Abstract
The antibacterial activities and membrane binding of nukacin ISK-1 and its fragments and mutants were evaluated to delineate the determinants governing structure-function relationships. The tail region (nukacin1-7) and ring region (nukacin7-27) were shown to have no antibacterial activity and also had no synergistic effect on each other or even on nukacin ISK-1. Both a fragment with three lysines in the N terminus deleted (nukacin4-27) and a mutant with three lysines in the N terminus replaced with alanine (K1-3A nukacin ISK-1) imparted very low activity (32-fold lower than nukacin ISK-1) and also exhibited a similar antagonistic effect on nukacin ISK-1. Addition of two lysine residues at the N terminus (+2K nukacin ISK-1) provided no further increased antibacterial activity. Surface plasmon resonance sensorgrams and kinetic rate constants determined by a BIAcore biosensor revealed that nukacin ISK-1 has remarkably higher binding affinity to anionic model membrane than to zwitterionic model membrane. Similar trends of strong binding responses and kinetics were indicated by the high affinities of nukacin ISK-1 and +2K nukacin ISK-1, but there was no binding of tail region, ring region, nukacin4-27, and K1-3A nukacin ISK-1 to the anionic model membrane. Our findings therefore suggest that the complete structure of nukacin ISK-1 is necessary for its full activity, in which the N-terminus three lysine residues play a crucial role in electrostatic binding to the target membrane and therefore nukacin ISK-1's ability to exert its potent antibacterial activity.
The emergence of resistance to traditional antibiotics among microorganisms has drawn the keen attention of researchers to ameliorate the antibiotic repertoires. In search of alternative antibacterial agents, lantibiotics seem to be one of the most promising candidates. Lantibiotics are synthesized on the ribosome as prepeptides that undergo several posttranslational modification events associated with several enzymes to form biologically active peptides (9, 25). Engineering of the naturally occurring novel lantibiotics might provide significant information for rational design of lantibiotics with improved activity and/or spectra that would be potent antimicrobials to augment, supplement, or replace the currently used antibiotics.
The antibacterial mode of actions of lantibiotics has been found to be diverse, and their activities pertain mostly to attacking the bacterial membrane, which eventually is induced to release ions, small molecules, and ATP from sensitive cells (16, 25, 27, 31). In addition, lantibiotics are reported to inhibit outgrowth of bacterial spores (22) and hinder cell wall biosynthesis (31). Among the structures/functions of lantibiotics, the type A(I) lantibiotic nisin has been explicitly studied to know well the structures responsible for its work as a potential antibiotic. The cationic nature of nisin allows it to bind to the phospholipid membrane by electrostatic interactions (4, 12, 33), and it exhibits higher affinity to anionic than to zwitterionic model membrane (8). N-terminal backbone amides of nisin interact with the pyrophosphate moiety of lipid II (14), and the transmembrane orientation of the molecule involves the insertion of its C-terminal part for pore formation (30). However, type A(II) lantibiotics that have two distinct regions (N-terminus tail and C-terminus ring) have not yet been studied well, and little is known about the structures of these peptides that are important for different steps necessary to function as antibiotics.
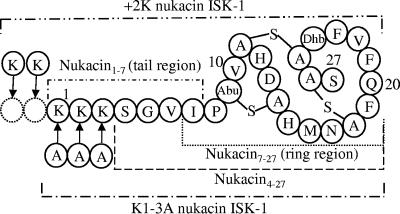
Nukacin ISK-1 is a novel type A(II) lantibiotic produced by Staphylococcus warneri ISK-1, isolated in our laboratory from well-aged Nukadoko, a bed of fermented rice bran (19, 20). It consists of 27 amino acids, including two molecules of lanthionine, one molecule of 3-metyllanthionine and one residue of dehydrobutyrine (Fig. 1) (1, 26). In this study, we generated and evaluated the fragments and mutants of nukacin ISK-1 to determine the roles of the structures involved in membrane binding and antibacterial activity. We used surface plasmon resonance (SPR) determined by BIAcore biosensor to investigate the binding behavior of nukacin ISK-1 and its fragments and mutants to model membrane. SPR is a fast and powerful tool for real-time monitoring of binding of the membrane-active peptides that supersedes the other relevant techniques (24). Our results suggested that the complete structure of nukacin ISK-1 is necessary for its full activity, and the first three lysine residues in the tail region play the vital role in its antibacterial activity, in which the positive charges are the key determinant for membrane binding of nukacin ISK-1. Since there is still little information on the mode of action of type A(II) lantibiotics, this study was designed to provide new insight into how type A(II) lantibiotics function in terms of the primary mode of action.
FIG. 1.
Proposed structure of nukacin ISK-1 showing its fragments and mutants used for this study. A-S-A, lanthionine; Abu-S-A, 3-methyllanthionine; Dhb, dehydrobutyrine. Tail region (nukacin1-7) was synthesized chemically, ring region (nukacin7-27) was generated by digestion with Pfu N-acetyl deblocking amino peptidase, and nukacin4-27 fragment was obtained by digestion with endoproteinase Lys-C. K1-3A nukacin ISK-1 and +2K nukacin ISK-1 were generated by genetic engineering.
MATERIALS AND METHODS
Synthesis and purification of nukacin ISK-1 and its fragments.
Nukacin ISK-1 was purified in accordance with the protocol described by Aso et al. (2).
The tail region (nukacin1-7) of nukacin ISK-1 was synthesized chemically by a solid-phase method using a 9-fluorenylmethoxy carbonyl (Fmoc) strategy with p-alkoxybenzyl alcohol resin (0.1 mmol; Kokusan Chemical, Tokyo, Japan) as the solid support. After chain assembly, the peptide was cleaved off from the support and the side chain-protecting groups were also deprotected by treatment with a trifluoroacetic acid (TFA)-triisopropylsilane-water mixture (95:2.5:2.5 [vol/vol]) for 90 min. The reaction mixture was passed over an HLC-DISK 13 (Kanto Chemical, Tokyo, Japan), and cold diethyl ether was added. The mixture was kept standing overnight at −20°C. The precipitant was collected on a polytetrafluoroethylene membrane filter (ADVANTEC, Ehime, Japan). The crude peptide on the filter was dissolved in 20% acetonitrile-0.1% TFA and then purified by reverse-phase (RP) column (PepRPC HR 5/5; Amersham Bioscience, Uppsala, Sweden) integrated in an LC-10A high-performance liquid chromatography (HPLC) system (Shimadzu, Kyoto, Japan). Peptides were eluted with a linear gradient of 15 to 80% acetonitrile-0.1% TFA at a flow rate of 1 ml/min and were analyzed by electrospray ionization-mass spectrometry (ESI-MS) (Accutof T100LC; JEOL, Tokyo, Japan).
The ring region (nukacin7-27) was obtained by digestion of nukacin ISK-1 (180 μg) with Pfu N-acetyl deblocking aminopeptidase (Takara, Shiga, Japan) (30 μg) in the buffer supplied by the manufacturer (250 mM N-ethylmorpholine-AcOH buffer, pH 8.0, containing 0.5 mM CoCl2) to make a 1-ml reaction volume, and the final reaction mixture was incubated at 50°C for 72 h. The nukacin4-27 fragment was generated by deletion of three lysine residues from the N terminus of nukacin ISK-1 (42 μg) by digestion with endoproteinase Lys-C (0.5 μg) (Sigma-Aldrich, St. Louis, MO) dissolved in 100 mM NH4HCO3 buffer (pH 8.5) to make a 1-ml reaction mixture and was incubated at 25°C for 24 h. After purification of the fragments by RP-HPLC, masses of the peptides were determined by ESI-MS.
Generation of nukacin ISK-1 mutants.
K1-3A nukacin ISK-1 and +2K nukacin ISK-1 were generated by amplification of the nukacin ISK-1 structural gene (nukA) by inverse PCR using plasmid pNZA (1) as a template with the following sets of primers: 5′-GCTGCTGCTTCAGGAGTAATCCCAACTGTG-3′ and 5′-AGCTCCTAAGACTTCATTCAATTCA-3′ and 5′-AAGAAAAAGAAAAAGTCAGGAGTAATCCCA-3′ and 5′-AGCTCCTAAGACTTCATTCAATTCA-3′, respectively. PCR was performed with KOD plus DNA polymerase (Toyobo, Osaka, Japan). The PCR products were purified with the Qiaquick PCR purification kit (QIAGEN, West Sussex, United Kingdom) and self-ligated with Ligation High (Toyobo). Transformation of Lactococus lactis NZ9000 was done according to the method developed by Holo and Nes (13). The resultant plasmids, pNZAK1-3A and pNZA +2K, were extracted by the method of O'Sullivan and Klaenhammer (23) and were then introduced into L. lactis NZ9000 harboring plasmid pInukdA, which contains all of the nukacin ISK-1 biosynthetic genes except for the nukA gene (1). Nukacin ISK-1 mutants were expressed by a nisin-controlled expression system basically in accordance with the method of Aso et al. (1). The recombinant strains were grown in chemically defined medium (18) with 5 μg/ml each chloramphenicol and erythromycin at 30°C. Nisin solution (crude nisin) (Sigma-Aldrich) was added to 10-ng/ml final concentrations to the culture at an optical density at 600 nm (OD600) of 0.6, and incubation continued for another 5 h.
For purification of expressed K1-3A nukacin ISK-1 and +2K nukacin ISK-1, culture supernatant was collected by centrifugation of the culture at 6,000 × g at 4°C for 15 min. Ten milliliters of culture supernatant was loaded onto a Sep-Pak C18 cartridge column (100 mg; Waters, Milford, MA), washed with 2 ml of 0.1% TFA, and eluted with 3 ml of 50% acetonitrile-0.1% TFA. Concentrated elutes were injected directly into the RP-HPLC column, and the mass of the peptides was determined by ESI-MS. The N-terminal amino acid sequences of the mutants were obtained by Edman degradation performed on a PPSQ-21 gas-phase automatic protein sequence analyzer (Shimadzu).
Antibacterial activity.
Antibacterial activities of purified nukacin ISK-1 and its fragments and mutants dissolved in water (pH 7.0) were determined by the spot-on-lawn method (20). Lactobacillus agar (Becton Dickinson, Sparks, MD) was overlaid on MRS agar medium with the specific indicator strains, and serial twofold-diluted peptides with water (pH 7.0) in 96-well plates (F96 microtiter plates, Nunc, Roskilde, Denmark) were spotted onto the surface of the medium. By observation of the clear zone of inhibition, antibacterial activities were expressed as the MIC. Lactobacillus sakei subsp. sakei JCM 1157T and Leuconostoc mesenteroides subsp. mesenteroides JCM 6124T, grown in MRS broth medium at 30°C, and Pediococcus pentosaceus JCM 5885, grown in MRS broth medium at 37°C, were used as indicators to determine antibacterial activity. Inhibitory activities of the fragments/mutants were determined with equimolar concentrations of nukacin ISK-1 and the fragment/mutant.
Preparation of model membranes.
The phospholipids (Sigma-Aldrich) used were 1,2-dimyristoyl-sn-glycero-3-phospho-rac-(1-glycerol) sodium salt (anionic) and 1,2-dioleyol-sn-glycero-3-phosphocholine (zwitterionic). Small unilamellar vesicle was prepared separately by dissolving 3 mg of each of the phospholipids in 10 ml chloroform. The suspension was evaporated under N2 and then dissolved in 10 mM HEPES buffer (final liposome concentration, 0.5 mM). After sonication, the liposome was dispensed into a glass tube and stored at −30°C.
SPR biosensor.
SPR spectroscopy using a BIAcore biosensor (BIAcore AB, Uppsala, Sweden) was used to determine the interaction of nukacin ISK-1 and its fragments and mutants with the previously described model membrane by an HPA sensor chip (BIAcore AB). All of the experiments were performed at 25°C in HBS buffer (10 mM HEPES, 150 mM NaCl, pH 7.4). Anionic or zwitterionic vesicle (0.5 mM, 80 μl each) was applied to the flow cell of the HPA sensor chip surface at a flow rate of 2 μl/min. To remove the multilamellar lipid vesicles and also to regenerate the model membrane, 10 mM NaOH (25 μl) was injected at a flow rate of 5 μl/min to result in a stable baseline corresponding to the lipid monolayer. Nukacin ISK-1 and its fragments and mutants were injected (7 μM, 15 μl) onto the lipid surface at a flow rate of 5 μl/min. The bulk effect of buffer composition was subtracted from each set of experimental sensorgrams. The kinetic values, ka (association rate constant) and kd (dissociation rate constant) of the resultant sensorgrams were determined by the BIAevaluation 2.1 software (BIAcore AB). The dissociation constant (KD) was calculated as kd/ka. SPR spectroscopy was done at least twice for each experiment.
RESULTS
Comparison of antibacterial activities of nukacin ISK-1 and its fragments and mutants.
Chemically synthesized tail region (nukacin1-7), Pfu N-acetyl deblocking aminopeptidase-digested ring region (nukacin7-27), endoproteinase Lys-C-digested nukacin4-27 fragment, and genetically engineered K1-3A nukacin ISK-1 and +2K nukacin ISK-1 (Fig. 1) were successfully obtained, and the masses of each were determined exactly to their calculated values. RP-HPLC-purified fragments and mutants were used to determine antibacterial activities along with their synergistic effects on each other and also on nukacin ISK-1 against L. sakei subsp. sakei JCM 1157T, L. mesenteroides subsp. mesenteroides JCM 6124T, and P. pentosaceus JCM 5885 as indicators. No activity of the ring region and tail region alone or even in combination was observed. Nukacin ISK-1 with same molar concentrations of tail or ring region also had the same activity as nukacin ISK-1 alone. Nukacin4-27 and K1-3A nukacin ISK-1 imparted similar but very low activities (32-fold lower) compared to nukacin ISK-1, and both had an antagonistic effect (fourfold lower activity) on the activity of nukacin ISK-1. An increase of two lysine residues at the N terminus (+2K nukacin ISK-1) resulted in no further enhancement of antibacterial activity of nukacin ISK-1 against the tested indicator strains (Table 1).
TABLE 1.
Antibacterial activities of nukacin ISK-1 and its fragments and mutants along with their inhibitory effects on nukacin ISK-1 and each other
| Nukacin ISK-1 or derivative | MIC (μM)a
|
||
|---|---|---|---|
| L. sakei | P. pentosaceus | L. mesenteroides | |
| Nukacin ISK-1 (control) | 0.26 | 0.52 | 0.26 |
| +2K nukacin ISK-1 | 0.26 | 0.52 | 0.26 |
| Nukacin ISK-1 + tail region | 0.26 | 0.52 | 0.26 |
| Nukacin ISK-1 + ring region | 0.26 | 0.52 | 0.26 |
| Nukacin ISK-1 + K1-3A nukacin ISK-1 | 1.04 | 2.08 | 1.04 |
| Nukacin ISK-1 + nukacin4-27 | 1.04 | 2.08 | 1.04 |
| K1-3A nukacin ISK-1 | 8.32 | 16.64 | 8.32 |
| Nukacin4-27 | 8.32 | 16.64 | 8.32 |
| Tail region (nukacin1-7) | ND | ND | ND |
| Ring region (nukacin7-27) | ND | ND | ND |
| Tail region + ring region | ND | ND | ND |
Antibacterial activity was determined at least twice for each derivative. Results are shown for L. sakei subsp. sakei JCM 1157T, P. pentosaceus JCM 5885, and L. mesenteroides subsp. mesenteroides JCM 6124T. ND, not detected.
Binding affinity of nukacin ISK-1 to the model membranes.
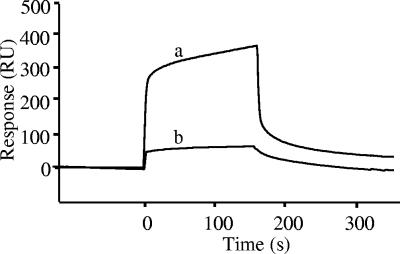
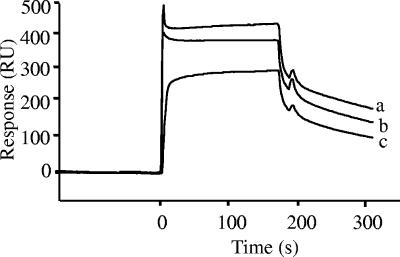
SPR determined by the BIAcore biosensor suggested strong binding responses of nukacin ISK-1 to anionic model membrane in contrast to the zwitterionic model membrane (Fig. 2). The integrity of the sensor chip HPA was also checked by allowing nukacin ISK-1 to bind to the free surface of its flow cells, and no affinity to the sensor chip was found (data not shown). Differences between the kinetics of each of the sensorgrams suggested different binding affinities towards the model membrane and provided further evidence for stronger binding of nukacin ISK-1 to anionic (KD, 5.15 μM) than to zwitterionic (KD, 32.5 μM) model membrane (Table 2). The variations in dissociation constant (KD) of nukacin ISK-1 to the anionic and zwitterionic model membranes were due to variations of both the association rate constant (ka) and dissociation rate constant (kd). These differences can be explained quantitatively to emphasize the binding behavior of nukacin ISK-1. The rate constants (ka and kd) distinctly indicate strong association and high stability of nukacin ISK-1 to anionic model membrane in contrast to zwitterionic model membrane, which were reflected similarly by the entire sensorgram showing high association (high number of resonance units [RU]) and low dissociation with anionic membrane and vice versa for zwitterionic membrane (Table 2). Kinetics data therefore strongly supported the sensorgrams and indicated high binding affinity of nukacin ISK-1 to anionic membrane in comparison with the zwitterionic model membrane. Variation of nukacin ISK-1 concentrations (5, 15, and 20 μM) allowed us to obtain high binding responses to anionic model membrane at higher concentrations (Fig. 3). Binding of nukacin ISK-1 to the model membrane thus correlated well with the amount of nukacin ISK-1.
FIG. 2.
Binding affinity of nukacin ISK-1 to anionic [1,2-dimyristoyl-sn-glycero-3-phospho-rac-(1-glycerol) sodium salt] and zwitterionic (1,2-dioleyol-sn-glycero-3-phosphocholine) model membranes determined by SPR biosensor. Sensorgrams for 7 μM nukacin ISK-1 bound to each of the anionic (a) and zwitterionic (b) model membranes are indicated.
TABLE 2.
Affinity kinetics of nukacin ISK-1 and +2K nukacin ISK-1 to model membrane
| Liposomea | Sample | ka (M−1 s−1) | kd (s−1) | KD (μM) |
|---|---|---|---|---|
| Anionic | Nukacin ISK-1 | 1.63 × 103 | 8.39 × 10−3 | 5.15 |
| +2K nukacin ISK-1 | 2.58 × 103 | 7.67 × 10−3 | 2.97 | |
| Zwitterionic | Nukacin ISK-1 | 5.29 × 102 | 1.72 × 10−2 | 32.5 |
Anionic, 1,2-dimyristoyl-sn-glycero-3-phospho-rac-(1-glycerol) sodium salt; zwitterionic, 1,2-dioleyol-sn-glycero-3-phosphocholine.
FIG. 3.
Dose response of nukacin ISK-1 towards the anionic model membrane. Nukacin ISK-1 concentrations were 20 (a), 15 (b), and 5 (c) μM.
Comparison of the binding affinities of nukacin ISK-1 and its fragments and mutants to the anionic model membrane.
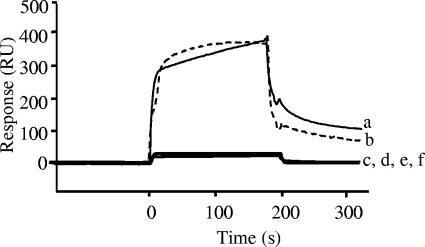
The SPR data detected as RU for the signal intensity obtained by the BIAcore biosensor were the functions of nukacin ISK-1 and its fragments and mutants for binding to the anionic model membrane (Fig. 4). The concentrations of nukacin ISK-1 and its fragments and mutants were 7 μM throughout this study. Strong binding responses of nukacin ISK-1 and +2K nukacin ISK-1 were detected by the entire sensorgrams that were further supported by the affinity rate constants for recognition and stability. Comparable but somewhat stronger (∼1.5-fold) affinity (KD, 5.15 μM and 2.97 μM for nukacin ISK-1 and +2K nukacin ISK-1, respectively) was observed for +2K nukacin ISK-1 (Table 2). No affinity of nukacin4-27, K1-3A nukacin ISK-1, tail region, and ring region to the anionic model membrane was detected by the SPR response under the specified conditions. The hypothesis that nukacin ISK-1 would have affinity towards negatively charged membrane was strongly supported by the binding of nukacin ISK-1 and +2K nukacin ISK-1 to anionic phospholipid membrane. The results therefore suggested that the association of nukacin ISK-1 with the model membrane is primarily by the electrostatic interactions for the presence of positively charged residues that dramatically increased the number of resonance units of SPR. The apparent argument for the possibility of the tail region to bind to the membrane was clarified by determining its secondary structure. Except for the tail region, nukacin ISK-1 and other fragments were shown to form a similar secondary structure (data not shown). The SPR results were thus quite robust for proving that the complete structure of nukacin ISK-1, especially the presence of the positively charged lysine residues in the tail region, was crucial for strong binding of nukacin ISK-1 to the anionic model membrane.
FIG. 4.
SPR sensorgrams denoting the binding of +2K nukacin ISK-1 (a), nukacin ISK-1 (b), tail region (c), ring region (d), nukacin4-27 (e), and K1-3A nukacin ISK-1 (f) to the anionic model membrane. The concentration of nukacin ISK-1 and its derivatives was 7 μM.
DISCUSSION
We tried to depict our understanding of which structure of nukacin ISK-1 is responsible for membrane binding and exerts its antibacterial activity. In this study, we used purified nukacin ISK-1, its tail region (nukacin1-7), ring region (nukacin7-27), nuakcin4-27 fragment, K1-3A nukacin ISK-1, and +2K nukacin ISK-1 (Fig. 1) to determine the antibacterial activity and their synergistic effects on nukacin ISK-1 as well as each other (Table 1). No activity of the ring or tail region was observed, these regions showed no synergistic effect in combination, and neither of them had any effect on the activity of nukacin ISK-1. Activities of nukacin4-27 and K1-3A nukacin ISK-1 were 32-fold lower than that of nukacin ISK-1, and both nukacin4-27 and K1-3A nukacin ISK-1 exhibited an inhibitory effect (fourfold lower) on the antibacterial activity of nukacin ISK-1. Our results are in good agreement with the findings that for the nisin1-12 fragment, deletion of five residues and deletion of a further nine residues from the C terminus of nisin resulted in no activity and 10-fold- and 100-fold-lower activities, respectively, along with the antagonistic effect of nisin1-12 on nisin activity (7). Truncation of five residues from the N terminus of lacticin 481 has also been reported to show 10-fold-lower activity (28).
Peptide antibiotics are thought primarily to be effective by their cationic and amphipathic nature. Nukacin ISK-1 has three net positive charges (determined by GENETYX-WIN; Software Development, Tokyo, Japan), so the anionic bacterial membrane would be the target for binding of nukacin ISK-1. However, we also tested and compared the zwitterionic vesicle to prove the fidelity of anionic vesicle as a model membrane. Stronger binding to anionic than to zwitterionic membrane (Table 2 and Fig. 2) and dose-dependent binding proved the electrostatic interactions of nukacin ISK-1 with anionic model membrane (Fig. 3). Though binding evaluation of lantibiotics by SPR has not yet been reported by others, our work was concurred with the binding responses of other antibiotic peptides to the model membrane (21, 24).
We used the affinity-related structure-function relationship as the parameter for biological activity. The binding of nukacin ISK-1 and its fragments and mutants to the model membrane was evaluated to determine the relationship between binding ability (Fig. 4) and antibacterial activity (Table 1). Binding affinities shown by SPR sensorgrams were significantly correlated with their observed antibacterial activities. High binding of nukacin ISK-1 and +2K nukacin ISK-1 to the anionic membrane reflected their high antibacterial activities. Increase of the cationic property of nukacin ISK-1 by two additional lysine residues at the N terminus (+2K nukacin ISK-1) provided a relatively stronger (∼1.5-fold) ability to bind to the model membrane (Table 2). However, this extra binding ability did not contribute further to enhancement of the antibacterial activity of +2K nukacin ISK-1. This was probably because the charges required for the electrostatic interactions to bind to the membrane would have already been satisfied by the three lysine residues to act as a novel antibacterial. Therefore, the increase in charges did not increase nukacin ISK-1's effectiveness as a potential antibiotic. Binding of nukacin ISK-1 to the membrane was thus proved to be primarily by the N terminus, and its antibacterial action might be dependent on the C terminus. No observable antibacterial activity of the tail and ring region was reflected, as expected by their inability to bind to the anionic model membrane. Many membrane-active peptides have been shown to form a secondary structure to undergo necessary alteration for various conformational changes needed to become an antibacterial (10, 11, 16, 17, 29). Our result also suggests that lack of secondary structure formation was the possible reason for the tail region not to bind to the anionic model membrane and not to show antibacterial activity, though the three lysine residues are present in the tail region. Low antibacterial activities of nukacin4-27 and K1-3A nukacin ISK-1 were detected, but none of them showed electrostatic interaction to bind to the anionic model membrane. Since the degrees of binding were found to be directly related to endowing nukacin ISK-1 and +2K nukacin ISK-1 with antibacterial activity, the low activities of nukacin4-27 and K1-3A nukacin ISK-1 might result from their binding by other weak associations (e.g., hydrophobic interaction) with the cytoplasmic membrane of an indicator strain. Nukacin4-27 and K1-3A nukacin ISK-1 showed inhibitory activities against nukacin ISK-1, though they did not bind to the model membrane. It has become well established that binding to the membrane is the primary step of a membrane-active antibacterial peptide, and in vivo activity depends on more than one factor. Existing structure-based antibacterial activity of nukacin ISK-1 might also be possible, as was discussed later for other lantibiotics. Therefore, the competition of nukacin4-27 and K1-3A nukacin ISK-1 against nukacin ISK-1 would probably be at the next steps of its activity (e.g., competition for docking molecule or a binding motif) or might even be against enzyme function inhibition.
The mode of actions of type A(I) (nisin type) lantibiotics has unambiguously been clarified. The overlapping killing activities of nisin have been elicited to be complex. Nisin and nisin-like (e.g., subtilin) lantibiotics use lipid II as a docking molecule for high-affinity binding that combines pore formation and inhibition of cell wall biosynthesis (3, 6, 31). The nisin-lipid II complex has a novel lipid II-binding motif in which the N-terminal backbone amides of nisin coordinate the pyrophosphate moiety of lipid II (14). Besides, some other lantibiotics have already been shown to target the peptidoglycan precursor lipid II for their potent mode of actions. Wiedemann et al. (32) recently found that the overall inhibitory features of plantaricin C are more similar to those of nisin, where it strongly inhibits in vitro lipid II synthesis and forms a stable complex with lipid II, indicating that both nisin and plantaricin C may target the same structures in lipid II. Mersacidin-like (type B, globular lantibiotics) lantibiotics block the precursor from incorporation into the cell wall to inhibit the transglycosylation step of cell wall biosynthesis (5). Hsu et al. (15) reported that electrostatic interactions play a central role in the mersacidin-lipid II interactions. However, information on the mode of action of type A(II) lantibiotics is still poor, and unraveling the details remains to be worked out. It might also be possible for the existence of a multiple-step mode of actions involved in antibacterial activity of nukacin ISK-1. We are now working on unraveling the detailed mode of actions of nukacin ISK-1. However, the present results clearly indicate that lysine-oriented charges crucially govern the binding of nukacin ISK-1 to the membrane through electrostatic interactions, which pave the primary way for nukacin ISK-1 to become a potent antibacterial.
Acknowledgments
We are grateful to M. Kimura of Kyushu University, Fukuoka, Japan, for his kind permission to get easy access to use the BIAcore biosensor. We are thankful to Yoshiko Morinaga for her sincere help with part of this study.
S.M.A. acknowledges a Monbukagakusho (MEXT, Japan) fellowship. This work was partially supported by grants from the Japan Society for the Promotion of Science (JSPS), the Japan Science Society, the Novartis Foundation (Japan) for the Promotion of Science, the Novozymes Japan Research Fund, and the Nagase Science and Technology Foundation.
REFERENCES
- 1.Aso, Y., J. Nagao, H. Koga, K. Okuda, Y. Kamenasa, T. Sashihara, J. Nakayama, and K. Sonomoto. 2004. Heterologous expression and functional analysis of the gene cluster for the biosynthesis of and immunity to the lantibiotic, nukacin ISK-1. J. Biosci. Bioeng. 98:429-436. [DOI] [PubMed] [Google Scholar]
- 2.Aso, Y., K. Okuda, J. Nagao, Y. Kamenasa, N. T. B. Phuong, H. Koga, K. Shioya, T. Sashihara, J. Nakayama, and K. Sonomoto. 2005. A novel type of immunity protein, NukH, for the lantibiotic nukacin ISK-1 produced by Staphylococcus warneri ISK-1. Biosci. Biotechnol. Biochem. 69:1403-1410. [DOI] [PubMed] [Google Scholar]
- 3.Breukink, E., I. Wiedemann, C. van Kraaij, O. P. Kuipers, H. -G. Sahl, and B. de Kruijff. 1999. Use of the cell wall precursor lipid II by a pore-forming peptide antibiotic. Science 286:2361-2364. [DOI] [PubMed] [Google Scholar]
- 4.Breukink, E., P. Ganz, B. de Kruijff, and J. Seelig. 2000. Binding of nisin Z to bilayer vesicles as determined with isothermal titration calorimetry. Biochemistry 39:10247-10254. [DOI] [PubMed] [Google Scholar]
- 5.Brotz, H., G. Bierbaum, P. E. Reynolds, and H. -G. Sahl. 1997. The lantibiotic mersacidin inhibits peptidoglycan biosynthesis at the level of transglycosylation. Eur. J. Biochem. 246:193-199. [DOI] [PubMed] [Google Scholar]
- 6.Brotz, H., M. Josten, I. Wiedemann, U. Schneider, F. Gotz, G. Bierbaum, and H. -G. Sahl. 1998. Role of lipid-bound peptidoglycan precursors in the formation of pores by nisin, epidermin and other lantibiotics. Mol. Microbiol. 30:317-327. [DOI] [PubMed] [Google Scholar]
- 7.Chan, W. C., M. Leyland, J. Clark, H. M. Dodd, L. Y. Lian, M. J. Gasson, B. W. Bycroft, and G. C. K. Roberts. 1996. Structure activity relationships in the peptide antibiotic nisin: antibacterial activity of fragments of nisin. FEBS Lett. 390:129-132. [DOI] [PubMed] [Google Scholar]
- 8.Demel, R. A., T. Peelen, R. J. Siezen, B. de Kruijff, and O. P. Kuipers. 1996. Nisin Z, mutant nisin Z and lacticin 481 interactions with anionic lipids correlate with antimicrobial activity. A monolayer study. Eur. J. Biochem. 235:267-274. [DOI] [PubMed] [Google Scholar]
- 9.de Vos, W. M., O. P. Kuipers, J. R. van der Meer, and R. J. Siezen. 1995. Maturation pathway of nisin and other lantibiotics: post-translationally modified antimicrobial peptides exported by Gram-positive bacteria. Mol. Microbiol. 17:427-437. [DOI] [PubMed] [Google Scholar]
- 10.Fimland, G., O. R. Blingsmo, K. Sletten, G. Jung, I. F. Nes, and J. Nissen-Meyer. 1996. New biologically active bacteriocins constructed by combining regions from various pediocin-like bacteriocins: the C-terminal region is important for determining specificity. Appl. Environ. Microbiol. 62:3313-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freund, S., G. Jung, W. A. Gibbons, and H. -G. Sahl. 1991. NMR and circular dichroism studies on Pep5, p. 103-112. In G. Jung and H. -G. Sahl (ed.), Nisin and novel lantibiotics. Escom, Leiden, The Netherlands.
- 12.Giffard, C. J., H. M. Dodd, N. Horn, S. Ladha, A. R. Machie, A. Parr, M. J. Gasson, and D. Sanders. 1997. Structure-function relations of variant and fragment nisins studied with model membrane systems. Biochemistry 36:3802-3810. [DOI] [PubMed] [Google Scholar]
- 13.Holo, H., and I. F. Nes. 1989. High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl. Environ. Microbiol. 55:3119-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsu, S.-T., E. Breukink, E. Tischenko, M. A. G. Lutters, B. de Kruijff, R. Kaptein, A. M. Bonvin, and N. A. van Nuland. 2004. The nisin-lipid II complex reveals a pyrophosphate cage that provides a blueprint for novel antibiotics. Nat. Struct. Mol. Biol. 11:963-967. [DOI] [PubMed] [Google Scholar]
- 15.Hsu, S.-T., E. Breukink, G. Bierbaum, H. -G. Sahl, B. de Kruijff, R. Kaptein, N. A. van Nuland, and A. M. Bonvin. 2003. NMR study of mersacidin and lipid II interaction in dodecylphosphocholine micelles. Conformational changes are a key to antimicrobial activity. J. Biol. Chem. 278:13110-13117. [DOI] [PubMed] [Google Scholar]
- 16.Jack, R., W. Benz, J. Tagg, and H.-G. Sahl. 1994. The mode of action of SA-FF22, a lantibiotic isolated from Streptococcus pyogenes strain FF22. Eur. J. Biochem. 219:699-705. [DOI] [PubMed] [Google Scholar]
- 17.Jack, R. W., A. Carne, J. Metzger, S. Stefanovic, H. G. Sahl, G. Jung, and J. Tagg. 1994. Elucidation of the structure of SA-FF22, a lanthionine-containing antibacterial peptide produced by Streptococcus pyogenes strain FF22. Eur. J. Biochem. 220:455-462. [DOI] [PubMed] [Google Scholar]
- 18.Jensen, P. R., and K. Hammer. 1993. Minimal requirements for exponential growth of Lactococcus lactis. Appl. Environ. Microbiol. 59:4363-4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kimura, H., H. Matsusaki, T. Sashihara, K. Sonomoto, and A. Ishizaki. 1998. Purification and partial identification of bacteriocin ISK-1, a new lantibiotic produced by Pediococcus sp. ISK-1. Biosci. Biotechnol. Biochem. 62:2341-2345. [DOI] [PubMed] [Google Scholar]
- 20.Kimura, H., R. Nagano, H. Matsusaki, K. Sonomoto, and A. Ishizaki. 1997. A bacteriocin of strain Pediococcus sp. ISK-1 isolated from Nukadoko, bed of fermented rice bran. Biosci. Biotechnol. Biochem. 61:1049-1051. [DOI] [PubMed] [Google Scholar]
- 21.Mangoni, M. L., N. Papo, G. Mignogna, D. Andreu, Y. Shai, D. Barra, and M. Simmaco. 2003. Ranacyclins, a new family of short cyclic antimicrobial peptides: biological function, mode of action, and parameters involved in target specificity. Biochemistry 42:14023-14035. [DOI] [PubMed] [Google Scholar]
- 22.Okereke, A., and T. J. Montville. 1992. Nisin dissipates the proton motive force of the obligate anaerobe Clostridium sporogenes PA 3679. Appl. Environ. Microbiol. 58:2463-2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O'Sullivan, D. J., and T. R. Klaenhammer. 1993. Rapid mini-prep isolation of high-quality plasmid DNA from Lactococcus and Lactobacillus spp. Appl. Environ. Microbiol. 59:2730-2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Papo, N., and Y. Shai. 2003. Exploring peptide membrane interaction using surface plasmon resonance: differentiation between pore formation versus membrane disruption by lytic peptides. Biochemistry 42:458-466. [DOI] [PubMed] [Google Scholar]
- 25.Sahl, H. -G., R. W. Jack, and G. Bierbaum. 1995. Biosynthesis and biological activities of lantibiotics with unique post-translational modifications. Eur. J. Biochem. 230:827-853. [DOI] [PubMed] [Google Scholar]
- 26.Sashihara, T., H. Kimura, T. Higuchi, A. Adachi, H. Matsusaki, K. Sonomoto, and A. Ishizaki. 2000. A novel lantibiotic, nukacin ISK-1, of Staphylococcus warneri ISK-1: cloning of the structural gene and identification of the structure. Biosci. Biotechnol. Biochem. 64:2420-2428. [DOI] [PubMed] [Google Scholar]
- 27.Schuller, F., R. Benz, and H. -G. Sahl. 1989. The peptide antibiotic subtilin acts by formation of voltage-dependent multi-state pores in bacterial and artificial membranes. Eur. J. Biochem. 182:181-186. [DOI] [PubMed] [Google Scholar]
- 28.Uguen, P., T. Hindre, S. Didelot, C. Marty, D. Haras, J.-P. Le Pennec, K. Vallée-Réhel, and A. Dufour. 2005. Maturation by LctT is required for biosynthesis of full-length lantibiotic lacticin 481. Appl. Environ. Microbiol. 71:562-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van de Ven, F. J. M., H. W. van den Hooven, R. N. H. Konings, and C. W. Hilbers. 1991. The spatial structure of nisin in aqueous solution, p. 35-42. In G. Jung and H. -G. Sahl (ed.), Nisin and novel lantibiotics. Escom, Leiden, The Netherlands.
- 30.van Heusden, H., B. de Kruijff, and E. Breukink. 2002. Lipid II induces an overall transmembrane orientation of the pore-forming peptide lantibiotic nisin. Biochemistry 41:12171-12178. [DOI] [PubMed] [Google Scholar]
- 31.Wiedemann, I., E. Breukink, C. van Kraij, O. P. Kuipers, G. Bierbaum, B. de Kruiff, and H.-G. Sahl. 2001. Specific binding of nisin to the peptidoglycan precursor lipid II combines pore formation and inhibition of cell wall biosynthesis for potent antibiotic activity. J. Biol. Chem. 276:1772-1779. [DOI] [PubMed] [Google Scholar]
- 32.Wiedemann, I., T. Böttiger, R. R. Bonelli, T. Schneider, H.-G. Sahl, and B. Martinez. 2006. Lipid II-based antimicrobial activity of the lantibiotic plantaricin C. Appl. Environ. Microbiol. 72:2809-2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Winkowski, K., R. D. Ludescher, and T. J. Montville. 1996. Physicochemical characterization of the nisin-membrane interaction with liposomes derived from Listeria monocytogenes. Appl. Environ. Microbiol. 62:323-327. [DOI] [PMC free article] [PubMed] [Google Scholar]