermented oligosaccharides most rapidly. Acetate was the principal fermentation product formed by biofilm bacteria, whereas higher levels of butyrate were produced by nonadherent populations, showing that the two communities were metabolically distinct.
The human colon is a highly specialized digestive organ that contains large numbers of bacteria that exist in stable complex communities. Bacteria are a major component of colonic material, comprising approximately 55% of solids in persons living on western-style diets (40), and the average daily fecal output is approximately 120 g (10). Many hundreds of different species and strains have been identified in intestinal contents (26) as well as on mucosal surfaces lining the bowel (30). The metabolic activities of the microbiota affect host physiology in a multiplicity of ways; through fermentation, for example, bacteria in the hindgut complete the digestive process and contribute to energy reclamation and host metabolism (11). The colonic epithelium has an obligate requirement for bacterial fermentation products (11), while the maintenance of colonization resistance to microbial pathogens (19), the activation or destruction of genotoxins and mutagens (33), and modulation of the immune system (39) are all dependent on the microbiota. Although host tissues and other endogenous substrates such as mucins, pancreatic enzymes, and other secretions are broken down and recycled by intestinal bacteria, the species composition and biochemical activities of the microbiota are determined primarily by diet and are strongly influenced by carbohydrate availability (26).
The large intestine is an open system in the sense that digestive materials from the small gut enter at one end and feces are periodically excreted at the other. Due to the anatomy of the large bowel and the mechanics of movement of particulate substances through the gut, bacteria that are able to colonize food residues in the cecum and maintain significant populations in the proximal bowel serve as inocula for new digestive materials entering the colon. These organisms may therefore be of particular ecological importance in maintaining the stability of the colonic microbiota. Little is known about how the colonization of particulate substances occurs in the large intestine, but it is likely that the organisms involved in the initial stages of this process form biofilms. Bacteria growing in these structures often behave differently from their nonadherent counterparts, and in particular, the nature and efficiency of their metabolism are changed, while many species exhibit greater resistance to antibiotics and other inhibitory factors that have deleterious effects on planktonic bacteria (3, 35, 42).
Microbial biofilms are ubiquitous, and they have been investigated in a variety of natural environments including sediments, soils, the oral cavity, and skin as well as in the gastrointestinal tracts of animals. However, while there is increasing interest in mucosal biofilms in the human colon (22, 37), particularly with respect to their role in disease processes (30, 41), study of the composition and ecological significance of these phenomena in the gut has generally been neglected. As a consequence, we know little about their ecology or physiological significance. The aims of this study, therefore, were to investigate the composition and activities of bacterial communities that colonize the surfaces of food residues in the gut lumen, with respect to their role in the breakdown of complex carbohydrates.
MATERIALS AND METHODS
Desorption of bacteria from food particles in feces.
Fresh stools were obtained from 15 healthy donors (eight males and seven females; age range, 23 to 54 years). The donors had not taken antibiotics for several months before the study commenced. Stools were homogenized in anaerobic sodium phosphate buffer (0.1 M, pH 6.5) to give 10% (wt/vol) slurries, which were passed sequentially through 500- and 250-μm-diameter sieves to separate large food particles. All samples were weighed and processed within 1 h to minimize the loss of cell viability. Filtrates containing nonattached bacteria were maintained under anaerobic conditions at 18°C for fermentation studies and bacteriological analyses. Material retained on the filters was washed twice with 500 ml of the anaerobic buffer to remove loosely adherent organisms. Washed particles were subsequently incubated for 30 min at 37°C under anaerobic conditions (O2-free N2 atmosphere) in phosphate buffer in the presence of 0.001% (wt/vol) cetyltrimethylammonium bromide (CTAB; BDH Ltd., Dagenham, United Kingdom) with mixing. Samples were then refiltered to separate food particles. Supernatants containing desorbed adherent bacteria were retained for further study.
Enumeration and identification of bacterial populations.
Suspensions (1.0 ml) of nonadherent bacteria and organisms desorbed from particulate material by CTAB were vortex mixed with sterile prereduced half-strength Wilkins-Chalgren anaerobe broth (9.0 ml) to form a 10-fold dilution series (10−1 to 10−9). Samples (0.1 ml) from the tubes (10−3 to 10−9) were then spread in triplicate onto a range of prereduced agar plates in an anaerobic chamber (atmosphere containing 10% H2, 10% CO2, and 80% CO2). Plates used for the isolation of aerobic and facultatively anaerobic organisms were as follows: nutrient agar for total aerobes and facultative anaerobes, MacConkey agar no. 2 for lactose fermenting and non-lactose-fermenting enterobacteria and enterococci, and azide blood agar base for facultatively anaerobic cocci. Isolation media used for strict anaerobes were Wilkins-Chalgren agar (WCA) for total anaerobes, anaerobic cocci, and clostridia; WCA with the addition of nonsporing anaerobe supplements (to prevent the growth of spore-forming species); WCA with gram-negative anaerobe supplements, which were selective for gram-negative organisms; Rogosa agar for lactobacilli; and Perfringens agar plus antibiotic supplements for Clostridium perfringens. Bacteria belonging to the Bacteroides fragilis group were isolated and enumerated using Bacteroides mineral salts agar (28), and bifidobacteria were counted using Beerens agar (4). Medium 10 (6) and YCFAG medium (13) were used for the isolation and cultivation of Faecalibacterium prausnitzii. Plates for aerobic incubation were removed from the anaerobic chamber and incubated at 37°C. Aerobic plates were incubated for 2 days and anaerobic plates were incubated for up to 5 days, with periodic examination, before colonies were counted. The bacteria were identified using a variety of techniques, including Gram stain, cell morphology, fermentation product analysis (see below), biochemical reactions in API (20A and 32A) tests (BioMerieux, Marcy l' Etoile, France), analysis of cellular fatty acid methyl esters (FAME), and 16S rRNA gene sequence analysis. FAME were extracted from bacterial pellets obtained from approximately 40 ml of culture in anaerobic peptone-yeast extract broth supplemented with glucose (10 g/liter) by saponification, methylation, and extraction as described previously (34). FAME were separated using a model 6890A microbial identification system (Microbial ID Inc., Newark, Del.), which consisted of a Hewlett-Packard (Palo Alto, CA) model 6890 gas chromatograph fitted with a 5% phenyl-methyl silicone capillary column (0.2 mm by 25 m), a flame ionization detector, a Hewlett-Packard model 7637A automatic sampler, and a Hewlett-Packard Vectra XM computer. The gas chromatography parameters were as follows: carrier gas, ultra-high-purity H2; column head pressure, 60 kPa; injection volume, 2 μl; column split ratio, 100:1; septum purge, 5 ml/min; column temperature, 170 to 270°C; injection port temperature, 300°C. Peaks were automatically integrated, and fatty acid names and percentages were calculated. Numerical analyses and predictions for bacterial identification were done using standard MIS library generation software. The system was calibrated using a standard MIDI FAME calibration mix before each operation and validated using the type strains Escherichia coli ATCC 11775, Bacteroides fragilis ATCC 25285, and Clostridium perfringens ATCC 13124. Yeast cells were identified using the API 20 C AUX biochemical identification system (bioMerieux, Basingstoke, England).
16S rRNA gene sequence analysis was used to confirm the identities of the isolates that could not be identified reliably by FAME analysis. DNA was extracted, purified, and amplified using universal primers (36) as described previously (21). Direct sequencing of the amplified DNA fragments was done using an automated ABI 3100 Genetic Analyzer capillary sequencer (Applied Biosystems, Foster City, CA).16S rRNA gene sequences were compared with all sequences in GenBank by using the BLAST algorithm (1).
SEM.
Samples of digestive residues for scanning electron microscopy (SEM) were placed in 3% (vol/vol) glutaraldehyde in PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)] buffer (100 mM, pH 7.4). The samples were then fixed with 4% (wt/vol) aqueous OsO4 and dehydrated stepwise in ethanol, with three changes (10 min) in each of 50%, 75%, 95%, and, finally, 100% ethanol. Samples were subsequently dried on a Poleron E 5000 critical-point drier, placed onto stubs, and gold coated to a depth of 30 nm. A Phillips XL 30 FEG scanning electron microscope was used to visualize the preparations.
Viability staining of bacteria growing in biofilms.
Food residues containing adherent bacteria were covered with 200 μl BacLight viability stain, comprised of 1.5 μl SYTO 9 and 1.5 μl propidium iodide, in 1 ml anaerobic distilled H2O (Molecular Probes Europe BV, Leiden, The Netherlands). Samples were then placed into an anaerobic chamber, in the dark, for 10 min to allow the stain to develop. Scans were taken using a Nikon Eclipse E800 upright microscope attached to a Nikon PCM 2000 CLSM system with a 488-nm argon laser (green fluorescence indicates live cells) and a 543-nm helium-neon laser (red fluorescence indicates dead cells). A 60× Plan Apo immersion lens with a numerical aperture of 1.4 was used for visualizing the bacteria, and images were captured and overlaid using C-Imaging software (Compix Inc., Cranberry Township, PA).
Oligonucleotide probes.
The range of fluorescent 16S rRNA gene oligonucleotide probes used in this study, which targeted all of the major populations of gut bacteria, has been described previously (7). In particular, probe Bif164 (5′-CATCCGGCATTACCACCC-3′) was used to identify bifidobacteria (24), probe Ent1 (5′-CCGCTTGCTCTCGCGAG-3′) was used for enterobacteria (25), probe Bac 303 (5′-CCAATGTGGGGGACCTT-3′) was used for Bacteroides and Prevotella (32), probe Erec 482 (5′-GCTTCTTAGTCAGGTACCG-3′) was used for the Eubacterium rectale-Clostridium coccoides group (17), and the universal eubacterial probe Eub338 (5′-GCTGCCTCCCGTAGGAGT-3′) was used for total eubacteria (2). Intestinal isolates and a range of culture collection type strains were used as controls for testing the specificities of the oligonucleotide probes (16). The organisms were cultured in Wilkins-Chalgren broth in an anaerobic chamber at 37°C. The bacteria were then fixed in fresh 4% paraformaldehyde, washed in phosphate-buffered saline (PBS), and stored in 50% (vol/vol) PBS-ethanol at −20°C (2). The probes were synthesized by Thermohybaid, Interactiva Division (Ulm, Germany), and 5′ labeled with the fluorochrome Cy3, Cy5, or fluorescein isothiocyanate (FITC).
Fluorescent in situ hybridization (FISH).
Food residues were fixed in 3 ml of 4% paraformaldehyde in PBS (pH 7.0) for 16 h at 4°C, gently washed with PBS (10 min) at room temperature, covered in 4 ml 50% (vol/vol) PBS-ethanol, and stored at 4°C until hybridization on the same day. Samples were then immersed in 4 ml of hybridization buffer (0.9 mol/liter NaCl, 20 mM Tris-HCl, 0.01% sodium dodecyl sulfate, pH 8.0) at room temperature for 10 min, removed, and covered with 100 μl hybridization buffer containing either FITC-, Cy3-, or Cy5-labeled probes at concentrations of 50 ng/μl, 30 ng/μl, and 30 ng/μl, respectively.
Hybridization was done at 45 or 50°C with the addition of formamide, depending on the type of probe (7), in a humid chamber for 4 h. Hybridization at 48°C was found to give optimum conditions for simultaneous visualization of the probes on the food residues. Unbound probe was subsequently removed with 5 ml wash buffer (0.9 mol/liter NaCl, 20 mM Tris-HCl), and the food residues were placed in 50 ml wash buffer for 20 min.
Food residues were gently rinsed with distilled H2O and mounted onto Teflon-coated eight-well glass slides (VWR; Merck Eurolab Ltd., Poole, United Kingdom). Citifluor medium (Citifluor Ltd., London, United Kingdom) was used as a mounting medium, and the slides were visualized as described above for viability staining. The three-dimensional characteristics of the biofilms were studied using the Nikon PCM 2000 CLSM system as described above. Images were captured and overlaid using C-Imaging software (Compix Inc., Cranberry Township, PA). Individual scans of 0.5 μm through the biofilms were combined using Autovisualize 5.5 3D visualization software (Autoquant Imaging Inc., Troy, NY). Images were taken from different volunteers.
Fermentation studies of strongly adherent and nonadherent bacteria.
Ten milliliters of desorbed adherent bacteria and nonadherent fecal bacteria was incubated in an orbital shaker at 37°C under O2-free N2 gas in sodium phosphate buffer (0.1 M, pH 6.5) in sealed 70-ml serum bottles (Wheaton) with 40 ml of either of the following substrates (20-g/liter final concentration): pectin (citrus), Lintner's starch (BDH), xylan (oatspelt), arabinogalactan (larchwood), porcine gastric mucin (type III; Sigma), fructo-oligosaccharides (Orafti, Tienen, Belgium), xylo-oligosaccharides (XOS; Suntory, Japan), soya-oligosaccharides (SOS; Calpis, Japan), and galacto-oligosaccharides (GOS; Yakult, Japan). The structure and composition of the oligosaccharides are shown in Table 1. Samples (2 ml) were taken periodically over a 6-h time course and frozen (−20°C) for subsequent analysis of fermentation products.
TABLE 1.
Structure and composition of oligosaccharides used in the studya
| Substrate | Chemical compositionb | Purity |
|---|---|---|
| Fructo-oligosaccharides (Raftilose P95) | β(2-1)-fructan; 60% Gfn; 40% Fn (d.p., 2-8; avg, 4-5) | 87% oligosaccharides |
| Xylo-oligosaccharides | β(1-4)-linked xylose; d.p. of oligosaccharide fraction, 2-4 | 29% oligosaccharides |
| Soya-oligosaccharides | Stachyose (F, Gal, Gal, Glu) and raffinose (F, Gal, Glu); d.p., 3-4 | 25% stachyose, 10% raffinose, 50% sucrose, 15% monosaccharides |
| Galacto-oligosaccharides | Small amounts of G, Gal, and Lac | 85% oligosaccharides |
F, fructose; G, glucose; Gal, galactose; Lac, lactose.
d.p., degree of polymerization.
SCFA measurements.
Short-chain fatty acid (SCFA) samples were determined by gas chromatography as follows. Culture samples (1 ml) were acidified with 50 μl 50% (vol/vol) H2SO4. One hundred microliters of internal standard (tert-butylacetate) was added to give a concentration of 30 mM. After mixing, the free fatty acids were extracted into ether and dried using anhydrous CaCl2. SCFA were separated using Unicam 10% FFAP, 100/120-mesh Chromosorb WAW-DMCS in a 1.8-m by 2-mm (internal diameter) glass column. Injector, detector, and column temperatures were 200, 300, and 155°C, respectively. Flow rates of the N2 carrier gas, H2, and air were set at 50, 30, and 370 ml/min, respectively. All samples were quantitated by comparison of sample peak heights with those of authentic standards. Lactate and succinate were measured using the same analytical system and operating conditions after methylation of samples and extraction into chloroform (20).
Dry weight measurements.
Dry weight measurements were done as described previously (12).
Statistical analysis.
Statistical analyses were performed using the Graphpad Prism 4 Statistics Package for Macintosh (Graphpad Software, Inc., San Diego, CA). Data relating to the biofilm and nonadherent populations were found to be not normally distributed and were compared by the Mann-Whitney U test. Probability values of <0.05 were considered statistically significant.
Chemicals.
Bacteriological supplies were obtained from Oxoid Ltd. (Basingstoke, Hamps, United Kingdom). Unless otherwise stated, all other chemicals were purchased from Sigma Chemical Co. (Poole, Dorset, United Kingdom).
RESULTS
Desorption of intestinal bacteria from food residues in feces.
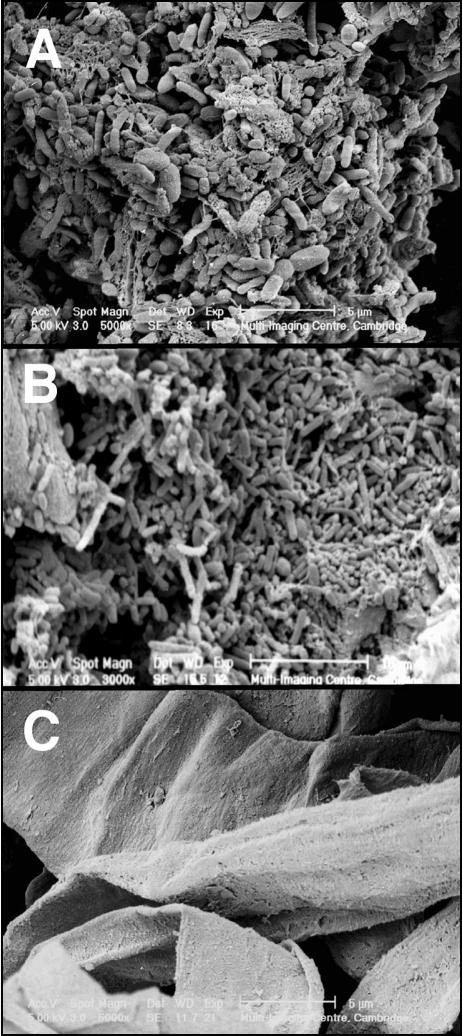
Approximately 5% of the bacterial cell mass in the lumen of the large intestine was found to be strongly adherent to the surface of food particles, although a considerably higher percentage was more loosely attached. A variety of bacterial morphologies was visualized in these biofilms using SEM (Fig. 1A). Washing with buffer removed part of the biofilm (Fig. 1B); however, surfactant treatment with CTAB was required for the complete desorption of bacteria from the surfaces of food particles (Fig. 1C).
FIG. 1.
SEM micrographs of food particles. (A) Intestinal bacteria growing on the surface before washing. (B) Bacteria remaining after several washes with buffer. (C) Complete removal of adherent bacteria following surfactant treatment with 0.001% CTAB.
Bacteriological analysis of biofilm communities.
Identification of bacteria growing on the surfaces of food particles in the large gut showed that the predominant biofilm communities were structurally similar to nonadherent populations (Table 2) and that there were no significant differences between them. Strict anaerobes belonging to the genera Bacteroides (seven species) and Bifidobacterium (six species) were the principal culturable anaerobes. Bacteroides thetaiotaomicron and Bacteroides vulgatus were the most prevalent bacteroides, and these species were also present in the highest numbers. Bacteroides caccae occurred in the biofilms but not in the nonadherent population in four people. Bifidobacterium adolescentis and Bifidobacterium angulatum were the most prevalent bifidobacteria; however, Bifidobacterium bifidum, when present, occurred in higher numbers. A variety of other anaerobes, including eubacteria, clostridia, peptostreptococci, actinomyces, faecalibacteria, and colinsella were present in biofilms; however, species such as Eubacteria rectale, C. perfringens, and Clostridium innocuum were not detected. Numbers of Bifidobacterium angulatum were higher than those in the nonadherent population, but this was not significant. Escherichia coli and Enterococcus faecalis were the predominant facultative anaerobes and occurred in cell population densities that were similar to those of their nonadherent counterparts; however, microorganisms considered to be nonautochthonous in the healthy gut, such as Bacillus subtilis, Staphylococcus aureus, and Candida albicans, were not found in the biofilms.
TABLE 2.
Predominant culturable anaerobes and facultative anaerobes isolated from particulate material in feces obtained from 15 healthy donors
| Organism (no. of positive subjects) | Biofilm bacteria
|
Nonadherent bacteria
|
||||
|---|---|---|---|---|---|---|
| No. of isolates | Log10 CFU/g (wet wt)
|
No. of isolates | Log10 CFU/g (wet wt)
|
|||
| Mean ± SEMa | Rangeb | Mean ± SEMa | Rangeb | |||
| Anaerobes | ||||||
| Bacteroides thetaiotaomicron (8)c | 8 | 5.1 ± 1.29 | 9.2-10.0 | 8 | 5.2 ± 1.31 | 9.3-10.3 |
| Bacteroides splanchnicus (6) | 6 | 3.5 ± 1.15 | 8.3-9.3 | 6 | 3.6 ± 1.18 | 8.7-9.3 |
| Bacteroides ovatus (7) | 7 | 4.1 ± 1.16 | 8.4-9.0 | 7 | 4.2 ± 1.2 | 8.4-9.6 |
| Bacteroides fragilis (5) | 5 | 3.0 ± 1.13 | 8.5-9.5 | 5 | 3.2 ± 1.2 | 9.4-10.0 |
| Bacteroides caccae (4) | 4 | 2.3 ± 1.0 | 8.3-9.7 | NDd | ||
| Bacteroides vulgatus (9) | 9 | 5.5 ± 1.2 | 8.6-9.8 | 9 | 5.2 ± 1.31 | 9.1-10.5 |
| Bacteroides distasonis (6) | 7 | 3.4 ± 1.1 | 7.5-9.3 | 7 | 3.4 ± 1.31 | 7.8-9.4 |
| Bifidobacterium adolescentis (9) | 10 | 5.6 ± 1.24 | 8.2-10.6 | 10 | 5.4 ± 1.19 | 8.2-9.8 |
| Bifidobacterium angulatum (8) | 8 | 5.0 ± 1.26 | 8.6-10.2 | 8 | 4.4 ± 1.1 | 7.6-8.8 |
| Bifidobacterium bifidum (6) | 6 | 4.0 ± 1.32 | 9.9-10.3 | 6 | 4.3 ± 1.1 | 10.4-11.0 |
| Bifidobacterium pseudolongum (5) | 6 | 3.3 ± 1.26 | 9.6-10.4 | 6 | 3.4 ± 1.27 | 9.8-10.4 |
| Bifidobacterium longum (7) | 7 | 4.5 ± 1.29 | 8.7-10.5 | 7 | 4.6 ± 1.31 | 9.1-10.5 |
| Bifidobacterium pseudocatenulatum (4) | 4 | 2.4 ± 1.07 | 8.2-9.8 | 4 | 2.3 ± 1.0 | 7.9-9.1 |
| Eubacterium limosume (6) | 6 | 3.3 ± 1.16 | 7.4-9.0 | 6 | 3.1 ± 1.02 | 7.5-8.1 |
| Eubacterium rectalee (5) | ND | 5 | 3.4 ± 1.29 | 9.7-10.7 | ||
| Unclassified eubacteriae (6) | 6 | 3.6 ± 1.17 | 8.2-9.6 | 6 | 3.8 ± 1.26 | 9.0-10.2 |
| Lactobacillus plantarumc (4) | 4 | 2.0 ± 0.89 | 7.1-7.9 | 4 | 2.2 ± 0.97 | 8.0-8.4 |
| Lactobacillus acidophilus (6) | 6 | 3.3 ± 1.09 | 8.0-8.6 | 6 | 3.6 ± 1.18 | 8.5-9.5 |
| Lactobacillus paracasei (5) | 5 | 2.8 ± 1.06 | 8.2-8.6 | 5 | 2.7 ± 1.02 | 7.8-8.4 |
| Unclassified lactobacillus type 1e (4) | 4 | 2.5 ± 1.1 | 8.5-10.1 | 4 | 2.8 ± 1.22 | 9.5-11.3 |
| Unclassified lactobacillus type 2e (2) | 2 | 1.0 ± 0.70 | 6.9-8.5 | 2 | 1.0 ± 0.66 | 6.9-7.7 |
| Peptostreptococcus asaccharolyticus (4) | 4 | 2.0 ± 0.88 | 7.0-7.8 | 4 | 2.5 ± 1.12 | 9.0-10.0 |
| Peptostreptococcus anaerobius (5) | 5 | 2.9 ± 1.14 | 7.9-9.7 | 5 | 3.2 ± 1.22 | 8.8-10.2 |
| Unclassified peptostreptococcus type 1e (3) | 3 | 1.8 ± 0.97 | 8.2-9.8 | 3 | 1.9 ± 1.0 | 8.8-10.0 |
| Unclassified peptostreptococcus type 2e (2) | 2 | 1.0 ± 0.68 | 6.9-8.1 | 2 | 1.0 ± 0.71 | 7.5-8.1 |
| Clostridium innocuum (2) | ND | 2 | 1.0 ± 0.67 | 6.7-7.9 | ||
| Clostridium clostridioforme (7) | 7 | 4.0 ± 1.13 | 8.1-8.9 | 7 | 4.2 ± 1.19 | 8.2-9.6 |
| Clostridium malenominatum (4) | 4 | 2.0 ± 0.92 | 7.0-8.0 | 4 | 1.1 ± 0.98 | 8.0-8.6 |
| Clostridium perfringens (5) | 5 | ND | 7.7-8.5 | 5 | 2.5 ± 0.95 | 7.1-7.9 |
| Unclassified clostridium type 1e (3) | 3 | 1.6 ± 0.87 | 7.0-8.2 | 3 | 1.8 ± 0.94 | 8.5-9.1 |
| Unclassified clostridium type 2e (2) | 2 | 1.0 ± 0.69 | 2 | 1.0 ± 0.66 | 6.8-7.6 | |
| Actinomyces spp. (2) | 2 | 1.3 ± 0.87 | 9.4-9.8 | 2 | 1.3 ± 0.89 | 9.5-10.1 |
| Faecalibacterium prausnitziie (5) | 5 | 3.1 ± 1.2 | 7.6-11.2 | 5 | 3.2 ± 1.28 | 8.2-10.6 |
| Colinsella aerofaciensense (6) | 6 | 3.1 ± 1.05 | 6.6-9.0 | 6 | 3.3 ± 1.11 | 6.8-9.8 |
| Aerobes and facultative anaerobes | ||||||
| Escherichia coli (9) | 11 | 5.8 ± 1.1 | 8.2-9.3 | 11 | 5.3 ± 1.17 | 8.6-9.2 |
| Klebsiella pneumoniae (3) | ND | 3 | 1.0 ± 0.56 | 4.5-5.9 | ||
| Enterococcus faecalis (8) | 9 | 4.1 ± 1.05 | 5.7-9.5 | 9 | 4.2 ± 1.06 | 7.1-8.7 |
| Bacillus subtilis (2) | ND | 2 | 1.0 ± 0.66 | 7.2-7.4 | ||
| Staphylococcus aureus (3) | ND | 3 | 1.3 ± 0.68 | 6.2-6.6 | ||
| Candida albicans (3) | ND | 3 | 1.7 ± 0.88 | 7.7-8.9 | ||
Results are expressed as the population mean log10 CFU/gram (wet weight) ± standard error of the mean for each group.
Results show the range of log10 CFU/gram (wet weight) of feces from subjects in which the bacteria were isolated.
Values in parentheses show the number of subjects positive for carriage of the bacteria.
ND, not detected.
Isolates were identified by BLAST search; all other bacteria were identified by FAME analysis, and yeasts were identified by API 20C AUX.
Visualization of biofilms using FISH.
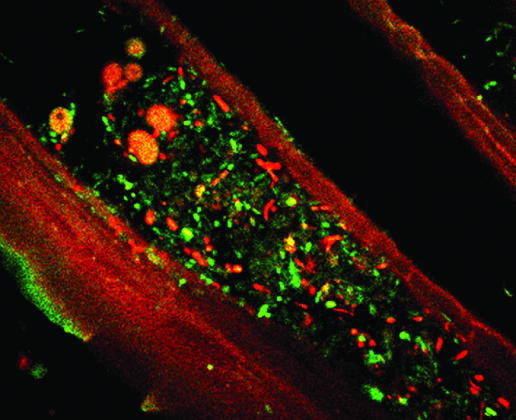
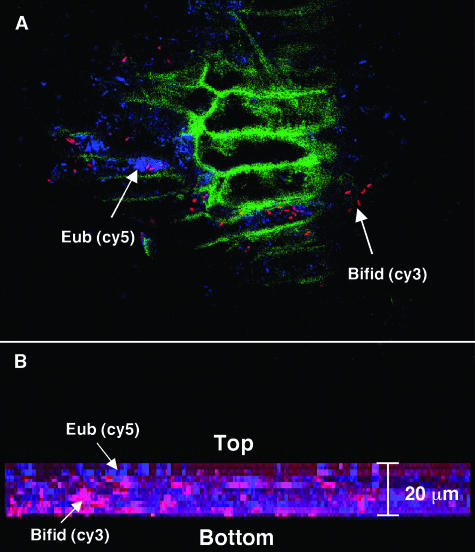
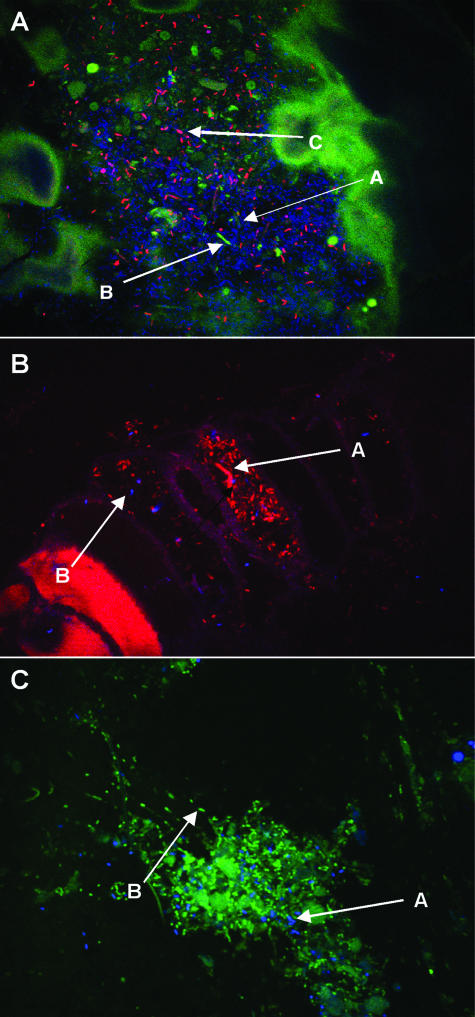
Figures 2 to 4 show representative biofilm populations from different healthy donors. In Fig. 2, the biofilm covering the surface of the food particle was comprised of a mixture of living and dead bacteria, while Fig. 3A shows a dense biofilm stained with a mixture of Cy3-, Cy5-, and FITC-labeled oligonucleotide probes specific for bifidobacteria, total eubacteria, and enterobacteria, respectively. Very few enterobacteria can be seen; however, the biofilm contained large numbers of dispersed bifidobacteria. In Fig. 3B, the food material was stained with a Cy3-labeled group-specific probe to detect the presence of bacteria belonging to the Eubacterium rectale-Clostridium coccoides group and a Cy5-labeled probe to detect the presence of bifidobacteria. Large numbers of bacteria belonging to members of the Eubacterium rectale-C. coccoides group can be seen in segments of the food material along with smaller numbers of bifidobacteria. Group-specific probes for the Bacteroides-Prevotella group and bifidobacteria labeled with FITC and Cy5, respectively, were used to stain the food particles shown in Fig. 3C. The bacteroides can be seen covering the food material in a biofilm interspersed with bifidobacterial colonies. Figure 4A shows plant material that is autofluorescing and that is stained with Cy5 (total eubacteria) and Cy3 (bifidobacteria). The bifidobacteria appear to be present in low numbers; however, the combination of confocal sections taken through the biofilm (Fig. 4B) shows that the structure was approximately 20 μm in depth and that large numbers of bifidobacteria were present, many of which were present in microcolonies growing below the surface, close to the substratum.
FIG. 2.
Light micrograph (magnification, ×100) of adherent bacteria on the surface of food particles stained with a live/dead stain. Yellow cells are living, and red cells are dead.
FIG. 4.
Fluorescent light micrograph (CLSM) of bacteria on plant cells in feces (magnification, ×60) stained with 16S rRNA oligonucleotide probes. (A) Total eubacteria (Eub) and bifidobacteria (Bifid) stained with Cy5 (blue)- and Cy3 (red)-labeled probes, respectively, are shown adhering to autofluorescent plant material (green). (B) Composite image of 0.5-μm confocal z sections through the food particle (total depth, 20 μm) showing the presence of bifidobacteria growing in microcolonies below the surface, close to the substratum.
FIG. 3.
Fluorescent light micrograph (magnification, ×100) of intestinal bacteria on food particles labeled with 16S rRNA gene oligonucleotide probes. (A) Total eubacteria are stained with Cy5 (blue, A), enterobacteria are stained with FITC (green, B), and bifidobacteria are stained with Cy3 (red, C). (B) Members of the Eubacterium rectale-C. coccoides group are stained with Cy3 (red, A), and bifidobacteria are stained with Cy5 (blue, B). (C) Large numbers of bacteroides stained with FITC (green, B) interspersed with bifidobacteria stained with Cy5 (blue, A) can be seen.
Fermentation of complex carbohydrates in adherent and nonadherent microbiotas.
Short-term fermentations were done with strongly adherent and nonadherent bacteria to investigate how different complex carbohydrates were fermented. Results in Table 3 show specific rates of SCFA production from various polysaccharides and oligosaccharides. Overall rates of fermentation of pectin and xylan were similar in the two communities. However, arabinogalactan digestion was considerably more rapid in the biofilm organisms, whereas the reverse was true with starch and mucin. Fermentation of small, highly soluble oligosaccharide molecules was invariably more rapid in nonadherent bacteria. GOS preparations were the most rapidly fermented by both bacterial communities, while XOS and SOS were slowly utilized by bacteria from the biofilms. Metabolism of the different carbohydrates gave rise to distinct patterns of fermentation product formation. SCFA molar ratios show that with the exceptions of SOS, adherent cells produced markedly higher levels of acetate than nonadherent bacteria. In contrast, these nonadherent organisms always formed proportionately more butyrate than the biofilm communities. Propionate production from the different carbohydrates was highly variable in both bacterial populations.
TABLE 3.
Specific rates of SCFA formation and molar ratios of acetate, propionate, and butyrate produced by biofilm and nonadherent bacteria in short-term fermentation experimentsa
| Carbohydrate | mmol SCFA produced/h/g (dry wt) bacteria ± SD (final SCFA molar ratio)c
|
|||||
|---|---|---|---|---|---|---|
| Biofilm bacteria
|
Nonadherent bacteria
|
|||||
| Acetate | Propionate | Butyrate | Acetate | Propionate | Butyrate | |
| Polysaccharides | ||||||
| Starch | 0.62 ± 0.12b (88) | 0.05 ± 0.03 (4) | 0.08 ± 0.02b (8) | 0.99 ± 0.12 (72) | 0.09 ± 0.03 (6) | 0.19 ± 0.05 (22) |
| Pectin | 0.58 ± 0.13 (80) | 0.12 ± 0.03 (17) | 0.02 ± 0.02b (3) | 0.62 ± 0.11 (64) | 0.21 ± 0.05 (28) | 0.07 ± 0.04 (8) |
| Arabinogalactan | 1.20 ± 0.09b (82) | 0.18 ± 0.02 (12) | 0.06 ± 0.02b (6) | 0.65 ± 0.10 (73) | 0.12 ± 0.04 (14) | 0.14 ± 0.03 (13) |
| Xylan | 0.89 ± 0.14 (89) | 0.10 ± 0.02 (9) | 0.05 ± 0.02 (2) | 0.70 ± 0.14 (70) | 0.17 ± 0.04) (16) | 0.15 ± 0.07 (14) |
| Mucin | 0.88 ± 0.14 (84) | 0.08 ± 0.02b (10) | 0.09 ± 0.03b (6) | 0.98 ± 0.13 (64) | 0.28 ± 0.10 (20) | 0.25 ± 0.03 (16) |
| Oligosaccharides | ||||||
| Fructo-oligosaccharides | 0.51 ± 0.05b (78) | 0.12 ± 0.03 (12) | 0.09 ± 0.03 (10) | 0.78 ± 0.15 (70) | 0.08 ± 0.02 (10) | 0.15 ± 0.03 (20) |
| Xylo-oligosaccharides | 0.46 ± 0.03b (90) | 0.04 ± 0.02 (4) | 0.09 ± 0.01 (6) | 0.74 ± 0.14 (78) | 0.06 ± 0.01 (8) | 0.14 ± 0.02 (14) |
| Galacto-oligosaccharides | 0.64 ± 0.15b (89) | 0.06 ± 0.01b (4) | 0.10 ± 0.10 (7) | 0.98 ± 0.28 (75) | 0.12 ± 0.03 (8) | 0.22 ± 0.04 (17) |
| Soya-oligosaccharides | 0.35 ± 0.03b (71) | 0.10 ± 0.02 (18) | 0.08 ± 0.02b (11) | 0.75 ± 0.04 (78) | 0.12 ± 0.03 (9) | 0.15 ± 0.02 (13) |
Bacteria were incubated with each substrate for 6 h under conditions of nitrogen limitation. Results show mean values obtained from fecal material obtained from 10 donors ± standard deviations.
Significant difference (P < 0.05) compared to the nonadherent group.
Values in parentheses are final SCFA molar ratios calculated from individual SCFA concentrations at the end of the fermentation.
DISCUSSION
The human colonic microbiota is generally perceived as being a homogeneous entity, but intestinal microorganisms exist in a multiplicity of microhabitats and metabolic niches on the bowel wall and on the surfaces of dietary residues in the gut lumen. These microenvironments constantly change as resources are consumed or produced. Molecular analysis using FISH has demonstrated that bacterial cells in the large intestine often exist as discrete entities, but they can also occur in microcolonies (30, 31) and, as shown in this study, in complex associations with other species on the surfaces of particulate materials.
SEM showed a variety of morphological forms in biofilms occurring on the surfaces of food residues in fecal material (Fig. 1). While washing with buffer removed part of the biofilm, treatment with the surfactant CTAB was required to completely desorb strongly adhering bacteria from food surfaces. Microbiological analysis of partially digested food particles in feces showed that the biofilm communities were members of complex multispecies consortia, and at the genus level, at least, biofilm populations were shown to be broadly similar to nonadherent microbiotas, with bacteroides and bifidobacteria predominating (Table 2). Interestingly, staphylococci and bacilli, which are not indigenous organisms in the colon, were found only in the nonadherent population and were not detected on the food particles. This inability to colonize may limit their ability to permanently establish in the large gut. The methods used to enumerate these communities are innately destructive and do not provide information on the organization and community structure in biofilms. However, CLSM sectioning of biofilms on food particles in combination with 16S rRNA gene oligonucleotide probes (Fig. 4) showed that bifidobacteria in the biofilms were present in microcolonies, as well as being dispersed with other organisms. Bifidobacteria have also been shown to occur in microcolonies in biofilms on the gut mucosa (30).
Bacteria first need to colonize food particles in the gut as a prelude to their digestion, which leads to the formation of biofilms. An important facet of bacterial growth in biofilms in the large gut is that species colonizing the surfaces of food particles are likely to be more directly involved in the breakdown of complex insoluble polymeric substances than nonadherent organisms, giving them an advantage in competing for nutrients in the ecosystem. Close spatial relationships between bacterial cells growing on surfaces are important in relation to metabolic communication and are ecologically significant in that they minimize potential growth-limiting effects on secondary cross-feeding populations that are associated with mass transfer resistance, for example, between H2-producing bacteria and H2-consuming syntrophs (8).
The majority of human intestinal bacteria are saccharolytic, and carbohydrate availability is an important factor regulating the composition and metabolic activities of the colonic microbiota (27). A wide range of different carbohydrates is potentially available for fermentation in the large bowel (11), and one of the main determinants of species diversity in the colonic ecosystem is the multiplicity of different carbon sources to which intestinal microorganisms potentially have access. Varying the availability of polysaccharide substrates has been shown to cause significant shifts in luminal anaerobic populations in a gut fermentor system (14). A variety of nutritional, host, and dietary factors affect the outcome of carbohydrate fermentation reactions in the large intestine, and because the majority of carbohydrates entering the colon is in the form of polysaccharides, the rate at which these substances can be depolymerized controls the rate at which fermentable carbohydrates become available for assimilation by the bacteria.
Starches and nonstarch polysaccharides (dietary fiber) are the principal sources of carbohydrates in the human colon (9), although nondigestible oligosaccharides such as fructo-oligosaccharides are increasingly being introduced into the western diet (18). A wide range of mucins from the upper gastrointestinal tract enter the large bowel in effluent from the small intestine, while more mucus is formed by goblet cells in the colonic mucosa. In small intestinal effluent, particulate substances such as partially digested plant cell materials are entrapped in a viscoelastic mucus gel, which must be broken down by bacteria in the colon to facilitate access to the food residues.
The chemical composition of the growth substrate markedly influences the fermentation products that can be formed by bacteria. This was demonstrated previously by Englyst et al. (15), who originally showed that acetate and butyrate were the principal SCFA produced from starch by fecal bacteria, whereas acetate was the main fermentation product from both pectin and xylan. Similar results were obtained in the present study, when different carbohydrates were incubated with biofilm and nonadherent fecal bacteria in fermentation experiments (Table 3). These short-term incubations were done under nitrogen limitation to restrict bacterial growth and changes in community structure.
Qualitative and quantitative differences in biofilm metabolism were seen in SCFA production rates, particularly when oligosaccharides served as substrates; previous studies have shown that these bacteria are also different with respect to their polysaccharidase and glycosidase activities (29). These results clearly showed that nonadherent bacteria fermented oligosaccharides considerably more rapidly than the biofilm communities, whereas this was only true with highly soluble polymers such as starch and, to a lesser degree, mucin. The relatively insoluble polysaccharide arabinogalactan associated with cell wall material was digested more rapidly by bacteria desorbed from the biofilms, reflecting their adaptation to this substrate. Interestingly, butyrate, which is used as an energy source by the colonic mucosa (38) and which has a number of anticancer properties (23), was formed primarily by nonadherent fecal communities, irrespective of the fermentation substrate, indicating that these bacteria are of physiological importance to the host.
An interesting facet of the results on carbohydrate metabolism obtained in this investigation was the data concerning oligosaccharide fermentation. Fructo-oligosaccharides, GOS, XOS, and SOS are all potential prebiotic substances. Prebiotics are food ingredients that are not hydrolyzed by human digestive enzymes and that selectively stimulate the growth and activities of specific groups of bacteria in the gut, usually bifidobacteria and lactobacilli, with health benefits (18). Surprisingly, fermentation of oligosaccharides was in some cases slower than the breakdown of their more complex polysaccharide counterparts. Indeed, in biofilm bacteria, SCFA generation from xylan and arabinogalactan was faster than that with the corresponding oligosaccharides.
However, the molar ratios of acetate, propionate, and butyrate produced from polysaccharides and chemically similar oligosaccharides were not sufficiently distinct to suggest that different groups of bacteria were involved in their fermentation. This indicates that substrate uptake was a significant factor affecting fermentation rate. Similar observations have been made in pure culture studies with Bacteroides ovatus (28).
In conclusion, while we know little about the fine structure of biofilm communities in the lumen of the human colon, it is clear that these microbiotas are heterogeneous assemblages that must form rapidly due to the relatively short retention time of digestive materials in the cecum and ascending colon, which act as a mixing chamber in the large bowel. However, it is unclear whether intestinal biofilms in the colon are comparable to the highly evolved assemblages seen in oral biofilm communities (5). More detailed studies, in combination with molecular methods of analysis, are needed to assess the ecological importance of these structures in the colonic ecosystem as a whole and, more importantly, to determine their significance with respect to host metabolism.
Acknowledgments
This research was funded by the Biotechnology and Biological Sciences Research Council.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anwar, H., M. K. Dasgupta, and J. W. Costerton. 1990. Testing the susceptibility of bacteria in biofilms to antibacterial agents. Antimicrob. Agents Chemother. 34:2043-2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beerens, H. 1990. An elective and selective isolation medium for Bifidobacterium spp. Lett. Appl. Bacteriol. 11:155-157. [Google Scholar]
- 5.Bradshaw, D. J., and P. D. Marsh. 2003. Novel microscopic methods to study the structure and metabolism of oral biofilms, p. 173-188. In M. Wilson and D. Devine (ed.), Medical implications of biofilms. Cambridge University Press, Cambridge, United Kingdom.
- 6.Caldwell, D. R., and M. P. Bryant. 1966. Medium without rumen fluid for nonselective enumeration and isolation of rumen bacteria. Appl. Microbiol. 14:794-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Child, M. W., A. Kennedy, B. Bahrami, S. Macfarlane, and G. T. Macfarlane. 2006. Studies on the effect of system retention time on bacterial populations colonising a three-stage continuous culture model of the human large gut using fluorescent in situ hybridisation techniques. FEMS Microbiol. Ecol. 55:299-310. [DOI] [PubMed] [Google Scholar]
- 8.Conrad, R., T. J. Phelps, and J. G. Zeikus. 1985. Gas metabolism evidence in support of the juxtaposition of hydrogen-producing and methanogenic bacteria in sewage sludge and lake sediments. Appl. Environ. Microbiol. 50:595-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cummings, J. H., E. R. Beatty, S. M. Kingman, and H. N. Englyst. 1996. Digestion and physiological properties of resistant starch in the human large bowel. Br. J. Nutr. 75:733-747. [DOI] [PubMed] [Google Scholar]
- 10.Cummings, J. H., S. A. Bingham, K. W. Heaton, and M. A. Eastwood. 1993. Fecal weight, colon cancer risk and dietary intake of non-starch polysaccharides (dietary fiber). Gastroenterology 103:1783-1789. [DOI] [PubMed] [Google Scholar]
- 11.Cummings, J. H., and G. T. Macfarlane. 1991. The control and consequences of bacterial fermentation in the human colon—a review. J. Appl. Bacteriol. 70:443-459. [DOI] [PubMed] [Google Scholar]
- 12.Degnan, B. A., and G. T. Macfarlane. 1995. Arabinogalactan utilization in continuous cultures of Bifidobacterium longum: effect of co-culture with Bacteroides thetaiotaomicron. Anaerobe 1:103-112. [DOI] [PubMed] [Google Scholar]
- 13.Duncan, S. H., G. L. Hold, H. J. M. Harmsen, C. S. Stewart, and H. J. Flint. 2002. Growth requirements and fermentation products of Fusobacterium prausnitzii, and a proposal to reclassify it as Faecalibacterium prausnitzii gen. nov., comb. nov. Int. J. Syst. Evol. Microbiol. 52:2141-2146. [DOI] [PubMed] [Google Scholar]
- 14.Duncan, S. H., K. P. Scott, A. G. Ramsey, H. J. M. Harsen, G. W. Welling, C. S. Stewart, and H. J. Flint. 2003. Effects of alternative dietary substrates on competition between human colonic bacteria in an anaerobic fermentor system. Appl. Environ. Microbiol. 69:1136-1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Englyst, H. N., S. Hay, and G. T. Macfarlane. 1987. Breakdown of resistant and readily digestible starch by human gut bacteria. J. Sci. Food Agric. 37:699-706. [Google Scholar]
- 16.Fite, A., G. T. Macfarlane, J. H. Cummings, M. J. Hopkins, S. C. Kong, E. Furrie, and S. Macfarlane. 2004. Identification and quantitation of mucosal and faecal desulfovibrios using real time PCR. Gut 53:523-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Franks, A. H., H. J. Harmsen, G. C. Raangs, G. J. Jansen, F. Schut, and G. W. Welling. 1998. Variations of bacterial populations in human feces quantified by fluorescence in situ hybridization with group-specific 16S rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol. 64:3336-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gibson, G. R., R. A. Rastall, and R. Fuller. 2003. The health benefits of probiotics and prebiotics, p. 52-76. In R. Fuller and G. Perdigon (ed.), Gut flora, nutrition immunity and health. Blackwell Publishing, Oxford, United Kingdom.
- 19.Hentges, D. J. 1983. Role of the intestinal flora in host defense against infection, p. 311-332. In D. J. Hentges (ed.), Human intestinal microflora in health and disease. Academic Press, London, United Kingdom.
- 20.Holdeman, L. V., E. P. Cato, and W. E. C. Moore. 1977. Anaerobe laboratory manual, 4th ed. Virginia Polytechnic Institute and State University, Blacksburg, Va.
- 21.Hopkins, M. J., G. T. Macfarlane, E. Furrie, A. Fite, and S. Macfarlane. 2005. Characterisation of bacteria in infant's stools using real-time PCR and Northern hybridisation analyses. FEMS Microbiol. Ecol. 54:77-85. [DOI] [PubMed] [Google Scholar]
- 22.Huijsdens, X. W., R. K. Linskens, M. Mak, S. G. Meuwissen, C. M. Vandenbroucke-Grauls, and P. H. Savelkoul. 2002. Quantification of bacteria adherent to gastrointestinal mucosa by real-time PCR. J. Clin. Microbiol. 40:4423-4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kruh, J., N. Defer, and L. Tichonicky. 1995. Effects of butyrate on cell proliferation and gene expression, p. 275-288. In J. H. Cummings, J. L. Rombeau, and T. Sakata (ed.), Physiological and clinical aspects of short chain fatty acids. Cambridge University Press, Cambridge, United Kingdom.
- 24.Langendijk, P. S., F. Schut, G. J. Jansen, G. C. Raangs, G. R. Kamphuis, M. H. F. Wilkinson, and G. W. Welling. 1995. Quantitative fluorescence in situ hybridization of Bifidobacterium spp. with genus-specific 16S rRNA-targeted probes and its application in fecal samples. Appl. Environ. Microbiol. 61:3069-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loge, F. J., R. W. Emerick, D. E. Thompson, D. C. Nelson, and J. L. Darby. 1999. Development of a fluorescent 16S rRNA oligonucleotide probe specific to the family Enterobacteriaceae. Water Environ. Res. 71:75-83. [Google Scholar]
- 26.Macfarlane, G. T., and J. H. Cummings. 1991. The colonic flora, fermentation and large bowel digestive function, p. 51-92. In S. F. Phillips, J. H. Pemberton, and R. G. Shorter (ed.), The large intestine: physiology, pathophysiology and disease. Raven Press, New York, N.Y.
- 27.Macfarlane, G. T., and G. R. Gibson. 1996. Carbohydrate fermentation, energy transduction and gas metabolism in the human large intestine, p. 269-318. In R. I. Mackie and B. A. White (ed.), Ecology and physiology of gastrointestinal microbes, vol. 1. Gastrointestinal fermentations and ecosystem. Chapman & Hall, New York, N.Y. [Google Scholar]
- 28.Macfarlane, G. T., S. Hay, S. Macfarlane, and G. R. Gibson. 1990. Effect of different carbohydrates on growth, polysaccharidase and glycosidase production by Bacteroides ovatus, in batch and continuous culture. J. Appl. Bacteriol. 68:179-187. [DOI] [PubMed] [Google Scholar]
- 29.Macfarlane, G. T., and S. Macfarlane. 1995. Human intestinal biofilm communities, p. 83-89. In J. Wimpenny, P. Handley, P. Gilbert, and H. Lappin-Scott (ed.), The life and death of biofilms. Bioline, University of Wales College of Cardiff, Cardiff, United Kingdom.
- 30.Macfarlane, S., E. Furrie, J. H. Cummings, and G. T. Macfarlane. 2004. Chemotaxonomic analysis of bacterial populations colonizing the rectal mucosa in patients with ulcerative colitis. Clin. Infect. Dis. 38:1690-1699. [DOI] [PubMed] [Google Scholar]
- 31.Macfarlane, S., M. J. Hopkins, and G. T. Macfarlane. 2000. Bacterial growth and metabolism on surfaces in the large intestine. Microb. Ecol. Health Dis. 2:64-72. [Google Scholar]
- 32.Manz, W., R. Amann, W. Ludwig, M. Vancanneyt, and K. H. Schleifer. 1996. Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum Cytophaga-Flavobacter-Bacteroides in the natural environment. Microbiology 142:1097-1106. [DOI] [PubMed] [Google Scholar]
- 33.McBain, A. J., and G. T. Macfarlane. 1998. Ecological and physiological studies on large intestinal bacteria in relation to production of hydrolytic and reductive enzymes involved in formation of genotoxic metabolites. J. Med. Microbiol. 47:407-416. [DOI] [PubMed] [Google Scholar]
- 34.Microbial ID Inc. 1992. Microbial Identification system operational manual. Microbial ID Inc., Newark, Del.
- 35.Mozes, N., and P. G. Rouxhet. 1992. Influence of surfaces on microbial activity, p. 125-136. In L. Melo, F. T. R. Bott, and B. Capdeville (ed.), Biofilms—science and technology. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 36.Nadkarni, M. A., F. E. Martin, N. A. Jacques, and N. Hunter. 2002. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology 148:257-266. [DOI] [PubMed] [Google Scholar]
- 37.Poxton, I. R., R. Brown, A. Sawyer, and A. Ferguson. 1997. Mucosa-associated bacterial flora of the human colon. J. Med. Microbiol. 46:85-91. [DOI] [PubMed] [Google Scholar]
- 38.Roediger, W. E. W. 1980. The colonic epithelium in ulcerative colitis: an energy-deficiency disease? Lancet ii:712-715. [DOI] [PubMed] [Google Scholar]
- 39.Schiffrin, E. J., D. Brassart, A. L. Servin, F. Rochart, and A. Donnet-Hughes. 1997. Immune modulation of blood leukocytes in humans by lactic acid bacteria: criteria for strain selection. Am. J. Clin. Nutr. 66:515S-520S. [DOI] [PubMed] [Google Scholar]
- 40.Stephen, A. M., and J. H. Cummings. 1980. The microbial contribution to human faecal mass. J. Med. Microbiol. 13:45-56. [DOI] [PubMed] [Google Scholar]
- 41.Swidsinski, A., A. Ladhoff, A. Pernthaler, S. Swidsinski, V. Loening-Baucke, M. Ortner, J. Weber, U. Hoffmann, S. Screiber, M. Dietel, and H. Lochs. 2002. Mucosal flora in inflammatory bowel disease. Gastroenterology 122:44-54. [DOI] [PubMed] [Google Scholar]
- 42.Van Loosdrecht, M. C. M., J. Lyklema, W. Norde, and A. J. B. Zehnder. 1990. Influence of interfaces on microbial activity. Microbiol. Rev. 54:75-87. [DOI] [PMC free article] [PubMed] [Google Scholar]