Abstract
We studied the frequency and composition of potential microcystin (MC) producers in 70 Finnish lakes with general and genus-specific microcystin synthetase gene E (mcyE) PCR. Potential MC-producing Microcystis, Planktothrixand Anabaena spp. existed in 70%, 63%, and 37% of the lake samples, respectively. Approximately two-thirds of the lake samples contained one or two potential MC producers, while all three genera existed in 24% of the samples. In oligotrophic lakes, the occurrence of only one MC producer was most common. The combination of Microcystis and Planktothrix was slightly more prevalent than others in mesotrophic lakes, and the cooccurrence of all three MC producers was most widespread in both eutrophic and hypertrophic lakes. The proportion of the three-producer lakes increased with the trophic status of the lakes. In correlation analysis, the presence of multiple MC-producing genera was associated with higher cyanobacterial and phytoplankton biomass, pH, chlorophyll a, total nitrogen, and MC concentrations. Total nitrogen, pH, and the surface area of the lake predicted the occurrence probability of mcyE genes, whereas total phosphorus alone accounted for MC concentrations in the samples by logistic and linear regression analyses. In conclusion, the results suggested that eutrophication increased the cooccurrence of potentially MC-producing cyanobacterial genera, raising the risk of toxic-bloom formation.
Cyanobacterial mass occurrences are a frequent phenomenon worldwide. A survey of the blooms in freshwaters has shown that on average, 59% contain toxins, with hepatotoxic blooms being more common than neurotoxic blooms (45). Toxic blooms expose water users to health risks and prevent the recreational use of water (19).
Microcystins (MCs) are the most prevalent cyanobacterial hepatotoxins in freshwaters, where they are produced mainly by strains of the genera Anabaena, Microcystis, Planktothrix, and occasionally Nostoc (45). The toxicity of MCs is due to the inhibition of eukaryotic proteinphosphatases 1 and 2A (11, 25) in liver cells, where MCs enter via the bile acid transport system (1). MCs are cyclic heptapeptides with a general structure of cyclo(-d-Ala-X-d-erythro-β-methylaspartic acid-Z-Adda-d-Glu-N-methyldehydroalanine), where X and Z are various l-amino acids and Adda is 3-amino-9-methoxy-2,6,8-trimethyl-10-phenyldeca-4,6-dienoic acid. d-Glu and Adda form the part of the molecule that interacts with the protein phosphatases and thus are the crucial amino acids for the toxicity of MCs (8).
MCs are produced by nonribosomal enzyme complexes. Adda is synthesized and integrated into the MC molecule by the enzymes McyG, McyD, and McyE. McyE also incorporates d-Glu, the other crucial amino acid for toxicity. Microcystin synthetase (mcy) gene clusters that encode these biosynthetic enzymes have now been characterized from all the main MC-producing genera (2, 30, 43, 48). The presence of biosynthetic genes has also been proven a prerequisite for MC production (5). Although intensively studied, only a few strains of Microcystis (16, 27, 29, 49) that contain mcy genes have been shown not to produce MCs. Recent results, however, indicate that this may be more frequent among Planktothrix strains (3, 21).
MC-producing genera include both toxic strains (with the mcy genes) and nontoxic strains (without the mcy genes). Toxic and nontoxic strains, sometimes of more than one genus, can coexist even in the same bloom (21, 53, 54). These strains cannot be separated from each other by microscopy. The revelation of the genetic basis for MC production has enabled the development of molecular methods for the detection and identification of MC producers. Most of these methods are based on PCR using primers designed to recognize the mcy genes. Many studies have concentrated on detecting either solelyMicrocystis, the most common and important MC producer throughout the world (12, 20, 31, 38, 49, 55, 58), or Planktothrix (21, 26). Often the target gene has been either mcyA or mcyB (20, 21, 31, 49, 55, 58). Some studies have used a combination of several biosynthetic genes (12, 38) but again concentrated only on Microcystis. Mbedi et al. (26) used eight different genes (mcyA, mcyB, mcyE, and mcyT) and intergenic regions (mcyCJ, mcyEG, mcyHA, and mcyTD) to validate their usability in detecting MC-producing Planktothrix strains. Both MC-producing Anabaena and Microcystis strains were detected and quantified with genus-specific mcyE primers (51) in two Finnish lakes. MC-producing strains of Anabaena, Microcystis, and Planktothrix were differentiated by a restriction fragment length polymorphism analysis of a general (non-genus-specific) mcyA PCR product in a German lake (10). However, studies of larger sample sets with genus-specific detection of all principal MC-producing genera are lacking.
We studied samples from 70 Finnish lakes with general and genus-specific PCR amplification of the mcyE gene to reveal the frequency of occurrence and compositions of potential microcystin-producing cyanobacterial genera: Anabaena, Microcystis, and Planktothrix. PCR results were compared to results of microcystin analyses and correlated to environmental variables. Genus-specific PCRs were efficient in detecting MC producers in samples taken before bloom season. Interestingly, we observed the simultaneous occurrence of the main MC producers most frequently in eutrophic and hypertrophic lakes.
MATERIALS AND METHODS
Environmental samples.
Samples from 70 lakes located in Southern and Central Finland were collected in summer (mainly in July) 2002 as described by Rajaniemi-Wacklin et al. (39). For DNA extraction, cells were collected from 75 ml to 1,000 ml of lake water by filtration. Depending on the concentration of humic or other substances clogging the filter, a series of filters with various pore sizes (10, 5, and 1 μm [Osmonics Inc.] and 0.2 μm [Pall Corporation]) were used to maximize the sample volume filtered. Filters were stored in 2 ml of lysis buffer (40 mM EDTA, 400 mM NaCl, 0.75 M sucrose, 50 mM Tris-HCl, pH 8.3) at −20°C until use. To measure MC concentration, an unconcentrated 5-ml aliquot was taken from each water sample for enzyme-linked immunosorbent analysis (ELISA). For measuring cell-bound MC concentration with ELISA, 2 to 18.5 liters of lake water was concentrated with a net (<25 μm pore size) and filtered through glass fiber filters (GF 52; Schleicher & Schuell) to collect the cells.
Environmental variables.
Samples were taken from a depth of 1 meter to determine temperature (°C), total phosphorus (TP; μg/liter), PO4-P (dissolved phosphorus [DIP]; μg/liter), total nitrogen (TN; μg/liter), NH4-N (μg/liter), NO3-N plus NO2-N (μg/liter), water color (mg/liter Pt), pH, chlorophyll a (chl-a; μg/liter), and Secchi depth (m) using standard methods (28). Dissolved nitrogen (DIN) was calculated as the sum of NH4-N and NO3-N plus NO2-N. Phytoplankton and cyanobacterial biomasses (mg/liter) and species composition were analyzed by microscopy using a Nordic variant of the Utermöhl technique (36, 50) and phase-contrast illumination at ×200 and ×800 to ×1,200 magnifications by a trained group of investigators. Cell counts were converted to biovolumes using the cell volumes of the phytoplankton database of the Finnish Environment Institute (www.environment.fi). Environmental data from the sampling date were available for all but four lakes, for which data from the nearest available date (within 2 to 6 weeks' time) were used.
Toxin analyses.
For MC measurements by ELISA, unconcentrated 5-ml water samples were sonicated (Labsonic U; Braun) for 2 min with a 0.5-s repeating duty cycle and filtered through a 0.2-μm polyethersulfone membrane (Puradisc 25 AS; Whatman). MCs were detected with an EnviroGard microcystins plate kit (Strategic Diagnostics Inc.) according to the manufacturer's instructions. The detection limit of MCs was 0.1 μg/liter. Absorbances were measured with an iEMS Reader MF (Labsystems) at wavelengths of 450 nm and 620 nm.
MCs from the concentrated water samples were extracted by ultrasonication from glass fiber filters in 75% aqueous methanol, according to Jurczak et al. (15). MCs were dissolved in 1 ml 75% aqueous methanol before analysis with ELISA. The MC concentration was determined as an equivalent to MC-LR with a QuantiPlate kit for microcystins (EnviroLogix) according to the manufacturer's instructions. The detection range of MCs was from 0.16 to 2.5 μg/liter. Absorbances were measured with a Multiscan RC microplate analyzer (Labsystems) at a wavelength of 450 nm.
DNA extraction and purification.
DNA was extracted from filters with a modified hot phenol method (7). For the lysis of cyanobacterial cells, higher concentrations of lysozyme (1.25 mg/ml final concentration) and proteinase K (300 μg/ml final concentration) were used. After extraction of the DNA-containing aqueous phase with phenol-chloroform-isoamyl alcohol (25:24:1, vol/vol/vol) and chloroform-isoamyl alcohol (24:1, vol/vol), DNA was precipitated with sodium acetate and ethanol at −20°C overnight. Precipitated DNA was washed with 70% ethanol and resuspended in Tris-EDTA (10:1, vol/vol) buffer. Extracted DNA was further purified with NucleoTrap PCR purification (BD Biosciences) and QuickStep PCR purification (Edge BioSystems) kits according to the manufacturers' instructions. For PCRs with mcyE gene-targeted primers, DNAs extracted from different filters were combined in equal amounts and DNA concentrations measured with a BioPhotometer (Eppendorf).
Design and testing of a Planktothrix-specific reverse primer.
A Planktothrix-specific reverse primer (mcyE-plaR3) was manually designed based on the alignment of mcyE gene sequences from 30 MC- or nodularin-producing Anabaena, Microcystis, Nostoc, Planktothrix, and Nodularia strains (40) for use with the mcyE-general forward primer mcyE-F2 (51). The specificity of the resulting primer pair was tested with 63 MC- or nodularin-producing and non-MC- or non-nodularin-producing Anabaena, Aphanizomenon, Hapalosiphon, Limnothrix, Microcystis, Nodularia, Nostoc, Phormidium, Planktothrix, and Synechococcus strains maintained in the culture collection of K. Sivonen, University of Helsinki (Table 1). PCR was performed in a total volume of 20 μl of 1× DyNAzyme PCR buffer (Finnzymes), including 1 μl of DNA, 250 μM each deoxynucleotide (Finnzymes), a 0.5 μM concentration of both primers (Sigma Genosys Ltd.), and 0.5 U of DyNAzyme II DNA polymerase (Finnzymes). PCR amplification consisted of an initial denaturation for 3 min at 95°C, 30 cycles of 30 s at 94°C, 30 s at 57°C, and 60 s at 72°C, with a final extension of 10 min at 72°C. The whole PCR mixture was loaded into a 1.5% agarose gel dyed with ethidium bromide (0.15 μg/ml) to ensure the detection of even the faintest amplification products. Gels were documented with a Kodak DC290 camera and the Kodak 1D v 3.5.0 imaging program. Images were visually inspected for amplification products.
TABLE 1.
Cyanobacterial strains used to test the specificity of the Planktothrix-specific primer pair mcyE-F2/plaR3
| Organism(s) and strain(s) | No. of strains | Specificity of primersa |
|---|---|---|
| Anabaena spp. | ||
| MC-producing strains: 90, 202A1, 202A2, NIVA-CYA83/1, 66A, PH256, 315, 318, 299B | 9 | − |
| Non-MC-producing strains: 299A, 141, 86, 123, 14, 37, IC-1, PH133, 277, PCC6309, PCC7108, PCC73105, PCC9208 | 13 | − |
| Aphanizomenon spp. | ||
| Non-MC-producing strains: TR183, 202, PCC7905 | 3 | − |
| Hapalosiphon sp. | ||
| MC-producing strain BZ31 | 1 | − |
| Limnothrix spp. | ||
| Non-MC-producing strains: 165-C, 007-A | 2 | − |
| Microcystis spp. | ||
| MC-producing strains: 205, 98, GL260735, Izancya 25, NIES 102, NIES A89, PCC7941, PCC7806, GL280646 | 9 | − |
| Non-MC-producing strains: PCC7820, 130, 269, GL060916 | 4 | − |
| Nodularia spp. | ||
| Nodularin-producing strains: HEM, AV3, F81, PCC7804, BY1, GR8b | 6 | − |
| Non-nodularin-producing strain HKVV | 1 | − |
| Nostoc spp. | ||
| MC-producing strain 152 | 1 | − |
| Non-MC-producing strain 159 | 1 | − |
| Phormidium sp. | ||
| MC-producing strain DVL1003 | 1 | − |
| Planktothrix spp. | ||
| MC-producing strains: 49, 97, NIVA-CYA126/8, NIVA-CYA127, NIVA-CYA128/R, 213, 226 | 7 | + |
| Non-MC-producing strains: 18, 214, 45, PCC6304 | 4 | −/(+)b |
| Synechococcus sp. | ||
| Non-MC-producing strain GL150636 | 1 | − |
+, amplification product with mcyE-F2/plaR3 primer pair; (+), weak amplification; −, no amplification.
Weak amplification with strain PCC6304.
PCR with lake water samples.
To exclude the possibility of PCR inhibition causing negative mcyE PCR results, a PCR with cyanobacterium-specific 16S rRNA gene-targeted primers (32) was performed with DNA extracted from different-cell-size fractions. If there was no amplification, an additional PCR was carried out with eubacterium-specific 16S rRNA primers (6) in case the sample contained no cyanobacteria. To identify potential MC producers in lake samples, DNA extractions from fractions of different cell sizes were combined before amplification to decrease the number of samples. PCR was performed with four primer pairs designed to amplify regions of the mcyE gene. In all reaction mixtures, the same forward primer, mcyE-F2, was used (51). All potential MC-producing genera were targeted with the use of a general reverse primer, mcyE-R4 (40), and MC-producing Anabaena, Microcystis, and Planktothrix spp. were targeted with the genus-specific reverse primers mcyE-12R, mcyE-R8 (51), and mcyE-plaR3 (5′-CTCAATCTGAGGATAACGAT-3′), respectively. All reaction mixtures were prepared in a 20-μl total volume containing 30 ng of extracted lake DNA as a template, a 250 μM concentration of each deoxynucleotide (Finnzymes), and a 0.5 μM concentration of both primers (Sigma Genosys Ltd.). Anabaena- and Microcystis-specific PCRs were performed with 1× DyNAzyme PCR buffer (Finnzymes) containing 1 U of DyNAzyme II DNA polymerase (Finnzymes). General and Planktothrix-specific PCRs took place in 1× Super Taq Plus PCR buffer (HT Biotechnology Ltd.) with 1 U of Super Taq Plus polymerase (HT Biotechnology Ltd.) and 1.25 μg/μl of bovine serum albumin (Promega). PCR protocols involved an initial denaturation for 3 min at 95°C; 35 cycles of 30 s at 94°C, 30 s at 56°C (mcyE-R4), 57°C (mcyE-plaR3), or 58°C (mcyE-12R, mcyE-R8), and 60 s at 68°C (Super Taq Plus) or 72°C (DyNAzyme II); and a final extension of 10 min at 68°C or 72°C. The whole PCR was loaded into a 1.5% agarose gel dyed with ethidium bromide (0.15 μg/ml). Gels were documented with a Kodak DC290 camera and the Kodak 1D v 3.5.0 imaging program. Images were analyzed with Bionumerics v 4.0 (Applied Maths BVBA). The spectral analysis feature of the program was used to determine optimal settings for background subtraction (disk size) and least-square filtering (cutoff value). The presence or absence of the bands was determined with the automatic band search feature using 5% minimum profiling; the bands were checked manually.
PCA and correlation analysis.
A principal component analysis (PCA) ordination was performed to group the lakes on the basis of the genus-specific mcyE PCR results. The presence or absence of Anabaena-, Microcystis-, and Planktothrix-specific PCR amplification was used as the input variable. Correlations between the resulting PC axes 1 and 2 and environmental variables were then calculated using an R statistical package (41). To find out which environmental variables were significant, both analyses were first done with a set of 48 lake samples, since for these lakes, data were available from all of the following environmental variables: latitude, longitude, surface area (km2), mean depth (m), temperature (°C), TP (μg/liter), DIP (μg/liter), TN (μg/liter), NH4-N (μg/liter), NO3 plus NO2-N (μg/liter), DIN (μg/liter), the ratio of TN to TP (TN/TP), the ratio of DIN to DIP (DIN/DIP), water color (mg/1 part), pH, cyanobacterial biomass (mg/liter), phytoplankton biomass (mg/liter), chl-a (μg/liter), Secchi depth (m), Anabaena biomass (mg/liter), Microcystis biomass (mg/liter), Planktothrix biomass (mg/liter), Oscillatoriales biomass (combined Planktothrix agardhii, Planktolyngbya subtilis, and Oscillatoriales biomasses; mg/liter), Aphanizomenon biomass (mg/liter), and the microcystin concentration (μg/liter) of unconcentrated and concentrated water samples. For the final PCA, all the lakes (n = 58) that had data from every statistically significantly correlated (P < 0.05) environmental variable were selected. The final correlation analysis was performed with environmental variables significant in the first analysis complemented with all the variables available for the 58 lakes selected (Table 2).
TABLE 2.
Environmental variables used in the correlation analysis with principal components 1 and 2c
| Environmental variable | PC 1
|
PC 2
|
||
|---|---|---|---|---|
| Ra | Pb | R | P | |
| Latitude north | 0.06 | 0.655 | 0.03 | 0.828 |
| Longitude east | 0.08 | 0.536 | −0.05 | 0.737 |
| Surface area | −0.23 | 0.082 | −0.03 | 0.827 |
| TP | −0.25 | 0.060 | −0.17 | 0.200 |
| DIP | −0.25 | 0.054 | −0.10 | 0.449 |
| TN | −0.39 | 0.002 | −0.12 | 0.360 |
| TN/TP ratio | 0.12 | 0.369 | 0.02 | 0.890 |
| Water color | −0.17 | 0.214 | −0.13 | 0.329 |
| pH | −0.45 | 0.000 | −0.06 | 0.668 |
| Cyanobacterial biomass | −0.42 | 0.001 | 0.04 | 0.791 |
| Phytoplankton biomass | −0.46 | 0.000 | −0.09 | 0.514 |
| Chlorophyll a | −0.39 | 0.002 | −0.16 | 0.226 |
| Secchi depth | 0.18 | 0.181 | 0.22 | 0.095 |
| Anabaena biomass | −0.22 | 0.092 | 0.11 | 0.391 |
| Microcystis biomass | −0.39 | 0.003 | 0.06 | 0.682 |
| Planktothrix biomass | −0.21 | 0.107 | 0.17 | 0.198 |
| Oscillatoriales biomass | −0.26 | 0.047 | 0.06 | 0.626 |
| Aphanizomenon biomass | −0.31 | 0.020 | 0.02 | 0.861 |
| Microcystin concn (concentrated samples) | −0.45 | 0.000 | −0.04 | 0.553 |
| Microcystin concn (unconcentrated samples) | −0.41 | 0.001 | 0.08 | 0.556 |
Strength of the correlations.
Significance of the correlations.
Principal components 1 and 2 represented variation in the presence of the three genus-specific mcyE genes in water samples (n = 58). Statistically significant P values (P < 0.05) are marked in bold.
Regression analyses.
Regression analyses were performed to determine whether variation in the response variables, the presence or absence of the three genus-specific mcyE genes, and MC concentrations (log transformed) of the concentrated water samples could be explained by environmental variables (scaled to an average of 0 and unit variance). The same explanatory variables, i.e., surface area, TP, DIP, TN, TN/TP, water color, pH, and Secchi depth, were used for both analyses. Nevertheless, it was impossible to consider genus-level factors in the analysis of MC concentrations, because MC analysis does not reveal the producer organisms, such as genus-specific mcyE PCR. Biomasses of cyanobacteria and phytoplankton, chl-a, and MC concentrations (the response variable in linear regression analysis) were not used as explanatory variables, because it was assumed that these variables could have been partially affected by the same environmental factors as response variables in the following regression analyses. Potential correlations among the environmental variables used (colinearity) were measured with the diagnostic “perturb” package functioning in R (41). Because of the binomial (presence-absence) response of the mcyE gene, a logistic regression model was used for the PCR data, whereas a linear regression model was used for the analysis of the continuous-response variable (i.e., MC concentrations). A forward stepwise selection of parameters was performed using the stepAIC procedure in the R statistical package (R library MASS) (52) for both analyses. The starting point was a model with no parameter effects (except intercept). Every possible term (interactions between the categorical terms measuring the differences in the occurrence probabilities of the mcyE genes of the three genera only in the logistic model) was included in the model, in turn, and their effect was proved with Akaike's information criterion. At each step, the best explanatory (i.e., most informative) variable was selected. During the stepwise process, the effect of the elimination of previously selected parameters and the inclusion of a new one was valued, and the action that most improved the model was performed. This was continued until the most optimal model, comprising the best explanatory variables, was found. Finally, the significance of stepwise-model parameters was tested (deviance and F statistics for logistic and linear regressions, respectively) and insignificant parameters (P > 0.05) were removed. The Kolmogorov-Smirnow and Shapiro-Wilk normality tests and visual diagnostics of the residuals (41) were used to validate the assumptions of the regression theory of the linear regression analysis on MC concentrations. The effect of overdispersion on the logistic regression was measured.
RESULTS
Specificity of Planktothrix mcyE PCR.
To test whether the mcyE-F2/plaR3 primer pair specifically amplified the mcyE gene region from MC-producing Planktothrix strains, PCRs were performed with 63 MC- or nodularin-producing and nonproducing cyanobacterial strains (Table 1). Amplification was detected only when DNA from an MC-producing Planktothrix strain was used as a template, except with one non-MC-producing Planktothrix strain (PCC 6304), which produced a faint amplification product. Results showed that the mcyE-F2/plaR3 primer pair was specific for MC-producing Planktothrix strains and could serve to monitor the environmental water samples for the presence of Planktothrix-specific mcyE genes.
Occurrence of mcyE genes in water samples.
To reveal how frequently potential MC producers appeared in the 70 lake samples, PCR with general and Anabaena-, Microcystis-, and Planktothrix-specific mcyE primers was performed. When studied with general mcyE primers, 59 of the lake samples (84%) showed the presence of a potential MC producer. Potential MC-producing Anabaena, Microcystis, and Planktothrix strains appeared in 26 (37%), 49 (70%), and 44 (63%) lake samples, respectively. This suggested that, at the time of sampling, Microcystis was the most common MC producer in Finnish lakes, followed by Planktothrix and Anabaena (Fig. 1). In seven lake samples, the general primer pair detected no potential MC producers, although amplification was positive with at least one of the genus-specific primers. This could imply that genus-specific primer pairs are more sensitive than the general primer pair. On the contrary, in two lake samples, no genus-specific amplification was detected despite positive amplification with general primers (Fig. 1). However, these amplification products were extremely faint.
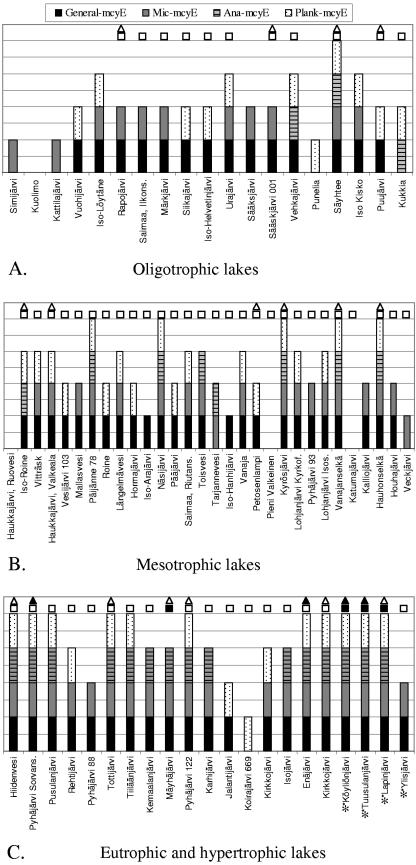
FIG. 1.
PCR results of 70 Finnish lakes with general mcyE and Microcystis-, Anabaena-, and Planktothrix-specific mcyE primer pairs. (A) Oligotrophic lakes (n = 19), with a TP concentration of <10 μg/liter. (B) Mesotrophic lakes (n = 30), with a TP concentration 10 to 34 μg/liter. (C) Eutrophic (n = 17) and hypertrophic (n = 4) lakes, with TP concentrations of 35 to 100 μg/liter and >100 μg/liter, respectively. Hypertrophic lakes are marked with an asterisk. Lakes are marked with white squares and triangles if MCs were detected with ELISA in concentrated and unconcentrated samples and with black squares and triangles if MC concentrations of concentrated and unconcentrated samples exceeded 1 μg/liter, respectively.
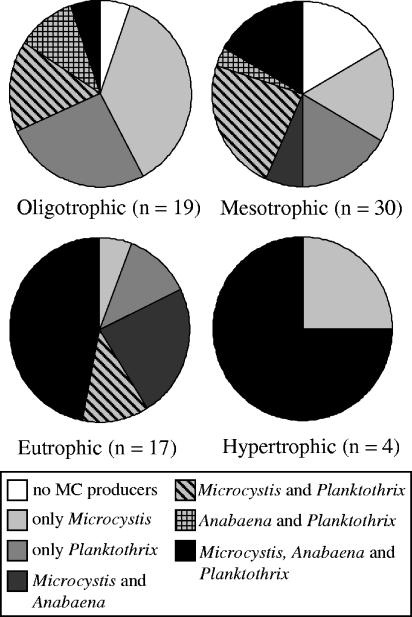
The composition of potential MC producers in lake samples was analyzed with three genus-specific mcyE primers. Of these lakes sampled, 26 (37%) indicated the presence of only one MC-producing genus, 21 (30%) contained two genera, and 17 (24%) contained all three genera (Fig. 1). To see how the trophic level of the lake influenced the composition of potential MC producers, lakes were grouped according to their TP concentration into oligotrophic (<10 μg/liter), mesotrophic, (10 to 34 μg/liter), eutrophic (35 to 100 μg/liter), and hypertrophic (>100 μg/liter) lakes (33). In oligotrophic lakes, the occurrence of only one MC producer, either Microcystis or Planktothrix, was most common. No lakes with Anabaena as a sole producer were found at any trophic level. Mesotrophic lakes had the widest distribution of different MC producer combinations, but lakes with Microcystis and Planktothrix were slightly more prevalent than others. The combination of Microcystis, Anabaena, and Planktothrix was the most widespread in both eutrophic and hypertrophic lakes (Fig. 2). The proportion of these three-producer lakes clearly increased under more-nutrient-rich conditions (Fig. 2). Eutrophic and hypertrophic lakes always contained potential MC producers, and the combination of all main producers was common.
FIG. 2.
Proportion of lakes with different combinations of potential MC producers, Anabaena, Microcystis, and Planktothrix, based on the presence of genus-specific mcyE genes in oligotrophic (TP, <10 μg/liter), mesotrophic (TP, 10 to 34 μg/liter), eutrophic (TP, 35 to 100 μg/liter), and hypertrophic (TP, >100 μg/liter) lakes.
Detection of MCs in water samples.
MC concentrations were measured with ELISA from unconcentrated and concentrated water samples. MCs were detected in only 20 of the unconcentrated water samples. The concentrations varied from 0.110 to 3.297 μg/liter (mean, 0.671 μg/liter; median, 0.302 μg/liter; standard deviation, 0.851 μg/liter). Of the measurements with concentrated water samples, MC concentrations of 11 samples were under the detection limit (0.16 μg/liter), while 24 samples exceeded the upper limit of detection (2.5 μg/liter). Because the original amount of filtered water differed from sample to sample, the lower and upper limits of detection corresponded to different MC concentrations in the original water samples. The lower detection limit ranged from <0.009 to <0.055 μg/liter, and the upper limit ranged from >0.135 to >1.765 μg/liter of lake water. The MC concentration of the concentrated water samples varied from 0.012 to 1.765 μg/liter (n = 59; mean, 0.306 μg/liter; median, 0.22 μg/liter; standard deviation, 0.364 μg/liter). The majority of the samples showed undetectable or small amounts of MCs due most likely to sampling before bloom season. In six samples, however, the MC concentration exceeded the WHO guideline value of 1 μg/liter for drinking water (57) (Fig. 1). All these samples came either from eutrophic or from hypertrophic lakes and contained two or three MC producers.
MCs were detected in 88% (58/66) of the lakes that showed the presence of MC producers by any primer pair. In addition, MCs were also detected in two lake samples with no PCR products. In both of these lakes, MCs were detected in concentrated samples but not in unconcentrated water samples. A low MC concentration, possibly resulting from a low number of MC producers, may explain the absence of PCR products in these two samples. Of the 10 lakes in which ELISA detected no MCs, 2 produced no PCR products with any primer pair, while 8 bore a positive PCR amplification with at least one primer pair, possibly representing inactive mcyE genotypes incapable of producing MCs.
Environmental variables associated with multiple MC producers by correlation analysis.
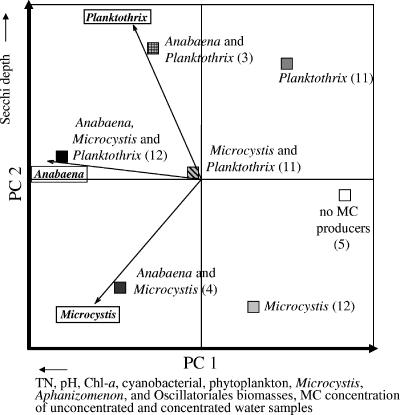
PCA was used to group the lake samples on the basis of the PCR results of Anabaena-, Microcystis-, and Planktothrix-specific primer pairs, and correlation analysis was used to determine environmental factors that significantly (P < 0.05) correlated to PC axes 1 and 2. Seven groups were formed: lakes with no PCR amplification, lakes with only Microcystis, those with only Planktothrix, those with Microcystis and Anabaena, those with Microcystis and Planktothrix, those with Anabaena and Planktothrix, and those with Anabaena, Microcystis, and Planktothrix (Fig. 3). PC axis 1 separated lakes with no or only one MC producer from lakes with multiple producers. Of the environmental variables phytoplankton and cyanobacterial biomasses, MC concentrations of concentrated and unconcentrated water samples, pH, TN, and chl-a concentration, Microcystis, Oscillatoriales, and Aphanizomenon biomasses correlated significantly (P < 0.05) with PC axis 1 (Table 2). Thus, the presence of several MC-producing genera at the same time seemed to be associated with greater cyanobacterial and phytoplankton biomasses, higher MC concentrations, a more alkaline pH, and growing concentrations of TN and chl-a (Fig. 3.). In contrast, the locations and sizes of the lakes, TP, DIP, TN/TP ratio, water color, Secchi depth, and Anabaena and Planktothrix biomasses did not significantly correlate with PC axis 1 and hence were likely unassociated with the presence of multiple MC producers. Secchi depth correlated most strongly (P = 0.095) with PC axis 2 (Table 2), which separated lakes with Planktothrix from those with no Planktothrix (Fig. 3), suggesting that MC-producing Planktothrix was more frequently present in lakes with greater transparency. PC axis 1 explained 42.4% and PC axis 2 explained 38.1% of the variation.
FIG. 3.
PCA ordination of the lake samples (n = 58) based on the mcyE PCR results (presence/absence) with Anabaena-, Microcystis-, and Planktothrix-specific primers. The different PCRs are indicated with string vectors, and the specificity of the PCR is indicated with a boxed genus name. Squares indicate positions of the lake groups, which have different combinations of mcyE genes. The number of lakes belonging to each group is in parentheses. Environmental variables correlating significantly with PC axis 1 and the most significant variable correlating with PC axis 2 are indicated with arrows showing the direction of correlation below or beside the corresponding axis. PC axes 1 and 2 explained 42.4% and 38.1% of the variation, respectively.
pH and TN: main explanatory variables of the presence of mcyE genes in the logistic regression model.
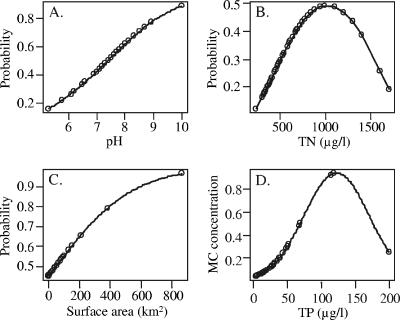
Logistic regression analysis was performed to determine whether environmental variables could explain the presence of the mcyE genes in water samples. The diagnostic inspection of model parameters did not suggest that colinearity among the explanatory variables would cause problems in stepwise regressions. The final model included the following variables: pH, TN, TN2 (second-order orthogonal polynomial term), and surface area (Table 3). Thus, the model predicted that the occurrence probability of the mcyE genes rises with a higher pH, a higher TN concentration, or a larger lake surface area (Fig. 4A to C). The effect of the term TN2, however, changes the otherwise linear response to a parabolic response (Fig. 4B), where the occurrence probability begins to drop with the highest TN concentrations. The parameter effects were similar for the three genera, as no significant interaction terms were detected (i.e., no statistically significant interaction terms appeared during the stepwise model selection). This suggested that the environmental variables affected potential MC-producing Anabaena, Microcystis, and Planktothrix organisms in similar manners. The results of genus-specific PCR amplifications were also seen in the logistic model. The Microcystis and Planktothrix mcyE genes were significantly more common than the Anabaena mcyE gene, since the model's Microcystis and Planktothrix estimates were significantly higher than 0 (Table 3). The logistic regression model was well suited to explain the occurrence of mcyE genes in the lakes studied, since the dispersion parameter (1.16) was close to 1. The slight overdispersion (residual deviance, 193.45 on 167 degrees of freedom) of the model, however, did not weaken the significance of the estimates or argue for modifications of the final model.
TABLE 3.
Parameters of a logistic regression model explaining the occurrence probability of the mcyE genes among the 58 lake samples
| Parameter | Estimate | SE | z test value | P |
|---|---|---|---|---|
| Intercepta | −0.85 | 0.31 | −2.74 | 0.006 |
| pH | 0.59 | 0.23 | 2.51 | 0.012 |
| TN | 0.32 | 0.21 | 1.54 | 0.124 |
| TN2 | −0.41 | 0.18 | −2.35 | 0.019 |
| Surface area | 0.51 | 0.28 | 1.78 | 0.075 |
| Microcystis mcyE geneb | 1.72 | 0.44 | 3.89 | <0.001 |
| Planktothrix mcyE geneb | 1.54 | 0.44 | 3.53 | <0.001 |
The probability level of Anabaena is adjusted via Intercept.
Categorical term measuring the differences in the occurrence probabilities of the mcyE genes of Microcystis and Planktothrix.
FIG. 4.
Estimated occurrence probability of the mcyE genes based on the logistic regression model parameters pH (A), TN (B), and the surface area of a lake (C) and on the estimated MC concentrations (μg/liter) based on linear regression model parameter TP (D). The effects of the other parameters were removed in the case of the logistic regression model (A to C) to reveal the subjective forms of response.
MC concentrations are explained mainly by TP in the linear regression model.
A linear regression model for the MC concentration (log transformed) was constructed in the same way as the logistic regression model for the presence of the mcyE genes. According to the final model, TP explained the MC concentration (log) of the sample (Table 4). The effect of the second-order (orthogonal) polynomial term TP2 again resulted in a parabolic response curve, where MC concentrations began to drop with the highest TP concentrations (Fig. 4D). Although reduced to include only one environmental variable, the model explained over a third of the variation of MC concentrations (R2, 0.34; adjusted R2, 0.31; residual standard error, 1.111 on 55 degrees of freedom; F statistic, 13.96 on 2 and 55 degrees of freedom; P < 0.001). Normality tests (Kolmogorov-Smirnow, D = 0.11, P = 0.525; Shapiro-Wilk, W = 0.96, P = 0.057) (41) permit us to assume that the residuals of the model were normally distributed. The visual diagnostics of residuals revealed no serious flaws with respect to assumptions of the regression theory.
TABLE 4.
Parameters of a linear regression model explaining the MC concentrations (log) measured from the 58 concentrated water samples
| Parameter | Estimate | SE | t test value | P |
|---|---|---|---|---|
| Intercept | −2.29 | 0.15 | −15.67 | <0.001 |
| TP | 0.62 | 0.15 | 4.20 | <0.001 |
| TP2 | −0.47 | 0.15 | −3.21 | 0.002 |
DISCUSSION
PCR results revealed that the cooccurrence of the three MC-producing genera, Anabaena, Microcystis, and Planktothrix, was more prevalent when the P concentration of the lakes increased but that oligotrophic lakes most commonly contained only one MC producer (Fig. 1 and 2). In the correlation analysis, the higher concentration of TN was instead significantly associated with multiple MC producer genera (Fig. 3). These results implied that nutrient-rich waters offered suitable conditions for the existence of multiple toxin producers and suggested that lake restoration efforts to reduce nutrient loading of the lakes could be beneficial in lowering the occurrence probability of MC-producing cyanobacterial genera. Additionally, we found more than one MC producer genus in over half of the samples, which further emphasizes the importance of studying the cooccurrence of several MC producers. Thus far, a majority of the studies have concentrated on the detection of only one producer genus (4). If more than one MC-producing genus exists in a lake, any one of them has the potential to become dominant or to form blooms in response to changed conditions (13, 23). Traditionally, microscopy has been used to determine the cyanobacterial composition of the sample. It cannot, however, differentiate MC-producing strains from the nonproducing ones, and previously, the only way to identify the toxin producer of the bloom was to isolate strains from the sample and prove their ability to produce MCs. Not only is such a procedure very time-consuming, but the result depends on the success of isolation (35, 46, 54).
The troublesome testing for the ability of strains to produce toxins is most probably the reason why correlations between environmental variables and MC producers have previously remained unreported. Instead, associations between environmental variables and MC concentrations have been studied earlier. In these studies, the role of the producing organism has been assigned to the dominant species present in the samples (9, 17, 18, 22, 34, 42). However, the existence of both toxic and nontoxic strains of the MC-producing genera and various amounts of MCs produced by a strain make it possible that the main MC producer of the lake is not necessarily the dominant species but could be the one that exists in smaller amounts, producing large amounts of MCs (45). Our method enabled us both to reveal the cooccurrence and to identify the potential MC producers, which offered a clear advantage over MC analyses. Thus, we could study which environmental variables were associated with the presence of MC producers and whether they were the same as variables correlated to MC concentrations. Lakes containing all three MC-producing genera were clearly separated from those lakes with no detected MC producers in PCA (Fig. 3). In addition to a higher TN concentration, known to promote the growth of cyanobacteria, factors such as a more alkaline pH, a higher chl-a concentration, and cyanobacterial and phytoplankton biomasses that usually prevail during blooms associated significantly with the presence of multiple MC-producing genera in the subsequent correlation analysis. This is in accordance with the hypotheses that MCs are produced under favorable growth conditions (45) and that production is associated with growth (37). The same variables have also positively correlated with higher MC concentrations in environmental studies (9, 17, 18, 22, 34, 42, 56). The correlation analysis also suggested that the Secchi depth separated lakes with MC-producing Planktothrix organisms and greater transparency from lakes with no MC-producing Planktothrix organisms and lower transparency along PC axis 2 (Fig. 3). This could reflect the ability of Planktothrix to grow and to form blooms in deep water layers (24). The relationship between multiple MC producers and higher MC concentrations that is anticipated based on the assumption of constitutive expression of the mcy genes (although it is equally possible that high MC concentrations in a sample are produced by a single genus or by many genera) was also seen in the correlation analysis (Fig. 3).
The environmental variables significant in the logistic regression model (pH, TN, surface area) for the occurrence probability of the mcyE genes agreed with the correlation analysis. The logistic regression also showed that both the nitrogen-fixing and non-nitrogen-fixing MC-producing genera were similarly associated with the environmental variables pH and nitrogen level but that Anabaena mcyE genes were generally present at levels lower than those of Microcystis and Planktothrix. At the time of the sampling, the majority of the lakes were not nitrogen limited (TN/TP < 10), a situation thought to select for non-nitrogen-fixing Microcystis and Planktothrix (45). If the model included more samples from August and September, when the amount of nitrogen begins to decline, the proportion of Anabaena could have been greater (47; P. Rajaniemi-Wacklin, A. Rantala, P. Kuuppo, K. Haukka, and K. Sivonen, unpublished data). The assumption that the same environmental variables would explain the presence of the mcyE genes and MC concentrations, however, was not seen in the linear regression model for MC concentrations. According to that model, only TP was needed to predict the MC concentration of the lakes. The difference between results could be due to the different characteristics of response variables, the other being presence and absence data and the other, quantitative data. However, the result itself was unsurprising, since TP is commonly associated positively with MC concentrations (9, 17, 18, 22) and is the key limiting nutrient for phytoplankton growth in fresh waters (44). Although different variables were found to explain the variation in response variables, they showed a similar trend. At the highest values of TN and TP, both the occurrence probability of the mcyE genes and the MC concentrations began to decline (Fig. 4B and D). This result is in accordance with the observation of Graham et al. (9) that maximal particulate MC values did not occur at the highest TN and TP values but reached the maxima at lower nutrient concentrations.
Potential microcystin producers were present in 84% of the lakes when studied with general mcyE primers and in 91% when studied with three genus-specific primer pairs. The results are in accordance with previous studies, where on average 59% (ranging from 25% to 92%) of the cyanobacterial blooms were hepatotoxic (45). In a previous survey of Finnish lakes, however, only 29% (54/188) of the blooms were hepatotoxic (46). Our present study revealed that potential toxin producers were over three times more frequent, although the analysis used samples collected before bloom season. At the time of sampling, only two of the lakes had blooms. The difference between our study and the previous survey is that the previous study analyzed the hepatotoxicity of apparent blooms with a nonsensitive mouse bioassay.
In our study, the most prevalent MC producer was Microcystis, which appeared in 70% of the lakes. This was not surprising considering that Microcystis is the most important MC producer throughout the world. Unexpectedly, however, our study revealed potential MC-producing Anabaena organisms in only 37% of the lakes. In a previous survey of Finnish lakes, both Anabaena (78%) and Microcystis (69%) were commonly found in hepatotoxic blooms by microscopy (46). The difference could again be explained by the sampling time. Most (60/70) of the samples studied were collected in mid-July. In Finnish lakes, however, Anabaena becomes more common only in late August and September, when nitrogen depletion starts to limit the growth of the non-nitrogen-fixing cyanobacterial genera (47; P. Rajaniemi-Wacklin, A. Rantala, P. Kuuppo, K. Haukka, and K. Sivonen, unpublished data). This could also explain why lakes with Anabaena as the sole MC producer went undetected.
Contrary to a previous survey, in which 25% of hepatotoxic blooms contained Oscillatoria (Planktothrix) (46), 63% of the lakes studied here contained potentially MC-producing Planktothrix. Since Planktothrix does not generally form surface blooms, its prevalence could have been underestimated in the previous survey, which studied only surface bloom samples. We had a better chance of harvesting Planktothrix cells in the integrate samples collected from 0 to 2 m, although the depth maximum of Planktothrix can occur even in deeper water layers (24). However, our study may also have overestimated the frequency of MC-producing Planktothrix. The faint amplification product from one nontoxic Planktothrix strain could mean that some of the PCR results with environmental lake samples were false positive. False positives could also indicate representatives of strains with mcy genes unable to produce MCs. Such inactive microcystin genotypes of Planktothrix strains appear to be quite common (3), and their proportions were estimated to be relatively high (5% and 21%) in two Alpine lakes (21). Whether this is also the case in Finnish lakes remains to be determined.
Of the many potentially usable mcy genes, we chose to use mcyE. This gene encodes McyE, a mixed polyketide peptide synthetase involved in the synthesis of Adda and the activation and addition of d-glutamate into the MC molecule. These two amino acids are crucial to toxicity and vary less than do the other amino acids of the molecule (45), and thus, this gene region was thought to be a reliable molecular marker for the detection of MC producers. In addition, we have shown in a phylogenetic study that mcyE sequences from different producer genera form their own clusters and remain excluded from horizontal gene transfer (40). Thus, primers and probes designed for this region are suitable not only for the detection but also for the identification of MC producers. Recently, other regions of the mcyE gene have also been found suitable for the detection of MC-producing Planktothrix strains in environmental samples (26) and MC- or nodularin-producing cyanobacteria (14).
In conclusion, the PCR method presented here is very sensitive and suitable for the detection of potential MC producers in environmental samples. Its ability to reveal the toxic potential of the lakes could be utilized as an early warning method for toxic blooms. Because of a very high similarity (97% to 100%) of genus-specific mcyE sequences used to design the primers, the method is not restricted to Finnish lake samples but may be used with any fresh water samples worldwide. In addition, this method can identify all the principal MC producers in the sample and thus can serve to evaluate the community composition of MC producers in a lake. Significance of the knowledge of all the producers was accentuated by the genus-specific PCR results, according to which over half of the lake samples contained at least two MC-producing genera and nearly a fourth contained all three genera studied. The importance of reducing nutrients as part of lake restoration and protecting waters from eutrophication was underlined by the results. The frequency of the lakes with three MC producers increased along with the trophic status (TP concentration) of the lakes. Statistical analyses revealed that TN significantly correlated to and explained the presence of multiple MC-producing genera. In the future, the primers now used in conventional PCR could be optimized for use in reverse transcriptase and quantitative real-time PCR applications with environmental RNA as a template. These methods would enable the determination of active populations of MC producers, the quantification of different producers, and, thus, the revelation of dominant producer genera.
Acknowledgments
This work was supported financially by the Academy of Finland (grants 53305 and 214457), EU projects MIDI-CHIP (EVK2-CT-1999-00026) and PEPCY (QLK4-CT-2002-02634) (K.S.), and the Viikki Graduate School of Biosciences (A.R.).
We thank Regional Environment Centres of Uusimaa, southwest Finland; Häme, Pirkanmaa, southeast Finland; and North Savo for collecting and sending the water samples. We are grateful to Lyudmila Saari and Matti Wahlsten for their valuable help in handling the samples.
REFERENCES
- 1.Carmichael, W. W. 1994. The toxins of cyanobacteria. Sci. Am. 270:78-86. [DOI] [PubMed] [Google Scholar]
- 2.Christiansen, G., J. Fastner, M. Erhard, T. Börner, and E. Dittmann. 2003. Microcystin biosynthesis in Planktothrix: genes, evolution, and manipulation. J. Bacteriol. 185:564-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Christiansen, G., R. Kurmayer, Q. Liu, and T. Börner. 2006. Transposons inactivate biosynthesis of the nonribosomal peptide microcystin in naturally occurring Planktothrix spp. Appl. Environ. Microbiol. 72:117-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dittmann, E., and T. Börner. 2005. Genetic contributions to the risk assessment of microcystin in the environment. Toxicol. Appl. Pharmacol. 203:192-200. [DOI] [PubMed] [Google Scholar]
- 5.Dittmann, E., B. A. Neilan, M. Erhard, H. von Döhren, and T. Börner. 1997. Insertional mutagenesis of a peptide synthetase gene that is responsible for microcystin production in the cyanobacterium Microcystis aeruginosa PCC 7806. Mol. Microbiol. 26:779-787. [DOI] [PubMed] [Google Scholar]
- 6.Edwards, U., T. Rogall, H. Blöcker, M. Emde, and E. C. Böttger. 1989. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 17:7843-7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giovannoni, S. J., E. F. DeLong, T. M. Schmidt, and N. R. Pace. 1990. Tangential flow filtration and preliminary phylogenetic analysis of marine picoplankton. Appl. Environ. Microbiol. 56:2572-2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goldberg, J., H.-B. Huang, Y.-G. Kwon, P. Greengard, A. C. Nairn, and J. Kuriyan. 1995. Three-dimensional structure of the catalytic subunit of protein serine/threonine phosphatase-1. Nature 376:745-753. [DOI] [PubMed] [Google Scholar]
- 9.Graham, J. L., J. R. Jones, S. B. Jones, J. A. Downing, and T. E. Clevenger. 2004. Environmental factors influencing microcystin distribution and concentration in the Midwestern United States. Water Res. 38:4395-4404. [DOI] [PubMed] [Google Scholar]
- 10.Hisbergues, M., G. Christiansen, L. Rouhiainen, K. Sivonen, and T. Börner. 2003. PCR-based identification of microcystin-producing genotypes of different cyanobacterial genera. Arch. Microbiol. 180:402-410. [DOI] [PubMed] [Google Scholar]
- 11.Honkanen, R. E., J. Zwiller, R. E. Moore, S. L. Daily, B. S. Khatra, M. Dukelow, and A. L. Boynton. 1990. Characterization of microcystin-LR, a potent inhibitor of type 1 and type 2A protein phosphatases. J. Biol. Chem. 265:19401-19404. [PubMed] [Google Scholar]
- 12.Hotto, A., M. Satchwell, and G. Boyer. 2005. Seasonal production and molecular characterization of microcystins in Oneida Lake, New York, USA. Environ. Toxicol. 20:243-248. [DOI] [PubMed] [Google Scholar]
- 13.Jacquet, S., J.-F. Briand, C. Leboulanger, C. Avois-Jacquet, L. Oberhaus, B. Tassin, B. Vinçon-Leite, G. Paolini, J.-C. Druart, O. Anneville, and J.-F. Humbert. 2005. The proliferation of the toxic cyanobacterium Planktothrix rubescens following restoration of the largest natural French lake (Lac du Bourget). Harmful Algae 4:651-672. [Google Scholar]
- 14.Jungblut, A.-D., and B. A. Neilan. 2006. Molecular identification and evolution of the cyclic peptide hepatotoxins, microcystin and nodularin, synthetase genes in three orders of cyanobacteria. Arch. Microbiol. 185:107-114. [DOI] [PubMed] [Google Scholar]
- 15.Jurczak, T., M. Tarczynska, K. Izydorczyk, J. Mankiewicz, M. Zalewski, and J. Meriluoto. 2005. Elimination of microcystins by water treatment processes—examples from Sulejow Reservoir, Poland. Water Res. 39:2394-2406. [DOI] [PubMed] [Google Scholar]
- 16.Kaebernick, M., T. Rohrlack, K. Christoffersen, and B. A. Neilan. 2001. A spontaneous mutant of microcystin biosynthesis: genetic characterization and effect on Daphnia. Environ. Microbiol. 3:669-679. [DOI] [PubMed] [Google Scholar]
- 17.Kotak, B. G., A. K.-Y. Lam, E. E. Prepas, and S. E. Hrudey. 2000. Role of chemical and physical variables in regulating microcystin-LR concentration in phytoplankton of eutrophic lakes. Can. J. Fish. Aquat. Sci. 57:1584-1593. [Google Scholar]
- 18.Kotak, B. G., A. K.-Y. Lam, E. E. Prepas, S. L. Kenefick, and S. E. Hrudey. 1995. Variability of the hepatotoxin microcystin-LR in hypereutrophic drinking water lakes. J. Phycol. 31:248-263. [Google Scholar]
- 19.Kuiper-Goodman, T., I. Falconer, and J. Fitzgerald. 1999. Human health aspects, p. 113-153. In I. Chorus and J. Bartram (ed.), Toxic cyanobacteria in water: a guide to their public health consequences, monitoring and management. E & FN Spon, London, United Kingdom.
- 20.Kurmayer, R., E. Dittmann, J. Fastner, and I. Chorus. 2002. Diversity of microcystin genes within a population of the toxic cyanobacterium Microcystis spp. in Lake Wannsee (Berlin, Germany). Microb. Ecol. 43:107-118. [DOI] [PubMed] [Google Scholar]
- 21.Kurmayer, R., G. Christiansen, J. Fastner, and T. Börner. 2004. Abundance of active and inactive microcystin genotypes in populations of the toxic cyanobacterium Planktothrix spp. Environ. Microbiol. 6:831-841. [DOI] [PubMed] [Google Scholar]
- 22.Lahti, K., J. Rapala, M. Färdig, M. Niemelä, and K. Sivonen. 1997. Persistence of cyanobacterial hepatotoxin, microcystin-LR in particulate material and dissolved in lake water. Water Res. 31:1005-1012. [Google Scholar]
- 23.Lepistö, L., A. Räike, and O.-P. Pietiläinen. 1999. Long-term changes of phytoplankton in a eutrophicated boreal lake during the past one hundred years (1893-1998). Algol. Stud. 94:223-244. [Google Scholar]
- 24.Lindholm, T. 1992. Ecological role of depth maxima of phytoplankton. Arch. Hydrobiol. Beih. Ergebn. Limnol. 35:33-45. [Google Scholar]
- 25.MacKintosh, C., K. A. Beattie, S. Klumpp, P. Cohen, and G. A. Codd. 1990. Cyanobacterial microcystin-LR is a potent and specific inhibitor of protein phosphatases 1 and 2A from both mammals and higher plants. FEBS Lett. 264:187-192. [DOI] [PubMed] [Google Scholar]
- 26.Mbedi, S., M. Welker, J. Fastner, and C. Wiedner. 2005. Variability of the microcystin synthetase gene cluster in the genus Planktothrix (Oscillatoriales, Cyanobacteria). FEMS Microbiol. Lett. 245:299-306. [DOI] [PubMed] [Google Scholar]
- 27.Mikalsen, B., G. Boison, O. M. Skulberg, J. Fastner, W. Davies, T. M. Gabrielsen, K. Rudi, and K. S. Jakobsen. 2003. Natural variation in the microcystin synthetase operon mcyABC and impact on microcystin production in Microcystis strains. J. Bacteriol. 185:2774-2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Niemi, J., P. Heinonen, S. Mitikka, H. Vuoristo, O.-P. Pietiläinen, M. Puupponen, and E. Rönkä (ed.). 2001. The Finnish Environment 445. The Finnish Eurowaternet—with information about Finnish water resources and monitoring strategies. Finnish Environment Institute, Edita Ltd., Helsinki.
- 29.Nishizawa, T., M. Asayama, K. Fuji, K.-I. Harada, and M. Shirai. 1999. Genetic analysis of the peptide synthetase genes for a cyclic heptapeptide microcystin in Microcystis spp. J. Biochem. 126:520-529. [DOI] [PubMed] [Google Scholar]
- 30.Nishizawa, T., A. Ueda, M. Asayama, K. Fuji, K.-I. Harada, K. Ochi, and M. Shirai. 2000. Polyketide synthase gene coupled to the peptide synthetase module involved in the biosynthesis of the cyclic heptapeptide microcystin. J. Biochem. 127:779-789. [DOI] [PubMed] [Google Scholar]
- 31.Nonneman, D., and P. V. Zimba. 2002. A PCR-based test to assess the potential for microcystin occurrence in channel catfish production ponds. J. Phycol. 38:230-233. [Google Scholar]
- 32.Nübel, U., F. Garcia-Pichel, and G. Muyzer. 1997. PCR primers to amplify 16R rRNA genes from cyanobacteria. Appl. Environ. Microbiol. 63:3327-3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.OECD. 1982. Eutrophication of water, monitoring, assessment and control. Organization for economic cooperation and development, Paris, France.
- 34.Oh, H.-M., S. J. Lee, J.-H. Kim, H.-S. Kim, and B.-D. Yoon. 2001. Seasonal variation and indirect monitoring of microcystin concentrations in Daechung Reservoir, Korea. Appl. Environ. Microbiol. 67:1484-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ohtake, A., M. Shirai, T. Aida, N. Mori, K.-I. Harada, K. Matsuura, M. Suzuki, and M. Nakano. 1989. Toxicity of Microcystis species isolated from natural blooms and purification of the toxin. Appl. Environ. Microbiol. 55:3202-3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Olrik, K., P. Blomqvist, P. Brettum, G. Cronberg, and P. Eloranta. 1998. Swedish Environmental Protection Agency Report 4860. Methods for quantitative assessment of phytoplankton in freshwaters, part I: sampling, processing and application in freshwater environmental monitoring programmes. Swedish Environmental Protection Agency, Stockholm.
- 37.Orr, P. T., and G. J. Jones. 1998. Relationship between microcystin production and cell division rates in nitrogen-limited Microcystis aeruginosa cultures. Limnol. Oceanogr. 43:1604-1614. [Google Scholar]
- 38.Ouahid, Y., G. Pérez-Silva, and F. F. del Campo. 2005. Identification of potentially toxic environmental Microcystis by individual and multiple PCR amplifications of specific microcystin synthetase gene regions. Environ. Toxicol. 20:235-242. [DOI] [PubMed] [Google Scholar]
- 39.Rajaniemi-Wacklin, P., A. Rantala, M. A. Mugnai, S. Turicchia, S. Ventura, J. Komarkova, L. Lepistö, and K. Sivonen. 2005. Correspondence between phylogeny and morphology of Snowella spp. and Woronichinia naegeliana, cyanobacteria commonly occurring in lakes. J. Phycol. 42:226-232. [Google Scholar]
- 40.Rantala, A., D. P. Fewer, M. Hisbergues, L. Rouhiainen, J. Vaitomaa, T. Börner, and K. Sivonen. 2004. Phylogenetic evidence for the early evolution of microcystin synthesis. Proc. Natl. Acad. Sci. USA 101:568-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.R Development Core Team, R Foundation for Statistical Computing. 2005. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Online.] http://www.R-project.org.
- 42.Rolland, A., D. F. Bird, and A. Giani. 2005. Seasonal changes in composition of the cyanobacterial community and the occurrence of hepatotoxic blooms in the eastern townships, Québec, Canada. J. Plankton Res. 27:683-694. [Google Scholar]
- 43.Rouhiainen, L., T. Vakkilainen, B. L. Siemer, W. Buikema, R. Haselkorn, and K. Sivonen. 2004. Genes coding for hepatotoxic heptapeptides (microcystins) in the cyanobacterium Anabaena strain 90. Appl. Environ. Microbiol. 70:686-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schindler, D. W. 1977. Evolution of phosphorus in lakes. Science 195:260-262. [DOI] [PubMed] [Google Scholar]
- 45.Sivonen, K., and G. Jones. 1999. Cyanobacterial toxins, p. 41-111. In I. Chorus and J. Bartram (ed.), Toxic cyanobacteria in water: a guide to their public health consequences, monitoring, and management. E & FN Spon, London, United Kingdom.
- 46.Sivonen, K., S. I. Niemelä, R. M. Niemi, L. Lepistö, T. H. Luoma, and L. A. Räsänen. 1990. Toxic cyanobacteria (blue-green algae) in Finnish fresh and coastal waters. Hydrobiology 190:267-275. [Google Scholar]
- 47.Sommer, U., Z. M. Glliwicz, W. Lampert, and A. Duncan. 1986. The PEG*-model of seasonal succession of planktonic events in fresh waters. Arch. Hydrobiol. 106:433-471. [Google Scholar]
- 48.Tillett, D., E. Dittmann, M. Erhard, H. von Döhren, T. Börner, and B. A. Neilan. 2000. Structural organization of microcystin biosynthesis in Microcystis aeruginosa PCC7806: an integrated peptide-polyketide synthetase system. Chem. Biol. 7:753-764. [DOI] [PubMed] [Google Scholar]
- 49.Tillett, D., D. L. Parker, and B. A. Neilan. 2001. Detection of toxigenicity by a probe for the microcystin synthetase A gene (mcyA) of the cyanobacterial genus Microcystis: comparison of toxicities with 16S rRNA and phycocyanin operon (phycocyanin intergenic spacer) phylogenies. Appl. Environ. Microbiol. 67:2810-2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Utermöhl, H. 1958. Zur Vervollkommnung der quantitativen Phytoplankton-Methodik. Mitt. Int. Ver. Limnol. 9:1-38. [Google Scholar]
- 51.Vaitomaa, J., A. Rantala, K. Halinen, L. Rouhiainen, P. Tallberg, L. Mokelke, and K. Sivonen. 2003. Quantitative real-time PCR for determination of microcystin synthetase gene E copy numbers for Microcystis and Anabaena in lakes. Appl. Environ. Microbiol. 69:7289-7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Venables, W. N., and B. D. Ripley. 2002. Modern applied statistics with S. Springer-Verlag, New York, N.Y.
- 53.Vezie, C., L. Brient, K. Sivonen, G. Bertru, J.-C. Lefeuvre, and M. Salkinoja-Salonen. 1997. Occurrence of microcystin-containing cyanobacterial blooms in freshwaters of Brittany (France). Arch. Hydrobiol. 139:401-413. [Google Scholar]
- 54.Vezie, C., L. Brient, K. Sivonen, G. Bertru, J.-C. Lefeuvre, and M. Salkinoja-Salonen. 1998. Variation of microcystin content of cyanobacterial blooms and isolated strains in Lake Grand-Lieu (France). Microb. Ecol. 35:126-135. [DOI] [PubMed] [Google Scholar]
- 55.Via-Ordorika, L., J. Fastner, R. Kurmayer, M. Hisbergues, E. Dittmann, J. Komarek, M. Erhard, and I. Chorus. 2004. Distribution of microcystin-producing and non-microcystin-producing Microcystis sp. in European freshwater bodies: detection of microcystins and microcystin genes in individual colonies. Syst. Appl. Microbiol. 27:592-602. [DOI] [PubMed] [Google Scholar]
- 56.Wicks, R. J., and P. G. Thiel. 1990. Environmental factors affecting the production of peptide toxins in floating scums of the cyanobacterium Microcystis aeruginosa in a hypertrophic African reservoir. Environ. Sci. Technol. 24:1413-1418. [Google Scholar]
- 57.World Health Organization. 2004. Guidelines for drinking water quality, 3rd ed., vol. 1. Recommendations. World Health Organization, Geneva, Switzerland.
- 58.Yoshida, M., T. Yoshida, Y. Takashima, R. Kondo, and S. Hiroishi. 2005. Genetic diversity of the toxic cyanobacterium Microcystis in Lake Mikata. Environ. Toxicol. 20:229-234. [DOI] [PubMed] [Google Scholar]