Abstract
Autolysis is self-degradation of the bacterial cell wall that results in the release of enzymes and DNA. Autolysis of starter bacteria, such as lactococci and propionibacteria, is essential for cheese ripening, but our understanding of this important process is limited. This is mainly because the current tools for measuring autolysis cannot readily be used for analysis of bacteria in mixed populations. We have now addressed this problem by species-specific detection and quantification of free DNA released during autolysis. This was done by use of 16S rRNA gene single-nucleotide extension probes in combination with competitive PCR. We analyzed pure and mixed populations of Lactococcus lactis subsp. lactis and three different species of Propionibacterium. Results showed that L. lactis subsp. lactis INF L2 autolyzed first, followed by Propionibacterium acidipropionici ATCC 4965, Propionibacterium freudenreichii ISU P59, and then Propionibacterium jensenii INF P303. We also investigated the autolytic effect of rennet (commonly used in cheese production). We found that the effect was highly strain specific, with all the strains responding differently. Finally, autolysis of L. lactis subsp. lactis INF L2 and P. freudenreichii ISU P59 was analyzed in a liquid cheese model. Autolysis was detected later in this cheese model system than in broth media. A challenge with DNA, however, is DNA degradation. We addressed this challenge by using a DNA degradation marker. We obtained a good correlation between the degradation of the marker and the target in a model experiment. We conclude that our DNA approach will be a valuable tool for use in future analyses and for understanding autolysis in mixed bacterial populations.
The present tools for measuring autolysis are not easily adaptable to analysis of mixed populations. This severely limits our understanding of autolysis in microbial populations, such as in cheese. Autolysis is self-degradation of the bacterial cell wall due to hydrolysis of the peptidoglycan network by autolysins (32, 34, 36, 37), and the result is leakage of intracellular components (9, 27). Lactococcus lactis subsp. lactis and Propionibacterium freudenreichii are bacteria essential to the ripening of Norwegian Jarlsberg cheese (17). The propionibacteria produce propionic acid, acetic acid, and CO2, with the latter responsible for the typical eye formation. In addition, this secondary starter contributes to the characteristic sweet, nutty flavor, provided mainly by free proline (20-22). Autolysis of lactococci is important for cheese ripening, in which the release of intracellular enzymes plays an important role in cheese flavor development (9, 40, 48). The role and contribution of intracellular enzymes from propionibacteria in the ripening process are poorly understood (11, 12, 23, 27, 40). Proteolytic enzymes, such as chymotrypsin and pepsin, are reported to modify autolytic activity (28, 29, 38, 39). Rennet, consisting of chymosin and bovine pepsin, hydrolyzes the κ-casein in milk and thus is a vital factor in cheese production (40, 48).
Several different methods are currently used to monitor bacterial autolysis. Electron microscopy and detection of differently released intracellular components are methods commonly used for milk and model systems and in cheese experiments, whereas absorbance measurements are applied in broth and buffer approaches (6, 8, 9, 12, 18, 26, 27, 32, 47, 49) (among other approaches). The intracellular metabolic enzyme lactate dehydrogenase (LDH), which is widely distributed among lactic acid bacteria, is commonly used as an enzymatic marker for autolysis (9, 30). Østlie et al. (32) reported that RNA and DNA were good markers for measuring initiation of autolysis. Measurement of CFU is not a good marker for autolysis since it demonstrates only decrease in culturability and not lysis (5, 9, 45). CFU measurements, however, are often used in combination with other approaches (4, 49). A fluorescence method introduced by Bunthof and coworkers (5) uses dual staining with a LIVE/DEAD BacLight viability kit to measure lysis of L. lactis subsp. lactis.
A limitation with the above-mentioned methods is that they do not discriminate between different bacterial species. Recently, however, an immunoassay for the detection of two bacterial species was developed. Valence et al. (46) developed a species-specific assay in which autolysis of Lactobacillus helveticus and P. freudenreichii was distinguished and monitored using immunoblotting analysis with Emmental cheese. Moreover, Deutsch et al. (10) have developed an immunological method for differentiating between lysis of L. helveticus and Streptococcus thermophilus based on different isomers of LDH. The challenge with these approaches lies in the difficulty in identifying species-specific immunological markers. Furthermore, it is difficult to obtain quantitative results by using immunological methods.
The objective of the present work was to develop and evaluate a DNA-based assay for detection and quantification of growth and autolysis of lactococci and propionibacteria in mixed populations. We applied species-specific detection (44) to quantify free DNA released during autolysis. The concept was demonstrated in pure and mixed sample cultures. We included rennet-treated samples to examine the influence of rennet on autolysis. Finally, autolysis in a liquid cheese model was evaluated to better simulate the situation in real cheese production. The challenge with DNA instability was addressed using a DNA degradation marker.
MATERIALS AND METHODS
Organisms and cultivation procedure.
The experimental series were conducted with Lactococcus lactis subsp. lactis INF L2 (Department of Chemistry, Biotechnology and Food Science, Norwegian University of Life Sciences), Propionibacterium jensenii INF P303, P. freudenreichii ISU P59 (Department of Food Science and Human Nutrition, Iowa State University, Ames), and Propionibacterium acidipropionici ATCC 4965 (American Type Culture Collection, Rockville, Maryland). Lactococci were grown in M17 broth (Oxoid) (42) with the addition of 0.5% (wt/vol) lactose (LM17), and propionibacteria were grown in sodium lactate broth (SLB) (31), with pH adjusted to 6.8. The impact of rennet on autolysis was analyzed with the addition of 1% rennet (Standard 190; Chr. Hansen, Hørsholm, Denmark). All strains were cultivated at 30°C in 500-ml screw cap bottles containing 200 ml media. Exponentially growing cells were used as an inoculum (1%). Samples were taken regularly during a 5-week period. The mixed sample was prepared by mixing the cultivated pure cultures in equal proportions. Viable-cell counts (CFU/ml), absorbance (A600), and pH measurements were determined, and the experiments were repeated at least three times.
Liquid cheese model.
The liquid cheese model system was recently developed by Treimo et al. (44) in order to imitate the lactose content and pH of the Jarlsberg cheese environment. The liquid cheese model system consists of 3% sodium caseinate (wt/vol) (Tine Norwegian Dairies BA, Norway), 0.25% lactose (wt/vol), 1.9% β-glycerophosphate (wt/vol) (Sigma), and 2.5% yeast extract (wt/vol) (Oxoid) dissolved in a simulated milk ultrafiltrate buffer (15). Growth of L. lactis subsp. lactis INF L2 and P. freudenreichii ISU P59 was measured in pure cultures and in cocultures, and the cultivation was carried out as previously described. Samples were analyzed after 0 h, 24 h, 48 h, 7 days, and 5 weeks.
DNA purification.
Cell DNA was retrieved by centrifugation of 1-ml samples for 5 min at 10,000 rpm. The supernatant was discharged, and the pellet was resuspended in 1 ml Tris-EDTA (TE) buffer (pH 7.8). The cells were disrupted mechanically in a Fast-Prep bead beater (Bio 101, La Jolla, California) (44). Free DNA was obtained by centrifugation of 5-ml samples for 5 min at 10,000 rpm (Beckman J2-MC centrifuge, JA-17 rotor) and at 4°C. The supernatant was then passed through a sterile filter (0.22 μm pore size; Millex-GP; Millipore, Bedford, Massachusetts) to secure a cell-free fraction and immediately frozen by liquid nitrogen. DNA was purified from both the supernatant and the pellet fractions with a DNeasy tissue kit, following the recommendations of the manufacturer (QIAGEN, Hilden, Germany). Each DNA purification was done in triplicate, adding a competitor in three different concentrations (10-fold dilutions) prior to the purifications (44).
DNA degradation assay.
The potential of using an internal control to measure DNA degradation activity was evaluated by use of DNase I (Roche Diagnostics, Mannheim, Germany) in different concentrations (200, 20, 2, and 0.2 U/ml). DNA degradation was analyzed after 0, 10, 20, 30, 40, 50, and 60 min. The samples evaluated contained both the competitor and L. lactis subsp. lactis INF L2 DNA. Separate detection of the two targets was done using real-time PCR (ABI 7900 HT; Applied Biosystems) with the Fast Start SYBR green Master (Rox) reagents (Roche). We used a primer pair equal to the universal 16S rRNA gene primer pair (44), with three nucleotides unique to the two amplicons for both the forward and the reverse primers. The real-time PCR was initiated with a denaturation step at 95°C for 10 min. The PCR was followed by 40 cycles with the following denaturation, annealing, and synthesis parameters: 95°C for 15 s, 61°C for 30 s, and 72°C for 45 s. An extension step at 95°C for 15 s, 60°C for 15 s, and 95°C for 15 s was included at the end of the PCR.
DNA quantification.
We used competitive PCR for quantification (35), following the protocol developed by Treimo et al. (44). The genes encoding the 16S rRNA gene product were coamplified with the competitor through application of a primer pair targeted toward a universally conserved region of the 16S rRNA gene. The competitive PCR was carried out for the three different dilutions (10-fold) of the competitor, followed by fragment separation on agarose electrophoresis gel. Samples with band intensities corresponding approximately to those of the competitor and target were selected and analyzed further using the DNA microarray approach.
The probes used in this assay were previously identified and constructed by Treimo et al. (44). The universal probe was constructed from a conserved region of the 16S rRNA gene, whereas the specific probes were constructed from informative nucleotide positions in the alignment from regions with relatively high variability. The probes were constructed so that the nucleotide following at the 3′ end was a discriminatory cytosine when using sequence-specific labeling (the mini-sequencing principle) (41).
The nucleotides from the competitive PCR were dephosphorylated and subsequently used in the cyclic labeling reaction with 6-carboxy-N,N,N′,N′-tetramethylrhodamine (TAMRA)-dideoxycytosine triphosphate (NEN Life Science Products, Inc., Boston, Massachusetts).
The hybridization experiments were carried out with CMT-GAPS coated slides (Corning, Inc, Corning, New York) for microarrays. Each slide was divided into 24 independent reaction chambers by using a homemade silicone rubber mask. Complementary probes containing a 15-mer poly(T) tail at the 5′ end were spotted on the slides in triplicate at a concentration of 40 pmol/ml in 50% dimethyl sulfoxide (Norwegian Radium Hospital, Oslo, Norway) and then UV cross linked (200 mJ) and baked for 3 h at 80°C. The slides were pretreated with succinyl anhydride and kept dry in the dark. Before use, the slides were prehybridized, washed in distilled water and subsequently in isopropanol, dried, and used within 1 hour. ArrayHyb LowTemp hybridization buffer (Sigma) was added to the sample mixture, heated, and then applied to the hybridization chamber. The hybridizations were done overnight at 30°C. Then, the slides were washed with different dilutions of SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and sodium dodecyl sulfate to increase the stringency of the environment and eventually in distilled water. The slides were then transferred to ethyl alcohol (100%) before being dried in a centrifuge. All wash solutions were passed through a filter (0.22 μm pore size) before use.
Fluorescence was detected by a Scan Array Express microarray scanner (Perkin Elmer, Boston, Massachusetts), and Scan Array Express version 2.0 software was used. Data were analyzed with Quant Array version 3.0 microarray analysis software.
Normalization.
Cyanine-5 (Cy5)-prelabeled probes, corresponding to the sequence-specifically TAMRA-labeled probes, were added to each sample before hybridization as an internal control. An internal control was included to make the assay hybridization independent (35). An additional internal control probe prelabeled with both Cy5 and TAMRA was included as a labeling control as well as for normalization since two different lasers were used to scan TAMRA and Cy5 (50).
Statistical analyses.
The results are presented as the means for two or three replicates ± the standard deviations. The significance of the differences detected for the species-specific DNA composition was determined by a two-sample t test provided in the Excel software package (Microsoft Excel 2000, Microsoft Corp., Redmond, Washington).
RESULTS AND DISCUSSION
DNA as a marker of autolysis.
The amount of free DNA is a result of the balance between DNA release and degradation. We determined whether shifts in this balance could be used as an indicator of growth and autolysis in mixed populations by evaluating whether the accumulation of total free DNA could be correlated to autolysis. Østlie et al. (32) reported that total DNA content was a good marker for measuring initiation of autolysis. Our comparison between free DNA accumulation and A600 measurements confirmed these observations and showed that accumulation of free DNA corresponded to initiation of autolysis for all strains evaluated (results not shown). In particular, the L. lactis subsp. lactis INF L2 strain showed a rapid release of free DNA. The relative changes in DNA were shifting in the supernatant and pellet fractions throughout the incubation period, probably due to continuous DNA release and degradation.
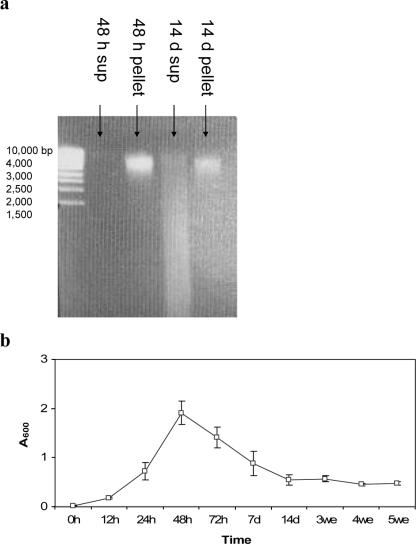
Figure 1 illustrates a comparison between DNA accumulation and absorbance measurements. We have shown DNA from supernatant and pellet fractions of P. freudenreichii ISU P59 on agarose gel (Fig. 1a) at maximum and minimum A600 (Fig. 1b). We observed an increase in free DNA content in the supernatant between 2 and 14 days, while the amount of pellet-bound DNA decreased during this period. The corresponding decrease determined by absorbance measurements is shown in Fig. 1b. All the strains included in our work showed the same main pattern, but the kinetics in DNA release was different.
FIG. 1.
Purified DNA on agarose gel from supernatant (free DNA) (sup) and pellet (cell DNA) fractions of P. freudenreichii ISU P59 cultivated in SLB broth at 30°C at 2 and 14 days (a) and absorbance measurements (A600) for the 5-week incubation period (b). The samples were separated for 1 h (90 V) on an agarose gel (0.8%) containing ethidium bromide (1 μg/ml). A DNA ladder (MBI Fermentas) ranging from 1,500 bp to 10,000 bp is shown. Error bars represent standard deviations for at least four replicates.
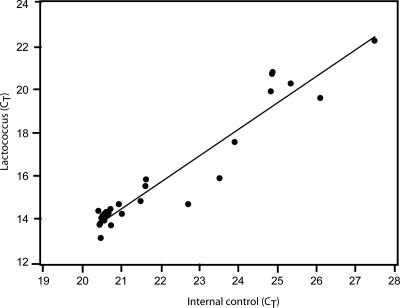
Previous work using DNA as a marker to estimate autolysis documented that DNA degradation needs to be taken into account, as DNA liberated from the cells by autolysis degenerates at different rates (2, 3, 25, 31, 32). We have addressed this problem by evaluating a model assay where an internal DNA degradation standard was included. We obtained a high correlation between the target signal and the standard (Fig. 2), indicating that it should be feasible to include such standards to improve autolysis detection in the future.
FIG. 2.
Comparison of Lactococcus lactis subsp. lactis INF L2 and internal control DNA after degradation by DNase I. DNA was degraded with DNase I as described in Materials and Methods. A scatter plot and a regression line are shown for the corresponding real-time PCR cycle threshold (CT) values. The squared regression coefficient (R2) for the regression line was 0.92. There was no detectable cross-reactivity between the two primer pairs for the two targets analyzed.
Measurements of species-specific DNA release.
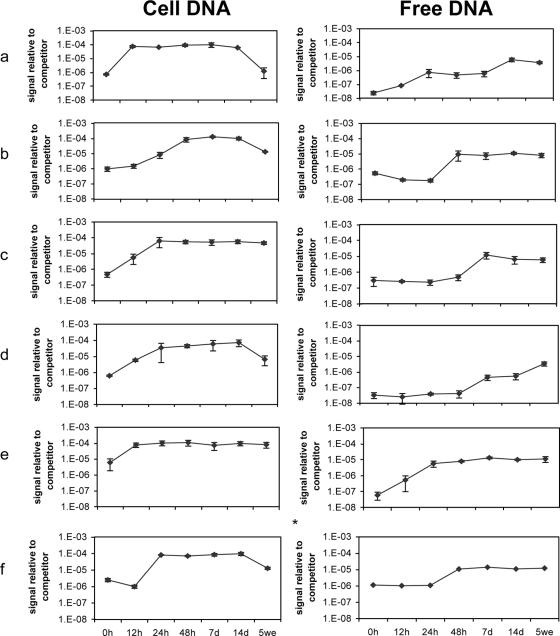
We determined the species-specific DNA release from bacteria in both pure and mixed samples. This was done by use of competitive PCR in combination with microarrays (44). Shifts in both free and cell DNA were analyzed. The general pattern for the cell DNA fraction was that the total bacterial 16S rRNA gene content increased initially, shifted to a constant level, and then decreased during the incubation period (Fig. 3). The decrease, however, was less profound for P. freudenreichii ISU P59 and the mixed sample. Lactococcus lactis subsp. lactis INF L2 showed maximum DNA content in the cell fraction after 12 h, followed by the propionibacteria, with a maximum between 24 and 48 h. This corresponds to a slower growth rate for propionibacteria than for lactococci (1).
FIG. 3.
16S rRNA gene microarray quantification of total cell and free bacterial DNA in Lactococcus lactis subsp. lactis INF L2 (a), Propionibacterium acidipropionici ATCC 4965 (b), P. freudenreichii ISU P59 (c), P. jensenii INF P303 (d), a mixed sample of the above-mentioned strains (e), and a coculture of the above-mentioned propionibacteria (f). The total bacterial DNA content was calculated as previously described (44). The 10−7 dilution of the competitor corresponds to approximately 107 DNA copies per ml in the original sample. The error bars for all samples indicate standard deviations for two or three independent quantifications. *, no parallels are available; d, days; we, weeks.
The species-specific 16S rRNA gene content in the mixed samples showed that L. lactis subsp. lactis INF L2 dominated after 12 h (Table 1). The relative DNA content of P. acidipropionici ATCC 4965 increased significantly between 12 and 24 h (P = 0.03) and further to a maximum after 48 h, while there was a relative maximum for P. freudenreichii ISU P59 after 48 h (P = 0.01) followed by a decrease. The relative amount of P. jensenii INF 303 DNA increased significantly from 24 h to 5 weeks (P = 0.02). The cell fraction of the coculture of propionibacteria was dominated by P. acidipropionici ATCC 4965. A difference between the coculture and the mixed samples was that P. acidipropionici ATCC 4965 reached a relative maximum DNA content earlier in the coculture (Table 1).
TABLE 1.
Species-specific DNA compositiona
| Bacterial strain and probe | Incubation time for indicated DNA type
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 h
|
12 h
|
24 h
|
48 h
|
7 d
|
14 d
|
5 wk
|
||||||||
| Cellc | Free | Cell | Free | Cell | Free | Cell | Free | Cell | Free | Cell | Free | Cellc | Free | |
| L. lactis subsp. lactis INF L2 | ||||||||||||||
| LCL | 71 ± 8b | - | 77 ± 30b | 70 ± 30 | 77 ± 29b | 73 ± 29 | 77 ± 30b | 73 ± 30 | 80 ± 20b | 76 ± 20 | 80 ± 27b | 77 ± 27 | 66 ± 20 | 72 ± 20 |
| PAB#3 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| PAB#4 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| PAB#5 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| P. acidipropionici ATCC 4965 | ||||||||||||||
| LCL | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| PAB#3 | 69 ± 19b | - | 79 ± 14b | - | 81 ± 12b | - | 81 ± 6b | 75 ± 7 | 80 ± 9b | 67 ± 11 | 87 ± 22b | 73 ± 19 | 75 ± 14 | 81 ± 4 |
| PAB#4 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| PAB#5 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| P. freudenreichii ISU P59 | ||||||||||||||
| LCL | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| PAB#3 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| PAB#4 | 52 ± 5b | - | 70 ± 13b | - | 73 ± 18b | - | 73 ± 3b | 57 ± 15 | 72 ± 2b | 66 ± 1 | 49 ± 7b,c | 72 ± 22 | 50 ± 7 | 68 ± 10 |
| PAB#5 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| P. jensenii INF P303 | ||||||||||||||
| LCL | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| PAB#3 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| PAB#4 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| PAB#5 | 37 ± 10b | - | 54 ± 13b | - | 62 ± 4b | - | 65 ± 9b | - | 59 ± 4b | 49 ± 6 | 62 ± 19b | 54 ± 8 | 52 ± 6 | 50 ± 8 |
| MIX | ||||||||||||||
| LCL | 34 ± 4b | - | 66 ± 6b | 34 ± 3 | 48 ± 12b | 51 ± 7 | 29 ± 2b | 18 ± 2 | 43 ± 6b | 20 ± 4 | 43 ± 9b | 20 ± 4 | 28 ± 6 | 20 ± 1 |
| PAB#3 | 27 ± 6b | - | 18 ± 1b | - | 23 ± 2b | - | 35 ± 8b | 44 ± 1 | 33 ± 8b | 18 ± 3 | 26 ± 6b | 25 ± 5 | 25 ± 4 | 34 ± 13 |
| PAB#4 | - | - | - | - | 21 ± 9b | - | 24 ± 2b | 21 ± 8 | 11 ± 8b | 49 ± 15 | 13 ± 0b | 40 ± 14 | 15 ± 4 | 35 ± 10 |
| PAB#5 | - | - | - | - | - | - | 13 ± 8b | - | 13 ± 4b | 15 ± 7 | 18 ± 4b | 15 ± 7 | 32 ± 4 | 12 ± 4 |
| PAB | NA | NA | NA | NA | ||||||||||
| LCL | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| PAB#3 | 48 ± 1 | - | 56 ± 10 | - | 58 ± 10 | - | 48 ± 14 | 34 | 50 ± 13 | 33 | 53 ± 12 | 40 | 55 ± 9 | 51 |
| PAB#4 | - | - | - | - | 21 ± 3 | - | 22 ± 6 | 31 | 19 ± 3 | 43 | 18 ± 1 | 32 | 23 ± 8 | 22 |
| PAB#5 | - | - | - | - | - | - | 22 ± 4 | - | 22 ± 1 | 15 | 21 ± 4 | 22 | 18 ± 6 | 20 |
Relative DNA fractions for the different bacteria are shown. Calculations were done as previously described (44). Probe abbreviations: LCL, L. lactis; PAB#3, P. acidipropionici; PAB#4, P. freudenreichii; PAB#5, P. jensenii. Standard deviations for two or three independent replicates are shown. Abbreviations: cell, cell DNA; free, free DNA; MIX, mixed sample of all the strains; PAB, coculture of the propionibacteria; -, signal less than background threshold; NA, no parallels are available.
Data from Treimo et al. (44).
Weak signals due to low cell content.
Measurement of total free DNA showed that L. lactis subsp. lactis INF L2 accumulated DNA after 12 h, P. acidipropionici ATCC 4965 accumulated DNA mainly from 24 to 48 h, P. freudenreichii ISU P59 accumulated DNA slightly from 24 to 48 h and more extensively after 48 h, and P. jensenii INF P303 accumulated DNA after 48 h and throughout the incubation period (Fig. 3). The mixed sample showed rapid accumulation of free DNA during the first 24 h mainly due to the lactococcal strain, followed by a slow increase for up to 7 days. Autolysis was observed mainly from 24 to 48 h in the coculture of propionibacteria. Analyses of the species-specific 16S rRNA gene content (Table 1) showed that the times/points for maximum DNA accumulation for the different bacteria in the mixed sample were comparable to those observed for pure cultures (compare Fig. 3 and Table 1). Maximum accumulation of DNA from L. lactis subsp. lactis INF L2 was observed after 24 h, followed by a decrease for up to 5 weeks (P = 0.02). Propionibacterium acidipropionici ATCC 4965 showed the highest accumulation after 48 h and P. freudenreichii ISU P59 after 7 days, the latter in accordance with the maximum release of the intracellular enzyme proline iminopeptidase as measured by Østlie et al. (31). Free DNA for P. jensenii INF 303 was detected after 7 days in the mixed sample, and the same relative level was obtained for the rest of the incubation period. DNA release from P. acidipropionici ATCC 4965 and P. freudenreichii ISU P59 was detected after 48 h in the coculture, whereas the latter strain showed maximum DNA release after 7 days. A slower DNA release was observed for P. jensenii INF 303, in which free DNA was detected after 7 days.
Effect of rennet on growth and autolysis.
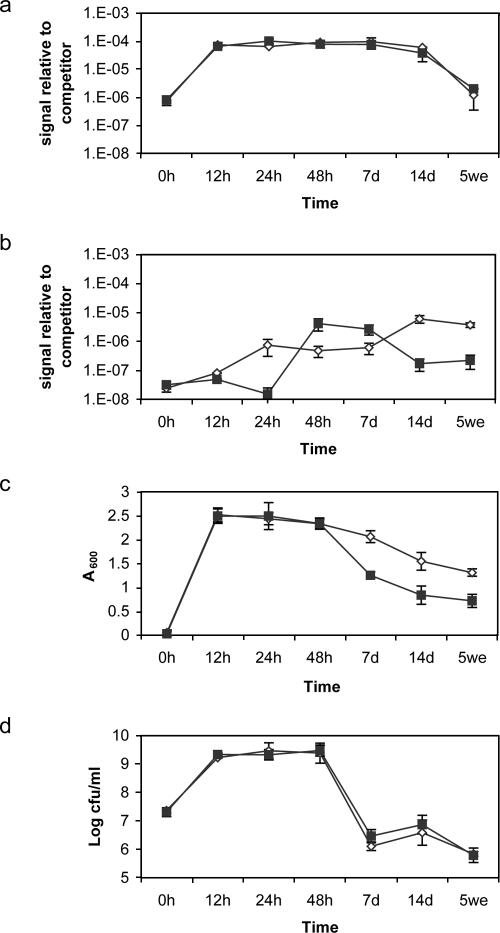
We compared our novel DNA approach to A600 and CFU measurements in the evaluation of the effect of rennet on growth and autolysis of L. lactis subsp. lactis INF L2 (Fig. 4). Maximum total bacterial L. lactis subsp. lactis INF L2 cell DNA content was observed after 12 h (Fig. 4a), and comparably, maximum turbidity and viable-cell counts were obtained after 12 h (Fig. 4c and d). Langsrud et al. (19) reported maximum cellular DNA content after 9 h with the same strain.
FIG. 4.
Influence of rennet on growth and autolysis as determined by measurement of total cell bacterial DNA (a), free DNA (b), A600 (c), and CFU/ml (d) for Lactococcus lactis subsp. lactis INF L2. Symbols: ⋄, sample without rennet; ▪, rennet-treated sample. The error bars for all samples indicate standard deviations for at least three independent quantifications.
Free DNA was detected after 12 h (Fig. 4b), as was a simultaneous decrease in A600 growth curve initiate (Fig. 4c). Comparably, the number of CFU (Fig. 4d) decreased rapidly after 48 h, as observed somewhat later with our approach by a shift in cell DNA (Fig. 4a) after 7 days.
DNA accumulation was faster between 24 and 48 h (approximately 2-log10 increase) in the presence of rennet than without it (Fig. 4b). DNA turnover, however, seemed higher in the presence of rennet than without it. This is seen as a decrease in the amount of free DNA particularly between 7 and 14 days. Rennet, which is a crude extract from the calf stomach, may contain DNases with the ability to degrade free DNA. The effect of rennet on autolysis was also observed by absorbance measurements (Fig. 4c). Autolytic activity is described as a decrease in absorbance (19), and previous work supported autolysis after 12 h for the same strain (19, 47). The presence of rennet resulted in a decrease in absorbance and hence an increased occurrence of autolysis of L. lactis subsp. lactis INF L2 after 48 h. The ability to form colonies, however, was not affected by rennet, as determined by the CFU measurements (Fig. 4d). There was a major CFU decrease in both treated and untreated samples between 48 h and 7 days. However, enumerations of viable CFU are not suited to detect autolysis, since CFU measurement demonstrates only decreases in culturability and not lysis. Some cells may be nonculturable yet still metabolically active or may be dead (i.e., permeabilized with a damaged membrane) but still not completely lysed, as was clearly observed by confocal microscopy (5).
Growth and autolysis results were obtained for CFU, A600, and cell DNA for the tested propionibacteria strains with respect to measurement of maximum cell density (A600 and CFU results not shown). Maximum total bacterial cell DNA content and maximum viable-cell counts were observed from 24 to 48 h for the different propionibacteria analyzed. Free DNA was also detected at different times (from 24 to 48 h), consistent with initiation of autolysis after 48 h for all the propionibacteria as measured by A600. However, the extent and rate of the decrease were strain dependent. Propionibacterium freudenreichii ISU P59 revealed the highest and fastest autolysis after 5 weeks by A600 measurements, followed by P. acidipropionici ATCC 4965 and finally P. jensenii INF P303. The autolysis measurements (A600) showed the same pattern for P. freudenreichii ISU P59 and P. acidipropionici ATCC 4965 as that previously described by Østlie et al. (31).
We also conducted a DNA-based evaluation of the autolytic effect of rennet on the other bacteria included in this work. All the strains responded differently to rennet. DNA release of P. acidipropionici ATCC 4965 was slightly later (between 48 h and 14 days) in the presence of rennet than without rennet (mainly between 24 and 48 h, but the accumulation increased slightly for up to 14 days). In addition, DNA accumulation showed nearly a 3-log10 increase after 5 weeks compared to a 2-log10 total increase in the sample without rennet (Fig. 3b). The rennet-treated P. freudenreichii ISU P59 sample showed a 1.5-log10 increase in free DNA between 12 h and 7 days, while DNA release occurred between 24 h and 7 days in the sample without rennet, with the same log10 change. However, we could not detect an effect of rennet on the DNA release for P. jensenii INF P303. The 16S rRNA gene method appeared to be more sensitive than previously described absorbance measurements for evaluating the influence of rennet. Our conclusion is that the DNA method is suitable for early detection of autolysis, while measurements of progressed autolysis are influenced by DNA degradation. Inclusion of a DNA degradation marker may also make the method more suitable to measure the progression of autolysis. A brief summary of the comparison of our DNA method for measurement of autolysis in mixed populations to some alternative methods is shown in Table 2.
TABLE 2.
Comparison of autolysis detection methods
| Detection method | Potential of mixed-population analyses | Sensitivity to autolysis | Considerations |
|---|---|---|---|
| CFU measurement | Low | Not sensitive (5) | Cells may form aggregates; cells may be nonculturable but still not lysed (5) |
| A600 measurement | Not possible | Medium | Influenced by extracellular material |
| Electron microscopy (26) | Low | Medium | No quantification can be made |
| BacLight (5) | Not possible | Medium | Quantification is difficult (26) |
| Intracellular metabolic enzyme, e.g., LDH (9, 30) | Mediuma | High | Sensitive to enzyme activity |
| Immunological (10, 46) | Medium | High | Difficulty in identifying species-specific immunological markers and in obtaining quantitative results |
| DNA array | High | High | Sensitive to DNA degradation |
Applications in a liquid cheese model.
The potential use of the 16S rRNA gene microarray method was further demonstrated with a liquid cheese model by L. lactis subsp. lactis INF L2 and P. freudenreichii ISU P59 in pure cultures and in a coculture. Generally, growth and autolysis occurred later in this medium than in broth. Maximum total bacterial DNA content was observed in the pure L. lactis subsp. lactis INF L2 cultures after 24 h and in P. freudenreichii ISU P59 after 7 days, compared to 12 and 48 h, respectively, in broth studies. DNA release increased in the first 48 h for L. lactis subsp. lactis INF L2 and then stayed at a constant level. Accumulation of free DNA of P. freudenreichii ISU P59 increased rapidly from 48 h to 7 days and increased slightly throughout the 5 weeks of the experimental period. Viable-cell counts of L. lactis subsp. lactis INF L2 developed comparably to those in the broth studies; however, a decrease (3 log10) was observed after 2 weeks. Propionibacterium freudenreichii ISU P59 reached a maximum number of CFU/ml (9.8 log10 ± 0.7 log10) after nearly 2 weeks, and an approximately 1-log10 decrease was detected after 5 weeks. Comparably, maximum viable-cell counts were observed after 48 h in SLB broth, and an approximately 1.5-log10 decrease was detected.
Coculture of L. lactis subsp. lactis INF L2 and P. freudenreichii ISU P59 in the liquid cheese model showed an accumulation of free DNA during the first 48 h, followed by a slightly accelerated accumulation for up to 7 days, and then there was a constant level of free DNA during the rest of the incubation period. The extent of DNA release, however, was in the same range as that observed for broth studies. Species-specific DNA detection of the coculture is shown in Table 3. L. lactis subsp. lactis INF L2 DNA showed maximum accumulation of free DNA after 24 to 48 h, followed by a decrease after 5 weeks. Autolysis of P. freudenreichii ISU P59 increased after 7 days and slightly more to a measured maximum after 5 weeks.
TABLE 3.
Summary of species-specific DNA detection of autolysis in a liquid cheese modela
| Cocultureb | Incubation time for indicated DNA typec
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 0 h
|
24 h
|
48 h
|
7 d
|
5 wk
|
||||||
| Cell | Free | Cell | Free | Cell | Free | Cell | Free | Cell | Free | |
| LCL | ++ | - | ++ | +++ | +++ | +++ | ++ | +++ | ++ | + |
| PAB#4 | - | - | + | - | + | - | + | + | + | + |
Relative DNA fractions for the different bacteria are shown. Calculations were done as previously described (44). Probe abbreviations: LCL, L. lactis; PAB#4, P. freudenreichii.
Coculture consists of a mixture of L. lactis subsp. lactis INF L2 and P. freudenreichii ISU P59.
Abbreviations: cell, cell DNA; free, free DNA; -, signal less than background threshold; +, relative signal of 15 to 25%; ++, relative signal of 40 to 55%; +++, relative signal of 65 to 75%.
Future understanding of autolysis in cheese ripening.
Occurrence of autolysis of propionibacteria in cheese and their contribution to proteolysis are poorly understood (11, 16, 40), and more knowledge is clearly needed. Damaged P. freudenreichii cells were visualized in Grana cheese by scanning electron microscopy, indicating the occurrence of autolysis after 8 months of ripening (7). These observations are in contrast to recent work by Valence et al. (46), in which autolysis of P. freudenreichii was reported to occur to a limited extent and very late during cold storage of Swiss cheese. Research on proteolysis by propionibacteria has mainly focused on free proline and intracellular proline-specific peptidase activity, and release of peptidases into cheese through autolysis has been assumed (11, 23, 27, 33, 43). Langsrud et al. (21, 22) observed a correlation between autolysis of propionibacteria in milk and production of free proline, and Østlie et al. (31) confirmed this correlation for broth media. Gagnaire et al. (13) showed that strains of P. freudenreichii did not contribute to proteolysis in an aqueous phase in Emmental cheese and that propionibacteria were not significantly involved in the production of proline. Few peptidases from P. freudenreichii were also found in Emmental cheese by a proteomic approach (12). However, the extent of autolysis is considered to be a strain-dependent phenomenon in the genera Propionibacterium and Lactococcus (8, 23, 24, 47), and this might be the situation for P. freudenreichii strains in cheese. However, we detected autolysis of P. freudenreichii in our broth and liquid cheese model analyses. The presence of different environmental factors, such as rennet, will in addition have an influence on autolysis, making the process complex to understand. The lack of proper tools to analyze complex samples is the main factor that limits our understanding of autolysis in cheese.
The use of model systems overcomes some of the complexities and difficulties in interpretations associated with cheese studies, although autolysis in a model does not entirely represent autolysis in cheese. Model systems, however, can be used to acquire general knowledge about autolysis that is relevant for real cheese systems. We have now shown that it is possible to analyze autolysis in mixed populations. Ultimately, with technological developments, we should also be able to generate accurate information from real cheese systems in the future and thereby increase our understanding of the underlying mechanisms of autolysis in the cheese ecosystem.
Acknowledgments
We thank the Norwegian Radium Hospital, Oslo, Norway, for printing the slides and LMGT, Laboratory of Microbial Gene Technology, Norwegian University of Life Sciences, for use of the scanner. We also thank S. M. Grønhovd for technical assistance with the growth experiments and Silje Marki for the liquid cheese model experiments.
This work was supported by MATFORSK Norwegian Food Research Institute, Hedmark Sparebank, and UMB, Norwegian University of Life Sciences.
REFERENCES
- 1.Beresford, T. P., N. A. Fitzsimons, N. L. Brennnan, and T. M. Cogan. 2001. Recent advances in cheese microbiology. Int. Dairy J. 11:259-274. [Google Scholar]
- 2.Bie, R., and G. Sjöström. 1975. Autolytic properties of some lactic acid bacteria used in cheese production. I. Materials and methods. Milchwissenschaft 30:653-657. [Google Scholar]
- 3.Bie, R., and G. Sjöström. 1975. Autolytic properties of some lactic acid bacteria used in cheese production. II. Experiments with fluid substrates and cheese. Milchwissenschaft 30:739-747. [Google Scholar]
- 4.Boutrou, R., A. Sepulchre, J. C. Gripon, and V. Monnet. 1998. Simple tests for predicting the lytic behavior and proteolytic activity of lactococcal strains in cheese. J. Dairy Sci. 81:2321-2328. [Google Scholar]
- 5.Bunthof, C. J., S. van Schalkwijk, W. Meijer, T. Abee, and J. Hugenholtz. 2001. Fluorescent method for monitoring cheese starter permeabilization and lysis. Appl. Environ. Microbiol. 67:4264-4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burton, K. 1956. A study of the conditions and mechanism of the diphenylamine reaction for the colorimetric estimation of deoxyribonucleic acid. Biochem. J. 62:315-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cappa, F., V. Bottazzi, F. Bosi, and M. G. Parisi. 1997. Characterization of propionibacteria in Grana cheese. Sci. Tech. Latt. Caesaria 47:405-414. [Google Scholar]
- 8.Chapot-Chartier, M.-P., C. Deniel, M. Rousseau, L. Vassal, and J.-C. Gripon. 1994. Autolysis of two strains of Lactococcus lactis during cheese ripening. Int. Dairy J. 4:251-269. [Google Scholar]
- 9.Crow, V. L., T. Coolbear, P. K. Gopal, F. G. Martley, L. L. McKay, and H. Riepe. 1995. The role of autolysis of lactic acid bacteria in the ripening of cheese. Int. Dairy J. 5:855-875. [Google Scholar]
- 10.Deutsch, S.-M., T. Ferain, J. Delcour, and S. Lortal. 2002. Lysis of lysogenic strains of Lactobacillus helveticus in Swiss cheeses and first evidence of concomitant Streptococcus thermophilus lysis. Int. Dairy J. 12:591-600. [Google Scholar]
- 11.Gagnaire, V., D. Molle, T. Sorhaug, and J. Leonil. 1999. Peptidases of dairy propionic acid bacteria. Lait 79:43-57. [Google Scholar]
- 12.Gagnaire, V., M. Piot, B. Camier, J. P. Vissers, G. Jan, and J. Leonil. 2004. Survey of bacterial proteins released in cheese: a proteomic approach. Int. J. Food Microbiol. 94:185-201. [DOI] [PubMed] [Google Scholar]
- 13.Gagnaire, V., A. Thierry, and J. Leonil. 2001. Propionibacteria and facultatively heterofermentative lactobacilli weakly contribute to secondary proteolysis of Emmental cheese. Lait 81:339-353. [Google Scholar]
- 14.Hannon, J. A., M. G. Wilkinson, C. M. Delahunty, J. M. Wallace, P. A. Morrissey, and T. P. Beresford. 2003. Use of autolytic starter systems to accelerate the ripening of Cheddar cheese. Int. Dairy J. 13:313-323. [Google Scholar]
- 15.Jeness, R., and J. Koops. 1962. Preparation and properties of a salt solution which simulates milk ultrafiltrate. Neth. Milk Dairy J. 16:153-164. [Google Scholar]
- 16.Kerjean, J. R., S. Condon, R. Lodi, G. Kalantzopoulos, J. F. Chamba, T. Suomalainen, T. Cogan, and D. Moreau. 2000. Improving the quality of European hard-cheeses by controlling of interactions between lactic acid bacteria and propionibacteria. Food Res. Int. 33:281-287. [Google Scholar]
- 17.Kure, C. F., and I. Skaar. 2000. Mould growth on the Norwegian semi-hard cheeses Norvegia and Jarlsberg. Int. J. Food. Microbiol. 62:133-137. [DOI] [PubMed] [Google Scholar]
- 18.Laloy, E., J. C. Vuillemard, M. ElSoda, and R. E. Simard. 1996. Influence of the fat content of Cheddar cheese on retention and localization of starters. Int. Dairy J. 6:729-740. [Google Scholar]
- 19.Langsrud, T., A. Landaas, and H. B. Castberg. 1987. Autolytic properties of different strains of group-N streptococci. Milchwissenschaft 42:556-560. [Google Scholar]
- 20.Langsrud, T., and G. W. Reinbold. 1973. Flavor development and microbiology of Swiss cheese—a review. II. Starters, manufacturing processes and procedures. J. Milk Food Technol. 36:531-542. [Google Scholar]
- 21.Langsrud, T., G. W. Reinbold, and E. G. Hammond. 1978. Free proline production by strains of propionibacteria. J. Dairy Sci. 61:303-308. [Google Scholar]
- 22.Langsrud, T., G. W. Reinbold, and E. G. Hammond. 1977. Proline production by Propionibacterium shermanii P59. J. Dairy Sci. 60:16-23. [Google Scholar]
- 23.Langsrud, T., T. Sorhaug, and G. E. Vegarud. 1995. Protein degradation and amino acid metabolism by propionibacteria. Lait 75:325-330. [Google Scholar]
- 24.Lemee, R., A. Rouault, S. Guezenec, and S. Lortal. 1994. Autolysis of 57 strains of dairy propionibacteria. Lait 74:241-251. [Google Scholar]
- 25.Lindahl, T., and B. Nyberg. 1972. Rate of depurination of native deoxyribonucleic acid. Biochemistry 11:3610-3618. [DOI] [PubMed] [Google Scholar]
- 26.Lortal, S., and M. P. Chapot-Chartier. 2005. Role, mechanisms and control of lactic acid bacteria lysis in cheese. Int. Dairy J. 15:857-871. [Google Scholar]
- 27.Lortal, S., R. Lemee, and F. Valence. 1997. Autolysis of thermophilic lactobacilli and dairy propionibacteria: a review. Lait 77:133-150. [Google Scholar]
- 28.Niskasaari, K. 1989. Characteristics of the autolysis of variants of Lactococcus lactis subsp. cremoris J. Dairy Res. 56:639-649. [Google Scholar]
- 29.Ohmiya, K., and Y. Sato. 1970. Studies on the proteolytic action of dairy lactic acid bacteria. Part X. Autolysis of lactic acid bacterial cells in aseptic rennet curd. Agric. Biol. Chem. 34:457-463. [Google Scholar]
- 30.O'Reilly, C. E., P. M. O'Connor, P. M. Murphy, A. L. Kelly, and T. P. Beresford. 2002. Effects of high-pressure treatment on viability and autolysis of starter bacteria and proteolysis in Cheddar cheese. Int. Dairy J. 12:915-922. [Google Scholar]
- 31.Østlie, H., V. Floberghagen, G. Reinbold, E. G. Hammond, G. Vegarud, and T. Langsrud. 1995. Autolysis of dairy propionibacteria—growth-studies, peptidase activities, and proline production. J. Dairy Sci. 78:1224-1237. [Google Scholar]
- 32.Østlie, H. M., G. Vegarud, and T. Langsrud. 1999. Autolytic systems in propionic acid bacteria. Lait 79:105-112. [Google Scholar]
- 33.Quelen, L., C. Dupuis, and P. Boyaval. 1995. Proline-specific activities of Propionibacterium freudenreichii subsp. shermanii. J. Dairy Res. 62:661-666. [Google Scholar]
- 34.Rogers, H. J., J. B. Ward, and H. R. Perkins. 1980. The bacterial autolysins, p. 437-460. In H. J. Rogers, J. B. Ward, and H. R. Perkins. (ed.), Microbial cell walls and membranes. Chapman & Hall, London, United Kingdom.
- 35.Rudi, K., J. Treimo, B. Moen, I. Rud, and G. Vegarud. 2002. Internal controls for normalizing DNA arrays. BioTechniques 33:496-502. [DOI] [PubMed] [Google Scholar]
- 36.Shockman, G. D., C. P. Chu, R. Kariyama, L. K. Tepper, and L. Daneo-Moore. 1993. Peptidoglycan (murein) hydrolases: unusual enzymes for unusual substrates, p. 213-227. In M. A. de Pedro, J. V. Höltje, and W. Löffelhardt (ed.), Bacterial growth and lysis. Metabolism and structure of the bacterial sacculus. Plenum Press, New York, N.Y.
- 37.Shockman, G. D., and J. V. Höltje. 1994. Microbial peptidoglycan (murein) hydrolases, p. 131-166. In J. M. Ghuysen and R. Hakenbech (ed.), Bacterial cell wall. Elsevier Science B.V., Amsterdam, The Netherlands.
- 38.Shockman, G. D., H. M. Pooley, and J. S. Thompson. 1967. Autolytic enzyme system of Streptococcus faecalis. 3. Localization of the autolysin at the sites of cell wall synthesis. J. Bacteriol. 94:1525-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shockman, G. D., J. S. Thompson, and M. J. Conover. 1967. Autolytic enzyme system of Streptococcus faecalis. 2. Partial characterization of autolysin and its substrate. Biochemistry 6:1054-1065. [DOI] [PubMed] [Google Scholar]
- 40.Sousa, M. J., Y. Ardo, and P. L. H. McSweeney. 2001. Advances in the study of proteolysis during cheese ripening. Int. Dairy. J. 11:327-345. [Google Scholar]
- 41.Syvanen, A. C. 1994. Detection of point mutations in human genes by the solid-phase minisequencing method. Clin. Chim. Acta 226:225-236. [DOI] [PubMed] [Google Scholar]
- 42.Terzaghi, B. E., and W. E. Sandine. 1975. Improved medium for lactic streptococci and their bacteriophages. Appl. Microbiol. 29:807-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tobiassen, R. O., L. Stepaniak, and T. Sorhaug. 1997. Screening for differences in the proteolytic systems of Lactococcus, Lactobacillus and Propionibacterium. Z. Lebensm. Unters. Forsch. A 204:273-278. [Google Scholar]
- 44.Treimo, J., G. Vegarud, T. Langsrud, S. Marki, and K. Rudi. 2006. Total bacterial and species-specific 16S rDNA micro-array quantification of complex samples. J. Appl. Microbiol. 100:985-998. [DOI] [PubMed] [Google Scholar]
- 45.Valence, F., S. M. Deutsch, R. Richoux, V. Gagnaire, and S. Lortal. 2000. Autolysis and related proteolysis in Swiss cheese for two Lactobacillus helveticus strains. J. Dairy Res. 67:261-271. [DOI] [PubMed] [Google Scholar]
- 46.Valence, F., R. Richoux, D. Thierry, A. Palva, and S. Lortal. 1998. Autolysis of Lactobacillus helveticus and Propionibacterium freudenreichii in Swiss cheeses: first evidence by using species-specific lysis markers. J. Dairy Res. 65:609-620. [Google Scholar]
- 47.Vegarud, G., H. B. Castberg, and T. Langsrud. 1983. Autolysis of group-N streptococci—effects of media composition modifications and temperature. J. Dairy Sci. 66:2294-2302. [Google Scholar]
- 48.Visser, S. 1993. Symposium—proteolytic-enzymes and cheese ripening—proteolytic-enzymes and their relation to cheese ripening and flavor—an overview. J. Dairy Sci. 76:329-350. [Google Scholar]
- 49.Wilkinson, M. G., T. P. Guinee, D. M. Ocallaghan, and P. F. Fox. 1994. Autolysis and proteolysis in different strains of starter bacteria during Cheddar cheese ripening. J. Dairy Res. 61:249-262. [Google Scholar]
- 50.Yang, Y. H., and T. Speed. 2003. Normalisation, p. 536-551. In D. Bowtell and J. Sambrook (ed.), DNA microarrays. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.