Abstract
Diet is a major factor in maintaining a healthy human gastrointestinal tract, and this has triggered the development of functional foods containing a probiotic and/or prebiotic component intended to improve the host's health via modulation of the intestinal microbiota. In this study, a long-term placebo-controlled crossover feeding study in which each subject received several treatments was performed to monitor the effect of a prebiotic substrate (i.e., lactulose), a probiotic organism (i.e., Saccharomyces boulardii), and their synbiotic combination on the fecal microbiota of three groups of 10 healthy human subjects differing in prebiotic dose and/or intake of placebo versus synbiotic. For this purpose, denaturing gradient gel electrophoresis (DGGE) analysis of 16S rRNA gene amplicons was used to detect possible changes in the overall bacterial composition using the universal V3 primer and to detect possible changes at the subpopulation level using group-specific primers targeting the Bacteroides fragilis subgroup, the genus Bifidobacterium, the Clostridium lituseburense group (cluster XI), and the Clostridium coccoides-Eubacterium rectale group (cluster XIVa). Although these populations remained fairly stable based on DGGE profiling, one pronounced change was observed in the universal fingerprint profiles after lactulose ingestion. Band position analysis and band sequencing revealed that a band appearing or intensifying following lactulose administration could be assigned to the species Bifidobacterium adolescentis. Subsequent analysis with real-time PCR (RT-PCR) indicated a statistically significant increase (P < 0.05) in total bifidobacteria in one of the three subject groups after lactulose administration, whereas a similar but nonsignificant trend was observed in the other two groups. Combined RT-PCR results from two subject groups indicated a borderline significant increase (P = 0.074) of B. adolescentis following lactulose intake. The probiotic yeast S. boulardii did not display any detectable universal changes in the DGGE profiles, nor did it influence the bifidobacterial levels. This study highlighted the capacity of an integrated approach consisting of DGGE analysis and RT-PCR to monitor and quantify pronounced changes in the fecal microbiota of healthy subjects upon functional food administration.
The human gastrointestinal (GI) tract harbors a complex community of microorganisms, with the largest concentration of bacteria and metabolic activity being found in the large intestine (2, 14, 32). According to current insights, the bacterial groups predominating the large bowel of human adults are facultative and obligate anaerobes mainly belonging to the genera Bacteroides, Eubacterium, Clostridium, Ruminococcus, Bifidobacterium, and Fusobacterium (23). Essentially, the role of these colonic organisms is confined to the fermentation of various substrates that escaped digestion in the upper GI tract. Whereas saccharolytic fermentation of carbohydrates leads to the production of short-chain fatty acids that provide additional energy to the host, the end products of proteolytic (protein) fermentation include various toxic substances such as phenolic compounds, amines, and ammonia. Colon bacteria are often classified as potentially harmful or potentially health promoting based on their fermentative features. In any given situation of intestinal balance, increased numbers of proteolytic clostridia and Bacteroides can be detrimental to health (11), while stimulation of bifidobacteria and lactobacilli is generally regarded as beneficial (10). The range of positive effects that have been linked to bifidobacteria and lactobacilli include stimulation of the immune system, production of vitamins, inhibition of intestinal pathogens, reduction of blood ammonia and cholesterol levels, and reduction of constipation (12, 33).
The fact that diet is a major factor controlling intestinal balance has triggered the development of so-called functional foods containing a probiotic and/or prebiotic component. Probiotics are commonly referred to as live microorganisms (bacteria or yeasts), which, when administered in adequate amounts, confer a health benefit on the host (8). However, other studies have suggested that inactivated microbes and their components can also exert probiotic effects (17, 43). Bacterial probiotic strains that are incorporated into commercial products worldwide most frequently belong to the genera Bifidobacterium and Lactobacillus. A prebiotic is a nondigestible, selectively fermented compound that induces specific changes in the composition and/or activity of the gastrointestinal microbiota that are beneficial for a host's well-being and health (13). Several oligosaccharides have been studied as potential prebiotics, including lactulose, galactooligosaccharides, fructooligosaccharides (oligofructose and inulin), and soybean oligosaccharides (6). Essentially, the aims of pro- and prebiotic supplementation are highly similar (i.e., to improve the host's health via modulation of the intestinal microbiota) but are achieved in different ways, namely, by introducing exogenous species (probiotics) or by stimulating indigenous bacteria (prebiotics), respectively.
The inadequacy of conventional culture techniques to reflect the microbial diversity of the intestinal ecosystem (20, 22, 35) has triggered the development of culture-independent techniques for the evaluation of pro- and prebiotic administration in humans. Commonly used molecular approaches to analyze the intestinal microbiota upon dietary intervention include population fingerprinting techniques like denaturing gradient gel electrophoresis (DGGE) or temperature gradient gel electrophoresis (34, 38) and terminal restriction fragment length polymorphism (19). These PCR-based tools allow the visualization of the predominant genetic diversity without prior knowledge of the composition or complexity of the microbial ecosystem present in the sample. Unlike population fingerprinting methods, fluorescent in situ hybridization (16, 42) and real-time PCR (RT-PCR) (15, 24) are able to generate more quantitative information for specific fecal bacterial groups. Although each of these techniques has been applied to human feeding trials, to our knowledge, no study so far has reported the integrated use of DGGE and RT-PCR to monitor the effects of pro-, pre-, and synbiotics on the fecal microbiota of healthy humans.
To demonstrate the efficacy of functional food components under in vivo conditions and to substantiate claims from studies in vitro and using experimental models, well-designed human dietary intervention studies are required (45). In the current study, a long-term placebo-controlled crossover feeding study was set up to monitor the effect of a prebiotic substrate (i.e., lactulose), a probiotic organism (i.e., Saccharomyces boulardii), and the synbiotic combination of the two on the predominant bacterial population of 30 healthy human subjects. Lactulose is a commercially available disaccharide that is used as a drug in the treatment of hepatic encephalopathy and chronic constipation (46), which has been shown to stimulate the growth of bifidobacteria (41). The probiotic yeast Saccharomyces boulardii is a biotherapeutic agent available as a registered pharmaceutical product and is used in the prevention and treatment of various types of diarrhea (9, 18, 36). For this purpose, modifications in the overall bacterial composition of fecal samples were monitored by population fingerprinting using DGGE analysis of 16S rRNA gene amplicons. In this way, DGGE analysis allowed the detection of pronounced changes in the predominant fecal microbiota following pro-, pre-, or synbiotic administration using the universal 16S rRNA gene V3 primer but also allowed the detection of changes at subpopulation level using group-specific primers targeting the Bacteroides fragilis subgroup, the genus Bifidobacterium, the Clostridium lituseburense group (cluster XI), and the Clostridium coccoides-Eubacterium rectale group (cluster XIVa). Pronounced changes revealed by DGGE analysis were further characterized by RT-PCR in order to obtain a quantitative estimate of the dietary intervention effect in healthy humans.
MATERIALS AND METHODS
Subjects.
Thirty healthy volunteers (11 women and 19 men) aged 23 ± 2 years (range, 20 to 26 years) participated in the study. None of the subjects had a history of gastrointestinal or metabolic disease or previous surgery (apart from appendectomy). The subjects did not receive antibiotic treatment or any other medical treatment influencing intestinal microbiota during the 3 months before the start of the study. Subjects were advised to maintain their usual diet during the study period and to avoid the intake of fermented milk products and food components containing high quantities of fermentable carbohydrates. The Ethics Committee of the University of Leuven (Belgium) approved the study, and all subjects gave informed consent.
Experimental design and sample collection.
Healthy volunteers were randomly assigned to three different treatment groups of a placebo-controlled crossover trial in which each subject participated in several treatments. The study was conducted over an 18-week period, which was divided into three ingestion periods of 4 weeks followed by a 4-week washout period, each separated by a 3-day sample collection interlude: (i) prebiotic period (day 1 to day 28), (ii) double placebo/synbiotic period (day 32 to day 60), (iii) probiotic period (day 64 to day 92), and (iv) washout period (day 96 to day 120) (Fig. 1). Lactulose (Duphalac; Solvay Pharma & Cie, Brussels, Belgium) and Saccharomyces boulardii (Enterol; Biomed, Dübendorf, Switzerland) were selected as prebiotic and probiotic compounds, respectively. The placebo consisted of maltodextrin (Paselli MD6; AVEBE B.A. Food, Foxhol, The Netherlands), an oligosaccharide that is obtained by enzymatic conversion of potato starch and that is completely digestible in the human small intestine. Twice a day, group 1 received 10 g lactulose together with 250 mg S. boulardii placebo in the first ingestion period (prebiotic), 10 g lactulose together with 250 mg S. boulardii placebo in the second ingestion period (placebo), 10 g lactulose placebo together with 250 mg S. boulardii in the third ingestion period (probiotic), and no intake during the final period (washout). According to the manufacturer, 250 mg of S. boulardii contains at least 2.5 × 109 viable lyophilized cells at the date of fabrication and 109 cells at the expiration date. However, the exact number of viable lyophilized cells at the moment of intake is unknown. Group 2 followed the same design except that higher doses of both active product and the respective placebo were administered: 15 g of lactulose and 500 mg of S. boulardii. Group 3 was analogous to group 1 except that the double-placebo period was replaced by the synbiotic combination of 10 g lactulose and 250 mg S. boulardii. The doses administered were chosen based on therapeutic recommendations in a way that subjects did not suffer from negative effects or discomfort. Throughout the study, the volunteers consumed their usual diet, taking care that the diet remained as stable as possible over the four periods. In addition, they were advised to avoid the intake of fermented milk products and food components containing high quantities of fermentable carbohydrates.
FIG. 1.
Schematic representation of the study design. The arrows (S1 to S5) indicate the time points of stool sample collection.
Before the start of the feeding study, at the end of each ingestion period, and at the end of the washout period, all stool samples produced during 72 h were collected. Because the mean transit takes between 60 and 72 h, samples were collected during three consecutive days. Each sample collected during 72 h was analyzed separately. Upon the collection of the fecal samples, 5 g (wet weight) was immediately frozen at −20°C for the purpose of DNA extraction.
Processing and total DNA extraction of the fecal samples.
Processing of the fecal samples and subsequent bacterial DNA extraction using a modified version of the method of Pitcher and coworkers (29) was performed as previously described (44).
Primer design and PCR program for DGGE.
Based on 16S rRNA gene sequences available from the EMBL database (http://srs6.ebi.ac.uk), specific PCR-DGGE primers for the Bacteroides fragilis subgroup and Clostridium clusters XI and XIVa were designed using Kodon (version 1.0) software (Applied Maths, St.-Martens-Latem, Belgium) as previously described (44) (Table 1). Validation of the developed primers was first performed in silico followed by in vitro specificity tests using type strains of species autochthonous to the human intestinal tract (Table 2). Other 16S rRNA gene primers used in this study targeting all (predominant) bacteria and the genus Bifidobacterium are listed in Table 1. The forward or reverse primer of each primer set was extended with a GC clamp at the 5′ end to allow the detection of all amplicons with DGGE.
TABLE 1.
Specifications of the 16S rRNA gene primers used in this study
| Target group | Primer | Primer sequencea (5′-3′) | Amplicon size (bp) | Annealing temp (°C) | Analysis used (gradient range [%]) | Reference or source |
|---|---|---|---|---|---|---|
| All bacteria | F357-GC | GC clamp-TACGGGAGGCAGCAG | 234 | 55 | DGGE (35-70) | 27 |
| R518 | ATTACCGCGGCTGCTGG | |||||
| Bacteroides fragilis | Bfra 531F | ATACGGAGGATCCGAGCGTTA | 293 | 65 | DGGE (35-60) | This study |
| subgroup | Bfra 766R-GC | GC clamp-CTGTTTGATACCCACACT | ||||
| Bifidobacterium | g-Bifid F | CTCCTGGAAACGGGTGG | 596 | 65 | DGGE (40-70) | 23 |
| g-Bifid R-GC | GC clamp-GGTGTTCTTCCCGATATCTACA | |||||
| Clostridium clusters | Erec 688F | GCGTAGATATTAGGAGGAAC | 211 | 60 | DGGE (35-70) | This study |
| XI and XIVa | Erec 841R-GC | GC clamp-TGCGTTWGCKRCGGCACCG | ||||
| Bifidobacterium | F_ado_IS | ATAGTGGACGCGAGCAAGAGA | 71 | 65 | RT-PCR | 15 |
| adolescentis | R_ado_IS | TTGAAGAGTTTGGCGAAATCG |
The GC clamp sequence is as follows: CGCCCGCCGCGCCCCGCGCCCGGCCCGCCGCCCCCGCCCC.
TABLE 2.
Results of 16S rRNA gene primer specificity tests
| Species | Strain | Amplicon obtained with primera:
|
||||
|---|---|---|---|---|---|---|
| V3 | Bfra | g-Bifid | Erec | Ado_IS | ||
| Bacillus cereus | LMG 6923T | + | − | − | − | − |
| Bacteroides distansonius | DSM 20701T | + | − | − | − | − |
| Bacteroides fragilis subgroupb | + | + | − | − | − | |
| Bifidobacterium speciesc | + | − | + | − | +g | |
| Clostridium clusters I and IId | + | − | − | − | − | |
| Clostridium cluster XIe | + | − | − | + | − | |
| Clostridium cluster XIVf | + | − | − | + | − | |
| Enterobacter aerogenes | LMG 2094T | + | − | − | − | − |
| Enterococcus solitarius | LMG 12890T | + | − | − | − | − |
| Escherichia coli | LMG 2092T | + | − | − | − | − |
| Faecalibacterium prausnitzii | A2-16S | + | − | − | − | − |
| Lactobacillus salivarius | LMG 9477T | + | − | − | − | − |
| Pediococcus pentosaceus | LMG 11488T | + | − | − | − | − |
| Prevotella melaninogenica | DSM 7089T | + | − | − | − | − |
| Proteus mirabilis | LMG 3257T | + | − | − | − | − |
| Staphylococcus aureus | LMG 8064T | + | − | − | − | − |
| Streptococcus salivarius | LMG 11489T | + | − | − | − | − |
| Veillonella parvula | DSM 2008T | + | − | − | − | − |
+, positive; −, negative.
The Bacteroides fragilis subgroup strains tested were Bacteroides fragilis LMG 10263T, Bacteroides eggerthii DSM 20697T, Bacteroides ovatus DSM 1896T, Bacteroides thetaiotaomicron DSM 2079T, and Bacteroides vulgatus LMG 17767T.
Bifidobacterium strains tested were B. adolescentis LMG 10502T, B. angulatum LMG 10503T, B. bifidum LMG 11041T, B. breve LMG 11042T, B. catenulatum LMG 11043T, B. dentium LMG 11045T, B. gallicum LMG 11596T, B. infantis LMG 8811T, B. longum LMG 13197T, B. pseudocatenulatum LMG 10505T, and B. ruminantium LMG 12588T.
Clostridium cluster I and II strains tested were C. beijerinckii LMG 5716T, C. butyricum LMG 1217T, C. perfringens LMG 11264T, C. sporogenes LMG 8421T, and C. tyrobutyricum LMG 1285T.
Clostridium cluster XI strains tested were C. bifermentans LMG 3029T, C. sordellii LMG 15708T, and Peptostreptococcus anaerobius LMG 15865T.
Clostridium cluster XIV strains tested were Anaerostipes caccae DSM 14662T, C. nexile DSM 1787T, Eubacterium hallii L2-7, Eubacterium rectale ATCC 33656T, Roseburia intestinalis DSM 14610T, and Ruminococcus productus LMG 21654T.
Positive for Bifidobacterium adolescentis only.
PCR assays were performed as previously described (44), using a single PCR core program for all primer pairs with primer-specific annealing temperatures (Table 1).
DGGE analysis and processing of the gels.
16S rRNA gene amplicons were analyzed with DGGE as previously described (44). In this study, different types of denaturing gradients were applied, depending on the primers used (Table 1). DGGE gels were stained for 30 min with 1× SYBR gold (catalog no. S-11494; Molecular Probes) in 1× TAE buffer (catalog no. 161-0773; Bio-Rad).
Inclusion of a standard reference every six lanes in each DGGE gel allowed the normalization of gel profiles using BioNumerics (BN) software, version 4.00 (Applied Maths, St.-Martens-Latem, Belgium). This normalization step enabled a comparison between DGGE profiles from different gels, provided that these gels were run under comparable denaturing and electrophoretic conditions. Cluster analysis of DGGE pattern profiles was performed using the unweighted-pair group method using arithmetic averages hierarchical clustering algorithm, and similarity between profiles was expressed by the curve-based Pearson product-moment correlation coefficient.
To perform band position analysis, a database containing the V3 primer amplicon DGGE band positions of all human GI tract-associated Bifidobacterium species was constructed using BN software. By comparing the V3 band position in the sample profiles with the BN database, a first tentative identification was obtained.
RT-PCR analysis.
Quantification of total bifidobacteria and Bifidobacterium adolescentis was performed with the LightCycler System I (Roche, Mannheim, Germany) using the FastStart DNA Master SYBR Green I kit and specific PCR primers (Table 1). To determine total bifidobacteria numbers present in the samples, g-Bifid primers (23) without a GC clamp were used. To determine the relative concentration of B. adolescentis, primers F_ado_IS and R_ado_IS targeting the intergenic spacer region of the 16S-23S rRNA gene described previously by Haarman and Knol (15) were used. The specificity of this primer set was confirmed against a panel of reference strains representing all human GI tract-associated Bifidobacterium species (Table 2) and Bifidobacterium ruminantium.
The efficiency of RT-PCR amplification was optimized for both primer sets. The highest efficiencies were obtained using 4 mM (final concentration) MgCl2 at an annealing temperature of 65°C for both primer sets. The 20-μl reaction mixture contained 4 mM MgCl2, 2 μl of 10× Mastermix (including FastStart enzyme, FastStart Taq DNA polymerase, reaction buffer, deoxynucleoside triphosphate mixture, MgCl2, and SYBR Green I dye), 2 μl of template DNA, and 1 mM of each primer. The temperature program for RT-PCR included one cycle at 95°C for 10 min for initial denaturation and activation of the FastStart Taq DNA polymerase followed by 40 cycles of denaturation at 95°C for 0 s followed by annealing at 65°C for 5 s and elongation at 72°C for 23 s and 5 s for g-Bifid and ado_IS primers, respectively. Detection of the fluorescent product was set at the end of the elongation step at each cycle. The melting curve was obtained by slow heating with a 0.1°C/s increment from 75°C to 95°C with continuous fluorescence measurement. Melting-point-determination analysis allowed the confirmation of the specificity of the amplification products. Calibration curves were constructed using dilutions of genomic DNA from a control strain (B. adolescentis LMG 10502) for which the number of bifidobacteria was determined by plate counting on MRS agar (Difco) incubated at 37°C under anaerobic conditions. The data presented are the mean values of duplicate RT-PCR analyses of the same DNA extracts in two independent runs.
Statistical analysis.
Results are expressed as mean values and standard deviations. Statistical analysis was performed with SPSS software (SPSS 12.0 for Windows; SPSS Inc., Chicago, IL). Given the low number of subjects in the treatment groups, nonparametric statistical analysis was used regardless of the distribution of results (Friedman analysis of variance [ANOVA]). The level for statistical significance was set at a P value of <0.05.
RESULTS
Population profiling with DGGE.
PCR-DGGE analysis with universal primers targeting the V3 region of the 16S rRNA gene was used to analyze the stability of the predominant fecal bacterial population of the samples across the different sampling periods (Fig. 2). Samples from all sampling points were pooled per person and analyzed on the same DGGE gel to rule out the possible influence of variations in electrophoretic conditions between different gels. Population fingerprint profiles were compared and analyzed both visually and numerically.
FIG. 2.
V3 16S rRNA gene DGGE profiles from all five sampling points (S1 to S5) of three individual subjects (I1 to I3), each representing one of the three test groups (G1 to G3). The day of sample collection at a given sampling point is indicated (day 1 [d1], d2, and d3). The square contains the band that appeared or intensified after lactulose intake (S2). There were three different scenarios regarding the presence of this band after lactulose ingestion: intensification (I1), appearance (I2), or remaining equally intense (I3).
Overall, DGGE band profiles of V3 16S rRNA gene amplicons displayed a relatively high complexity (mean of 20.4 bands per profile) and were relatively stable for each subject (Fig. 2). Between different subjects, considerable variation in the composition of the population fingerprints could be observed (data not shown). Although profiles were relatively stable for each subject, small qualitative (the presence or absence of a band) or quantitative (variable intensity of a band) variations did occur (even between two samples from the same day), most of which were subject specific (Fig. 2). However, one band fragment at a specific position in the V3 profiles appeared (n = 5) or intensified (n = 17) after lactulose ingestion in 22 of the 30 subjects. In five other subjects, the band was already present and remained equally intense (Fig. 2). In the three remaining subjects, the band was absent during the entire study. Repeated PCR-DGGE analyses of the same DNA extracts confirmed this observation (data not shown). This particular band was located in the high-percentage denaturant zone of the DGGE gel, indicating that it most probably represented a member of the genus Bifidobacterium, given the high percentage of G+C genome content of these organisms (55 to 67 mol%). After comparing the V3 band positions of all known Bifidobacterium species, only two species matched the particular band position, namely, B. adolescentis and B. ruminantium (Fig. 3). In contrast, the intake of placebo or S. boulardii with or without lactulose did not result in any uniform change in the DGGE band profiles (Fig. 2).
FIG. 3.
Comparison of V3 16S rRNA gene primer DGGE profile (50% to 70% gradient) of sample I2 G2 S2 d1 (see Fig. 2 legend) with the DGGE band positions of type and reference strains of Bifidobacterium species associated with the human GI tract and of B. ruminantium. The square contains the band that appeared or intensified after lactulose intake.
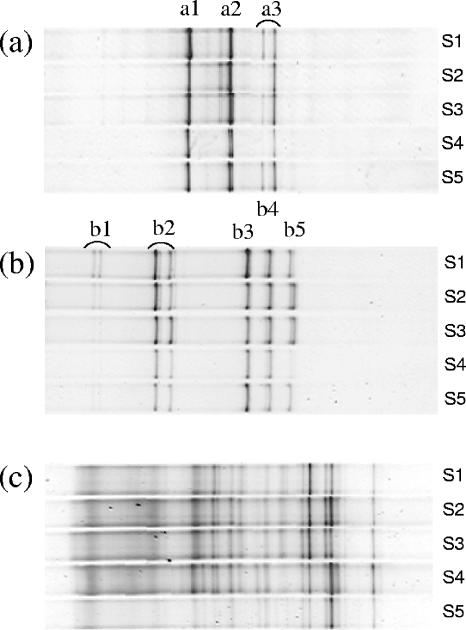
In addition to universal primers, a set of group-specific primers was applied to analyze the stability of certain fecal subpopulations during the intake periods. The Bacteroides fragilis (Fig. 4a) and Bifidobacterium (Fig. 4b) subpopulation, visualized by primers Bfra and g-Bifid, showed fairly stable profiles within each individual over the complete study period in all three test groups. These profiles were less complex than the profiles generated with the universal V3 primer and displayed less subject-specific variations. Based on band position analysis, profiles permitted the assignment of specific bands to known colonic species (Fig. 4a and b). In contrast, the complexity of the profiles of Clostridium clusters XI and XIVa (Fig. 4c), obtained with the Erec primer set, were comparable to the profiles obtained with the universal V3 primer. The profiles remained largely unchanged during the entire test period in all three test groups. Because a larger subgroup is targeted by this primer set, it was not possible to link band positions to specific species.
FIG. 4.
Clustering of group-specific DGGE band profiles from sampling points S1 to S5 of a representative subject from group 2: the Bacteroides fragilis subgroup (a), the genus Bifidobacterium (b), and Clostridium clusters XI and XIVa (c). By comparison to a DGGE database, band position analysis allowed the assignment of specific bands to the following species: Bacteroides vulgatus (a1), Bacteroides ovatus and/or Bacteroides fragilis and/or Bacteroides thetaiotaomicron (a2), Bacteroides uniformis (a3), B. adolescentis (b1), B. adolescentis (b2), B. longum/B. infantis (b3), B. breve (b4), and B. adolescentis (b5). Assignment of bands was not possible for the profiles for Clostridium clusters XI and XIVa because of the size of this group.
DGGE band sequence analysis.
The DGGE band fragment that appeared or intensified after lactulose ingestion was further characterized by sequencing analysis in order to link it to a specific Bifidobacterium species. This particular band was extracted from the V3 profiles of nine subjects from the three test groups and yielded identical 150-bp sequences in all these samples (data not shown). After comparison with the EMBL database, the sequence of the band in question exhibited the highest similarity (100%) with the 16S rRNA gene sequence of B. adolescentis and B. ruminantium followed by B. thermacidophylum, B. boum, B. catenulatum, and B. pseudocatenulatum, with a sequence similarity of 96.7%. Given the fact that B. ruminantium has never been reported to occur in humans and because of the documented predominance of B. adolescentis in the human gut, the band fragment was assigned to the latter species.
Quantification of fecal bifidobacteria using real-time PCR.
Since the Bifidobacterium population and particularly the species Bifidobacterium adolescentis appeared to be influenced by lactulose administration, real-time PCR was used to quantify the total fecal bifidobacteria (Table 3) and B. adolescentis (Table 4).
TABLE 3.
Results of RT-PCR-based quantification of total bifidobacteria in the three different treatment groupsa
| Group | Total bifidobacteria (log10 bifidobacteria/g [wet wt])
|
P value | ||||
|---|---|---|---|---|---|---|
| Baseline (S1) | Lactulose (S2) | Placebo/synbiotic (S3) | S. boulardii (S4) | Washout (S5) | ||
| 1 | 8.57 ± 0.67 | 8.95 ± 0.81 | 8.23 ± 0.73 | 8.20 ± 0.62 | 8.40 ± 0.78 | 0.371 (NS) |
| 2 | 8.61 ± 0.82 | 9.31 ± 0.72 | 8.53 ± 0.80 | 8.60 ± 1.08 | 8.62 ± 0.50 | 0.120 (NS) |
| 3 | 7.79 ± 1.27 | 8.98 ± 0.40b | 8.38 ± 0.50 | 8.13 ± 1.35 | 8.43 ± 1.41 | <0.05 |
α = 0.05 (Friedman ANOVA). S1, first time point of stool sample collection; NS, not significant.
Significantly different from baseline (P = 0.007) and synbiotic (P = 0.004) and borderline significantly different from S. boulardii cells (P = 0.061) and washout (P = 0.099).
TABLE 4.
Results of RT-PCR-based quantification of Bifidobacterium adolescentis in the three different treatment groupsa
| Group |
B. adolescentis level (log10B. adolescentis cells/g [wet wt])
|
P value | ||||
|---|---|---|---|---|---|---|
| Baseline (S1) | Lactulose (S2) | Placebo/synbiotic (S3) | S. boulardii (S4) | Washout (S5) | ||
| 1 | 7.76 ± 1.01 | 7.97 ± 0.94 | 7.95 ± 0.32 | 7.29 ± 1.14 | 7.41 ± 1.13 | 0.435 (NS) |
| 2 | 7.41 ± 1.22 | 8.14 ± 1.06 | 7.27 ± 0.98 | 7.52 ± 1.21 | 7.41 ± 0.93 | 0.164 (NS) |
| 3 | 7.14 ± 1.12 | 7.75 ± 1.16 | 7.35 ± 1.01 | 7.50 ± 0.96 | 7.68 ± 1.05 | 0.306 (NS) |
α = 0.05 (Friedman ANOVA). S1, first time point of stool sample collection; NS, not significant.
A statistically significant increase (P < 0.05) in the total level of fecal bifidobacteria was observed in group 3 after the intake of lactulose compared to the baseline conditions and synbiotic intake (Table 3). In groups 1 and 2, a nonsignificant tendency towards higher Bifidobacterium levels following lactulose intake was observed. A combination of the results of groups 1 and 3 (both of which received the same dose of lactulose during the first intake period) before and after lactulose intake resulted in a significant increase in the levels of fecal bifidobacteria, from 8.14 ± 1.09 to 8.99 ± 0.60 log10 bifidobacteria/g (wet weight) (P = 0.021). A mean standard deviation of 0.18 log10 bifidobacteria/g was obtained from duplicate RT-PCR runs, which is indicative of the good reproducibility of the method.
Nonsignificant tendencies indicating higher relative numbers after lactulose administration could be observed in the RT-PCR-based quantification of B. adolescentis (Table 4). A combination of the results from groups 1 and 3 before and after lactulose intake resulted in a borderline significant increase of the fecal B. adolescentis levels from 7.42 ± 1.09 to 7.84 ± 1.06 log10 bifidobacteria/g (wet weight) (P = 0.074). These results support the assumption that the DGGE band being discussed indeed represents a B. adolescentis species and not B. ruminantium, as the ado_IS primer set was shown to be specific for B. adolescentis in RT-PCR analyses (data not shown).
Although all subjects showed an overall increase in total levels of bifidobacteria and B. adolescentis after lactulose ingestion, these changes did not have the same proportional effect in all subjects. For instance, it was found that in one of the subjects, the DGGE band representing B. adolescentis was not present in the baseline sample but appeared as a very intense fragment after lactulose ingestion. In RT-PCR, this change was accompanied by a relative increase from 7.11 log10 to 9.26 log10 for the total bifidobacteria and from 5.41 log10 to 8.23 log10 for B. adolescentis. Likewise, two subjects for which no B. adolescentis cells could be detected in their fecal samples using RT-PCR lacked the DGGE band at the B. adolescentis position. A more detailed analysis of the RT-PCR data showed that the proportional increase in levels of total bifidobacteria and B. adolescentis was correlated to some extent with the initial level of bifidobacteria present (data not shown). Overall, subjects with lower initial bifidobacterial counts showed higher rates of response to lactulose administration than those exhibiting higher numbers under baseline conditions. No dose-effect relation was found for lactulose or S. boulardii.
DISCUSSION
In various studies that have analyzed the effect of pre-, pro-, and synbiotics on the human intestinal microbiota, it has been shown that bifidobacteria and lactobacilli can be stimulated, leading to a relative decrease of other organisms such as clostridia, streptococci, Bacteroides, and coliforms. Many of those studies still relied on conventional culture techniques (5, 21), which are less suitable for microbial monitoring and studying population dynamics than culture-independent approaches. In this study, the qualitative composition of the fecal microbiota was analyzed during the intake of a prebiotic and/or probiotic compound by DGGE using universal and group-specific primers. Subsequently, profound changes in DGGE patterns linked to prebiotic intake were further characterized quantitatively using two RT-PCR assays targeting total bifidobacteria and B. adolescentis. DGGE profiles of the predominant fecal microbiota using universal V3 16S rRNA gene primers generated complex but overall relatively stable and unique profiles in each of the three test groups. This confirms previous findings demonstrating the subject specificity of the predominant fecal microbiota in humans and its stability over a prolonged period of time (44, 47). Occasionally, subject-specific variations did occur during the intake period, most of which are probably due to variations in the daily diet of the subjects. One specific V3 DGGE band fragment that was observed in 90% of the subjects following lactulose intake was selected for further characterization. Based on the relative position of the band and sequence analysis of the extracted V3 amplicon, the band could be assigned to the species B. adolescentis and/or B. ruminantium. Because the two species display high 16S rRNA gene sequence similarities (up to 98.9%), reliable discrimination between both taxa was not possible. B. ruminantium is a typical rumen bacterium (3) that has so far never been reported in the human GI tract. Taken together with the fact that B. adolescentis is considered to be one of the most dominant Bifidobacterium species in the intestinal tract of human adults (24, 37), it can be concluded that the particular V3 band fragment represents B. adolescentis rather than B. ruminantium.
Because DGGE can, at its best, be considered as a semiquantitative tool for monitoring bacterial populations, additional analysis with RT-PCR was required to obtain a quantitative estimation of the stimulation of bifidobacteria following lactulose intake. In RT-PCR analyses, both the total bifidobacteria as well as B. adolescentis counts increased after lactulose intake independent of the administered dose, but this effect was found to be statistically significant only for total bifidobacteria. This finding may suggest that the increase in total bifidobacteria levels is not only due to the rise in B. adolescentis and also that other members of the genus Bifidobacterium, including B. catenulatum, B. pseudocatenulatum, B. longum, and B. infantis, may be influenced by the administration of lactulose. Most prebiotic feeding studies in which a bifidogenic effect was reported analyzed only total bifidobacteria levels. Tannock and coworkers (38) also demonstrated changes at the species level through the detection and increased staining intensity of RNA-DGGE bands assigned to B. adolescentis and Colinsella aerofaciens after the consumption of galactooligosaccharide- and fructooligosaccharide-containing biscuits.
The increase in fecal bifidobacteria after lactulose ingestion has been reported in various studies using culture-based methodologies (1, 3, 31, 39, 40) and fluorescent in situ hybridization analysis (41). To our knowledge, this is the first human study using DGGE and RT-PCR in an integrated approach to indicate an in vivo bifidogenic effect of lactulose. The stimulating effect of lactulose appeared to be restricted to the period of supplementation, as numbers of bifidobacteria returned to baseline levels after administration stopped. Also, in other prebiotic feeding studies (30, 42), increased numbers of fecal bifidobacteria returned to initial levels once prebiotic ingestion had ceased. This finding demonstrates the selective nature of prebiotic fermentation in the colon and supports the concept of beneficial modulation of the gut microbiota through dietary supplementation with specific oligosaccharides. Along with positive effects on the bacterial composition, prebiotic administration can also exert favorable effects on the intestinal metabolic activity, e.g., colonic NH3 metabolism. In this context, the colonic ammonia-nitrogen metabolism was investigated in the same volunteers by means of the biomarker lactose-[15N15N]-ureide and indicated a significant reduction of the urinary 15N excretion after the intake of lactulose, which was accompanied by a significant increase in the fecal 15N output, as was also demonstrated by higher 15N levels found in the fecal bacterial fractions (7). The observed decrease in levels of bifidobacteria after synbiotic treatment remains unclear. Possibly, consumption of lactulose by the probiotic yeast S. boulardii (26) and/or spatial competition between S. boulardii and the Bifidobacterium population may have contributed to this decrease.
From a methodological point of view, it is interesting that the intensity of the DGGE band representing B. adolescentis seemed proportional to the amount of total bifidobacteria and B. adolescentis determined by RT-PCR. Likewise, Bibiloni and coworkers (4) showed that an increased band intensity was correlated with relative abundance based on dot blot hybridization. In other studies, internal-standard systems were developed for DGGE analysis to conduct comparisons of relative fragment staining intensities (28, 38). As initially noticed by Roberfroid and colleagues (30) and as later confirmed in other studies (25, 42), the relative increase in levels of fecal bifidobacteria probably depends more on the baseline concentration of Bifidobacterium than on the prebiotic dose administered. A similar effect was also observed in the present study and could suggest that prebiotic intake may be particularly effective for subjects exhibiting low intrinsic numbers of bifidobacteria. On the other hand, it should be kept in mind that logarithmic values can give a biased view of absolute increases. For instance, an apparently small increase of 0.2 log10 starting from a baseline level of 9.3 log10 is comparable to an increase of 3.1 log10 starting from an initial concentration of 6.0 log10.
The observation that the administration of the probiotic yeast S. boulardii did not appear to cause profound changes in the intestinal microbiota of healthy subjects may not be entirely unexpected. Whereas S. boulardii is specifically used as a biotherapeutic agent for the prevention and treatment of different types of diarrhea (9, 18, 36), none of the subjects in this study had a disturbed intestinal balance or suffered from diarrhea during the sampling period. The therapeutic effects of S. boulardii in healthy subjects may be minimal compared to those in patients suffering from diarrhea.
Group-specific primers were used to allow a more in-depth analysis of three bacterial subpopulations of the human colon. However, DGGE profiles of the Bacteroides fragilis subgroup, the genus Bifidobacterium, and Clostridium clusters XI and XIVa were relatively stable and did not reveal significant temporal shifts during the feeding trial. These results are consistent with the findings of previous studies in which no major shifts of predominant autochthonous bacterial groups were observed (34, 44). In contrast to the observations from universal profiles, stimulation of B. adolescentis after lactulose ingestion could not be detected in the Bifidobacterium profiles, possibly because of the target concentration effect. It can be assumed that the use of universal primers will yield a DGGE band for only the most predominant species representing the largest fraction of the community DNA. Group-specific primers are directed to only a fraction of the total DNA pool, in which case concentration differences are expected to be less pronounced in the subpopulation DGGE profile.
In conclusion, this study has demonstrated that an integrated use of DGGE and RT-PCR has the potential to monitor the effects of pro-, pre-, and synbiotic intake on the predominant fecal microbiota of humans. Universal V3 primer DGGE fingerprint profiles from fecal samples revealed an effect of lactulose administration on fecal bifidobacteria and in particular on B. adolescentis. Subsequent quantification with RT-PCR confirmed the stimulation of total bifidobacteria and B. adolescentis. Because changes occurring mainly in the dominant bacterial populations can be detected, it cannot be excluded that subtle changes in less predominant species or groups of species may remain undetected by this approach. Although the sensitivity of this strategy needs to be elaborated further, the combination of DGGE analysis and RT-PCR quantification may be considered a promising approach in future studies, e.g., to analyze disturbed intestinal microbiota of specific patient groups. Especially in chronic intestinal disorders such as inflammatory bowel disease, in which the intestinal microbiota is believed to play a role in the (aethio)pathogenesis of the disease, this integrated approach could be useful to monitor potential indicator organisms and to assess the effects of new biotherapeutic agents during clinical intervention studies.
Acknowledgments
This work was supported by IWT-Vlaanderen, Brussels, Belgium (GBOU project no. 010054, “Development of a fast, non-invasive technological tool to investigate the functionality and effectivity of pro- and prebiotics in normal healthy humans: the use of a labeled biomarker”). G.H. is a postdoctoral fellow of the Fund for Scientific Research—Flanders (F.W.O.-Vlaanderen, Belgium).
L. De Vuyst and B. Pot are acknowledged for intellectual input. The dedication of the persons that participated in the study is also highly acknowledged.
REFERENCES
- 1.Ballongue, J., C. Schumann, and P. Quignon. 1997. Effects of lactulose and lactitol on colonic microflora and enzymatic activity. Scand. J. Gastroenterol. 222(Suppl.):41-44. [DOI] [PubMed] [Google Scholar]
- 2.Berg, R. D. 1996. The indigenous gastrointestinal microflora. Trends Microbiol. 4:430-435. [DOI] [PubMed] [Google Scholar]
- 3.Biavati, B. and P. Mattarelli. 1991. Bifidobacterium ruminantium sp. nov. and Bifidobacterium merycicum sp. nov. from the rumens of cattle. Int. J. Syst. Bacteriol. 41:163-168. [DOI] [PubMed] [Google Scholar]
- 4.Bibiloni, R., M. A. Simon, C. Albright, B. Sartor, and G. W. Tannock. 2005. Analysis of the large bowel microbiota of colitic mice using PCR/DGGE. Lett. Appl. Microbiol. 41:45-51. [DOI] [PubMed] [Google Scholar]
- 5.Bouhnik, Y., A. Attar, F. A. Joly, M. Riottot, F. Dyard, and B. Flourié. 2004. Lactulose ingestion increases faecal bifidobacterial counts: a randomised double-blind study in healthy humans. Eur. J. Clin. Nutr. 58:462-466. [DOI] [PubMed] [Google Scholar]
- 6.Cummings, J. H., G. T. Macfarlane, and H. N. Englyst. 2001. Prebiotic digestion and fermentation. Am. J. Clin. Nutr. 73:415S-420S. [DOI] [PubMed] [Google Scholar]
- 7.De Preter, V., T. Vanhoutte, G. Huys, J. Swings, P. Rutgeerts, and K. Verbeke. 2006. Effect of lactulose and Saccharomyces boulardii administration on the colonic urea-nitrogen metabolism and the bifidobacteria concentration in healthy human subjects. Aliment. Pharmacol. Ther. 23:963-974. [DOI] [PubMed] [Google Scholar]
- 8.FAO/WHO. 2001. Health and nutritional properties of probiotics in food including powder milk and live lactic acid bacteria. Report of a joint FAO/WHO expert consultation. [Online.] http://www.fao.org/es/ESN/Probio/report.pdf.
- 9.Fioramonti, J., V. Theodorou, and L. Bueno. 2003. Probiotics: what are they? What are their effects on gut physiology? Best Pract. Res. Clin. Gastroenterol. 17:711-724. [DOI] [PubMed] [Google Scholar]
- 10.Fuller, R., and G. R. Gibson. 1998. Probiotics and prebiotics: microflora management for improved gut health. Clin. Microbiol. Infect. 4:477-480. [Google Scholar]
- 11.Gibson, G. R., R. A. Rastall, and M. B. Roberfroid. 1999. Prebiotics, p. 101-124. In G. R. Gibson and M. B. Roberfroid (ed.), The colonic microbiota: nutrition and health. Kluwer Academic Publishers, London, United Kingdom.
- 12.Gibson, G. R., and M. B. Roberfroid. 1995. Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. J. Nutr. 125:1401-1412. [DOI] [PubMed] [Google Scholar]
- 13.Gibson, G. R., H. M. Probert, J. Van Loo, R. A. Rastall, and M. B. Roberfroid. 2004. Dietary modulation of the human colonic microbiota: updating the concept of prebiotics. Nutr. Res. Rev. 17:259-275. [DOI] [PubMed] [Google Scholar]
- 14.Guarner, F., and J. R. Malagelada. 2003. Gut flora in health and disease. Lancet 361:512-519. [DOI] [PubMed] [Google Scholar]
- 15.Haarman, M., and J. Knol. 2005. Quantitative real-time PCR assays to identify and quantify fecal Bifidobacterium species in infants receiving a prebiotic infant formula. Appl. Environ. Microbiol. 71:2318-2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harmsen, H. J., G. C. Raangs, A. Franks, A. C. Wildeboer-Veloo, and G. W. Welling. 2002. The effect of the prebiotic inulin and the probiotic Bifidobacterium longum on the faecal microflora of healthy volunteers measured by FISH and DGGE. Microb. Ecol. Health Dis. 14:219. [Google Scholar]
- 17.Isolauri, E., P. V. Kirjavainen, and S. Salminen. 2002. Probiotics—a role in the treatment of intestinal infection and inflammation. Gut 50(Suppl. III):54-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jahn, H. U., R. Ullrich, T. Schneider, R. M. Liehr, H. L. Schieferdecker, H. Holst, and M. Zeitz. 1996. Immunological and trophical effects of Saccharomyces boulardii on the small intestine in healthy human volunteers. Digestion 57:95-104. [DOI] [PubMed] [Google Scholar]
- 19.Jernberg, C., A. Sullivan, C. Edlund, and J. K. Jansson. 2005. Monitoring of antibiotic-induced alterations in the human intestinal microflora and detection of probiotic strains by use of terminal restriction fragment length polymorphism. Appl. Environ. Microbiol. 71:501-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Langendijk, P. S., F. Schut, G. J. Jansen, G. C. Raangs, G. R. Kamphuis, M. H. F. Wilkinson, and G. W. Welling. 1995. Quantitative fluorescence in situ hybridization of Bifidobacterium spp. with genus-specific 16S rRNA-targeted probes and its application in fecal samples. Appl. Environ. Microbiol. 61:3069-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Langlands, S. J., M. J. Hopkins, N. Coleman, and J. H. Cummings. 2004. Prebiotic carbohydrates modify the mucosa associated microflora of the human large bowel. Gut 53:1610-1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leser, T. D., J. Z. Amenuvor, T. K. Jensen, R. H. Lindecrona, M. Boye, and K. Møller. 2002. Culture-independent analysis of gut bacteria: the pig gastrointestinal tract microbiota revisited. Appl. Environ. Microbiol. 68:673-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsuki, T., K. Watanabe, J. Fujimoto, Y. Miyamoto, T. Takada, K. Matsumoto, H. Oyaizu, and R. Tanaka. 2002. Development of 16S rRNA-gene-targeted group-specific primers for the detection and identification of predominant bacteria in human feces. Appl. Environ. Microbiol. 68:5445-5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsuki., T., K. Watanabe, J. Fujimoto, Y. Kado, T. Takada, K. Matsumoto, and R. Tanaka. 2004. Quantitative PCR with 16S rRNA-gene-targeted species-specific primers for analysis of human intestinal bifidobacteria. Appl. Environ. Microbiol. 70:167-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matteuzzi, D., E. Swennen, M. Rossi, T. Hartman, and V. Lebet. 2004. Prebiotic effects of a wheat germ preparation in human healthy subjects. Food Microbiol. 21:119-124. [Google Scholar]
- 26.Mitterdorfer, G., W. Kniefel, and H. Viernstein. 2001. Utilization of prebiotic carbohydrates by yeasts of therapeutic relevance. Lett. Appl. Microbiol. 33:251-255. [DOI] [PubMed] [Google Scholar]
- 27.Muyzer, G., E. C. Dewaal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petersen, D. G., and I. Dahllöf. 2005. Improvements for comparative analysis of changes in diversity of microbial communities using internal standards in PCR-DGGE. FEMS Microbiol. Ecol. 53:339-348. [DOI] [PubMed] [Google Scholar]
- 29.Pitcher, D. G., N. A. Saunders, and R. J. Owen. 1989. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett. Appl. Microbiol. 8:151-156. [Google Scholar]
- 30.Roberfroid, M. B., J. A. E. Van Loo, and G. R. Gibson. 1998. The bifidogenic nature of chicory inulin and its hydrolysis products. J. Nutr. 128:11-19. [DOI] [PubMed] [Google Scholar]
- 31.Sahota, S. S., P. M. Bramley, and I. S. Menzies. 1982. The fermentation of lactulose by colonic bacteria. J. Gen. Microbiol. 128:319-325. [DOI] [PubMed] [Google Scholar]
- 32.Salminen, S., C. Bouley, M. C. Boutron-Ruault, J. H. Cummings, A. Franck, G. R. Gibson, E. Isolauri, M. C. Moreau, M. Roberfroid, and I. Rowland. 1998. Gastrointestinal physiology and function—targets for functional food development. Br. J. Nutr. 80(Suppl.):147-171. [DOI] [PubMed] [Google Scholar]
- 33.Salminen, S., P. Ramos, and R. Fonden. 1993. Substrates and lactic acid bacteria. In S. Salminen, and A. vonWright (ed.), Lactic acid bacteria. Marcel Dekker, New York, N.Y.
- 34.Satokari, R. M., E. E. Vaughan, A. D. L. Akkermans, M. Saarela, and W. M. de Vos. 2001. Bifidobacterial diversity in human feces detected by genus-specific PCR and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 67:504-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suau, A., R. Bonnet, M. Sutren, J. J. Godon, G. R. Gibson, M. D. Collins, and J. Doré. 1999. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl. Environ. Microbiol. 65:4799-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Surawicz, C. M., G. W. Elmer, P. Speelman, L. V. McFarland, J. Chinn, and G. Vanbelle. 1996. Prevention of antibiotic-associated diarrhea by Saccharomyces boulardii: a prospective study. Gastroenterology 96:981-988. [DOI] [PubMed] [Google Scholar]
- 37.Takada, T., K. Matsumoto, and K. Nomoto. 2004. Development of multi-color FISH method for analysis of seven Bifidobacterium species in human faeces. J. Microbiol. Methods 58:413-421. [DOI] [PubMed] [Google Scholar]
- 38.Tannock, G. W., K. Munro, R. Bibiloni, M. A. Simon, P. Hargreaves, P. Gopal, H. Harmsen, and G. Welling. 2004. Impact of consumption of oligosaccharide-containing biscuits on the fecal microbiota of humans. Appl. Environ. Microbiol. 70:2129-2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Terada, A., H. Hara, S. Kato, T. Kimura, I. Fujimori, K. Hara, T. Maruyama, and T. Mitsuoka. 1993. Effect of lactosucrose (4-beta-D-galactosylsucrose) on faecal flora and faecal putrefactive products of cats. J. Vet. Med. Sci. 55:291-295. [DOI] [PubMed] [Google Scholar]
- 40.Tomoda, T., Y. Nalano, and T. Kageyama. 1991. Effect of yogurt and yogurt supplemented with Bifidobacterium and/or lactulose in healthy persons: a comparative study. Bifdobacteria Microflora 10:123-130. [Google Scholar]
- 41.Tuohy, K. M., C. J. Ziemer, A. Klinder, Y. Knöbel, B. L. Pool-Zobel, and G. R. Gibson. 2002. A human volunteer study to determine the prebiotic effects of lactulose powder on human colonic microbiota. Microb. Ecol. Health Dis. 14:165-173. [Google Scholar]
- 42.Tuohy, K. M., S. Kolida, A. M. Lustenberger, and G. R. Gibson. 2001. The prebiotic effects of biscuits containing partially hydrolysed guar gum and fructo-oligosaccharides—a human volunteer study. Br. J. Nutr. 86:341-348. [DOI] [PubMed] [Google Scholar]
- 43.van der Aa Kühle, A., K. Skovgaardb, and L. Jespersena. 2005. In vitro screening of probiotic properties of Saccharomyces cerevisiae var.boulardii and food-borne Saccharomyces cerevisiae strains. Int. J. Food Microbiol. 101:29-39. [DOI] [PubMed] [Google Scholar]
- 44.Vanhoutte, T., G. Huys, E. De Brandt, and J. Swings. 2004. Temporal stability analysis of the microbiota in human faeces by denaturating gradient gel electrophoresis using universal and group-specific 16S rRNA gene primers. FEMS Microbiol. Ecol. 48:437-446. [DOI] [PubMed] [Google Scholar]
- 45.Van Loo, J. 2004. The specificity of the interaction with intestinal bacterial fermentation by prebiotics determines their physiological efficacy. Nutr. Res. Rev. 17:89-98. [DOI] [PubMed] [Google Scholar]
- 46.Weber, F. L. 1997. Effects of lactulose on nitrogen metabolism. Scand. J. Gastroenterol. 32:83-87. [DOI] [PubMed] [Google Scholar]
- 47.Zoetendal, E. G., A. D. L. Akkermans, and W. M. de Vos. 1998. Temperature gradient gel electrophoresis analysis of 16S rRNA from human fecal samples reveals stable and host-specific communities of active bacteria. Appl. Environ. Microbiol. 64:3854-3859. [DOI] [PMC free article] [PubMed] [Google Scholar]